Abstract
Background
Severe Acute Respiratory Syndrome (SARS) is a severe respiratory illness caused by a novel virus, the SARS coronavirus (SARS-CoV). 3C-like protease (3CLpro) of SARS-CoV plays a role in processing viral polypeptide precursors and is responsible of viral maturation. However, the function of 3CLpro in host cells remains unknown. This study investigated how the 3CLpro affected the secretion of cytokines in the gene-transfected cells.
Results
From immunofluorescence microscopy, the localization of c-myc tagged 3CLpro was detected both in the cytoplasm and nucleus of transfected A549 cells. Expression of granulocyte-macrophage colony-stimulating factor (GM-CSF) was significantly decreased in 3CLpro-transfected cells by both RT-PCR and ELISA, but without changes in other cytokines, i.e., IL-1β, IL-6, IL-8, IL12p40, TNF-α, and TGF-β. Furthermore, the protein levels of NF-kB decreased in 3CLpro-transfected A549 cells when compared to EGFP transfected cells.
Conclusions
Our results suggest that the 3CLpro may suppress expression of GM-CSF in transfected A549 cells through down-regulation of NF-kB production.
Background
Severe acute respiratory syndrome (SARS) emerged as a communicable human disease in November 2002 and rapidly spread throughout the world [1]. A coronavirus, called SARS-associated coronavirus (SARS-CoV), was identified as the causative agent [2,3]. SARS-CoV is a plus-strand RNA virus featuring a large single-stranded RNA genome of approximately 29.7 kb [4] that contains 16 non-structural proteins (nsps) with multiple enzymatic functions. These are known or are predicted to include types of enzymes that are common components of the replication machinery of plus-strand RNA viruses [5,6]. In these nsps, the Nsp5 [main protease (Mpro) or 3C-like protease (3CLpro)] is responsible for the proteolytic processing of viral polypeptides into functional proteins and 3CLpro mutant, C145A, blocks the maturation process [7]. This viral protease has been the target for design of the inhibitors [7-11]. Recently, SARS-3CLpro was found to induce cell apotosis and interact with cellular vacuolar-H+ ATPase [12,13]. Several human proteins might possess a similar structure of SARS-3CLpro cleavage sites using computational methods [14]. Theses studies implied the SARS-3CLpro could involve in virus-induced host pathology.
SARS-CoV causes a severe lung infection, and the epithelial cells of the upper respiratory tract are the primary targets [15-17]. SARS-CoV reproduces rapidly and causes massive cell damage. At the whole-body level, immune-mediated damage, due to activation of cytokines and/or chemokines [18-22] and, perhaps, autoimmunity [23,24], may play key roles in the clinical and pathological features of SARS. One of the major causes of host death is the induction of lung fibrosis. The mechanism of the lung fibrosis caused by SARS-CoV requires elucidation. Significant efforts have been made to analyze the function of SARS-3CLpro on viral replication, but the impact of SARS-3CLpro on cytokines of the host cell is still unclear. In this study, we constructed the pEGFP-C3 vector with SARS-3CLpro gene and transfected it into A549 lung epithelial cells. The expression profiles of cytokines were validated by RT-PCR and ELISA. The results indicated that the expression and secretion of granulocyte-macrophage colony-stimulating factor (GM-CSF) decrease in SARS-3CLpro transfected A549 cells. Furthermore, NF-kB, putative regulators of GM-CSF gene expression, was also decreased on its protein level in 3CLpro transfected cells.
Results
Construction of SARS-CoV 3CLpro wild type and a mutant C145A clone into pEGFP-C3 vector
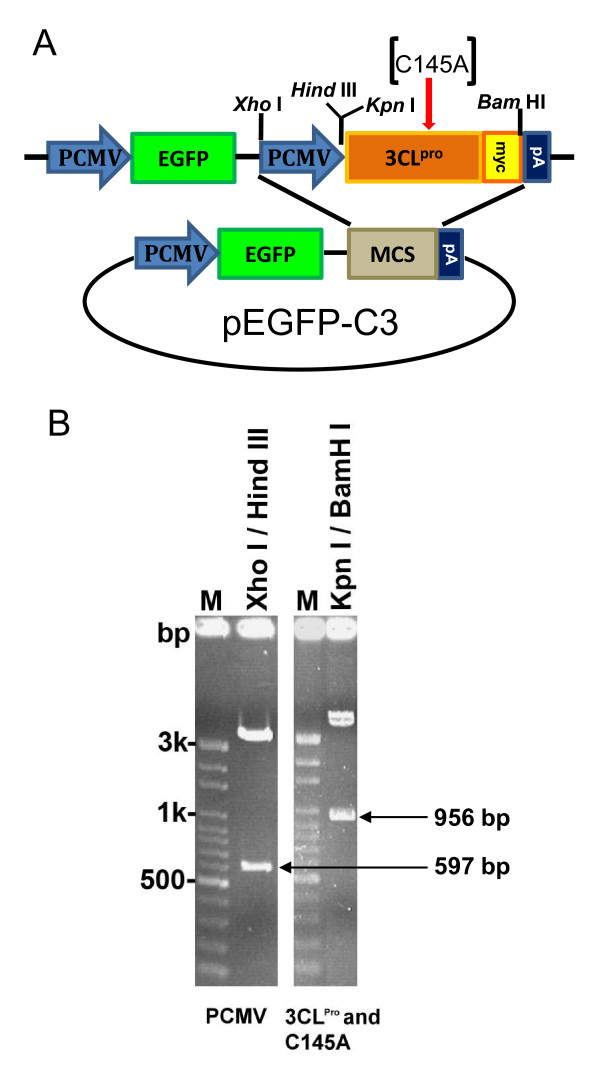
To optimize the efficiency of transfection and expression of functional 3CLpro in A549 lung cancer cell line, we constructed expression vector expressing SARS-CoV 3CLpro wild type gene into a pEGFP-C3 vector (Figure 1A). An inactive 3CLpro mutant, C145A (TGT→GCT), was also included in this study as a negative control [7]. Furthermore, the myc tag sequence was also introduced into the 3' end of the 3CLpro and C145A genes for immunofluorescence analysis. The DNA fragments of CMV promoter, 3CLpro wild type, and C145A 3CLpro mutant were amplified by PCR using primers containing restriction enzyme sites as shown in Figure 1A. The expression constructs were further verified by 1% agarose gel electrophoresis after restriction enzymes digestion (Figure 1B). As expected, the 597 bp of PCMV and 956 bp of 3CLpro (and C145A) fragments were excised from plasmids with appropriate restriction enzymes and were confirmed by DNA sequencing.
Figure 1.
Construction of pEGFP-3CLpro and pEGFP-C145A expression vectors. (A) Scheme of pEGFP-3CLpro and pEGFP-C145A construction. PCMV, human cytomegalovirus (CMV) immediate early promoter; EGFP, enhanced green fluorescent protein gene; MCS, multiple cloning sites; 3CLpro, SARS-CoV 3CLpro gene; C145A, 3CLpro C145A mutant gene; myc, myc-tag sequence; pA, SV40 early mRNA polyadenylation signal sequence. (B)The constructs were digested with restriction enzyme and separated on 1% agarose gel. M, DNA markers.
Expression of EGFP, 3CLpro, and C145A in A549 cells
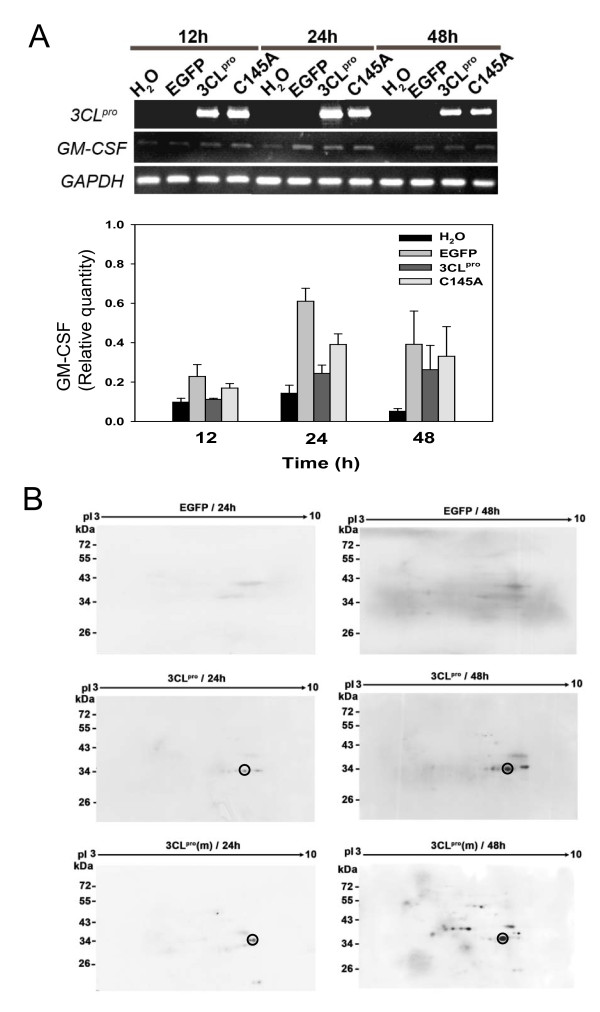
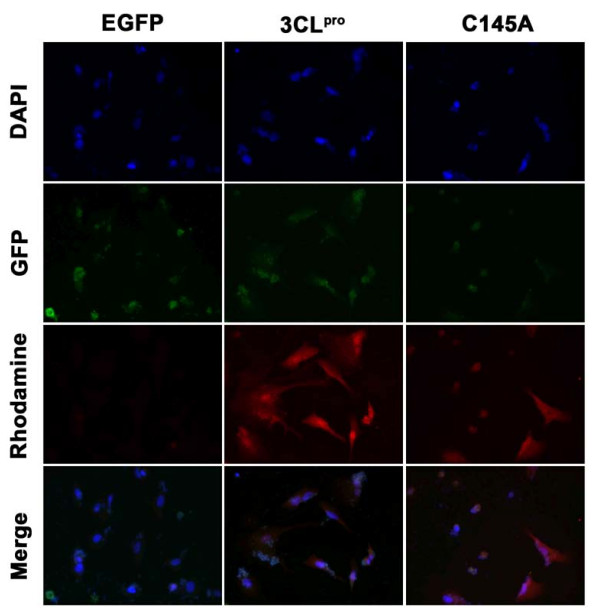
The plasmids of pEGFP-C3, pEGFP-3CLpro, and pEGFP-C145A were successfully transfected into A549 cells respectively. The expression levels of 3CLpro and C145A mRNA were determined by reverse transcription (RT)-PCR and the protein expression levels were analyzed using 2D western blots (Figure 2A and 2B). The results indicated that the expression of 3CLpro and C145A mRNA reached the highest level in A549 cells at 12 to 24 hrs and slightly decreased their expression at 48 h after transfection. However, 2D western blots results indicated that the protein expression levels of 3CLpro and C145A steadily increased with time up to 48 hrs after transfection. To track the locations of 3CLpro and C145A proteins in transfected A549 cells, Myc-tagged 3CLpro and C145A were detected using rhodamine-conjugated anti-Myc antibodies. The immunofluorescence microscopic images demonstrated 60-70% transfection efficiency (data not shown) and the localizations of c-myc tagged 3CLpro and C145A were detected both in the cytoplasm and nucleus of transfected A549 cells compared to control EGFP (Figure 3).
Figure 2.
RT-PCR and 2-D western blot analysis of EGFP, 3CLpro, and C145A transfected A549 cell line. (A) RT-PCR analysis of 3CLpro and GM-CSF genes expression at 12, 24, and 48 hrs in A549 cells which were transfected with EGFP, 3CLpro, and C145A respectively. The relative abundance compared with the expression level of GAPDH is depicted in the bar chart below. (B) 2-D western blots of 3CLpro and C145A expression patterns in A549 cells. A549 cells transfected with EGFP, 3CLpro and C145A were analyzed at 24 and 48 hrs after transfection. Proteins were resolved by 2-D gels and electroblotted onto PVDF membranes. Western blots were probed with anti-3CLpro specific antibody. The circle indicates the site of 3CLpro.
Figure 3.
Nuclear and cytoplasmic localization of 3CLpro and C145A in transfected A549 cells. EGFP, 3CLpro, and C145A transfected A549 cells were collected at 24 hrs after transfection. Cells were fixed and stained as described in Methods. Nuclei were stained with DAPI (blue fluorescence, obtained from the excitation of 359 nm and emission of 468 nm.). GFP panel shows the green fluorescence obtained from the excitation of 488 nm and emission of 509 nm. 3CLpro and C145A were detected with a rhodamine-conjugated secondary antibody (red fluorescence, obtained from the excitation of 496 nm and emission of 520 nm).
The expression of GM-CSF in SARS-3CLpro transfected A549 cells by semi-quantification RT-MPCR
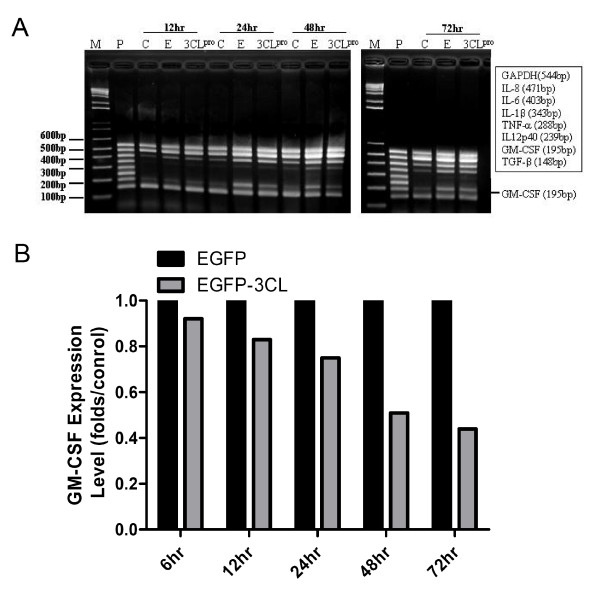
Total RNA from A549 cells (C), EGFP (E) and pEGFP-3CLpro (3CLpro) transfected cells were collected at 12, 24, 48, and 72 hrs. As shown in Figure 4, expression of GM-CSF was demonstrated in 3CLpro transfected cells, A549 cells and EGFP transfected cells (Figures 2A and 4A) by Multiplex PCR kits. The decreased percentage was observed in EGFP-3CLpro transfected A549 cells compared to the EGFP transfected A549 cells at 6, 12, 24, 48, and 72 hrs is 9%, 17%, 25%, 49%, and 56%, respectively. There was significant decrease of GM-CSF in 3CLpro transfected A549 cells after 24 hrs (p < 0.005). Additionally, there were no change to the mRNA expression of IL-1β, IL-6, IL-8, IL12p40, TNF-α, and TGF-β found in 3CLpro transfected cells. To confirm the mRNA expression of GM-CSF, other primer pairs were used to amplify GM-CSF by semi-quantification RT-MPCR. As shown in Figure 4B, decreased expression of GM-CSF was noted in 3CLpro transfected cells compared to GAPDH in A549 cells and EGFP transfected cells at 24, 48, and 72 hrs. A significant decrease of GM-CSF was observed in 3CLpro transfected A549 cells after 24, 48, and 72 hrs with maximal reduction folds of vector control about 0.75, 0.53 and 0.45, respectively (p < 0.005).
Figure 4.
Expression of various cytokines and chemokines in 3CLpro transfected A549 cells using RT-PCR. (A) Total RNAs were harvested at 12, 24, 48 and 72 hrs. RT-PCR was performed to detect the expression of various cytokines and chemokines including IL-1β (343 bp), IL-6 (403 bp), IL-8 (471 bp), IL12p40 (239 bp), TNF-α (288 bp), TGF-β (148 bp), GM-CSF (195 bp). GAPDH was used as control. A549 cells were used as the background. (B) Expression of GM-CSF in 3CLpro transfected A549 cells using RT-PCR. Total RNA was harvested at 6, 12, 24, 48 and 72 hrs. RT-PCR was performed to detect the mRNA expression of GM-CSF (277 bp). GAPDH was used to normalize the cell number (307 bp). There were marked decreases of GM-CSF in 3CLpro transfected A549 cells after 24 hrs (*; p < 0.05). A549 cells were used as the background. C, A549 cells; E, EGFP transfected cells; 3CLpro, 3CLpro transfected cells.
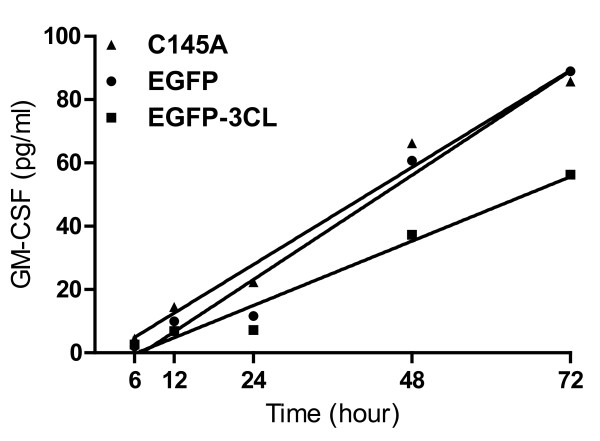
The secretion of GM-CSF in SARS-3CLpro transfected A549 cells by ELISA
As shown in Figure 5, the values of GM-CSF concentration in the culture mediums of EGFP transfected A549 cells at 6, 12, 24, 48, and 72 hrs were 2.047 ± 0.2(pg/ml), 9.928 ± 0.13(pg/ml), 11.657 ±.15 (pg/ml), 60.714 ±.0.18 (pg/ml), 88.926 ±.0.31 (pg/ml), respectively. The values of GM-CSF concentration in the culture mediums of 3CLpro transfected A549 cells at 6, 12, 24, 48, and 72 hrs were 2.624 ± 0.13(pg/ml), 6.852 ± 0.17(pg/ml), 7.237 ±.0.11 (pg/ml), 37.313 ±.0.18 (pg/ml), 56.25 ±.0.31 (pg/ml), respectively. The values of GM-CSF concentration in the culture mediums of C145A transfected A549 cells at 6, 12, 24, 48, and 72 hrs were 4.431 ± 0.11(pg/ml), 14.424 ± 0.21(pg/ml), 22.281 ±.0.54 (pg/ml), 66.235 ±.0.68 (pg/ml), 85.715 ±.0.43 (pg/ml), respectively. A significant decrease of GM-CSF was observed in 3CLpro transfected A549 cells compared to the EGFP-transfected and C145A-transfected A549 cells by an analysis of covariance (p = 0.0204 and 0.0236 respectively).
Figure 5.
Decreased secretion of GM-CSF in SARS-CoV-3CLpro transfected cells. A549 cells were transfected with EGFP or 3CLpro or C145A plasmids. The supernatants were collected from cell cultures at 6, 12, 24, 48 and 72 hrs and ELISA was performed to evaluate the amount of secreted GM-CSF. The supernatant from the cell culture of A549 cells was used as a control. A significant decrease of GM-CSF was observed in 3CLpro or C145A transfected A549 cells (p = 0.0204 and 0.0236 respectively).
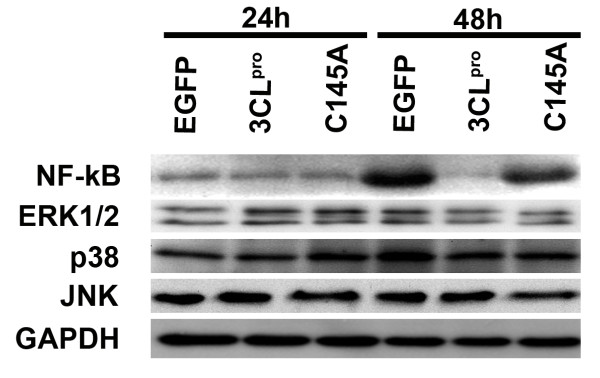
Involvement of NF-κB in 3CLpro-mediated GM-CSF expression in A549 cells
To investigate whether ERK1/2, p38, and JNK MAPK pathways and NF-κB were involved in 3CLpro-mediated GM-CSF production in A549 cells [25,26], we examined the expression levels of these MAP kinases and NF-κB by western blot analysis. As shown in Figure 6 (a bar graph including mean and standard deviation were provided in Additional File 1), the expression of NF-κB was reduced in 3CLpro transfected cells at 48 hrs after transfection compared to cells transfected with EGFP. In contrast, over expression of 3CLpro did not change the expression levels of ERK1/2, p38, and JNK in A549 cells significantly.
Figure 6.
Expression of NF-kB and MAPKs in EGFP, 3CLpro, and C145A tranfected A549 cells. A549 cells tranfected with EGFP, 3CLpro, and C145A were harvested at 24 and 48 hrs as indicated above and total cell lysate were resolved in 12.5% SDS-PAGE. Antibodies used in western blotting are indicated in left panel.
Discussion
Severe acute respiratory syndrome coronavirus (SARS-CoV) causes a severe lung infection [18-20]. It is important to elucidate the mechanism of the lung fibrosis caused by SARS-CoV. The expression of 3CLpro and C145A in A549 cells was established (Figures 2 and 3) to elucidate the pathogenesis of SARS-CoV in lung epithelial cells. The localization patterns of 3CLpro and C145A were observed both in the cytoplasm and nucleus of A549 cells by immunofluorescence (Figure 3). Decreased expression and levels of GM-CSF were found in 3CLpro-transfected cells using RT-PCR, but not in C145A transfected cells (Figure 2A). Down-regulation of GM-CSF expression by 3CLpro in A549 cells may be attributed in part to the reduced protein level of NF-kB (Figure 6). In addition, there were no significant decreases in the mRNA expression of IL-1β, IL-6, IL-8, IL12p40, TNF-α, TGF-β using RT-MPCR. This finding is consistent with that of Ziegler et al [27] who reported no induction of cytokines genes (IFN-α, IFN-β, IL-28, IL-29, TNF-α, CCLl5 or CXCL10) or IFN-α/β-induced MxA gene in SARS-CoV-infected A549 cells.
GM-CSF was originally defined by its ability to generate in vitro granulocyte and macrophage colonies from bone marrow precursor cells [28]. Apart from its physiological role in the control of alveolar macrophage development, it now appears more likely that its major role lies in its ability to govern the properties of the more mature myeloid cells of the granulocyte and macrophage lineages, particularly during host defense and inflammatory reactions. Recent evidence for the key role of GM-CSF in inflammatory and autoimmune diseases is summarized [29]. GM-CSF-deficient mice showed an increased susceptibility to pulmonary infections [30,31]. In addition, some studies also demonstrated that impaired production of GM-CSF played a role in the development of bleomycin-induced pulmonary fibrosis [32,33]. Together, there is growing evidence suggesting that GM-CSF is an important regulator of the alveolar epithelium, surfactant homeostasis, and lung host defense [31,34]. To support the above, our experimental results show decreased expression and secretion of GM-CSF in 3CLpro-transfected cells, and suggest that SARS-3CLpro will affect host defense.
Due to the functional importance of SARS-CoV 3CLpro in proteolytic recognition and processing of the virus polyproteins, the experimental approaches and computational prediction of substrate cleavage sites of 3CLpro become attractive topics in anti-SARS drug design [8-11,15,35]. It is clear that the 3CLpro cleavage sites are consistent preferentially with relative position of P1'-Ser/Ala P1-Gln P2-Leu [35,36]. In this study, 3CLpro transfected A549 cells decreased the protein levels of NF-kB with subsequent decline in GM-CSF mRNA and protein expression (Figures 4-6). The amino acid sequence analyses of NF-kB indicated that non-canonical 3CLpro cleavage sites (such as P1'-Tyr/Gln/Arg/Ile P1-Gln P2-Leu) commonly existed in NF-kB. Interestingly, NF-kB are associated in GM-CSF production in lung cancer cell lines [37]. In depth investigations are required to determine whether 3CLpro directly or indirectly affects the protein levels of NF-kB in transfected A549 cell.
Conclusions
The role of the viral cytopathic effect in the pathogenesis of SARS-CoV-associated lung diseases involvement remains unclear. Our results reveals that the 3CLpro -induced decreased GM-CSF herein may provide further information on the pathogenesis of lung diseases with associated SARS-CoV. Also, the protein levels of NF-kB decreased in 3CLpro transfected cells. Thus, we conclude that 3CLpro may mediate down-regulation of NF-kB production and subsequently suppress GM-CSF mRNA and protein expression in transfected A549 cells. Therefore, the SARS-CoV 3CLpro may play a role in the mechanism of lung fibrosis of SARS through the suppression of GM-CSF.
Methods
Plasmids
Plasmid pEGFP-C3 was obtained from CLONTECH (CLONTECH Laboratories, Palo Alto, CA, USA). This plasmid contains the EGFP variant and neomycin resistant genes under the control of the cytomegalovirus early gene promoter and the SV40 early gene promoter, respectively. Full-length 3CLpro versions (wild type and mutant, C145A) and CMV promoter were amplified by PCR using primers containing suitable restriction sites for subcloning into the pEGFP-C3 vector (3CLpro primers: 5' ATATGGTACCAgTGGTTTTAGGAAAATGGCATTC 3'/Kpn I, 5' GAGATGAGTTTCTGCTCTTGGAAGGTAACACCAGAGCATT 3', and ATATGGATCCCAGATCCTCTTCTGAGATGAGTTTCTGCTCTTGGAA 3'/BamH I; CMV promoter primers: 5' ATATCTCGAGTAGTTATTAATAGTAATCAATTAC 3'/XhoI and 5' ATATAAGCTTTAGCGCTAGCGGATCTGA 3'/Hind III). The PCR was performed with reagents containing a 0.2 μM primers mixture, 1.25 μM dNTP mixture, 1.5 μM MgCl2, 10 ng templates, and 2.5 U DNA polymerase (Takara, Tokyo, Japan). Amplified DNA fragments were then ligated into the cloning site of the pEGFP-C3 vector (EGFP) and transformed into Escherichia coli DH5α competent cells, which were obtained from Life Technologies (Carlsbad, California, USA). Restriction enzyme digestion, PCR and DNA sequencing analysis (Mission Biotech, Taipei, Taiwan) were used to verify the plasmids.
Cell culture and Transfection
The human lung carcinoma A549 cells were obtained from American type culture collection (ATCC) (Manassas, VA, USA) and were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (GIBCO-BRL, Carlsbad, California, USA) at 37°C in a 5% CO2 incubator. A total of 1 × 106 cells were grown to 70% confluence in 100 mm2 culture plates before transfection. The transfection reaction was performed by using Lipofectamine plus reagents (Invitrogen, California, USA) with 2 μg of each plasmid, pEGFP-C3, pEGFP-3CLpro, and pEGFP-C145A according to the manufacturer's instructions. The cells were then cultured in serum-free DMEM for 12 hrs at 37°C in a 5% CO2 incubator and subsequently in DMEM with 10% FBS.
Immunofluorescence cell staining and Fluorescence Microscopy
EGFP, 3CLpro and C145A transfected A549 cells were cultured on sterile cover slides for overnight and fixed by methanol. Samples were blocked with 10% blocking serum and incubated with 3CLpro antibody (Abcam Inc., Cambridge, MA) and rhodamine conjugated second antibody (Invitrogen Corporation; Grand Island, NY) separately. Nuclei were stained with DAPI (4',6-diamidino-2-phenylindole, Sigma, St. Louis, MO). All transfected A549 cells were observed with a Zeiss Axioplan-2 epifluorescence microscope equipped with appropriate fluorescence filters. Digital images of the cells were recorded by using a spot camera system.
mRNA analysis and RT-MPCR
All studies were carried out in a designated PCR-clean area. After treatments at 12-72 hrs, cell pellets were washed in DPBS twice and treated with Trizol mRNA lysis buffer for total RNA extraction. All samples were analyzed by reverse transcriptase Multiplex PCR (RT-MPCR) with messageScreenTM Human Inflammatory Cytokine Set 1 Multiplex PCR kits (Biosource, International, Inc., CA, USA) for a temporal and spatial distribution of mRNA expression of different cytokines. This method is an accurate and valid system to detect multiple gene expression by amplifying all the genes under the same conditions. The PCR derived DNA fragments, obtained by 25 PCR cycles, and was subjected to electrophoresis in a 1.7% agarose gel. The cDNAs encoding human granulocyte-macrophage-colony-stimulating factor (GM-CSF) and GAPDH were amplified by RT-PCR using the following primer pairs: 5'-TGAGTAGAGACACTGCTGCTG-3' (forward primer for GM-CSF cDNAs), 5'-TCAAAGGGGATGACAAGCAGAA-3' (reverse primer for GM-CSF cDNAs), 5'-CATGTTCGTCATGGGTGTGA-3' (forward primer for GAPDH cDNAs), and 5'-AGTGAGCTTCCCGTTCAGCTC-3' (reverse primer for GAPDH cDNAs). The amplification was performed in a 50 μl reaction volume containing 1× reaction buffer (Promega, Madison, Wisconsin, USA), 1.5 μM of MgCl2, 200 μM of dNTPs, 1 μM of each primer and 2.5 units of Taq DNA polymerase (Promega, Madison, Wisconsin, USA) using a Perkin-Elmer Gene Amp PCR system 2400. Each cycle consists of denaturation at 95°C for 1 min, annealing at 55°C for 45 sec, and amplification at 72°C for 45 sec. The RT-PCR derived DNA fragments obtained by 30 PCR cycles were subjected to electrophoresis on a 1.7% agarose gel. Following staining with ethidium bromide, the gels were photographed and band intensity was measured under UV light using the Alphaimager 2200 (Alphalnnotech, San Leandro, CA, USA). The specific RNA level of every sample was expressed as the product's intensity. cDNA encoding glyceraldehyde- 3-phosphate dehydrogenase (GAPDH) was quantified and used to normalize the expression of GA-CSF and each cytokine for each sample.
Cytokine ELISA
The quantification of cytokine levels from the cell culture mediums was performed by duplication using ELISA kits for GM-CSF (Biosource International, Inc, CA, USA), according to the manufacturer's instructions. In each case, the optical density of known standards was used to construct a calibration curve and the mean cytokine values ± SD were then calculated for each sample.
Statistical analysis
Statistical analysis was performed using the paired t-test. A P value less than 0.05 was considered significant. GraphPad Prism software v5.01 (GraphPad Software Inc., USA) was employed for the analysis of covariance by comparing linear regression lines. Because the slopes differ so much, it is not possible to test whether the intercepts differ significantly.
2-DE
The 2-DE procedure was performed as described elsewhere with minor modification [38]. Briefly, 100 μg of protein was loaded onto IPG strips (18 cm, pH 3-10, GE Healthcare) for analytic 2-DE analysis. The IPG strips were subsequently rehydrated on the IPGphor IEF system (GE Healthcare, Uppsala, Sweden) at 20°C and 30 V for 14 h. Subsequently, IEF was conducted at 8000 V for 56 kVh for analytic analysis. Strips were then treated with equilibration buffer containing dithiothreitol (Sigma, St. Louis, MO) and then alkylated with iodoacetate (Sigma, St. Louis, MO) for 15 min. The IPG strips were transferred onto 10-18% linear gradient polyacrylamide gels (18×18 cm) and the electrophoresis was performed at 45 mA per gel at 4°C.
Western blot
Proteins that had been resolved on 12.5% SDS-PAGE or 2-DE gels were electroblotted onto PVDF membranes using a semi-dry apparatus (GE Healthcare, Uppsala, Sweden). The membranes were blocked for 1 h with 5% non-fat milk and 0.1% Tween-20 in PBS, followed by incubation with primary antibody (3CLpro, NF-kB, ERK1/2, JNK, p38, GAPDH, Abcam Inc., Cambridge, MA) for 2 h at room temperature. After washing three times with PBS containing 0.05% Tween-20 (PBST), the primary antibody was detected with an anti-rabbit horseradish peroxidase-conjugated secondary antibody (Abcam Inc., Cambridge, MA) for 1 h at room temperature. Signals were developed with an enhanced chemiluminescence western blotting detection system (PerkinElmer, Boston, MA,).
List of abbreviations
SARS-CoV: Severe acute respiratory syndrome coronavirus; 3CLpro: 3C-like protease; GM-CSF: human granulocyte macrophage colony stimulating factor; ELISA: Enzyme-linked immunosorbent assay; RT-PCR: reverse transcription polymerase chain reaction; IL-1β: interleukin 1 beta; IL-6: interleukin 6; IL-8: interleukin 8; TNFα: tumour necrosis factor α; IL-12p40: interleukin 12 p40 subunit; TGF-β: transforming growth factor β; ERK1/2: extracellular-signal-regulated kinases 1 and 2; NF-kB: nuclear factor kappa B; JNK: c-Jun N-terminal kinases; p38 MAPK: P38 mitogen-activated protein kinases; EGFP: enhanced green fluorescent protein; GAPDH: Glyceraldehyde 3-phosphate dehydrogenase.
Authors' contributions
STC, GJT, and SLC participated in the design of study. MCMC, HYT, HHL, YCW, CHC, and TCH performed the experiments, collected and analyzed data. GJT, SLC, JL and MCMC wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
A bar graph of NF-kB and MAPKs expression in EGFP, 3CLpro, and C145A tranfected A549 cells. The relative expression level of NF-kB and MAPKs in EGFP, 3CLpro, and C145A tranfected A549 cells at 24 and 48 hrs in Figure 6 are compared with GAPDH and depicted in the bar graph including mean and standard deviation.
Contributor Information
Hsien-Hua Liao, Email: lhhy011@ms1.hinet.net.
Yao-Chen Wang, Email: wang5717@ms21.hinet.net.
Miles Chih-Ming Chen, Email: miles@gate.sinica.edu.tw.
Hsien-Yu Tsai, Email: suki@gate.sinica.edu.tw.
Johnson Lin, Email: linj@ukzn.ac.za.
Shui-Tein Chen, Email: bcchen@gate.sinica.edu.tw.
Gregory Jiazer Tsay, Email: gjt@csmu.edu.tw.
Sun-Long Cheng, Email: chuangsl@ms57.hinet.net.
Acknowledgements
This study was supported by grants 92-2751-B-040-002-Y from the National Science Council, Taiwan, R.O.C. We thank Dr. Liang, Po-Hung for generous contributions of 3CLpro and C145A mutant clone.
References
- Fouchier RA, Kuiken T, Schutten M, van Amerongen G, van Doornum GJ, van den Hoogen BG, Peiris M, Lim W, Stohr K, Osterhaus AD. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Penaranda S, Bankamp B, Maher K, Chen MH. et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–9. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W. et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–66. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Marra MA, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YS, Khattra J, Asano JK, Barber SA, Chan SY. et al. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Graham RL, Sparks JS, Eckerle LD, Sims AC, Denison MR. SARS coronavirus replicase proteins in pathogenesis. Virus Res. 2008;133:88–100. doi: 10.1016/j.virusres.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice E, McAuliffe J, Lu X, Subbarao K, Denison MR. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J Virol. 2004;78:9977–86. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MF, Kuo CJ, Chang KT, Chang HC, Chou CC, Ko TP, Shr HL, Chang GG, Wang AH, Liang PH. Mechanism of the maturation process of SARS-CoV 3CL protease. J Biol Chem. 2005;280:31257–66. doi: 10.1074/jbc.M502577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Luo H, Yu C, Sun T, Chen J, Peng S, Qin J, Shen J, Yang Y, Xie Y. et al. Molecular cloning, expression, purification, and mass spectrometric characterization of 3C-like protease of SARS coronavirus. Protein Expr Purif. 2003;32:302–8. doi: 10.1016/j.pep.2003.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan K, Wei P, Feng Q, Chen S, Huang C, Ma L, Lai B, Pei J, Liu Y, Chen J. et al. Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. J Biol Chem. 2004;279:1637–42. doi: 10.1074/jbc.M310875200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–7. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Huang C, Wei P, Fan K, Liu Y, Lai L. 3C-like proteinase from SARS coronavirus catalyzes substrate hydrolysis by a general base mechanism. Biochemistry. 2004;43:4568–74. doi: 10.1021/bi036022q. [DOI] [PubMed] [Google Scholar]
- Lin CW, Lin KH, Hsieh TH, Shiu SY, Li JY. Severe acute respiratory syndrome coronavirus 3C-like protease-induced apoptosis. FEMS Immunol Med Microbiol. 2006;46:375–80. doi: 10.1111/j.1574-695X.2006.00045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CW, Tsai FJ, Wan L, Lai CC, Lin KH, Hsieh TH, Shiu SY, Li JY. Binding interaction of SARS coronavirus 3CL(pro) protease with vacuolar-H+ ATPase G1 subunit. FEBS Lett. 2005;579:6089–94. doi: 10.1016/j.febslet.2005.09.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiemer L, Lund O, Brunak S, Blom N. Coronavirus 3CLpro proteinase cleavage sites: possible relevance to SARS virus pathology. BMC Bioinformatics. 2004;5:72. doi: 10.1186/1471-2105-5-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls JM, Poon LL, Lee KC, Ng WF, Lai ST, Leung CY, Chu CM, Hui PK, Mak KL, Lim W. et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–8. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CH, Liu CY, Wan YL, Chou CL, Huang KH, Lin HC, Lin SM, Lin TY, Chung KF, Kuo HP. Persistence of lung inflammation and lung cytokines with high-resolution CT abnormalities during recovery from SARS. Respir Res. 2005;6:42. doi: 10.1186/1465-9921-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo AW, Tang NL, To KF. How the SARS coronavirus causes disease: host or organism? J Pathol. 2006;208:142–51. doi: 10.1002/path.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CK, Lam CW, Wu AK, Ip WK, Lee NL, Chan IH, Lit LC, Hui DS, Chan MH, Chung SS. et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M. SARS Immunity and Vaccination. Cell Mol Immunol. 2004;1:193–8. [PubMed] [Google Scholar]
- Xu X, Gao X. Immunological responses against SARS-coronavirus infection in humans. Cell Mol Immunol. 2004;1:119–22. [PubMed] [Google Scholar]
- Cheung CY, Poon LL, Ng IH, Luk W, Sia SF, Wu MH, Chan KH, Yuen KY, Gordon S, Guan Y. et al. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J Virol. 2005;79:7819–26. doi: 10.1128/JVI.79.12.7819-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Xu J, Zhou C, Wu Z, Zhong S, Liu J, Luo W, Chen T, Qin Q, Deng P. Characterization of cytokine/chemokine profiles of severe acute respiratory syndrome. Am J Respir Crit Care Med. 2005;171:850–7. doi: 10.1164/rccm.200407-857OC. [DOI] [PubMed] [Google Scholar]
- Lin YS, Lin CF, Fang YT, Kuo YM, Liao PC, Yeh TM, Hwa KY, Shieh CC, Yen JH, Wang HJ. et al. Antibody to severe acute respiratory syndrome (SARS)-associated coronavirus spike protein domain 2 cross-reacts with lung epithelial cells and causes cytotoxicity. Clin Exp Immunol. 2005;141:500–8. doi: 10.1111/j.1365-2249.2005.02864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Huang YH, Chuang YH, Peng CM, Wang LC, Lin YT, Chiang BL. Autoantibodies against human epithelial cells and endothelial cells after severe acute respiratory syndrome (SARS)-associated coronavirus infection. J Med Virol. 2005;77:1–7. doi: 10.1002/jmv.20407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griego SD, Weston CB, Adams JL, Tal-Singer R, Dillon SB. Role of p38 mitogen-activated protein kinase in rhinovirus-induced cytokine production by bronchial epithelial cells. J Immunol. 2000;165:5211–20. doi: 10.4049/jimmunol.165.9.5211. [DOI] [PubMed] [Google Scholar]
- Munoz C, Pascual-Salcedo D, Castellanos MC, Alfranca A, Aragones J, Vara A, Redondo JM, de Landazuri MO. Pyrrolidine dithiocarbamate inhibits the production of interleukin-6, interleukin-8, and granulocyte-macrophage colony-stimulating factor by human endothelial cells in response to inflammatory mediators: modulation of NF-kappa B and AP-1 transcription factors activity. Blood. 1996;88:3482–90. [PubMed] [Google Scholar]
- Ziegler T, Matikainen S, Ronkko E, Osterlund P, Sillanpaa M, Siren J, Fagerlund R, Immonen M, Melen K, Julkunen I. Severe acute respiratory syndrome coronavirus fails to activate cytokine-mediated innate immune responses in cultured human monocyte-derived dendritic cells. J Virol. 2005;79:13800–5. doi: 10.1128/JVI.79.21.13800-13805.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltman AM, van der Kooij SW, Coffer PJ, Offringa R, Daha MR, van Kooten C. Rapamycin specifically interferes with GM-CSF signaling in human dendritic cells, leading to apoptosis via increased p27KIP1 expression. Blood. 2003;101:1439–45. doi: 10.1182/blood-2002-06-1688. [DOI] [PubMed] [Google Scholar]
- Eager R, Nemunaitis J. GM-CSF gene-transduced tumor vaccines. Mol Ther. 2005;12:18–27. doi: 10.1016/j.ymthe.2005.02.012. [DOI] [PubMed] [Google Scholar]
- LeVine AM, Reed JA, Kurak KE, Cianciolo E, Whitsett JA. GM-CSF-deficient mice are susceptible to pulmonary group B streptococcal infection. J Clin Invest. 1999;103:563–9. doi: 10.1172/JCI5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell BC, Whitsett JA. Gm-CSF regulates pulmonary surfactant homeostasis and alveolar macrophage-mediated innate host defense. Annu Rev Physiol. 2002;64:775–802. doi: 10.1146/annurev.physiol.64.090601.113847. [DOI] [PubMed] [Google Scholar]
- Moore BB, Coffey MJ, Christensen P, Sitterding S, Ngan R, Wilke CA, McDonald R, Phare SM, Peters-Golden M, Paine R. et al. GM-CSF regulates bleomycin-induced pulmonary fibrosis via a prostaglandin-dependent mechanism. J Immunol. 2000;165:4032–9. doi: 10.4049/jimmunol.165.7.4032. [DOI] [PubMed] [Google Scholar]
- Christensen PJ, Bailie MB, Goodman RE, O'Brien AD, Toews GB, Paine R. Role of diminished epithelial GM-CSF in the pathogenesis of bleomycin-induced pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2000;279:L487–95. doi: 10.1152/ajplung.2000.279.3.L487. [DOI] [PubMed] [Google Scholar]
- Trapnell BC, Whitsett JA, Nakata K. Pulmonary alveolar proteinosis. N Engl J Med. 2003;349:2527–39. doi: 10.1056/NEJMra023226. [DOI] [PubMed] [Google Scholar]
- Kiemer L, Lund O, Brunak S, Blom N. Coronavirus 3CLpro proteinase cleavage sites: Possible relevance to SARS virus pathology. BMC Bioinformatics. 2004;5:72. doi: 10.1186/1471-2105-5-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegyi A, Ziebuhr J. Conservation of substrate specificities among coronavirus main proteases. J Gen Virol. 2002;83:595–599. doi: 10.1099/0022-1317-83-3-595. [DOI] [PubMed] [Google Scholar]
- Hideshi N, Yoshiki U, Makoto K, Ryoji H, Hirokuni T. Cyclooxygenase-2 inhibitor NS-398 suppresses cell growth and constitutive production of granulocyte-colony stimulating factor and granulocyte macrophage-colony stimulating factor in lung cancer cells. Cancer Science. 2003;94:173–180. doi: 10.1111/j.1349-7006.2003.tb01415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongsachart P, Huang-Liu R, Sinchaikul S, Pan FM, Phutrakul S, Chuang YM, Yu CJ, Chen ST. Glycoproteomic analysis of WGA-bound glycoprotein biomarkers in sera from patients with lung adenocarcinoma. Electrophoresis. 2009;30:1206–20. doi: 10.1002/elps.200800405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A bar graph of NF-kB and MAPKs expression in EGFP, 3CLpro, and C145A tranfected A549 cells. The relative expression level of NF-kB and MAPKs in EGFP, 3CLpro, and C145A tranfected A549 cells at 24 and 48 hrs in Figure 6 are compared with GAPDH and depicted in the bar graph including mean and standard deviation.