Abstract
We have previously shown that human munc13 (hmunc13) is up-regulated by hyperglycemia under in vitro conditions in human mesangial cell cultures. The purpose of the present study was to determine the cellular function of hmunc13. To do this, we have investigated the subcellular localization of hmunc13 in a transiently transfected renal cell line, opossum kidney cells. We have found that hmunc13 is a cytoplasmic protein and is translocated to the Golgi apparatus after phorbol ester stimulation. In addition, cells transfected with hmunc13 demonstrate apoptosis after treatment with phorbol ester, but cells transfected with an hmunc13 deletion mutant in which the diacylglycerol (C1) binding domain is absent exhibit no change in intracellular distribution and no induction of apoptosis in the presence of phorbol ester stimulation. We conclude that both the diacylglycerol-induced translocation and the apoptosis represent functional activity of hmunc13. We have also demonstrated that munc13-1 and munc13-2 are localized mainly to cortical epithelial cells in rat kidney and both are overexpressed under conditions of hyperglycemia in a streptozotocin-treated diabetic rat model. Taken together, our data suggest that hmunc13 serves as a diacylglycerol-activated, PKC-independent signaling pathway capable of inducing apoptosis and that this pathway may contribute to the renal cell complications of hyperglycemia.
INTRODUCTION
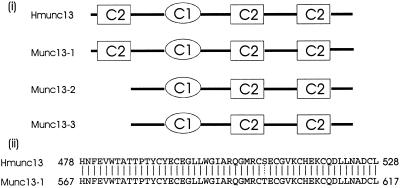
There has been growing interest in the functional characterization of a novel class of signaling proteins that belong to the same superfamily as protein kinase C (PKC) but lack its kinase activity. Unc-13, one of the members of this family, encodes a phorbol ester/diacylglycerol (DAG)-binding protein in Caenorhabditis elegans, and initial evaluation suggested it had a role in neurotransmitter release (Maruyama and Brenner, 1991; Brose et al., 1995). Mammalian homologues munc13-1, -2, and -3 (munc13s) were originally cloned from rat brain, and similar to Unc-13 they possess both DAG- and Ca2+-binding domains (Brose et al., 1995). Syntaxin, synaptobrevin, SNAP 25 (Betz et al., 1997), and Doc2 (Orita et al., 1997) were found to coimmunoprecipitate with munc13s, consistent with the suggestion that this new family of DAG-binding proteins is involved in vesicle trafficking and neurotransmitter release. Recently we cloned a human renal homologue, hmunc13, which is expressed in cultured renal mesangial and cortical epithelial cells (Song et al., 1998). As shown in Figure 1, hmunc13 is a differentially spliced isoform with partial identity to munc13-1, -2, and -3, having an N terminal similar to rat munc13-1 and a C terminal more similar to rat munc13-2 (Song et al., 1998). The overall structure of hmunc13 is closer to rat munc13-1, with three C2 (calcium binding) domains and one C1 (DAG) domain. The C1 domain is highly homologous between hmunc13 and munc13-1 (Figure 1), suggesting that both proteins have similar binding affinity to phorbol ester where the binding affinity (Kd) of munc13-1 C1 domain to phorbol ester has been reported as 5 nM (Betz et al., 1998). This binding affinity was found to be comparable with that of PKC (Betz et al., 1998). The accumulated structural information, therefore, suggests that unc-13 and its mammalian homologues serve as a DAG-activated alternative signaling pathway along with PKC and N-chimaerin (Areces et al., 1994; Betz et al., 1998); however, until recently, there has been little functional evidence to support this hypothesis. In an important new study, it was found that exposure of fibroblasts transfected with munc13-1 to phorbol esters causes translocation of munc13-1 to the plasma membrane. Furthermore, when overexpressed in the presynapses of Xenopus laevis neuromuscular junctions, munc13-1 acts as a phorbol ester-dependent enhancer of spontaneous and evoked neurotransmitter release (Betz et al., 1998). Taken together, these observations imply that phorbol esters and DAG mediate PKC-independent effects on membrane traffic and synaptic transmission. On the basis of these findings in neuronal tissue, it occurred to us that in kidney, hmunc13 might play a similar role in membrane trafficking in renal glomerular and tubular cells. In addition, because hmunc13 expression in cultured mesangial cells is up-regulated by hyperglycemia (Song et al., 1998), of interest to us was the possible role of hmunc13 activation in the development of microvascular and renal complications of hyperglycemia, especially because hyperglycemia increases intracellular DAG (King et al., 1997). Currently, PKC activation by DAG is believed to play a dominant role in the pathogenesis of the diabetic complications of hyperglycemia (H. Ishii et al., 1996; Schleicher and Nerlich, 1996), but in the light of the preceding discussion, it is possible that some of the effects previously attributed to DAG activation of PKC might instead be caused by DAG activation of hmunc13. To explore this question and also assess the functional role of hmunc13 in kidney, we first undertook to investigate the localization of hmunc13 and determine whether exposure to phorbol esters had any effect on its intracellular translocation. In the course of performing these studies, we observed that cells transfected with hmunc13 became rounded up and died after treatment with phorbol 12, 13-dibutyrate (PDBu), a phorbol ester analogue. This finding prompted us to explore the mechanism of phorbol ester-induced cell death in the transfected cells and in particular to test whether exposure to phorbol ester causes apoptosis through activation of hmunc13. If true, this might provide a connection between the diabetic state, activation of hmunc13, and cell damage. Such a hypothesis also seemed reasonable to us because apoptosis has been documented in the tubular epithelium of rat kidneys infused with high glucose solutions (N. Ishii et al., 1996), and also in a diabetic mouse model, accompanied by down-regulation of bcl-2 and bclx expression and a transient increase of bax expression (Ortiz et al., 1997).
Figure 1.
(i) Comparison of the structure of rat munc13s and hmunc13. C1 represents the DAG-binding (C1) domain; C2 represents the Ca2+-binding (C2) domain. (ii) Comparison of the sequence of the C1 domain of rat munc13-1 and hmunc13. Continuous lines indicate identical amino acids, and the dotted line indicates similar amino acids.
In the present investigation, we overexpressed an epitope-tagged hmunc13 in opossum kidney (OK) cells, a cell line of renal epithelial origin, and studied its intracellular localization by immunocytochemistry, using confocal microscopy. Under resting conditions hmunc13 was diffusely distributed in the cytoplasm. After exposure to phorbol ester, within 15–30 min we observed translocation of hmunc13 to the Golgi apparatus. This DAG-induced translocation did not occur when OK cells were transfected with hmunc13 mutated at the DAG binding site. An unexpected but novel finding was that OK cells transfected with hmunc13 demonstrated clear evidence of apoptotic changes within 8 h of exposure to phorbol esters, changes that did not occur when the cells were transfected with hmunc13 mutated at the DAG-binding site. An indication of the potential in vivo relevance of these findings is the fact that in situ hybridization experiments performed on normal rat kidney indicate that rat munc13-1 and munc13-2 are expressed mainly in cortical tubular epithelial cells and to a minor extent in the glomerulus; however, in kidney tissue from streptozotocin (STZ)-treated rats (a model of diabetes mellitus), under hyperglycemic conditions the expression of both genes is increased predominantly in tubule cells but also in the glomerulus.
Collectively these data imply that as a second messenger DAG can activate either a PKC (proliferative)-signaling pathway or, alternatively, a hmunc13 (apoptosis)-signaling pathway. The combined action of these two pathways may have implications in the functional responses of cells to various stimuli, such as hyperglycemia. Our results further suggest that hyperglycemic activation of hmunc13 and induction of apoptosis may be one of the factors contributing to cell injury in diabetic nephropathy.
MATERIALS AND METHODS
Materials and Animals
OK cells were obtained from American Type Cell Collection (Rockville, MD). FBS, MEM, penicillin, streptomycin, Lipofectamine Plus, Hanks’ solution, TRIzol reagent, SuperScript II RNase H− reverse transcriptase, dNTP, Taq DNA polymerase, DNA size markers, and T4 DNA ligase were obtained from Life Technologies (Burlington, Ontario, Canada). DNase I, yeast tRNA, poly(A), and T7Sequence kit were purchased from Pharmacia Biotech (Uppsala, Sweden). The TA cloning kit was from Invitrogen (San Diego, CA). T7 RNA polymerase with RNase inhibitor and its reaction buffer, 18S rRNA primers, and competimers were from Ambion (Austin, TX). Vent DNA polymerase and restriction enzymes were obtained from New England Biolab (Beverly, MA). Oligonucleotides were synthesized by Life Technologies or Biotech Service Center, Hospital for Sick Children (Toronto, Ontario, Canada). Mouse anti-hemagglutinin (HA) monoclonal antibody, an antibody against a peptide epitope derived from the HA protein of human influenza virus, goat anti-mouse immunoglobulin G (IgG) conjugated with rhodamine, complete mini protease inhibitor cocktail tablets, digoxigenin (DIG) RNA labeling mix, anti-DIG–rhodamine, protease K, RNase A, blocking reagent for DIG nucleic acid detection, and in situ cell death detection kit were purchased from Boehringer Mannheim (Mannheim, Germany). Wheat germ agglutinin (WGA) conjugated with FITC was purchased from EY Laboratories (San Mateo, CA). Midi plasmid preparation kits and RNeasy total RNA preparation kits were from Qiagen (Chatsworth, CA). Horse anti-mouse IgG conjugated with biotin and Vector ABC staining kits were from Vector Laboratory (Burlingame, CA). ECL was purchased from Amersham (Oakville, Ontario, Canada). DC Protein Assay kit was from Bio-Rad (Hercules, CA). STZ was purchased from ICN (Mississauga, Ontario, Canada). PDBu and nocodazole were from Sigma (St. Louis, MO). Other chemicals with cell culture or molecular biology grade were obtained from local suppliers.
Rats (Sprague Dawley, male, 250–280 g) were obtained from the Department of Comparative Medicine, University of Toronto, and maintained according to established guidelines.
Construction of HA-tagged hmunc13 and Truncated Mutant without C1 Domain
We constructed an HA-tagged hmunc13 by taking advantage of an EcoN I restriction site (nucleotide 3949) close to the 3′ end of the ORF of hmunc13 constructed in pCMV·SPORT (pCMV·SPORThmunc13; Life Technologies) and used PCR to introduce the HA tag at the C terminal of hmunc13. A PCR fragment was generated with Vent DNA polymerase, insert of pCMV·SPORThmunc13 as a template, and a pair of primers (5′-GAATACGGTTCTGGATGAGCT-3′ and 5′-gcggccgcTCAAGCGTAGTCTGGGACGTCGTATGGGTAGCTCCCCTCCTCCGTGGAACG-3′), where the HA tag sequence is underlined and a NotI site is shown in lower case. A stop code (5′-TCA-3′) was placed between the HA tag and the NotI site. The PCR product was then incubated with 2 U Taq DNA polymerase at 72°C for 15 min and extracted by phenol–chloroform and ethanol precipitation. The resulting pellet was resuspended and ligated to pCR2.1 by using a TA cloning kit. This plasmid was then digested with NotI and EcoN I, subjected to 1% agarose gel electrophoresis. The insert was purified and ligated to pCMV·SPORThmunc13 previously cut with NotI and EcoN I. The resulting construct (hmunc13-HA) was sequenced to confirm the addition of the HA tag.
To construct a deletion mutant lacking the C1 domain (C1-less mutant), we replaced the entire C1 domain (AA 478–528) with two residues, Ala and Arg. Primers 5′-CGTTGGCGCGCCAGCGGGCTGCAGAAAAGAGC-3′ (Asc I site is underlined) and 5′-CTGTCTCATCAAAGTACACC-3′ were used to generate a PCR fragment with Vent DNA polymerase and pCMV·SPORThmunc13 as a template. Another piece of PCR fragment was generated by primers of Sp6 promoter (5′-AGCTATTTAGGTGACACTATAG-3′) and 5′-GCTAGGCGCGCCGGAGTGGTGCACGAAATGG-3′ (Asc I site is underlined). The two PCR fragments were digested with Asc I and ligated with T4 DNA ligase, and the ligated product was subjected to 1% agarose gel electrophoresis to check the size and purification. The gel-purified ligated piece was further digested with KpnI and BstZ17 I and ligated to KpnI- and BstZ17 I-digested pCMV·SPORThmunc13-HA.
Plasmids for cell transfection were prepared using a Midi plasmid preparation kit according to manufacturer’s instructions.
Cells and Transfection
OK cells were grown in MEM supplemented with 10% FBS and 100 U/ml penicillin and 100 μg/ml streptomycin, and plated in 60- or 100-mm culture dishes or on glass coverslips placed in 24-well culture plates. Cells were transiently transfected (transfected rate 30–50%) with hmunc13-HA or C1-less mutant by using Lipofectamine Plus according to the manufacturer’s instructions and maintained in serum-free MEM overnight (3 h for apoptotic experiments) after 24 h of transfection. Cell monolayers were washed, and fresh medium containing PDBu or the same amount of vehicle (DMSO at a final concentration of 0.0001%) was added to the culture medium at a final concentration of 0.1 μM, and cells were analyzed at different time points as indicated. For nocodazole treatment experiments, nocodazole in DMSO was added to the medium at a final concentration of 4 μM for 1 h and followed by addition of PDBu at a final concentration of 0.1 μM. Cells were subjected to immunostaining after 3 h of PDBu treatment. An identical quantity of DMSO was added to control cells.
Immunocytochemistry
Cells grown on coverslips were washed three times with ice-cold Hanks’ solution, fixed, and permeabilized with 100% methanol at −20°C for 5 min. The coverslips were then air-dried, washed three times with PBS, pH 7.4, and incubated in blocking solution (PBS + 0.2% Tween 20) containing 10% no-fat dry milk. Cells were then incubated with 0.02 mg/ml anti-HA for 30 min at room temperature followed by 0.02 mg/ml anti-mouse IgG–rhodamine for 30 min. Cells were washed at least eight times with PBS + 0.2% Tween 20 between incubation of anti-HA and anti-mouse IgG–rhodamine or after anti-mouse IgG–rhodamine. Coverslips were then mounted on a glass slide and observed under a confocal scanning microscope. For labeling of the Golgi apparatus, 0.05 mg/ml WGA–FITC was added to the anti-mouse IgG–rhodamine.
Immunoblot Analysis and Preparation of Crude Golgi Membrane
Cells grown on culture plates were washed three times with ice-cold Hanks’ solution, scraped into 0.5 ml cell lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 0.25% sodium deoxycholate, 1% NP-40, 1 mM EDTA, and protease inhibitor cocktail, pH 7.5), and then rocked at 4°C for 45 min. The insoluble fraction was removed by centrifugation at 14,000 × g for 5 min. Supernatants were subjected to 6% SDS-PAGE and transferred to nitrocellulose. The membrane was washed twice with TBS, blocked with TBS containing 0.1% Tween 20 (TBST) and 1% normal horse serum for 30 min, and then incubated with 0.5 μg/ml anti-HA in TBST. After washing with TBST at least four times, the membrane fraction was incubated with 0.2 μg/ml anti-mouse IgG–biotin, washed with TBST, and then incubated with the A and B reagent mix in a Vector ABC staining kit according to manufacturer’s instructions. The blot was detected by ECL according to the manufacturer’s instruction.
Golgi membranes were prepared by a sucrose density method reported previously (Balch et al., 1994), with a protease inhibitor cocktail presented in all buffer solution. The band at the interface of 0.8 and 1.2 M sucrose was collected and subjected to 6% SDS PAGE and immunoblotting as described above. Protein concentration was determined by Lowry assay with BSA as standard using a DC Protein Assay kit following its instructions.
Detection of Apoptosis by DNA Fragmentation
Cleaved genomic DNA during apoptosis for cells grown on coverslips was detected by terminal deoxynucleotidyl transferase (TdT)-mediated dUTP nick end labeling (TUNEL) using an in situ cell death detection kit following manufacturer’s directions. Fluorescein labels were incorporated in nucleotide polymers. Negative controls were obtained by incubating label solution without TdT under the same conditions. After labeling for apoptosis, cells were further subjected to immunocytochemistry as described above to detect expression of hmunc13 or its C1-less mutant.
Genomic DNA fragmentation of cells grown on 60 mm culture dishes was analyzed by 2% agarose gel electrophoresis using the procedure described elsewhere (Eastman, 1995).
STZ-treated Diabetic Rat Model
Rats received a single, intraperitoneal injection of STZ (65 mg/kg body weight) dissolved in 20 mM citric acid, pH 4.5. Blood glucose was monitored daily by tail blood sampling with a Medisense blood glucose sensor (Medisense Canada, Mississauga, ON, Canada). Blood glucose was maintained at a concentration of 15–20 mM with 2 U NPH insulin daily (subcutaneous) after diabetes was confirmed by elevated blood and urinary glucose. Rats were killed after 1 or 11 d of diabetes. Rat kidneys were collected as soon as possible, usually within 3–5 min, and processed for total RNA preparation or tissue preparation for in situ hybridization as described below. Control rats were injected intraperitoneally with the same amount of 20 mM citric acid, and their blood glucose levels were also tested daily (<5 mM).
Relative RT-PCR
Total RNA from rat kidney cortex was prepared using a TRIzol reagent according to instructions provided by the manufacturer, and then treated with DNase I. Confirmation of no genomic DNA contamination in RNA preparations and relative RT-PCR were performed as described elsewhere (Song et al., 1998). Primers for amplification of rat munc13-1 are 5′-CGTGACCAAGATGAGTACTCC-3′ (sense) and 5′-CGAAGTCGTGTAGTAAGGCG-3′ (antisense); they yielded a fragment of 195 bp. Primers for rat munc13-2 are 5′-GAGTCCTGAAGGAGCTCTGG-3′ (sense) and 5′-AGGACAGCACACTGCTTTGG-3′ (antisense); they yielded a fragment of 193 bp. Primers for rat munc13-2 are 5′-AGATGACCTTGGCAAGTGC-3′ (sense) and 5′-CGATACATCATGGATGGATGG-3′ (antisense); they yielded a fragment of 198 bp. The sequence of PCR products was confirmed by cloning PCR fragments into pCR2.1 using a TA cloning kit and by sequencing using a T7Sequencing kit with T7 promoter as a primer.
In Situ Hybridization
Templates for in vitro transcription were generated by PCR with primers described above for three different isoforms, except that for antisense cRNA, addition of T7 promoter (5′-TAATACGACTCACTATAGGGA-3′) was present in the sense strain, and for sense cRNA, T7 promoter was present in the antisense strain. Antisense and sense cRNA for different isoforms were obtained by in vitro transcription. PCR templates (200 ng) were incubated with T7 RNA polymerase (40 U) and DIG RNA labeling mix in a total volume of 40 μl at 37°C for 90 min. Recombinant RNA (20 μl) was purified by using an RNeasy total RNA preparation kit, and its yield was estimated by A260. The remaining cRNA was subjected to ethanol precipitation and resuspended in nuclease-free water.
All solutions used before the posthybridization step were diethylpyrocarbonate-treated or prepared in diethylpyrocarbonate-treated water. Kidneys were quickly cut into 2-mm-thick blocks after dissection, then put in PBS containing 4% paraformaldehyde for 4 h at 4°C. The tissue was soaked in PBS containing 30% sucrose overnight at 4°C and then stored in liquid nitrogen. Frozen tissues were sectioned (10 μm) and placed on a poly-l-lysine–coated glass slides. To ensure the same experimental conditions, kidney sections from control and diabetic rats were placed on the same slide. Tissue slides were then dried at 40°C overnight and stored at −80°C for <1 wk. On the day of hybridization, slides with tissue sections were dried at 40°C for 2 h, then washed twice with PBS. Slides were then incubated with 0.3% Triton X-100 in PBS for 15 min at room temperature and washed twice with PBS afterward. Sections were incubated with 1 μg/ml RNase-free proteinase K in TE buffer (100 mM Tris-HCl, 50 mM EDTA, pH 8.0) for 30 min at 37°C and then fixed by incubating with PBS containing 4% paraformaldehyde for 5 min at 4°C. Sections were then washed twice with PBS and acetylated with freshly prepared 0.1 M triethanolamine buffer, pH 8.0, containing 0.25% acetic anhydride. Slides were then incubated first with 4× SSPE (1× SSPE containing 150 mM NaCl, 20 mM NaH2PO4, and 1 mM EDTA, pH 7.4) containing 50% formamide at 37°C for 20 min and then overlaid with 75 μl hybridization buffer (40% formamide, 10% dextran sulfate, 0.02% Ficoll, 0.02% polyvinylpyrolidone, 10 mg/ml BSA, 4× SSPE, 10 mM DTT, 0.4 mg/ml yeast t-RNA, and 0.1 mg/ml poly(A)) containing 50 ng of denatured DIG-labeled cRNA probe. Slides were incubated in a humid chamber at 42°C overnight. After hybridization, slides were washed at least four times in 1× SSPE at 37°C. Sections were incubated with 20 μg/ml RNase A in NTE buffer (500 mM NaCl, 10 mM Tris-HCl, 1 mM EDTA, pH 8.0) at 37°C for 30 min and washed twice with 0.1× SSPE. Slides were washed and blocked in TBS (100 mM Tris-HCl and 150 mM NaCl, pH 7.5) containing 1% blocking reagent and then incubated with 0.02 mg/ml anti-DIG–rhodamine for 1 h. Slides were washed at least five times with TBS. Staining was assessed by a confocal scanning microscopy.
RESULTS
Translocation of hmunc13 to Golgi Apparatus after DAG Treatment
To study its cellular function, we elected to overexpress hmunc13 in OK cells, a cell line of renal epithelia origin, and compare two constructs—an HA-tagged hmunc13 and an HA-tagged hmunc13 deletion mutant lacking the C1 domain (C1-less mutant). Cells used in the present study were grown on glass coverslips under growth-arrested conditions with serum starvation. Transient transfection of OK cells was confirmed by Western blot analysis (Figure 2). As shown in Figure 2, an ∼180-kDa protein was expressed in the hmunc13-HA–transfected cells and a ∼175-kDa protein was detected in the C1-less mutant-transfected cells. No band was detected in cells transfected with empty plasmid pCMV·SPORT.
Figure 2.
Immunoblot of hmunc13 and the C1-less mutant. hmunc13-HA (Hmunc13), C1-less mutant (C1 less), or empty plasmid pCMV·SPORT (PCMV) were transiently transfected into OK cells. Whole-cell lysates were prepared and subjected to 6% SDS-PAGE. The blot was detected by anti-HA. Note the slightly decreased molecular weight of the C1-less mutant.
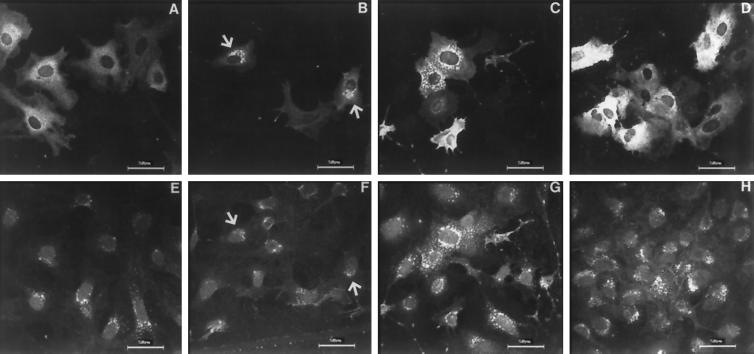
Intracellular localization of hmunc13-HA in transfected OK cells was monitored by immunocytochemistry using cells doubly labeled with anti-HA antibody (Figure 3, top panels) and WGA (Figure 3, bottom panels). As indicated in Figure 3, inspection of A reveals that hmunc13 exhibits a cytosolic distribution compared with the Golgi apparatus stained with WGA shown in E, but after exposure to 0.1 μM PDBu, a DAG analogue, hmunc13 is translocated to the perinuclear area (B) and colocalizes with WGA at the Golgi apparatus (Figure 3, compare B and F; Figure 4). Translocation of hmunc13 to the Golgi after PDBu treatment occurred in 15–30 min and became more obvious in 2–3 h. By contrast, when cells were transfected with the C1-less mutant, which lacks a DAG-binding domain, there was no translocation after PDBu treatment (refer to Figure 3, D and H), and hmunc13 staining remained cytosolic.
Figure 3.
Immunostaining of OK cells transiently transfected with hmunc13-HA (A–C and E–G) and C1-less mutant (D and H). Cells were stained with anti-HA, then probed with anti-mouse IgG–rhodamine for detection of hmunc13 (A–C) and C1-less mutant (D). The Golgi apparatus was detected by staining with WGA–FITC (E–H). Slides were observed by confocal microscopy using a laser scanning microscope with excitation wavelength at 568 nm for detecting rhodamine (A–D) and at 488 nm for detecting FITC (E–H). Cells were treated with vehicle (A and E), 0.1 μM PDBu for 3 h (B, D, F and H), 4 μM nocodazole + PDBu (C and G) as described in MATERIALS AND METHODS. Negative controls obtained by incubating with normal mouse IgG or immunostaining of cells transfected with empty plasmid (pCMV·SPORT) yielded very little or no staining (our unpublished results). Arrowheads indicate colocalization of anti-HA and WGA staining. Note: top and bottom panel pairs, i.e., A and E, B and F, etc., represent anti-HA and WGA-FITC staining, respectively, of identical fields.
Figure 4.
Localization of hmunc13 to Golgi in hmunc13-HA transiently transfected OK cells in response to PDBu treatment for 2 h. Cells were treated and detected as in Figure 3. Note the colocalization (orange with the mix of red and green) of hmunc13 (red) and WGA (green) staining. Bar, 10 μm.
When cells were treated with nocodazole, a drug that depolymerizes microtubules (Morris and Yu-Lee, 1998), after PDBu treatment the patterns of WGA and hmunc13 staining became identical, and both revealed a dispersed Golgi pattern (Figure 3, compare C and E).
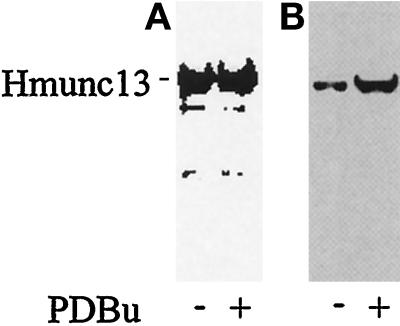
Translocation of hmunc13 from cytosol to the Golgi apparatus after PDBu treatment was also confirmed by immunoblot analysis of a Golgi membrane preparation, after subcellular fractionation. As shown in Figure 5, after PDBU treatment, hmunc13 is enriched in Golgi membranes compared with whole-cell lysates.
Figure 5.
Immunoblots of whole-cell lysates (A) and Golgi membrane preparations (B) from hmunc13-transfected OK cells with (+) or without (−) PDBu treatment for 3 h. The whole-cell lysates represent small aliquots of cells for Golgi membrane preparations. Equal amounts of protein were loaded onto each lane of A or B. The blots were then detected by anti-HA antibody.
Hmunc13-overexpressed Cells Are Apoptotic after DAG Treatment
The PDBu-induced translocation from cytosol to Golgi suggests that hmunc13, like rat munc13s, is activated by DAG and has functional implications. While attempting to study the effect of prolonged exposure to DAG activation on hmunc13-transfected cells, we noticed that the cells rounded up and died; however, hmunc13-transfected cells without PDBu treatment and cells transfected with the C1-less mutant, with or without PDBu treatment, were relatively healthy. This finding was somewhat unexpected because DAG has long been known as a carcinogen and a promoter of cell growth, and it led us to investigate the possibility that treatment with phorbol ester is inducing apoptosis in cells transfected with hmunc13.
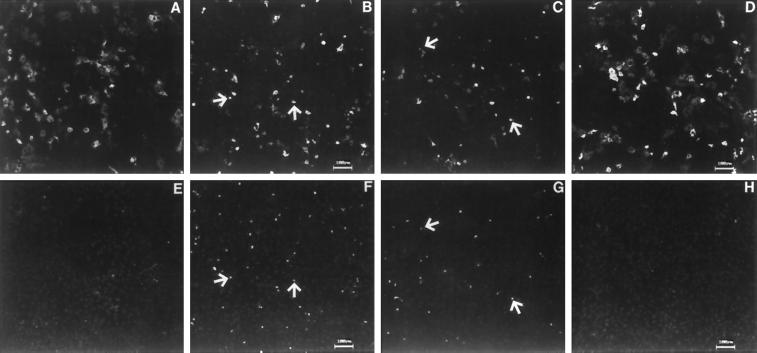
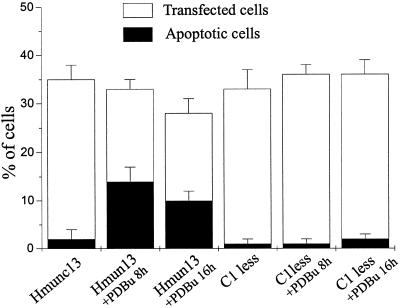
Using the TUNEL assay, we found that the number of apoptotic cells was significantly increased in hmunc13-transfected OK cells after 8 and 16 h of PDBu treatment. These results are displayed in Figure 6 and summarized in Figure 7. The top panels of Figure 6 show the expression of hmunc13 in OK cells, and the bottom panels demonstrate the presence of fluorescein-labeled TUNEL on the same cells. Inspection of F (8 h of PDBu treatment) and G (16 h of PDBu treatment) compared with E (treatment with vehicle control) reveals evidence of DAG-induced increase in TUNEL-staining cells. The reason that the number of hmunc13-transfected and TUNEL-positive cells appears to be less in C and G compared with G and F is because the apoptotic cells lift off and are washed away during the staining process after 16 h of PDBu treatment. The conclusion that there is DAG-induced apoptosis in hmunc13-transfected cells is further supported by the fact that cells transfected with the C1-less mutant exhibit almost no labeling with TUNEL after exposure to PDBu for 8 h (compare H with F and G). In Figure 6, some TUNEL-staining cells do not appear to exhibit expression of hmunc13. The reason for this is that these cells are expressing low levels of hmunc13 that are not detected at low power. By contrast, the TUNEL assay results in a strong fluorescence emission and can easily be detected under the same conditions. Therefore under high power almost all TUNEL-staining cells (>90%) also express hmunc13 (our unpublished results). Finally, cells transfected with empty plasmid also showed almost no TUNEL labeling with or without PDBu treatment (our unpublished results).
Figure 6.
Double labeling of apoptotic cells and expression of hmunc13 or C1-less mutant. hmunc13 (A–C and E–G) and C1-less mutant (D and H) transiently transfected cells were subjected to TUNEL labeled with fluorescein (E–H) and then subjected to anti-HA and anti-mouse IgG–rhodamine labeling for expression of hmunc13 and C1-less mutant (A–D). Cells were treated with vehicle (A and E) or 0.1 μM PDBu for 8 h (B, D, F and H) or 16 h (C and G). C1-less mutant-transfected cells treated with vehicle exhibit an image similar to D and H (our unpublished results). Negative controls of TUNEL by incubating cells with labeling mix and no TdT yielded no staining of fluorescein (our unpublished results). Arrowheads indicate representative cells costained with anti-HA (A–D) and TUNEL (E–H) from identical fields.
Figure 7.
Graphic representation of the percentage of transfected (immunostaining-positive) and apoptotic (TUNEL-positive) cells in hmunc13-transfected cells or C1-less mutant-transfected (C1 less) cells treated with or without PDBu for 8 or 16 h. Cell numbers were counted with an average of three low-power views under the confocal microscope. Bars are representations of means ± SD of three experiments.
To further confirm this, a DNA fragmentation assay was used. Further evidence of a breakdown in genomic DNA is revealed by the “laddering” pattern shown in Figure 8, obtained after 8 and 16 h of PDBu treatment in hmunc13-transfected cells.
Figure 8.
Genomic DNA breakdown in hmunc13-transfected cells by PDBu treatment. Genomic DNA obtained from empty plasmid (PCMV), hmunc13-transfected cells (Hmunc13), or C1-less mutant-transfected cells (C1 less) treated with vehicle (−) or 0.1 μM PDBu for 8 or 16 h was subjected to 2% agarose gel electrophoresis. Molecular size marker (M) is shown.
Expression of munc13s in Normal and STZ-treated Diabetic Rat Kidney
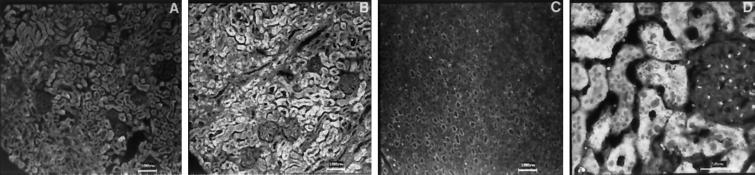
We have previously demonstrated that hmunc13 is up-regulated by high-glucose treatment in cultured human mesangial (Song et al., 1998). Because the main thrust of the present study was to investigate the functional role of hmunc13, we thought it was important to document its in vivo expression. Furthermore, confirmation of up-regulation of hmunc13 by hyperglycemia in an in vivo state is necessary to establish a role for this gene in diabetic nephropathy. Ideally, because hmunc13 was cloned from a human cDNA library, we would have preferred to characterize its expression in human kidney; however, because of difficulty in obtaining suitable human tissue, we elected instead to use an animal model of diabetes—the STZ-treated rat (the relevant isoforms being munc13-1, -2, and -3). As shown in Figure 9, munc13-1 is expressed mainly in cortical tubular epithelial cells of both normal and STZ-treated diabetic rats; however, the expression level of munc13-1 was higher in STZ-treated diabetic rat after 11 d of hyperglycemia. Expression of munc13-1 was significantly higher in certain glomerular cells of diabetic animals, but it is impossible to identify these cells with any certainty at the resolution of confocal microscopy; however, because of our previous in vitro results (Song et al., 1998), we suspect that munc13-1 is up-regulated in the mesangial cells. An increased expression level of munc13-2 was also detected in diabetic rats with an expression pattern similar to that of munc13-1 (our unpublished results). Possibly because of low basal expression, we could not obtain satisfactory in situ hybridization data for munc13-3 in rat kidney.
Figure 9.
Expression of rat munc13-1 in kidney of normal (A) or STZ-treated diabetic (B–D) rat detected by in situ hybridization. Outer cortex (A and B), medulla (C), and a higher power view of outer cortex (D) from diabetic rat kidney are shown. Similar to diabetic rats, staining in the renal medulla for normal rat kidney is less than the cortex (our unpublished results). Note the increased expression of munc13-1 in the tubular epithelial cells as well as in certain glomerular cells. Negative controls with sense cRNA showed little staining in both normal and diabetic rat sections (our unpublished results).
To confirm the overexpression of munc13-1 and munc13-2 in diabetic rat kidney, we performed relative RT-PCR on renal cortical RNA preparation. Relative RT-PCR was chosen because low expression of munc13s in the rat kidney and a very low signal were detected in Northern blot analysis (our unpublished observation; Brose et al., 1995). As shown in Figure 10, compared with the housekeeping gene, 18S ribosome RNA, expression of munc13-1 is overexpressed in the renal cortex of the STZ-treated diabetic rat after only 1 d of hyperglycemia, whereas expression of munc13-2 is increased to a much lesser extent. Interestingly, munc13-3 is down-regulated in the same animal model. So far, we have failed to detect a human homologue of rat munc13-3 in a commercial human kidney cDNA library (Life Technologies) using PCR with primers targeted to different regions of munc13-3. Therefore, the role of munc13-3 in diabetic nephropathy remains to be determined.
Figure 10.
Expression of munc13-1, munc13-2, and munc13-3 in the renal cortex of the normal rat and after 1 d (1d) and 11 d (11d) of hyperglycemia in STZ-treated rats. 18S ribosome RNA (18S) served as a housekeeping gene.
DISCUSSION
The results of the present study provide the first direct evidence of the subcellular localization of hmunc13 in vitro. Expression of epitope-tagged hmunc13 in OK cells indicates that this protein has a cytoplasmic distribution under basal conditions, but with PDBu stimulation, hmunc13 is translocated to the Golgi apparatus. This effect is unlikely to have taken place through activation of PKC, because the deletion mutant C1-less mutant (without the DAG-binding domain) showed no translocation.
In a recent study reported by Betz et al. (1998), munc13-1 was localized to the presynaptic region in rat brain by immunocytochemistry. In transfected HEK 293 cells, green fluorescent protein-tagged munc13-1, -2, and -3 are all translocated to plasma membrane after phorbol ester stimulation. We believe that the fact that hmunc13 is translocated to the Golgi apparatus in response to phorbol ester activation compared with translocation of munc13-1, -2, and -3 to the plasma membrane is because hmunc13 is a unique isoform of munc13s-containing domains not present in rat munc13s. Other reasons for the different localization could be that different gene constructs (green fluorescent protein-tagged versus HA-tagged) were used or that there were differences in cell types and experimental culture conditions.
This brings up the relationship of the DAG-activated signaling pathways of munc13s and PKC. The multiplicity of PKC isoforms and the tissue specificity of PKC functional expression are well known (Blobe et al., 1998). The picture that is emerging for the munc13 pathways is that it also will be composed of tissue-specific, functionally different isoforms; however, unlike PKC, the munc13 proteins have no kinase domain (Brose et al., 1995; Song et al., 1998). The question that arises is how they will affect their functional role. The Golgi apparatus is involved in vesicular traffic. A number of SNARE proteins, such as yeast Sed5p (Banfield et al., 1994) and mVps45 (Tellam et al., 1997), mammalian syntaxin 6 (Bock et al., 1997), VAMP4, Syntaxin 13, and mVtib (Advani et al., 1998), have all been reported to be localized to the Golgi. Rat munc13-1 has been shown to interact with a number of proteins involved in vesicle docking and trafficking, such as syntaxin (Betz et al., 1997) and Doc2 (Orita et al., 1997). Interaction of munc13-1 and Doc2 was stimulated by DAG and has been suggested to be involved in Ca2+-dependent exocytosis (Orita et al., 1997). The finding in the present study that translocation of hmunc13 to the Golgi after DAG stimulation is another indication that hmunc13 is a protein that participates in DAG-regulated vesicle trafficking and exocytosis. Further studies are required to investigate whether hmunc13 interacts with other Golgi-localized SNARE proteins or whether some SNARE proteins cotranslocate to the Golgi with hmunc13 after DAG stimulation. It has also been suggested that PKC plays a role in Golgi budding (for review see Martin, 1997). For example, a study in Saccharomyces cerevisiae implicated DAG as playing an important role in the formation of Golgi budding involving Sec14 (Kearns et al., 1997). Because hmunc13 translocates to the Golgi after DAG stimulation, it would also be of interest to determine whether hmunc13 is involved in Golgi budding and interacts with Sec14L, the partial mammalian homologue of yeast Sec14 (Chinen et al., 1996).
One of the unexpected but intriguing results of the present study is that hmunc13-transfected cells are apoptotic after PDBu stimulation. This effect is unlikely to have occurred through other DAG-activated pathways such as PKC because the C1-less mutant transfected cells were not apoptotic after PDBu treatment, and cells transfected with empty plasmid were not apoptotic without or with PDBu stimulation. PDBu is a reagent known to be a tumor promoter capable of stimulating cell proliferation through PKC activation (Mochly-Rosen and Kauvar, 1998). Although the role of PKC in apoptosis is not consistent in the literature (Lavin et al., 1996; Deacon et al., 1997), the bulk of evidence suggests that PKC, especially PKCα, activated by phorbol esters such as phorbol 12-myristate 13-acetate and PDBu, inhibits apoptosis (Lavin et al., 1996; Deacon et al., 1997; Whitman et al., 1997; Mochly-Rosen and Kauvar, 1998). In the case of PKC-induced apoptosis, there is evidence suggesting that down-regulation rather than DAG activation of PKC is responsible for this effect, because prolonged exposure to a stable phorbol ester form could result in down-regulation of PKC expression (Leszczynski, 1996; Deacon et al., 1997). A reasonable possibility is that signaling through hmunc13 becomes dominant after PKC activity is decreased. The effect of hmunc13 on apoptosis may not be limited to the kidney because clones with homology to munc13-1 or munc13-2 have been identified by the expression sequencing tag project in mouse mammalian gland (GenBank accession no. AI156592), heart (GenBank accession no. AA855543), and human testis (GenBank accession no. AA620907 and AA412093). Because of the lack of a specific hmunc13 activator or inhibitor compound, the function of hmunc13 could not be assessed in vivo. Nevertheless we believe that hmunc13-induced apoptosis is not simply because of the overexpression of this protein, because overexpression of its truncated mutant without DAG-binding (C1-less) domain does not produce apoptosis. Therefore both up-regulation and activation of hmunc13 with DAG are necessary for this effect.
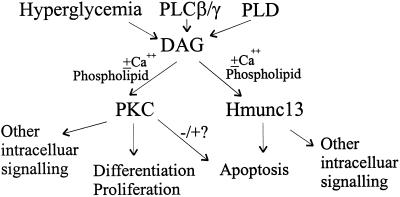
With the perspective of the functional characteristics of hmunc13 as revealed in the present study, combined with the known behavior of munc13-1, -2, and -3 in rat brain, it is possible to suggest a model for the cellular activation of hmunc13 and PKC isoforms. Because, as noted in the INTRODUCTION, both munc13s and PKC have similar binding affinity for phorbol esters, our results showing that cells transfected with hmunc13 become apoptotic after DAG treatment could mean that hmunc13 participates in a signaling pathway that serves to counterbalance DAG-activated PKC. This concept is illustrated schematically in Figure 11. We propose that DAG acts as a secondary messenger to activate two alternate pathways. One pathway effected through PKC results in kinase activation and serine–threonine phosphorylation of downstream targets leading to cell proliferation, whereas the other pathway effected through hmunc13 induces apoptosis, possibly through interaction involving vesicle trafficking.
Figure 11.
Schematic representation of DAG-activated, branched signaling pathways involving PKC and hmunc13. DAG levels are increased by such factors as hyperglycemia, phospholipase C (PLC) β/γ, and phospholipase D (PLD), resulting in activation of PKC and hmunc13 and leading to two separate signaling pathways, resulting in proliferation and differentiation or apoptosis.
This model may help to explain the pathogenesis of renal complications of diabetes. In the present study we have shown that in rat kidney, mRNA of munc13-1 and munc13-2 are mainly expressed in cortical tubular epithelia cells. Preliminary data, using a peptide-specific antibody against all three isoforms of rat munc13s, also demonstrate that munc13s protein is localized in the same renal cortical tubular epithelia expressing rat munc13s mRNA (Song and Silverman, unpublished data). Furthermore, in the present study, using both in situ hybridization and relative RT-PCR, we have also shown that munc13-1 and munc13-2 are overexpressed in kidney of STZ-treated diabetic rats. This result in rat kidney is consistent with previous in vitro findings, demonstrating that expression of hmunc13 is up-regulated by high glucose treatment in cultured human mesangial cells (Song et al., 1998). It has been reported that an increase in intracellular DAG levels is only detectable after 2 d of high glucose treatment (Xia et al., 1994). The fact that expression of both rat munc13-1 and munc13-2 is found to be increased after only 1 d of hyperglycemia suggests that overexpression of these genes is a consequence of hyperglycemia and not secondary to stimulation by DAG. Therefore, in diabetes, there may be two mechanisms acting to increase activity of hmunc13: 1) hyperglycemia itself, and 2) hyperglycemia-induced increase in cellular DAG (Hise and Mehta, 1988; Derubertis and Craven, 1994; King et al., 1997). It is not our intention to provide a critical comparison of in vitro and in vivo manifestation. By using transient transfection, it is also difficult for us to determine the expression level of hmunc13 necessary to produce apoptotic response to DAG; however, after careful assessment of the immunocytochemical findings, it appears to us that cells begin to exhibit apoptosis at even very low levels of expression of hmunc13 (Figure 6, B and D). Our tentative conclusion is that overexpression of hmunc13 in diabetic renal cortical epithelia cells may make the cells more vulnerable to apoptosis and could be an important contributing factor in causing cell injury in diabetic nephropathy. In this regard, it is noteworthy that under hyperglycemic conditions, renal tubular cells exhibit evidence of apoptosis (N. Ishii et al., 1996; Ortiz et al., 1997). Finally, because PKC inhibitors have been developed to treat diabetic nephropathy (H. Ishii et al., 1996; King et al., 1997), a potential side effect of those inhibitors could result from overactivity of hmunc13. Elucidation of the detailed molecular pathway of the effect of hmunc13 on apoptosis will provide further insight into the pathogenesis of diabetic nephropathy.
In conclusion, our findings provide support for a model that incorporates two DAG-activated pathways: 1) PKC dependent and 2) hmunc13 dependent. Moreover, these two pathways seem to regulate two opposing cell phenotypes: PKC–proliferation and hmunc13–apoptosis. Overexpression of hmunc13 under hyperglycemic conditions and DAG-induced apoptosis implicate a role for hmunc13 in diabetic renal cell injury.
ACKNOWLEDGMENTS
Y.S. is the recipient of a postdoctoral fellowship from the Kidney Foundation of Canada. This study was supported by grants from the Canadian Diabetes Foundation and the Medical Research Council of Canada to M.S. The technical assistance of Mrs. P. Clayman is greatly appreciated.
REFERENCES
- Advani RJ, Bae H, Bock JB, Chao DS, Doung Y, Prekeris R, Yoo J, Scheller RH. Seven novel mammalian SNARE proteins localize to distinct membrane compartments. J Biol Chem. 1998;273:10371–10324. doi: 10.1074/jbc.273.17.10317. [DOI] [PubMed] [Google Scholar]
- Areces LB, Kazanietz MG, Blumberg PM. Close similarity of baculovirus-expressed n-chimaerin and protein kinase Cα as phorbol ester receptors. J Biol Chem. 1994;269:19553–19558. [PubMed] [Google Scholar]
- Balch WE, Bunphy WG, Braell WA, Rothman JE. Reconstitution of the transport of protein between successive compartments of the Golgi measured by the coupled incorporation of N-acetylglucosamine. Cell. 1994;39:405–416. doi: 10.1016/0092-8674(84)90019-9. [DOI] [PubMed] [Google Scholar]
- Banfield DK, Lewis MU, Rabouille C, Warren G, Pelham HRB. Localization of Sed5, a putative vesicle targeting molecule, to the cis-Golgi network involves both its transmembrane and cytoplasmic domains. J Cell Biol. 1994;127:357–371. doi: 10.1083/jcb.127.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz A, Ashery U, Rickmann M, Augustin I, Neher E, Sudhof TC, Rettig J, Brose N. Munc13 is a presynaptic phorbol ester receptor that enhances neurotransmitter release. Neuron. 1998;21:123–136. doi: 10.1016/s0896-6273(00)80520-6. [DOI] [PubMed] [Google Scholar]
- Betz A, Okamoto M, Benseler F, Brose N. Direct interaction of the rat unc-13 homologue Munc13–1 with the N terminus of syntaxin. J Biol Chem. 1997;272:2520–2526. doi: 10.1074/jbc.272.4.2520. [DOI] [PubMed] [Google Scholar]
- Blobe GC, Stribling S, Obeid LM, Hannum YA. Protein kinase C isoenzymes: regulation and function. Cancer Surv. 1998;27:213–248. [PubMed] [Google Scholar]
- Bock JB, Klumperman J, Davanger S, Scheller RH. Syntaxin 6 functions in trans-Golgi network vesicle trafficking. Mol Biol Cell. 1997;8:1261–1271. doi: 10.1091/mbc.8.7.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brose N, Hofmann K, Hata Y, Sudhof C. Mammalian homologues of Caenorhabditis elegans unc-13 gene define novel family of C2-domain proteins. J Biol Chem. 1995;270:25273–25280. doi: 10.1074/jbc.270.42.25273. [DOI] [PubMed] [Google Scholar]
- Chinen K, Takahashi E, Nakamura Y. Isolation and mapping of a human gene (SEC14L), partially homologous to yeast SEC14, that contains a variable number of tandem repeat (VNTR) sites in its 3′ untranslated region. Cytogenet Cell Genet. 1996;73:218–223. doi: 10.1159/000134342. [DOI] [PubMed] [Google Scholar]
- Deacon EM, Pongracz J, Griffiths G, Lord JM. Isoenzymes of protein kinase C: differential involvement in apoptosis and pathogenesis. Mol Pathol. 1997;50:124–131. doi: 10.1136/mp.50.3.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derubertis FR, Craven PA. Activation of protein kinase C in glomerular cells in diabetes: mechanisms and potential links to the pathogenesis of diabetic glomerulopathy. Diabetes. 1994;43:1–8. doi: 10.2337/diab.43.1.1. [DOI] [PubMed] [Google Scholar]
- Eastman A. Assays for DNA fragmentation, endonucleases, and intracellular pH and Ca2+ associated with apoptosis. In: Schwartz LM, Osborne BA, editors. Cell Death, Methods in Cell Biology. Vol. 46. San Diego: Academic Press; 1995. pp. 41–55. [DOI] [PubMed] [Google Scholar]
- Hise MK, Mehta PS. Characterization and localization of calcium/phospholipid-dependent protein kinase-C during diabetic renal growth. Endocrinology. 1988;123:1553–1558. doi: 10.1210/endo-123-3-1553. [DOI] [PubMed] [Google Scholar]
- Ishii H, et al. Amelioration of vascular dysfunctions in diabetic rats by an oral PKC β inhibitor. Science. 1996;272:728–731. doi: 10.1126/science.272.5262.728. [DOI] [PubMed] [Google Scholar]
- Ishii N, Ogawa Z, Suzuki K, Numakami K, Saruta T, Itoh H. Glucose loading induces DNA fragmentation in rat proximal tubular cells. Metabolism. 1996;45:1348–1353. doi: 10.1016/s0026-0495(96)90114-6. [DOI] [PubMed] [Google Scholar]
- Kearns BG, McGee TP, Mayinger P, Gedvilaite A, Phillips SE, Kagiwada S, Bankaitis VA. Essential role for diacylglycerol in protein transport from the yeast Golgi complex. Nature. 1997;387:101–105. doi: 10.1038/387101a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GL, Ishii H, Koya D. Diabetic vascular dysfunction: a model of excessive activation of protein kinase C. Kidney Int. 1997;52(suppl 60):S-77–S-85. [PubMed] [Google Scholar]
- Lavin MF, Watters D, Song Q. Role of protein kinase activity in apoptosis. Experientia. 1996;52:979–994. doi: 10.1007/BF01920107. [DOI] [PubMed] [Google Scholar]
- Leszczynski D. The role of protein kinase C in regulation of apoptosis: a brief overview of the controversy. Cancer J. 1996;9:308–313. [Google Scholar]
- Martin TFJ. Greasing the Golgi budding machine. Nature. 1997;387:21–22. doi: 10.1038/387021a0. [DOI] [PubMed] [Google Scholar]
- Maruyama IN, Brenner S. A phorbol ester/diacylglycerol-binding protein encoded by the unc-13 gene of Caenorhabditis elegans. Proc Natl Acad Sci USA. 1991;88:5729–5733. doi: 10.1073/pnas.88.13.5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochly-Rosen D, Kauvar LM. Modulating protein kinase C signal transduction. Adv Pharmacol. 1998;44:91–145. doi: 10.1016/s1054-3589(08)60126-x. [DOI] [PubMed] [Google Scholar]
- Morris SM, Yu-Lee L. Expression of RNUDC, a potential nuclear movement protein in mammalian cells: localization to the Golgi apparatus. Exp Cell Res. 1998;238:23–32. doi: 10.1006/excr.1997.3822. [DOI] [PubMed] [Google Scholar]
- Orita S, Naito A, Sakaguchi G, Maeda M, Igarashi H, Sasaki T, Takai Y. Physical and functional interactions of Doc2 and Munc13 in Ca2+-dependent exocytotic machinery. J Biol Chem. 1997;272:1681–1684. doi: 10.1074/jbc.272.26.16081. [DOI] [PubMed] [Google Scholar]
- Ortiz A, Ziyadeh FN, Neilson EG. Expression of apoptosis-regulatory genes in renal proximal tubular epithelia cells exposed to high ambient glucose and in diabetic kidneys. J Invest Med. 1997;45:50–56. [PubMed] [Google Scholar]
- Schleicher E, Nerlich A. The role of hyperglycemia in the development of diabetic complications. Horm Metab Res. 1996;28:367–373. doi: 10.1055/s-2007-979817. [DOI] [PubMed] [Google Scholar]
- Song Y, Ailenberg M, Silverman M. Cloning of a novel gene in the human kidney homologous to rat munc13s: its potential role in diabetic nephropathy. Kidney Int. 1998;53:1689–1695. doi: 10.1046/j.1523-1755.1998.00942.x. [DOI] [PubMed] [Google Scholar]
- Tellam JT, Jams DE, Stevens TH, Piper RC. Identification of a mammalian Golgi Sec1P-like protein, mVps45. J Biol Chem. 1997;272:6187–6193. doi: 10.1074/jbc.272.10.6187. [DOI] [PubMed] [Google Scholar]
- Whitman SP, Civoli F, Daniel LW. Protein kinase CβII activation by 1-(-d-arabinofuranosylcytosine is antagonistic to stimulation of apoptosis and bcl-2α down-regulation. J Biol Chem. 1997;272:23481–23484. doi: 10.1074/jbc.272.38.23481. [DOI] [PubMed] [Google Scholar]
- Xia P, Inoguchi T, Kern TS, Engerman RL, Oates PJ, King GL. Characterization of the mechanism for the chronic activation of diacyglycerol-protein kinase C pathway in diabetes and hypergalactosemia. Diabetes. 1994;43:1122–1129. doi: 10.2337/diab.43.9.1122. [DOI] [PubMed] [Google Scholar]