Abstract
Imaging of cell nuclei plays a critical role in cancer diagnosis and prognosis. In order to image noninvasively cell nuclei in vivo without staining, we developed ultraviolet photoacoustic microscopy (UV-PAM), in which 266-nm-wavelength ultraviolet light excites unlabeled DNA and RNA in cell nuclei to produce photoacoustic waves. We applied UV-PAM to ex vivo imaging of cell nuclei in a mouse lip and a mouse small intestine, and to in vivo imaging of the cell nuclei in the mouse skin. The UV-PAM images of unstained cell nuclei match the optical micrographs of the histologically stained cell nuclei. Given intrinsic optical contrast and high spatial resolution, in vivo label-free UV-PAM has potential for unique biological and clinical application.
Cell nuclei are organelles containing the DNA genome, in which major cell activities take place, such as DNA replication, RNA synthesis, and ribosome assembly. Since cancer cells lose their control of DNA replication, their nuclei are different from normal nuclei in morphology. For example, two morphological characteristics of the nuclei in cancer cells are their folded shape and enlarged size [1]. These characteristics are considered the hallmarks of cancer and are used by pathologists to determine cancer grade and evaluate prognosis [2]. Therefore, imaging of cell nuclei plays a critical role in cancer diagnosis. Classical imaging of cell nuclei in excised tissue goes through a complicated histological process—including tissue processing, embedding, sectioning, and staining before imaging with microscopy. Without sectioning, cell nuclei in tissue specimens have been imaged successfully by reflectance confocal microscopy [3], fluorescence confocal microscopy [4] and multiphoton microscopy [5]. However, confocal microscopy requires staining cell nuclei by citric acid, acridine orange, or methylene blue, and multiphoton microscopy provides nuclear images with perinuclear fluorescent speckles. Therefore, imaging of cell nuclei still falls short of a label-free imaging technique with high contrast and spatial resolution.
Photoacoustic microscopy (PAM) is a new imaging technique with rich optical contrast and high spatial resolution [6]. In PAM, a pulsed laser beam is focused into biological tissue. Once the light pulse is absorbed by the tissue and converted into heat, thermoelastic expansion of the tissue generates ultrasonic waves, which are detected by a focused ultrasonic transducer. By using light absorbed by the hemoglobin in red blood cells, PAM is capable of imaging the vasculature in various tissues, such as the human skin and the mouse brain [6,7]. Furthermore, PAM is capable of mapping the total concentration and the oxygen saturation of hemoglobin in single blood vessels by using two or more optical wavelengths in the visible spectral region [6,7]. So far, PAM has been developed to image single red blood cells [8], but has not been able to image cell nuclei.
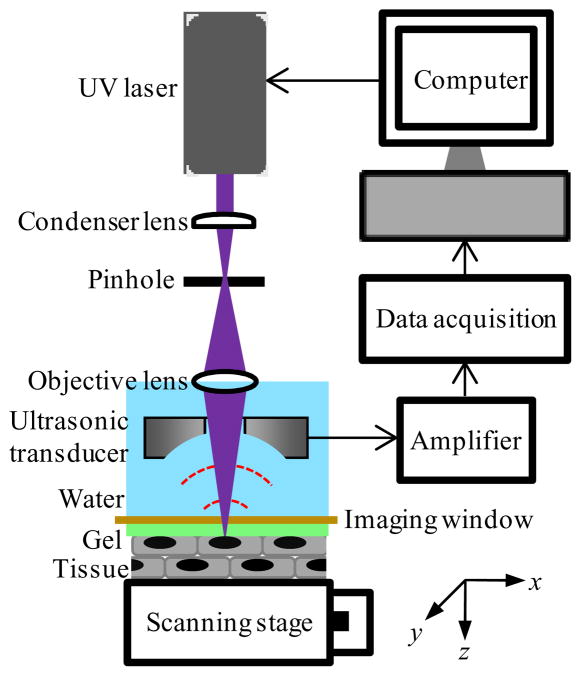
We have developed a special PAM called ultraviolet photoacoustic microscopy (UV-PAM) for imaging of cell nuclei in intact biological tissue. In UV-PAM, ultraviolet light (UV) is used in place of visible light. DNA and RNA—two major compounds in cell nuclei—strongly absorb UV light around a wavelength of 260 nm [9]. In contrast, the UV absorption of protein and lipids is weaker than that of DNA and RNA by one order of magnitude around 260 nm [9,10]. To take advantage of this high intrinsic absorption contrast of DNA and RNA, our UV-PAM system (Fig. 1) employs light at 266 nm emitted by a Nd:YLF Q-switched UV laser (QL266-010-O, Crystalaser; pulse width, 7 ns). The laser beam is spatially filtered by a 25-μm-diameter pinhole (910PH-25, Newport) and focused into a water tank by a water-immersion objective lens (LB4280, Thorlabs). Subsequently, the beam passes through a ring-shaped focused ultrasonic transducer (center frequency, 50 MHz; focal length, 7 mm) and penetrates a 25 μm thick polyethylene membrane before the beam focuses on the object to be imaged. The polyethylene membrane seals the bottom of the water tank to form an imaging window while maintaining acoustic coupling. The laser pulse energy behind the membrane is measured to be 35 nJ. The ultrasonic transducer is in coaxial alignment with the objective lens to a common focus. The specimen is mounted on a two-dimensional scanning stage with a minimal scan step size of 0.31 μm. Time-resolved photoacoustic signals are detected by the ultrasonic transducer during raster scanning to reconstruct tomographic images, which can be rendered in various forms such as cross sectional images and maximum amplitude projection (MAP) images.
Fig. 1.
Schematic of the ultraviolet photoacoustic microscopy system.
We imaged single sub-micron beads to measure the spatial resolution of the UV-PAM system. The beads were black polystyrene microspheres 0.2 μm in diameter (Polysciences), immobilized by adsorption on a quartz slide (Chemglass Life Sciences). Bead images were acquired by scanning with a 0.31 μm step size. We measured both the lateral full width at half maximum (FWHM) by fitting the Airy pattern to the amplitude profiles of the horizontal cross-sectional images of the single beads and the axial FWHM by Gaussian fitting to the axial amplitude profiles of bead images. By fitting the images of 15 beads, the lateral FWHM was found to be 0.70 ± 0.04 μm (mean ± standard error), and the axial FWHM to be 28.5 ± 0.8 μm.
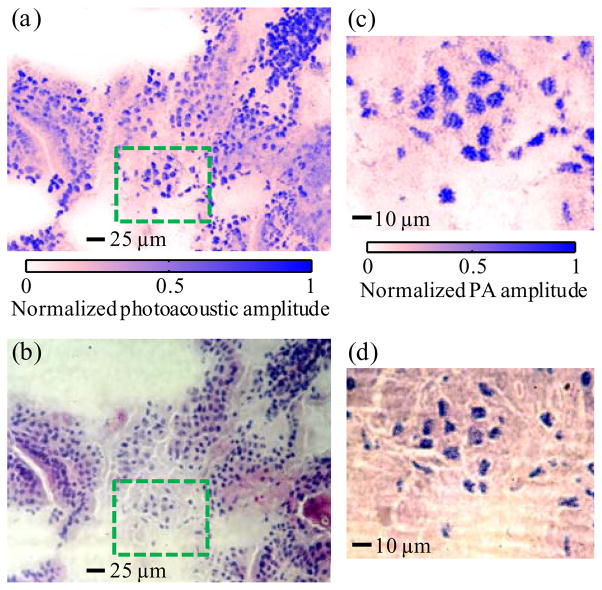
Then, we imaged cross sections of a mouse small intestine and compared the UV-PAM images of the sections with their histological micrographs. All experimental animal procedures were carried out in conformity with the laboratory animal protocol approved by the Animal Studies Committee of Washington University in St. Louis. The small intestine was excised from a sacrificed Swiss Webster mouse (Harlan Laboratories), and cut into 6 μm thick cross sections by a cryostat (CM1850; Leica Microsystems). We acquired a UV-PAM image of the cross section of the small intestine by scanning with a 0.62 μm step size (Fig. 2a). After the scanning, the section slide was stained by hematoxylin and eosin (Sigma-Aldrich). Hematoxylin stains the cell nucleus blue, and eosin stains the cytoplasm pink. Next, optical micrographs of the intestine section were obtained by using a microscope with a 20× objective (0.45 NA; Nikon). The cell nuclei are shown as dark-blue spots in Fig. 2b as expected in histology. Corresponding to each dark-blue spot in the histologic Fig. 2b, there is a light-blue spot in Fig. 2a, confirming that cell nuclei are imaged by UV-PAM. Because UV-PAM possesses background-free detection, the cell nuclei are shown in high positive contrast due to both the strong UV absorption of DNA and RNA and the weak absorption of cytoplasm. The signal-to-noise ratio was as high as 52 dB. In addition, Fig. 2a discloses the same spatial distribution of cell nuclei as Fig. 2b does. Furthermore, UV-PAM was able to resolve the shape and size of the cell nuclei as shown in a close-up in Fig. 2c. Figure 2d shows an optical micrograph of the same cell nuclei obtained with a 60× oil-immersion objective (1.4 NA; Nikon). The shape and size of the cell nuclei in Fig. 2c are consistent with those in Fig. 2d. Therefore, this comparative study shows the capability of UV-PAM for label-free histology.
Fig. 2.
Cell-nuclear images of a 6-μm thick cross section of mouse small intestine acquired with ultraviolet photoacoustic microscopy and histology. (a) Photoacoustic image (MAP) of the section without staining. (b) Histologic image of the section stained with hematoxylin and eosin. (c) Close-up of the area enclosed by dashed lines in (a). (d) Histologic image of the stained section enclosed by dashed lines in (b). The correlation coefficients are 0.88 between the images shown in panels a and b and 0.83 between the images shown in panels c and d. PA, photoacoustic.
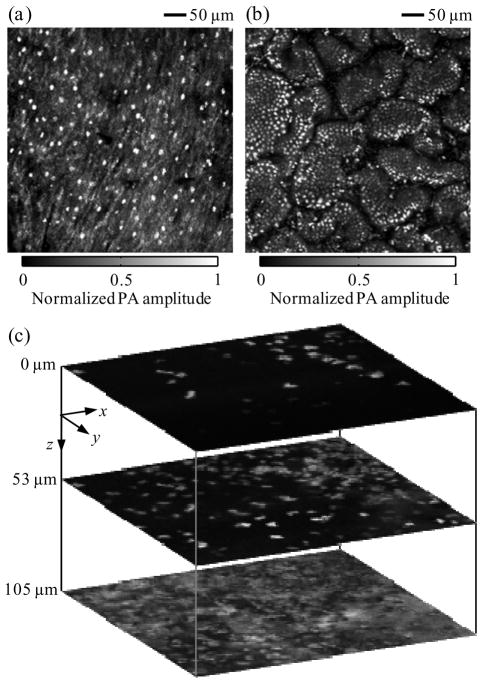
Next, we imaged cell nuclei in the epithelium of a mouse lip and in the intestinal villi of a mouse small intestine using UV-PAM ex vivo. Fresh specimens were taken from adult Swiss Webster mice and immersed in phosphate buffer solution (PBS; Sigma-Aldrich). After the small intestine was cut longitudinally and unfolded into a sheet, the specimens were mounted on the scanning stage with their inner surfaces in contact with the image window through PBS. By scanning with a 1.25 μm step size for 2.6 minutes, we imaged the nuclei of the epithelial cells in the mouse lip (Fig. 3a). The image shows a relatively homogeneous distribution of cell nuclei, each approximately 6 μm in diameter. The distance between the centers of neighboring cell nuclei ranges from 16 to 39 μm, suggesting that the stratified squamous epithelium on the lip was composed of cells with a lateral size of the same range. By scanning with a 0.62 μm step size for 7.4 minutes, we then imaged the nuclei of the epithelial cells on the mouse small intestine (Fig. 3b). By contrast, this image shows the tight arrangement of simple columnar epithelial cells into intestinal villi. The nuclear diameter was ~3 μm, and the cell width was ~6 μm. This study shows that UV-PAM is capable of imaging cell nuclei in non-sectioned tissues.
Fig. 3.
Cell-nuclear images of ex vivo and in vivo tissue acquired with ultraviolet photoacoustic microscopy. (a) Photoacoustic image (MAP) of epithelial cells in the ex vivo lip of a mouse. (b) Photoacoustic image (MAP) of epithelial cells in the ex vivo intestinal villi of a mouse. (c) In vivo photoacoustic images (dimensions: 250×250 μm2) of cells in the ear skin of a nude mouse at different depths (0, 53, and 105 μm)ss. PA, photoacoustic.
Finally, we imaged in vivo the cell nuclei in the ear skin of an athymic nude mouse using UV-PAM. The mouse (Harlan Laboratories) was anesthetized by isoflurane and held by a self-constructed stereotaxic imaging stage. After the imaging stage was mounted on the scanning stage, the image window was lowered to be in contact with the mouse ear through ultrasound gel (Parker Laboratories). After in vivo scanning with a 0.62 μm step size, we acquired a three-dimensional (3D) image of the cell nuclei in the mouse ear skin (Fig. 3c). Relative to the first slice in Fig. 3c, the second and third were 53 and 105 μm deep inside the mouse ear, respectively, suggesting that the penetration depth of UV-PAM is greater than 100 μm for in vivo imaging. Cell density varied with the depth: 45 cell nuclei are clearly visualized in the first slice, and 159 in the second. Although the lateral resolution of UV-PAM was expected to decrease with penetration depth, 76 cell nuclei could still be identified in the third slice. Therefore, UV-PAM is capable of 3D, noninvasive, in vivo imaging of cell nuclei without staining.
In summary, we developed a new photoacoustic imaging technology called UV-PAM to image cell nuclei ex vivo and in vivo by optical excitation of DNA and RNA. For the first time, label-free cell-nuclear images can be noninvasively acquired in vivo with strong optical contrast and high spatial resolution. UV-PAM images of unstained cell nuclei matched optical micrographs of the stained cell nuclei well in shape, size, and distribution. Therefore, UV-PAM has potentially broad applications in cancer studies and diagnosis.
Acknowledgments
The authors thank Ruimin Chen for ultrasonic transducer fabrication, Chris Favazza for data acquisition software, and Song Hu for data processing. This work was sponsored in part by National Institutes of Health grants R01 EB000712, R01 EB008085, R01 CA134539, and U54 CA136398. L.W. has a financial interest in Microphotoacoustics, Inc. and Endra, Inc., which, however, did not support this work.
References
- 1.Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat Rev Cancer. 2004;4:677. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
- 2.Molavi DW. The Practice of surgical pathology: a beginner’s guide to the diagnostic process. Springer; New York: 2008. [Google Scholar]
- 3.Kaeb S, Landthaler M, Hohenleutner U. Confocal laser scanning microscopy--evaluation of native tissue sections in micrographic surgery. Lasers Med Sci. 2009;24:819. doi: 10.1007/s10103-009-0660-9. [DOI] [PubMed] [Google Scholar]
- 4.Karen JK, Gareau DS, Dusza SW, Tudisco M, Rajadhyaksha M, Nehal KS. Detection of basal cell carcinomas in Mohs excisions with fluorescence confocal mosaicing microscopy. Br J Dermatol. 2009;160:1242. doi: 10.1111/j.1365-2133.2009.09141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paoli J, Smedh M, Wennberg AM, Ericson MB. Multiphoton Laser Scanning Microscopy on Non-Melanoma Skin Cancer: Morphologic Features for Future Non-Invasive Diagnostics. J Invest Dermatol. 2007;128:1248. doi: 10.1038/sj.jid.5701139. [DOI] [PubMed] [Google Scholar]
- 6.Wang LV. Multiscale photoacoustic microscopy and computed tomography. Nat Photonics. 2009;3:503. doi: 10.1038/nphoton.2009.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu S, Wang LV. Photoacoustic imaging and characterization of the microvasculature. J Biomed Opt. 2010;15:011101. doi: 10.1117/1.3281673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maslov K, Zhang HF, Hu S, Wang LV. Optical-resolution confocal photoacoustic microscopy. Proc SPIE. 2008;6856:68561I. [Google Scholar]
- 9.Stoscheck CM. Quantitation of protein. Methods Enzymol. 1990;182:50. doi: 10.1016/0076-6879(90)82008-p. [DOI] [PubMed] [Google Scholar]
- 10.McHowat J, Jones JH, Creer MH. Quantitation of individual phospholipid molecular species by UV absorption measurements. J Lipid Res. 1996;37:2450. [PubMed] [Google Scholar]