Abstract
Guanine nucleotide exchange factors (GEFs) activate Rho GTPases by catalyzing the exchange of bound GDP for GTP, thereby resulting in downstream effector recognition. Two metazoan families of GEFs have been described: Dbl-GEF family members that share conserved Dbl homology (DH) and Pleckstrin homology (PH) domains and the more recently described Dock180 family members that share little sequence homology with the Dbl family and are characterized by conserved Dock homology regions 1 and 2 (DHR-1 and -2). While extensive characterization of the Dbl family has been performed, less is known about how Dock180 family members act as GEFs, with only a single x-ray structure having recently been reported for the Dock9-Cdc42 complex. In order to learn more about the mechanisms used by the founding member of the family, Dock180, to act as a Rac-specific GEF, we set out to identify and characterize its limit functional GEF domain. A C-terminal portion of the DHR-2 domain, composed of approximately 300 residues (designated as Dock180DHR-2c), is shown to be necessary and sufficient for robust Rac-specific GEF activity both in vitro and in vivo. We further show that Dock180DHR-2c binds to Rac in a manner distinct from Rac-GEFs of the Dbl family. Specifically, Ala27 and Trp56 of Rac appear to provide a bipartite binding site for the specific recognition of Dock180DHR-2c, whereas, for Dbl family Rac-GEFs, Trp56 of Rac is the sole primary determinant of GEF specificity. Based on our findings, we are able to define the core of Dock180 responsible for its Rac-GEF activity as well as highlight key recognition sites that distinguish different Dock180 family members and determine their corresponding GTPase specificities.
Members of the Rho family of GTPases regulate a wide range of cellular activities including cell-cycle progression, gene transcription, cell migration, cell polarity, and vesicular trafficking through their abilities to bind to multiple downstream effectors (1-4). Rho GTPases switch between two states, the GDP-bound inactive state and the GTP-bound active state. Tight regulation of Rho GTPases and their nucleotide-bound state is important for mediating their different cellular functions (5). Three main classes of regulatory proteins for Rho GTPases have been identified and characterized. Guanine nucleotide exchange factors (GEFs) include ~70 mammalian/human proteins that promote the exchange of GDP for GTP on Rho GTPases. GTPase-activating proteins (GAPs) catalyze the hydrolysis of the bound GTP to GDP, and Rho GDP-dissociation inhibitors (GDIs) slow nucleotide exchange while serving to sequester Rho GTPases in the cytoplasm (6-8).
Two families of GEFs have been discovered for Rho GTPases, which we refer to here as the Dbl (9) and Dock180 superfamilies (10-12). The Dbl family members all possess two tandem conserved domains, the Dbl Homology (DH) and the Pleckstrin Homology (PH) domains (13-18). The DH domains of Dbl-GEFs are directly responsible for catalyzing GDP-GTP exchange activity while the PH domains are important for protein localization to the plasma membrane. More recently, Dock180-related proteins have been shown to catalyze nucleotide exchange on Rac and/or Cdc42 despite the absence of primary sequence homology to DH domains (19, 20). The functional domain for this unconventional class of GEFs was originally suggested to consist of ~500 residues located within the C-terminal half of Dock-180-related proteins and has been referred to as the Dock180 Homology Region 2 or DHR-2 domain.
Dock180 family members have been implicated in multiple biological pathways that include cell phagocytosis (21, 22), cell migration (21, 23-25), tumor suppression (26) and axonal outgrowth (27, 28). Dock180 is the founding member of the family and functions as a Rac-specific GEF. Full-length Dock180 shows much lower GEF activity compared to the isolated DHR-2 domain, apparently due to auto-inhibition (29, 30). Relief of the auto-inhibited state in cells has been suggested to result upon binding to the accessory protein Elmo (19, 31-34).
Recently, x-ray crystal structures were reported for the DHR-2 domain of one member of the Dock180 family, Dock9 (Zizimin 1), bound to different nucleotide-bound states of Cdc42 (35). This work has provided a first glimpse of how a Dock180 family member functions in catalyzing GDP-GTP exchange. However, as Dock9 is a Cdc42-specific GEF, we still do not have a detailed picture of how a Rac-specific GEF for the Dock180 family functions and confers specificity for Rac. Moreover, Dock180 exhibits much higher catalytic GEF activity, compared to Dock9, suggesting that the founding member of this GEF family may possess some important distinguishing features with regard to its mechanism of action.
In the present study, we set out to define a limit functional domain for Dock180 as an important first step for mechanistic studies and ultimately for high-resolution structural characterizations. During the course of our efforts to obtain an active limit functional domain for Dock180, we modeled the domain structure of the C-terminus of DHR-2 and isolated a stable region from Dock180 that can be expressed in high yield and exhibits full Rac-GEF activity. Here we show that this fragment, designated as Dock180DHR-2c (or sometimes simply DHR-2c), is fully active despite lacking the upstream helical domain that purportedly mediates dimerization of DHR-2 domains in Dock9 and has been suggested to be necessary for the full activation of its GEF activity (35). We provide evidence that this defined region of DHR-2 harbors specific recognition sites that allow for the discrimination between Rac and Cdc42 by Dock subfamilies A (Rac-specific) and D (Cdc42-specific) (20). Moreover, by specifically comparing the activity of the tandem DH-PH domains of the Rac-specific GEF Tiam-1 with that of the Rac-specific Dock180DHR-2c, we define a key contact made by Ala27 in Rac that is necessary together with Trp56 for Dock180DHR-2c- recognition and is absent in the functional coupling of DH-PH domains to Rac.
EXPERIMENTAL PROCEDURES
Plasmid constructs
The Dock180 plasmid was a gift from Dr. Michiyuki Matsuda (Kyoto University). To obtain the clone of the full length DHR-2 domain (amino acids 1178-1657), a polymerase chain reaction (PCR) was performed using the Dock180 plasmid as template DNA and primers 5′ -GCGGATCCATGGAAAGGC TTTTGGAT -3′ and 5′-CGGAATTCTCACGATGAG AGGGAAGAGA-3′. The PCR product was cloned into the pET28a plasmid (Novagen) using BamH I and EcoR I restriction sites. The constructs encoding the DHR-2n (1178-1334) and DHR-2c (1135-1657) sub-domains were generated by PCR using DHR-2 as the template and cloned into both pET28a and pKH-3 plasmids.
Protein expression and purification
Single colonies of E. coli BL21 (DE3) containing target plasmids were inoculated in 10 ml of LB medium with 50 μg/ml kanamycin or 100 μg/ml carbenicillin (RPI) and cultured overnight at 37°C. These cultures were subsequently used to inoculate 1 L LB medium with antibiotic in a shaking incubator at 37°C. The large-scale cultures were incubated to an OD600=0.6 and induced by IPTG (RPI) (final concentration = 200 μM) at room temperature overnight. Bacteria were harvested by centrifugation at 5000 rpm for 10 minutes and the pellets were re-suspended in lysis buffer (20 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 500 mM NaCl) with 10 μg/ml leupeptin and 10 μg/ml aprotinin. For DHR-2, DHR-2c and DHR-2n, the suspensions were sonicated on ice and the resulting lysates were cleared by centrifugation at 20,000 × g for 30 minutes at 4°C. The supernatants were collected and incubated with nickel-chelating beads (Amersham) for 30 minutes on ice. The beads were washed with 100 ml of lysis buffer containing 40 mM imidazole until no significant protein was detected in the wash buffer. The proteins were eluted with lysis buffer containing 200 mM imidazole, concentrated to ~200 μM, and stored at -80°C for further use. Rac and Cdc42 were expressed as GST-fusion proteins (36) using procedures similar to those described above and the supernatants were collected and applied to a glutathione-Sepharose column. After binding, the beads were washed extensively with lysis buffer and target proteins were then eluted using the same buffer with 10 mM glutathione adjusted to neutral pH. The eluted proteins were applied to PD-10 desalting columns (GE Healthcare) to remove glutathione and concentrated to ~300 μM. Mant-GDP-Rac and Mant-GDP-Cdc42 were prepared by mixing Rac or Cdc42 with a 10-fold excess of Mant-GDP in lysis buffer with 10 mM EDTA for 10 minutes followed by the addition of excess MgCl2 to quench the excess EDTA. The mixture was applied to a PD-10 desalting column equilibrated in lysis buffer in order to remove unbound Mant-GDP.
In vitro GEF assays
All fluorescence measurements were made using a Varian Eclipse Fluorescence Spectrophotometer. Samples were stirred continuously and thermostated at 25°C in HMA buffer (20 mM Hepes, pH 8.0, 5 mM MgCl2, 1 mM NaN3). In vitro GEF assays used Mant-GDP as a probe to monitor the extent of nucleotide exchange on GTPases. Mant-GDP was added to HMA buffer to a final concentration of 1 μM. Different concentrations of Rho GTPases (Rac, Cdc42) and their mutants were added into solution together with various concentrations of GEF proteins. The Mant-GDP fluorescence changes were monitored at 25°C using an excitation wavelength of 340 nm and an emission wavelength of 440 nm. All measurements were repeated at least three times. When measuring the turnover rates of the GEF proteins, Rac was preloaded with Mant-GDP and the decrease in fluorescence was detected as different concentrations of Rac were mixed with GEF proteins and excess GDP.
GST-Rac pull-down assays
To check the binding of DHR-2 and DHR-2c with Rac, GST-Rac (0.3 nmol) was prebound to 15 μl of glutathione-Sepharose beads while the same amount of DHR-2c was added to the beads in the presence of 5 mM EDTA. The negative control tube contained beads and DHR-2c (no GST-Rac). The mixtures were rotated at 4°C for 30 minutes and then centrifuged at 16,000 × g for 1 minute. The supernatant was discarded. The beads were washed (3X) with buffer and loaded onto an SDS-PAGE gel. To check the nucleotide-binding preference of different DHR-2c constructs, glutathione-Sepharose beads preloaded with GST-Rac were mixed with DHR-2c and excess GDP or GTPγS in EDTA (final concentration=10 mM)-containing buffer. Excess MgCl2 (20 mM) was added to the solution after 15 minutes and incubated for an additional 15 minutes. The beads were washed as described above and the binding was detected by Colloidal Blue-staining following SDS-PAGE.
Indirect immunofluorescence
The cells were transfected with the plasmids of interest and then plated on acid coverslips overnight. The cells were fixed on the coverslip in 4% formaldehyde in PBS for 20 minutes at room temperature and rinsed (3X) with PBS. Triton X-100 (0.1%) was added to permeabilize the cells, after which the cells were rinsed with PBS (3X). Incubations were performed with primary and secondary antibodies in PBS supplemented with 2% BSA. The cells were again washed with room temperature PBS (3X) after each incubation.
PBD assays
Cells transfected with plasmids of interest were harvested with MBL buffer (magnesium-containing lysis buffer) that contained 50 mM Hepes, pH 7.5, 150 mM NaCl, 1% Triton, 10% glycerol, 25 mM NaF, 10 mM MgCl2, 1 mM EDTA, 10 μg/ml leupeptin and 10 μg/ml aprotinin. Whole cell lysates were cleared by centrifugation, and the supernatants were used to assess the total amount of Rac. The remaining extract was combined with 2 volumes of lysis buffer and the recombinant Rac/Cdc42-binding domain of Pak (i.e. designated as GST-PBD (Upstate) (37)) coupled to glutathione beads and incubated for 30 minutes at 4°C. The beads were washed (3X) with lysis buffer and eluted in SDS-PAGE sample buffer. Aliquots of both total cell extracts and the eluents from the PBD beads were immunoblotted with anti-Myc monoclonal 9E10 (Covance) and visualized with ECL reagents from Amersham following the manufacturer's instructions.
RESULTS
Identifying a stable and functionally active GEF domain for Dock180
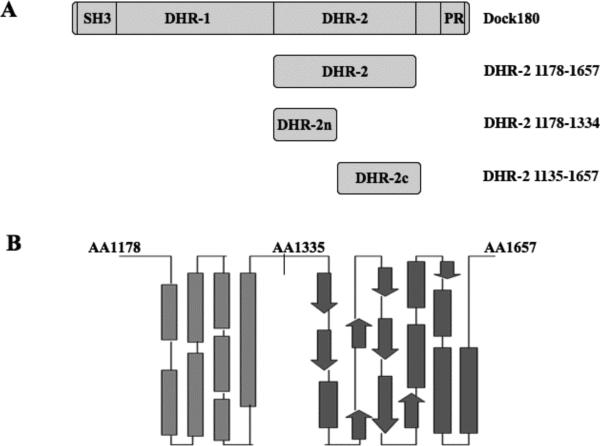
The functional DHR-2 domain was originally suggested to comprise a conserved region present in all Dock180 family members. It is composed of approximately 500 amino acids (residues 1111 to 1657 for Dock180) with sequence alignment of all the DHR-2 domains revealing a low degree of primary sequence identity among different members (i.e., 18% between Dock180 and Dock9) (20). Although we and others have found that the bacterially expressed, complete DHR-2 domain from Dock180 is active in vitro, it is relatively unstable and easily denatured whereas a shorter construct lacking the N-terminal 70 amino acids of DHR-2 yielded a more stable domain (amino acids 1178-1657; Figure 1A) that could be expressed in bacteria in good yield and possessed nucleotide exchange activity. However, further purification of this DHR-2 construct for biochemical and structure-function studies revealed that it exhibited a tendency to aggregate, especially in solutions containing less than 300 mM NaCl. Thus, it has been difficult to routinely use this domain in nucleotide exchange experiments for comparison with other known GEFs.
Figure 1.
Dock180 constructs examined in this study. (A) Schematic representation of the Dock180 and DHR-2 constructs that were used to study the in vivo and in vitro activation of Rac. (B) Secondary structural prediction of the DHR-2 domain of Dock180 using J-pred.
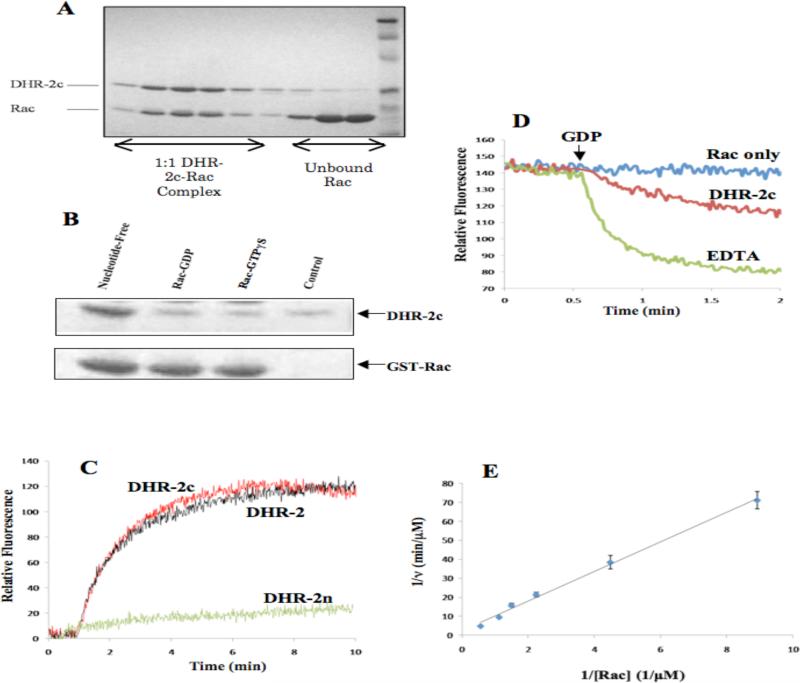
Further secondary structure analysis and homology modeling of DHR-2, which included our taking advantage of the recently published x-ray crystal structures for the DHR-2 domain of Dock-9 bound to Cdc42 (35), suggested that the organization of the entire DHR-2 domain can be resolved into two distinct regions, that we designate here as DHR-2n (i.e. the helix-rich N-terminus, amino acid residues 1178-1334) and DHR-2c (amino acid residues 1335-1657; Figures 1A and 1B). The expressed DHR-2c domain of Dock180 (Dock180DHR-2c) was purified to homogeneity and observed to bind to its cognate GTPase target, Rac, in a manner typical for a GEF, such that the nucleotide-free form of GST-Rac binds the recombinant Dock180DHR-2c much more tightly than either the GDP- or GTPγS-bound forms of Rac (Figures 2A and 2B). In addition, this ~37 kDa domain showed robust GEF activity towards purified Rac in fluorescence-based assays monitoring the exchange of GDP for the nucleotide analog Mant-GDP, and displayed a molar specific activity equal to that of the full length DHR-2 domain (Figure 2C). Thus, Dock180DHR-2c fully accounts for the GEF activity of the DHR-2 domain in Dock180. In our in vitro assays, we found no evidence that the DHR-2n domain of Dock180 contributes to the overall GEF activity in contrast to the Cdc42-specific GEF, Dock9, which was suggested to require the helix-rich DHR-2n domain for full activity (35). Further truncations at either end of Dock180DHR-2c totally abolished its GEF activity, and during the process of purifying each of the two sub-domains of DHR-2, we confirmed that DHR-2n is not stable in low salt solution and is responsible for the aggregation of the DHR-2 domain, while Dock180DHR-2c is stable and fully active under these same conditions. This may help explain our earlier observation regarding the labile nature of the full length DHR-2 domain of Dock180 and its tendency towards aggregation, as this is likely due to the rapid denaturation of the N-terminal half of DHR-2.
Figure 2.
Dock180DHR-2c activates Rac in vitro and in vivo. (A) The complex between the purified Dock180DHR-2c domain (designated DHR-2c in the figure) and Rac. Colloidal blue-stained SDS-PAGE was performed on fractions obtained from G75 gel-filtration chromatography. The samples shown were obtained from successive 4 ml fractions of the gel filtration elution profile. (B) Binding preference of DHR-2c to nucleotide-free Rac. Colloidal blue-stained SDS-PAGE of GST-Rac affinity precipitation of GDP- or GTPγS-bound Rac. Each of the GST-Rac bead samples was incubated with 1 μg of purified DHR-2c, washed and analyzed by SDS-PAGE and colloidal blue staining. (C) The C-terminal domain of the DHR-2 domain of Dock180 (DHR-2c) is sufficient to stimulate GDP-Mant-GDP nucleotide exchange on Rac. DHR-2c (Red) showed similar GEF activity as full-length DHR-2 (Black) while DHR-2n showed no activity. (D) Dissociation kinetics of pre-bound Mant-GDP in the presence of excess unlabeled GDP. Rac-Mant-GDP (1 μM) was stirred continuously, alone or in the presence of 100 nM DHR-2c or 10 mM EDTA. 1 mM GDP (final concentration) was added at the indicated time. (E) Kinetics of DHR-2c-catalyzed nucleotide exchange on Rac1. Each reaction contained 20 nM DHR-2c, 100 μM GDP and increasing concentrations of Rac preloaded with Mant-GDP. Initial rates were obtained by monitoring the decrease in fluorescence following the addition of GDP. Kinetic analysis of nucleotide exchange by DHR-2c yielded a Km of 3.3 μM and a kcat of 19.8/min.
Characterizing the highly specific GEF activity of Dock180DHR-2c towards Rac
In order to obtain catalytic activity and turnover rates for the GEF activity of Dock180DHR-2c, we assayed nucleotide exchange under conditions of excess Rac. Figure 2D shows an example of fluorescence experiments that were performed using recombinant Rac that had been preloaded with Mant-GDP and purified by gel filtration to remove any free nucleotide. Using this approach, different concentrations of the purified Mant-GDP-Rac complex were mixed with excess GDP and then 20 nM Dock180DHR-2c was added to initiate the exchange of the fluorescent nucleotide for unlabeled GDP. The concentration of Dock180DHR-2c in these assays was much lower than Rac in order to use the Michaelis-Menten approximation in subsequent analyses. The release of Mant-GDP from Rac (i.e. due to its exchange for excess unlabeled GDP) caused the fluorescence to decrease, as shown in Figure 2D, thus providing a real time measurement of the rate-limiting step for nucleotide exchange, i.e. the catalyzed dissociation of bound nucleotide. We obtained a series of fluorescence traces using this readout (not shown) and estimated the corresponding initial velocities (v) for each concentration of Rac. Figure 2E shows the resulting double reciprocal plot of 1/v vs. 1/[Rac-Mant-GDP], with the linear fit of the data yielding a Km value of 3.3 μM and a kcat value of 19.8/min (0.33/sec). As expected, fitting the same set of fluorescence traces individually with a single exponential gave a similar value for the time constant (not shown). We also calculated the intrinsic GDP-dissociation rate constant for Rac which was found to be 0.017/min, indicating that the dissociation rate of GDP from Rac is increased more than 1000-fold by the GEF activity of Dock180DHR-2c.
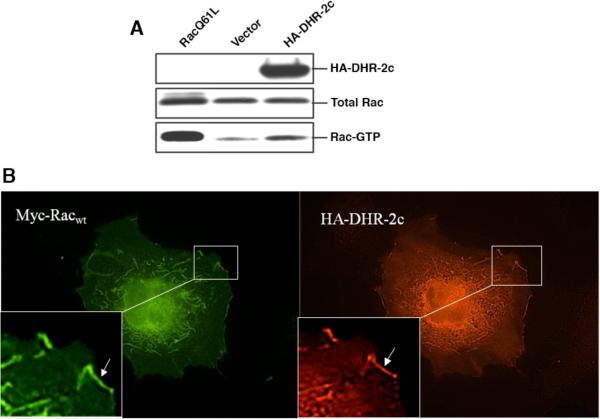
In order to demonstrate that Dock180DHR-2c represented an active GEF in vivo, we tested its ability to promote the activation of Rac in cells, i.e. following its transient transfection into Cos-7 cells. Figure 3A shows the results obtained when hemagglutinin (HA)-tagged Dock180DHR-2c was expressed in Cos-7 cells along with Myc epitope-tagged Rac and the lysates were tested for their relative amounts of activated GTP-bound Rac, following pull-downs using GST fused to the limit Rac-binding domain of its effector Pak3 (GST-PBD) (37). Figure 3A (bottom panel) shows that increased amounts of Myc-tagged Rac-GTP were pulled down from lysates that contained expressed HA-Dock180DHR-2c, compared to control lysates (in these experiments, the ability of GST-PBD to pull-down dominant-active Rac Q61L served as a positive control). Further evidence for the in vivo coupling of Rac and Dock180DHR-2c is provided by the co-localization of Myc-Rac and HA-Dock180DHR-2c observed in co-transfected cells as shown in Figure 3B. The incorporation of Dock180DHR-2c into membrane ruffles (lamellipodia) was striking and was somewhat unexpected, given that it had been suggested that the conserved DHR-1 domain, which contains a putative site for interaction with PtdIns(3,4,5)P3, might be necessary for the membrane binding of full-length Dock180 family members (38).
Figure 3.
Examining Dock180DHR-2c in cells. (A) PBD assay demonstrates activation of Rac in vivo by Dock180DHR-2c (DHR-2c). GTP-bound Rac was affinity precipitated with PBD beads from Cos-7 cell lysates that had been transiently transfected with vector plus Myc-RacWT (wild-type), HA-tagged DHR-2c plus Myc-RacWT, or vector plus Myc-Rac Q61L. The top and middle panels show the expression of HA-DHR-2c, and the relative expression of the Myc-Rac constructs. The bottom panel shows the relative amounts of activated Rac pulled down under the described conditions. (B) Rac and DHR-2c co-localize in membrane ruffles. Indirect immunofluoresence of Cos-7 cells cotransfected with Myc-RacWT (wild-type) and HA-DHR-2c.
Trp56 of Rac is required for Dock180DHR-2c binding and catalysis
In order to explore the factors underlying the selectivity of the GEF activity of Dock180DHR-2c towards Rac versus Cdc42, we began by evaluating the importance of position 56 which is a tryptophan residue in Rac and a phenylalanine residue in Cdc42. Starting with the first report of the x-ray crystal structure of the complex between the DH/PH domains of Tiam-1 and Rac (39), a wealth of crystallographic data has provided additional detail regarding how Rho GTPases bind to Dbl family GEFs and where in their primary structure the specificity for GEFs resides. In the case of Rac, these critical contact sites are primarily located in switch I and switch II, as well as in the β2-β3 strands that lie in between the switch regions. Tyr32, Asp38, and Asn39 of switch I, together with Gln61 and Gln64 of switch II, are all critical either for GEF binding or for activation, as substitutions for any of these residues severely impair the ability of Dbl family members to catalyze nucleotide exchange (40).
The β3 strand of Rac, that is just upstream of Trp56 (Phe56 in Cdc42) in switch II, is particularly important, as it is this residue that is primarily responsible for Rac specificity in DH-domain-mediated nucleotide exchange. Thus, the W56F mutant of Rac is not activated by Rac-GEFs of the Dbl family while, conversely, the F56W mutant of Cdc42 enables it to be recognized by these same Rac-specific GEFs (40, 41). However, the primary structures of members of Dock180 family members bear little similarity to Dbl family GEFs, thus raising the question of whether the DHR-2 domain of Dock180 binds to a similar region on Rac as the DH domain of Dbl-GEFs. In addition, whether Trp56 of Rac is equally critical for Dock180 recognition is controversial, as it has been reported that the W56F mutant of Rac can still interact with the DHR-2 domain of Dock180 (19) as well as to be partially activated by the DHR-2 domain of Dock2 (42). In contrast, when Trp56 of Rac is mutated to phenylalanine, which is the corresponding residue in Cdc42, we find that Rac W56F is activated to <10% of the level of wild-type Rac by DHR-2c (Figure 4A). We confirmed that this Rac mutant can still fully load Mant-GDP when treated with EDTA, demonstrating that this substitution did not compromise its GTP-binding capability (not shown). As will be described further below, our data clearly demonstrate the important role that Trp56 in Rac plays in the specificity of DHR-2 binding and activation, similar to what has been observed for the DH domain of the Dbl family GEF Tiam-1.
Figure 4.
Met1524 of Dock180DHR-2c (DHR-2c) specifically recognizes Trp56 of Rac. (A) The Rac W56F mutant is ineffective in coupling to DHR-2c. (B) Sequence comparisons of key residues within the DHR-2 domains of Dock subfamilies A and D. (C) Mutation of methionine 1524 of DHR-2c reduces its GEF activity. DHR-2c and the DHR-2c M1524L mutant (60 nM, each) were mixed with 600 nM Rac and 1 μM Mant-GDP in HMA buffer. The relative rate of nucleotide exchange by wild-type DHR-2c or the DHR-2c M1524L mutant with RacWT or Rac W56F is shown by changes in Mant fluorescence in A and B, respectively. The GEF activity of DHR-2c M1524L mutant was significantly reduced (~25% of wild-type Rac). (D) DHR-2c and DHR-2c M1524L (240 nM, each) were mixed with 600 nM Rac and 1 μM Mant-GDP in HMA buffer. Only DHR-2c Met1524L showed significant GEF activity toward Rac W56, while wild-type DHR-2c did not catalyze nucleotide exchange on Rac W56F.
Using the Dock9-Cdc42 x-ray crystal structure as a guide (35), we set out to identify critical residues within Dock180DHR-2c that make contact with Trp56 of Rac. The candidate residues from the DHR-2 domain of Dock9 that interact with Cdc42 are Leu1940 and Gln1943. Specifically, the methyl group of Leu1940 in Dock9 interacts with the phenyl ring of Phe56. This leucine residue is conserved in members of Dock subfamily D (Docks 9-11, which are specific GEFs for Cdc42), while in the Rac-specific subfamily A (Docks 1, 2, and 5; Dock1 being Dock180), the corresponding residue is Met1524 (Figure 4B). In order to test the degree to which the leucine-phenylalanine interaction in the Dock9-Cdc42 complex, and the methionine-tryptophan contact in the Dock180-Rac complex, encode the specificity for these interactions, we mutated the corresponding residue Met1524 to leucine in Dock180DHR-2c. As shown in Figure 4C, changing the methionine to leucine decreased the GEF activity of Dock180DHR-2c by ~80%. Conversely, the methionine-to-leucine substitution at 1524 in Dock180DHR-2c resulted in a five-fold increase in GEF activity toward the Rac W56F mutant (Figure 4D). Taken together, these results confirmed a point of specific discrimination between Rac versus Cdc42 toward Dock180 (versus Dock9): the indole ring of Trp56 of Rac interacts with the methyl thio-ether side chain of Met1524 of Dock180 while the extra methyl group of the leucine residue at the corresponding position in Dock9 is a key hydrophobic site of interaction for the phenyl ring of Phe56 in Cdc42.
A comparison of Rac specific-GEF activity by Tiam-1 and Dock180DHR-2c
Although there are similarities between the set of residues in Rac that are important for binding DHR-2 and DH domains, we wanted to further define these residues and investigate their relative contribution to the overall GEF activity. The data in Figures 5A and 5B again show that Trp56 of Rac is necessary for Dock180DHR-2c binding and activation, similar to what has been observed for the DH-domain of Tiam-1. However, the complementary substitution (i.e. Phe to Trp) at position 56 in Cdc42 is not sufficient to allow for GEF activity by Dock180DHR-2c, although this Cdc42 mutant can be strongly activated by the DH-PH domains of Tiam-1 (Figures 5C and 5D). Moreover, although the M1524L mutant of Dock180DHR-2c can activate the W56F mutant of Rac, the introduction of this Cdc42-specific residue into Dock180DHR-2c conferred no detectable GEF activity when it was tested with wild-type Cdc42 (data not shown). Thus, Trp56 is necessary but not sufficient for the specific binding and GEF activity of Dock180 towards Rac, indicating that additional contacts are necessary for full GEF activity.
Figure 5.
Comparisons of the abilities of the Rac-specific Dock180DHR-2c and Tiam-1DH-PH to activate Rac and Cdc42 mutants. 180 nM Dock180DHR-2c and 15 μM Tiam-1DH-PH have similar catalytic properties towards the same concentration of RacWT and were used in all subsequent experiments. The concentration of Rac, Cdc42 or their mutants was kept at 600 nM. Bars show the relative nucleotide exchange activity of the Rac and Cdc42 mutants compared to the wild-type proteins and the standard error of three trials. Note that the A27K and G30S mutants of Rac show reduced capabilities for being activated by Dock180DHR-2c while they still can be fully activated by Tiam-1DH-PH (A and B). Residues 27 and 56 of Rac or Cdc42 are both critical for specific Dock180 GEF recognition (C) while the single Cdc42 F56W mutant can be nearly fully activated by the DH-PH domain of Tiam-1 (D). Note that the Cdc42 K27A-F56W double-mutant can be partially activated by Dock180DHR-2c.
Structural comparisons and primary sequence alignments between Cdc42 and Rac highlight other candidate residues, which may play critical roles in mediating Dock180DHR-2c-catalyzed nucleotide exchange on Rac. These residues, which are shown in the Dock9-Cdc42 structure to be sites of contact in Cdc42 that are not conserved in Rac, and to interact with non-conserved residues in the respective DHR-2 domains, are Ala27, Gly30, Ser41 and Asn52 in Rac. To test whether these residues played a significant role in the selectivity of the GEF, Ser41 and Asn52 in Rac were mutated to their corresponding residues Ala41 and Thr52 in Cdc42. We found that neither residue had a detectable effect on the ability of Dock180DHR-2c to catalyze nucleotide exchange (see Supplementary Table 1). Residues 27 and 30 of Rac or Cdc42 have been suggested to mediate selective recognition of Rac by the DHR-2 domain of Dock2 (42). To test the importance of these residues for the recognition of Rac by Dock180DHR-2c, we introduced the substitutions A27K and G30S (i.e. Rac-to-Cdc42 changes at these positions) into Rac. Figure 5A shows the results of fluorescence nucleotide exchange assays demonstrating that both of these Rac mutants were impaired in their ability to functionally couple to Dock180DHR-2c. The Rac A27K mutant was ~20% effective, and thus only marginally better than the Rac W56F mutant, whereas the G30S mutant was about 30% as effective as wild-type Rac in coupling to Dock180DHR-2c. In contrast to their essential role in Dock180DHR-2c-mediated nucleotide exchange, when we tested the Rac A27K and Rac G30S mutants with the DH-PH domains of Tiam-1, we found that both Rac mutants were activated to a degree comparable to wild-type Rac (Figure 5B). Taken together, these data confirm a key contact site in Rac for Dock180DHR-2c that does not play a significant role in binding Dbl family GEFs.
To determine whether these two residues (i.e. at positions 27 and 30), along with position 56, are sufficient for specific recognition by Dock180DHR-2c, we mutated these sites in Cdc42 either individually or in combination in order to provide Rac contact points in a Cdc42 background. Figure 5C shows that the double-mutant S30G-F56W of Cdc42 was incapable of being activated by Dock180DHR-2c. However, the K27A-F56W mutant of Cdc42 showed a partial ability to recognize Dock180DHR-2c (Figure 5C). Unlike the Cdc42 F56W mutant, which was nearly 80% effective compared to wild-type Rac in coupling to Tiam-1 (Figure 5D), the K27A-F56W mutant of Cdc42 was only about 25% as effective as wild-type Rac in its interaction with Dock180DHR-2c. Thus far, we have not found any other residue that contributes to the specific recognition of Dock180DHR-2c.
In addition to these residues that are divergent between Cdc42 and Rac, we also mutated several residues common to Cdc42 and Rac that have been shown to be necessary for binding the DH-PH domain of Tiam-1. In the β2-β3 and switch II regions, the G54A and Q61L mutants of Rac are unable to be activated by Dock180DHR-2c (Supplementary Table 1). Together with the results obtained for the Rac W56F mutant of Rac, we conclude that the β3 and switch II regions of Rho GTPases are the critical area for the binding of both the Dock180 and Dbl families of GEFs. However, in the switch I region, the binding site for Rac in Dock180DHR-2c appears to be different from its binding site on the DH domain of Tiam-1. In addition to Ala27 and Gly30 of Rac, the Rac Y32A mutant also responds differently to the DHR-2c and DH domains, as the nucleotide exchange activity of this mutant is not responsive to DH-PH domains but it is still able to be activated by Dock180DHR-2c. The Rac E39A mutant, which contains another substitution at the end of switch I, is unable to be activated by either Dock180DHR-2c or the DH-PH domain of Tiam-1 (Supplementary Table 1). Taken together, these data suggest that the switch I region in GTPases plays an important role in the specific binding of both Dock180 and Dbl family GEFs but significantly, the residues which make up the contact site are different for these two families of GEFs.
DISCUSSION
The Rho family GEFs are critical cellular regulatory proteins, given the important roles that their target GTPases play in a wide variety of cellular and biological functions ranging from actin cytoskeletal changes, to cell migration and invasion, to cell-cycle progression, differentiation and developmental events. Two classes of upstream activators (GEFs) for the Rho GTPases have been identified. One family of GEFs, for which the founding member is the oncogenic Dbl protein, has been extensively characterized and a good deal of structure-function information is now available regarding how different members of the Dbl family functionally engage their target Rho GTPases. Less is known, at least from a biochemical and structural perspective, about the second family of Rho-GEFs for which Dock180 is the prototype.
The recent x-ray crystal structure for the DHR-2 domain from one member of the Dock180 family, Dock9, bound to its GTPase target, Cdc42, was recently determined and has begun to shed some light on how this Cdc42-specific GEF couples to different nucleotide-bound states of Cdc42. However, some important questions have remained unanswered regarding whether the Rac-specific GEF Dock180 uses a similar mechanism to activate Rac as that used by Dock9 to activate Cdc42, as well as those used by Rac-specific GEFs from the Dbl family, such as Tiam-1.
As a means for developing biochemical strategies to address these mechanistic issues, we set out to delineate a limit functional domain for Dock180 and examine whether its ability to catalyze nucleotide exchange is mechanistically comparable to that of the DH-PH domains of Dbl GEFs. Our earlier modeling based on secondary structure predictions led us, in a trial and error manner, to the discovery of a limit functional sub-domain of DHR-2 (Dock180DHR-2c). This fully active sub-domain is unexpectedly small in light of the recently published x-ray structures for the Dock9-Cdc42 complexes, as it consists of only the C-terminal 300 residues of DHR-2. The limit domain, Dock180DHR-2c, can bind and activate Rac in vitro with a rate of catalysis that is essentially identical to that for the full-length DHR-2 domain of Dock180. It also is able to activate Rac in cells, while maintaining specificity as it is completely incapable of coupling to Cdc42. This differs from the findings regarding the regulation of Dock9 activity, as the N-terminal helix-rich domain is necessary for the full activation of Dock9 by providing the binding site for homodimerization with a second Dock9 DHR-2 domain. Interestingly, even under optimal dimerization conditions, the nucleotide exchange activity of Dock9 for Cdc42 is ~30 times less than that measured for either the DHR-2 domain of Dock180 or Dock180DHR-2c (i.e. the rate for catalyzed nucleotide exchange is 0.01 s-1 for the DHR-2 domain of Dock9 compared to 0.33 s-1 for the corresponding domain of Dock180), indicating a significant difference in catalytic efficiency. This intrinsically higher turnover of Dock180-Rac complexes could have important implications for the full-length Dock180 protein in vivo and a significant influence on the rate of phagocytosis and cell engulfment, and thus will be an interesting subject for further study.
Mutagenesis of Dock180DHR-2c has enabled us to identify several residues in Rac that are essential for Dock180 recognition. In addition, we found that the selective nature of Rac recognition of Dock180 is mediated by Trp56 of Rac, as is the case for Rac specificity for the Dbl family GEF Tiam-1, therefore indicating a similarity in their use of position 56 as a means of GTPase discrimination. While the importance of position 56 in Rac and Cdc42 is common to both the Dbl and Dock180 families of GEFs, we also demonstrate here the critical role of a second contact site on Rac at Ala27 and to a lesser extent, Gly30. Replacement with either residue in Rac with the corresponding residue in Cdc42 dramatically abrogates the nucleotide exchange activity of Dock180DHR-2c for Rac (Figure 5A). Using Cdc42 as a GTPase scaffold, we found that replacement of Cdc42 residues 27 and 56 (lysine and phenylalanine, respectively), but not 30 and 56, was able to restore a modest degree of coupling to Dock180DHR-2c, as this Cdc42 double mutant showed ~25% of the nucleotide exchange activity of wild-type Rac (Figure 5C). Using the crystallographic data provided by the structure for the DHR-2 domain of Dock9 bound to Cdc42, we examined the importance of the residue from Dock180DHR-2c that makes contact with residue 56 of Rac and found that Met1524 of Dock180 is critical for Rac recognition, as it is for Cdc42 recognition by Dock9, where the methionine is replaced with leucine at residue 1941.
In an attempt to rationalize the discrimination of Dock180 between Rac1 and Cdc42, we homology modeled the complex between Dock180DHR-2c and Rac1, and their mutants, using the x-ray structure of Dock9-Cdc42 as a template. Within the modeled complex of wild-type Dock180DHR-2c and wild-type Rac, there is a strong interaction between the aromatic ring of Trp56 in Rac (Figure 6, top-left; green ring) and the sulfur of Met1524 in Dock180DHR-2c (Figure 6, top-left; yellow), with the distance between the ring centroid and the sulfur being 4.2 Å. This interaction is weakened in the complex between wild-type Dock180DHR-2c and the Rac W56F mutant (Figure 6, top-right, i.e. the distance between the ring centroid (orange) and the sulfur (yellow) is 4.7 Å), which might account for the loss of GEF activity by wild-type Dock180DHR-2c upon the Rac W56F mutant. Also, within the complex between wild-type Dock180DHR-2c and wild-type Rac, there are considerable hydrophobic contacts between the sulfur atom of Met1524 and the indole ring of Trp56. When Met1524 in Dock180DHR-2c is changed to leucine, the strong interaction that occurred between the sulfur from the methionine and the tryptophan ring is lost while a few hydrophobic contacts are maintained between the leucine at position 1524 and Trp56 (Figure 6, bottom-left), which overall would account for this mutant having slightly less activity compared to wild-type Dock180DHR-2c. Surprisingly, the model for the complex between the Dock180DHR-2c M1524L mutant and the Rac W56F mutant (Figure 6, bottom-right) is very similar to that of the complex between Dock180DHR-2c M1524L and wild-type Rac, making it difficult to rationalize why the exchange activity for the former complex is higher. In this regard, we are in the process of obtaining structural information for the complex between Dock180DHR-2c and wild-type Rac using x-ray crystallography which should shed light on this question as well as provide further insights into the mechanism of GEF activity by Dock180.
Figure 6.
Structure of the modeled complexes showing the interaction between residues 1524 of Dock180DHR-2c and 56 of Rac1. The side-chains are shown as both sticks and dots. Met1524 (blue), Leu1524 (red), Trp56 (green) and Phe56 (orange). The sulfur of Met1524 is shown in yellow. The homology model for the Dock180DHR-2c domain (residues 1336 to 1615) and wild-type Rac1 (residues 1-177) was obtained using the structures of the Dock9-DHR-2 domain and Cdc42 from the structure of their complex (PDB ID 1WM9) as a template using the Swiss-model server (43-45). The homology-modeled structures were superimposed on the structure of the complex of Dock9-DHR-2 and Cdc42 using PyMOL (DeLano Scientific LLC). The structure of this complex was energy-minimized first by steepest-descent and then by conjugate-gradient methods using Gromacs (ver 4.0) (46) to remove any steric clashes. Mutations were made using the mutagenesis module on PyMOL, keeping the side-chain dihedral angles the same as in the wild-type complexes. The structures for the mutant complexes were again energy minimized to obtain the final structures. The interactions between residues were calculated either using the PIC server (47) or the CSU software (48).
In conclusion, we have been able to define a minimal region harboring full nucleotide exchange activity within the DHR-2 domain of Dock180 and provide the molecular basis by which the founding member of the Dock180 family of GEFs specifically couples with the Rac GTPase. The establishment of a minimal functional domain for the Dock180 protein now sets the stage for structural and biochemical studies to further probe the mechanism by which this GEF shows its extraordinary catalytic activity as well as to learn more about how other parts of the Dock180 protein are used to regulate this activity in cells in order to initiate Rac-mediated signal transduction pathways.
Supplementary Material
ACKNOWLEDGMENT
The authors wish to acknowledge Miao-chong Joy Lin for help with the PBD assays and Cindy Westmiller for help with the manuscript preparation.
Abbreviations
- DHR-
Dock180 Homology Domain 2
- Dock180DHR-2c
C-terminus of DHR-2 in Dock180, residues 1335-1657
- DH
Dbl Homology
- PH
Pleckstrin Homology
- GEF
Guanine Nucleotide Exchange Factor
- Mant-GDP
2′-3′-O-(N′-Methylanthraniloyl)guanosine-5′-O-diphosphate
Footnotes
This work was supported by National Institutes of Health R01 Grants GM40654 and GM47458.
REFERENCES
- 1.Cerione RA. Cdc42: new roads to travel. Trends Cell Biol. 2004;14:127–132. doi: 10.1016/j.tcb.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 2.Chardin P. The ras superfamily proteins. Biochimie. 1988;70:865–868. doi: 10.1016/0300-9084(88)90226-x. [DOI] [PubMed] [Google Scholar]
- 3.Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 4.Tapon N, Hall A. Rho, Rac and Cdc42 GTPases regulate the organization of the actin cytoskeleton. Curr. Opin. Cell Biol. 1997;9:86–92. doi: 10.1016/s0955-0674(97)80156-1. [DOI] [PubMed] [Google Scholar]
- 5.Malliri A, Collard JG. Role of Rho-family proteins in cell adhesion and cancer. Curr. Opin. Cell Biol. 2003;15:583–589. doi: 10.1016/s0955-0674(03)00098-x. [DOI] [PubMed] [Google Scholar]
- 6.Nassar N, Hoffman GR, Manor D, Clardy JC, Cerione RA. Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP. Nat. Struct. Biol. 1998;5:1047–1052. doi: 10.1038/4156. [DOI] [PubMed] [Google Scholar]
- 7.Hoffman GR, Cerione RA. Signaling to the Rho GTPases: networking with the DH domain. FEBS Lett. 2002;513:85–91. doi: 10.1016/s0014-5793(01)03310-5. [DOI] [PubMed] [Google Scholar]
- 8.Johnson JL, Erickson JW, Cerione RA. New insights into how The Rho guanine nucleotide dissociation inhibitor regulates the interaction of Cdc42 with membranes. J. Biol. Chem. 2009;284:23860–23871. doi: 10.1074/jbc.M109.031815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hart MJ, Eva A, Evans T, Aaronson SA, Cerione RA. Catalysis of guanine nucleotide exchange on the CDC42Hs protein by the dbl oncogene product. Nature. 1991;354:311–314. doi: 10.1038/354311a0. [DOI] [PubMed] [Google Scholar]
- 10.Takai S, Hasegawa H, Kiyokawa E, Yamada K, Kurata T, Matsuda M. Chromosomal mapping of the gene encoding DOCK180, a major Crk-binding protein, to 10q26.13-q26.3 by fluorescence in situ hybridization. Genomics. 1996;35:403–404. doi: 10.1006/geno.1996.0378. [DOI] [PubMed] [Google Scholar]
- 11.Hasegawa H, Kiyokawa E, Tanaka S, Nagashima K, Gotoh N, Shibuya M, Kurata T, Matsuda M. DOCK180, a major CRK-binding protein, alters cell morphology upon translocation to the cell membrane. Mol. Cell. Biol. 1996;16:1770–1776. doi: 10.1128/mcb.16.4.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erickson JW, Cerione RA. Structural elements, mechanism, and evolutionary convergence of Rho protein-guanine nucleotide exchange factor complexes. Biochemistry. 2004;43:837–842. doi: 10.1021/bi036026v. [DOI] [PubMed] [Google Scholar]
- 13.Klinger MB, Guilbault B, Kay RJ. The RhoA- and CDC42-specific exchange factor Dbs promotes expansion of immature thymocytes and deletion of double-positive and single-positive thymocytes. Eur. J. Immunol. 2004;34:806–816. doi: 10.1002/eji.200324400. [DOI] [PubMed] [Google Scholar]
- 14.Srivastava SK, Wheelock RH, Aaronson SA, Eva A. Identification of the protein encoded by the human diffuse B-cell lymphoma (dbl) oncogene. Proc. Natl. Acad. Sci. U.S.A. 1986;83:8868–8872. doi: 10.1073/pnas.83.23.8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Worthylake DK, Rossman KL, Sondek J. Crystal structure of the DH/PH fragment of Dbs without bound GTPase. Structure. 2004;12:1078–1086. doi: 10.1016/j.str.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 16.Feng Q, Albeck JG, Cerione RA, Yang W. Regulation of the Cool/Pix proteins: key binding partners of the Cdc42/Rac targets, the p21-activated kinases. J. Biol. Chem. 2002;277:5644–5650. doi: 10.1074/jbc.M107704200. [DOI] [PubMed] [Google Scholar]
- 17.Feng Q, Baird D, Cerione RA. Novel regulatory mechanisms for the Dbl family guanine nucleotide exchange factor Cool-2/alpha-Pix. EMBO J. 2004;23:3492–3504. doi: 10.1038/sj.emboj.7600331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baird D, Feng Q, Cerione RA. The Cool-2/alpha-Pix protein mediates a Cdc42-Rac signaling cascade. Curr. Biol. 2005;15:1–10. doi: 10.1016/j.cub.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 19.Brugnera E, Haney L, Grimsley C, Lu M, Walk SF, Tosello-Trampont AC, Macara IG, Madhani H, Fink GR, Ravichandran KS. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 2002;4:574–582. doi: 10.1038/ncb824. [DOI] [PubMed] [Google Scholar]
- 20.Cote JF, Vuori K. Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J. Cell Sci. 2002;115:4901–4913. doi: 10.1242/jcs.00219. [DOI] [PubMed] [Google Scholar]
- 21.Park D, Tosello-Trampont AC, Elliott MR, Lu M, Haney LB, Ma Z, Klibanov AL, Mandell JW, Ravichandran KS. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007;450:430–434. doi: 10.1038/nature06329. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Wu YC, Fadok VA, Lee MC, Gengyo-Ando K, Cheng LC, Ledwich D, Hsu PK, Chen JY, Chou BK, Henson P, Mitani S, Xue D. Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12. Science. 2003;302:1563–1566. doi: 10.1126/science.1087641. [DOI] [PubMed] [Google Scholar]
- 23.Wu YC, Horvitz HR. C. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180. Nature. 1998;392:501–504. doi: 10.1038/33163. [DOI] [PubMed] [Google Scholar]
- 24.Wu YC, Tsai MC, Cheng LC, Chou CJ, Weng NY. C. elegans CED-12 acts in the conserved crkII/DOCK180/Rac pathway to control cell migration and cell corpse engulfment. Dev. Cell. 2001;1:491–502. doi: 10.1016/s1534-5807(01)00056-9. [DOI] [PubMed] [Google Scholar]
- 25.Henson PM. Engulfment: ingestion and migration with Rac, Rho and TRIO. Curr. Biol. 2005;15:R29–30. doi: 10.1016/j.cub.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 26.Yajnik V, Paulding C, Sordella R, McClatchey AI, Saito M, Wahrer DC, Reynolds P, Bell DW, Lake R, van den Heuvel S, Settleman J, Haber DA. DOCK4, a GTPase activator, is disrupted during tumorigenesis. Cell. 2003;112:673–684. doi: 10.1016/s0092-8674(03)00155-7. [DOI] [PubMed] [Google Scholar]
- 27.Namekata K, Enokido Y, Iwasawa K, Kimura H. MOCA induces membrane spreading by activating Rac1. J. Biol. Chem. 2004;279:14331–14337. doi: 10.1074/jbc.M311275200. [DOI] [PubMed] [Google Scholar]
- 28.Miyamoto Y, Yamauchi J, Sanbe A, Tanoue A. Dock6, a Dock-C subfamily guanine nucleotide exchanger, has the dual specificity for Rac1 and Cdc42 and regulates neurite outgrowth. Exp. Cell Res. 2007;313:791–804. doi: 10.1016/j.yexcr.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 29.Lu M, Kinchen JM, Rossman KL, Grimsley C, Hall M, Sondek J, Hengartner MO, Yajnik V, Ravichandran KS. A Steric-inhibition model for regulation of nucleotide exchange via the Dock180 family of GEFs. Curr. Biol. 2005;15:371–377. doi: 10.1016/j.cub.2005.01.050. [DOI] [PubMed] [Google Scholar]
- 30.Cote JF, Vuori K. GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol. 2007;17:383–393. doi: 10.1016/j.tcb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gumienny TL, Brugnera E, Tosello-Trampont AC, Kinchen JM, Haney LB, Nishiwaki K, Walk SF, Nemergut ME, Macara IG, Francis R, Schedl T, Qin Y, Van Aelst L, Hengartner MO, Ravichandran KS. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 2001;107:27–41. doi: 10.1016/s0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- 32.Lu M, Kinchen JM, Rossman KL, Grimsley C, deBakker C, Brugnera E, Tosello-Trampont AC, Haney LB, Klingele D, Sondek J, Hengartner MO, Ravichandran KS. PH domain of ELMO functions in trans to regulate Rac activation via Dock180. Nat. Struct. Mol. Biol. 2004;11:756–762. doi: 10.1038/nsmb800. [DOI] [PubMed] [Google Scholar]
- 33.Santy LC, Ravichandran KS, Casanova JE. The DOCK180/Elmo complex couples ARNO-mediated Arf6 activation to the downstream activation of Rac1. Curr. Biol. 2005;15:1749–1754. doi: 10.1016/j.cub.2005.08.052. [DOI] [PubMed] [Google Scholar]
- 34.Grimsley CM, Kinchen JM, Tosello-Trampont AC, Brugnera E, Haney LB, Lu M, Chen Q, Klingele D, Hengartner MO, Ravichandran KS. Dock180 and ELMO1 proteins cooperate to promote evolutionarily conserved Rac-dependent cell migration. J. Biol. Chem. 2004;279:6087–6097. doi: 10.1074/jbc.M307087200. [DOI] [PubMed] [Google Scholar]
- 35.Yang J, Zhang Z, Roe SM, Marshall CJ, Barford D. Activation of Rho GTPases by DOCK exchange factors is mediated by a nucleotide sensor. Science. 2009;325:1398–1402. doi: 10.1126/science.1174468. [DOI] [PubMed] [Google Scholar]
- 36.Koland JG, O'Brien KM, Cerione RA. Expression of epidermal growth factor receptor sequences as E. coli fusion proteins: applications in the study of tyrosine kinase function. Biochem. Biophys. Res. Commun. 1990;166:90–100. doi: 10.1016/0006-291x(90)91915-f. [DOI] [PubMed] [Google Scholar]
- 37.Bagrodia S, Taylor SJ, Jordon KA, Van Aelst L, Cerione RA. A novel regulator of p21-activated kinases. J. Biol. Chem. 1998;273:23633–23636. doi: 10.1074/jbc.273.37.23633. [DOI] [PubMed] [Google Scholar]
- 38.Cote JF, Motoyama AB, Bush JA, Vuori K. A novel and evolutionarily conserved PtdIns(3,4,5)P3-binding domain is necessary for DOCK180 signalling. Nat. Cell Biol. 2005;7:797–807. doi: 10.1038/ncb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Worthylake DK, Rossman KL, Sondek J. Crystal structure of Rac1 in complex with the guanine nucleotide exchange region of Tiam1. Nature. 2000;408:682–688. doi: 10.1038/35047014. [DOI] [PubMed] [Google Scholar]
- 40.Gao Y, Xing J, Streuli M, Leto TL, Zheng Y. Trp(56) of rac1 specifies interaction with a subset of guanine nucleotide exchange factors. J. Biol. Chem. 2001;276:47530–47541. doi: 10.1074/jbc.M108865200. [DOI] [PubMed] [Google Scholar]
- 41.Karnoub AE, Worthylake DK, Rossman KL, Pruitt WM, Campbell SL, Sondek J, Der CJ. Molecular basis for Rac1 recognition by guanine nucleotide exchange factors. Nat. Struct. Biol. 2001;8:1037–1041. doi: 10.1038/nsb719. [DOI] [PubMed] [Google Scholar]
- 42.Kwofie MA, Skowronski J. Specific recognition of Rac2 and Cdc42 by DOCK2 and DOCK9 guanine nucleotide exchange factors. J. Biol. Chem. 2008;283:3088–3096. doi: 10.1074/jbc.M705170200. [DOI] [PubMed] [Google Scholar]
- 43.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL Workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 44.Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Research. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peitsch MC. Protein modeling by E-mail. BioTechnology. 1995;13:658–660. [Google Scholar]
- 46.Hess B, Kutzner C, van der spoel D, Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 47.Tina KG, Badra R, Srinivasan N. PIC: protein interactions calculator. Nucleic Acids Research. 2007;35(web-server issue):W473–476. doi: 10.1093/nar/gkm423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bionformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.