Abstract
A paramount objective of the eukaryotic cell division cycle is to overcome numerous internal and external insults to faithfully duplicate the genetic information once per every cycle. This is carried out by elaborate networks of genome surveillance signaling pathways, termed replication checkpoints. Central to replication checkpoints are two protein kinases, the upstream kinase ATR, and its downstream target kinase, Chk1. When the DNA replication process is interrupted, the ATR-Chk1 pathway transmits signals to delay cell cycle progression, and to maintain fork viability so that DNA duplication can resume after the initial damage is corrected. Previous studies showed that replicative stress not only activated Chk1, but also triggered the ubiquitin-dependent destruction of Chk1 in cultured human cells. In a recent study, we identified the F-box protein, Fbx6, as the mediator that regulates Chk1 ubiquitination and degradation in both normally cycling cells and during replication stress. We further showed that expression levels of Chk1 and Fbx6 exhibited an overall inverse correlation in both cultured cancer cell lines and in breast tumor tissues, and that defects in Chk1 degradation, for instance, due to reduced expression of Fbx6, rendered tumor cells resistant to anticancer treatment. Here we highlight those findings and their implications in the replication checkpoint and cellular sensitivity to cancer therapies.
Keywords: Chk1, Fbx6, replication checkpoint, cancer, therapy resistance
The replication checkpoint monitors the progress of DNA replication forks during S phase, and delays the firing of later replication origins when active replication forks are stalled due to collisions with damaged or abnormally structured DNA.1,2 Key components of the replication checkpoint are the apical protein kianse, ATR, and its downstream target kinase, Chk1. Activation of the ATR-Chk1 pathway is initiated by loading of replication protein A (RPA) onto single-stranded DNA (ssDNA) stretches generated either through the uncoupling between the active helicase and the stalled DNA polymerase or through the 5' to 3' end processing of double-stranded breaks (DSBs).3-6 These RPA-bound ssDNA attracts ATR and its partner protein ATRIP,7,8 together with a replication factor C (RFC)-like complex containing Rad17 that localizes specifically at ssDNA-DSB junctions. The Rad17-RFC, in turn, promotes loading of the PCNA-like Rad9-Rad1-Hus1 (9-1-1) complex at the DNA damage site.7,9 Phosphorylation of Rad17 and Rad9 by ATR facilitates the recruitment of adaptor proteins, including TopBP1 and Claspin, which significantly increase the catalytic activity of the ATR kinase to induce phosphorylation of the major ATR substrate, Chk1, at Ser-317 and Ser-345, leading to checkpoint activation.10-15
Multi-Faceted Regulation of Chk1
CHK1 encodes a serine/threonine protein kinase with a highly conserved amino-terminal catalytic domain and a regulatory carboxyl-terminus. Loss of CHK1 led to early embryonic lethality in mice,11,16 underscoring the importance of this protein kinase in the maintenance of cell viability even in the absence of external insults. Chk1 responds primarily to replication fork abnormalities via ATR-dependent phosphorylation at two sites, Ser-345 and Ser-317,10,11 through which it activates an array of downstream events to provoke cell cycle arrest, preserve replication fork viability, activate DNA repair mechanisms, and terminate the checkpoint to resume cell division cycle.
Recent studies revealed multi-faceted regulatory mechanisms of Chk1. First, our results and work of others indicated a spatiotemporal regulation of Chk1 upon DNA damage, in which phosphorylation of Chk1 by ATR triggers its release from the chromatin-enriched fraction into soluble nuclear, cytoplasmic and centrosome compartments,17-19 where it coordinates the activation of the cell cycle arrest or repair function, as well as being degraded to terminate the activated checkpoint. The chromatin-associated Chk1 probably not only monitors the movement of replicating forks, but may also regulate other chromosomal activities, such as transcriptional regulation.20 Second, evolution has overlaid Chk1 phosphorylation of different functions. For instance, while phosphorylation at Ser-317 is a prerequisite for phosphorylation at Ser-345, Ser-345 phosphorylation plays an essential role for mediating the replication checkpoint and cellular viability.21-23 Third, increasing evidence suggests that the carboxyl-terminal regulatory domain of Chk1 is not only required for Chk1 activation, but may also function as an auto-inhibition region in cells possibly through forming an intra-molecular interaction with the catalytic domain.24-28 Forth, a number of studies showed that Chk1 undergoes proteasome-dependent degradation under both normal and stressful environmental conditions.18,29-33 Further investigation indicates that this Chk1 degradation requires its ubiquitination by the Skp1-Cul1-Fbx6 and/or Cul4A-DDB1 ubiquitin E3 ligases.18,32,33 These studies propose a model, in which phosphorylation of Chk1 by ATR transits this protein kinase from a “closed” inactive conformation to an “open” active structure that both promotes Chk1 substrate phosphorylation and checkpoint activation, and renders the protein susceptible to proteasome-dependent degradation by ubiquitin ligases,32,33 and likely to de-phosphorylation by phosphatases as well,34-36 thereby limiting the duration of checkpoint signaling. This coupled activation-destruction mechanism prevents the accumulation of activated Chk1 as cells cope with transient replication stress encountered during a normal S phase; thus, it provides a negative feedback regulation to turn off the activated replication checkpoint and resume the normal cell cycle progression.
Targeted Degradation of Chk1 in Cancer Therapy
Numerous studies revealed that persistent exposure of cancer cell lines to a variety of replicative stresses, including deep hypoxia (O2 <0.1%), ionization radiation, the topoisomerase I inhibitor—CPT, methylmethanesulfonate and aphidicolin, provoked a significant downregulation of the Chk1 protein.10,18,29-33,35,37-43 This downregulation is mainly attributed to the ubiquitination and proteasomal-dependent degradation of Chk1 by the Skp1-Cul1-Fbx6 and/or Cul4A-DDB1 ubiquitin E3 ligases.18,30-33 CHK1 is an essential gene; even a 50% reduction in the level of Chk1 will lead to spontaneous cell death.44 In agreement with this, severe degradation of Chk1 by replicative stress treatment is counter-productive, presumably because when the level of Chk1 is reduced below a threshold, cells would not be able to maintain the replication forks and hold at S or G2 phase in the presence of replicative stress. As a result, the replication fork will collapse and cells will enter mitosis with significant amount of damaged DNA; ultimately, those cells will be cleared out through suicide mechanisms. In fact, this may well represent one of the molecular killing mechanisms for many anticancer drugs that target the replication checkpoint.
This led us to hypothesize that if cancer cells bear defects in downregulating Chk1, then they should show resistance to anti-cancer therapies, because maintaining a constant level of Chk1 will better protect them from the harsh anticancer treatment. Indeed, from the NCI 60 cell line database we found that two cell lines (the breast cancer cell line MDA-MB-231 and renal cancer cell line TK-10) that elicited extraordinary resistance to CPT did not degrade Chk1 upon replicative stress treatment, including CPT and IR. Similarly, an earlier study reported that a trinuclear platinum complex induced downregulation of Chk1 in the parent ovarian carcinoma cell line, but not in a drug resistant sub-clone.39 Depletion of Chk1 by RNA interference dramatically increased MDA-MB-231 cells’ sensitivity to CPT. Interestingly, inhibiting Chk1 suppressed the radio-resistance of glioblastoma cancer stem cells.45 These data suggest that Chk1 degradation defects may represent a relatively general mechanism by which cancer cells acquire therapy resistance, and ultimately facilitate the cell survival of those resistant cancer cells upon anticancer treatment. In consistent with this hypothesis, loss of function mutations of Chk1 is rarely observed in human cancers.46 Instead, Chk1 has been found to be overexpressed in various tumors compared to the adjacent normal tissues,40,47-49 and expression of Chk1 proteins positively correlated with the tumor grade and cancer cell proliferation of breast tumors.50 These lines of findings suggest that Chk1 plays an important role in regulating cellular response to anticancer treatment, and its expression in primary tumors might serve as a predictive biomarker of tumor responsiveness to these important anticancer drugs.
In order to understand the molecular mechanisms underlying the Chk1 degradation defect, we examined the expression level of Fbx6, the ubiquitin E3 ligase that regulates Chk1 protein stability, in both sensitive and resistant cancer cells. Remarkably, we found that the two CPT resistant lines (MDA-MB-231 and TK-10) displayed very low expression of Fbx6 compared to the sensitive cell line, A549; accordingly, the expression level of Chk1 was higher in these two resistant lines than in the sensitive line. To determine whether the relationship between Fbx6 and Chk1 expression levels extend beyond these three lines, we surveyed expression of Chk1 and Fbx6 in non-small cell lung carcinoma, glioblatoma and breast cancer cells. Indeed, we find that the majority of cell lines showed an inverse correlation between Chk1 and Fbx6, i.e., cells with high Fbx6 expression tend to express relatively low Chk1, and vice versa.
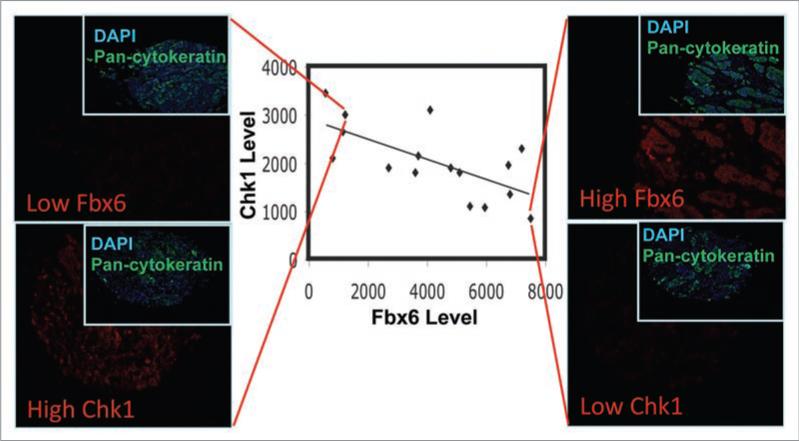
To further confirm this Fbx6-Chk1 correlation, we analyzed the expression of Fbx6 and Chk1 protein in chemotherapy-naïve human breast tumors. The results showed a significant inverse correlation between Chk1 and Fbx6 in a panel of 16 breast tumor tissues (Fig. 1). We recognize that the current tumor dataset, while suggestive, is not definitive, and that a much larger cohort of breast tumors, as well as other tumor types, will need to be evaluated before the true significance and scope of the Fbx6-Chk1 relationship is fully understood. However, we feel strongly that these preliminary results in the breast cancer tissue and cultured cell lines do suggest that a broader examination of human tumors (particularly the anticancer-resistant subset) is warranted, and believe that the current findings will provide the impetus to undertake a full investigation of the Fbx6-Chk1 relationship in tumor tissues with associated patient outcome data.
Figure 1.
Inverse correlation between Chk1 and Fbx6 in breast tumors. A panel of 16 breast tumor tissues were analyzed for expression of Fbx6 and Chk1 by quantitative immunostaining using the AQUA™ technology that allows reproducible measurements of proteins of interest in both the cytoplasmic and nuclear compartments of cells in fixed tissues. These tumor samples had not been categorized for cancer sub-type or stage at the time of excision. Expression of Fbx6 and Chk1 was plotted. The linear regression is well fitted, with an R-square of 0.351. The trend line is the result of linear regression between Fbx6 and Chk1 AQUA scores with a Pearson correlation coefficient of R = -0.59 (p = 0.016). Rank order correlation analysis of the data indicates a Spearman rho = -0.556 (p = 0.025). Two tumor staining representing high Chk1/low Fbx6 and low Chk1/high Fbx6 were shown on the left and right, respectively. DAPI and pan-cytokeratin were used to stain the nucleus and cytoplasm, respectively.
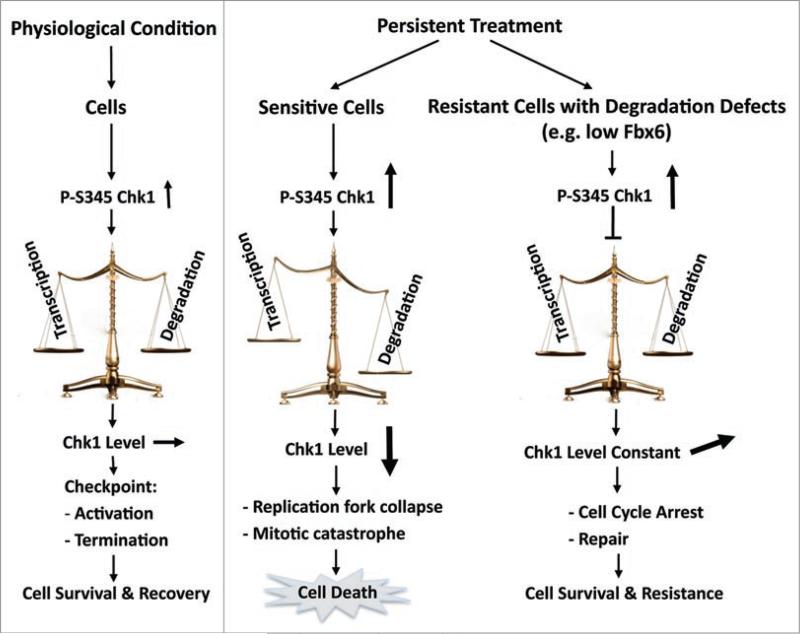
In addition to the reduced expression, deregulated function of Fbx6, for instance, abnormal localization or mutation that impairs its E3 ligase activity or interaction with Chk1, might also contribute to Chk1 degradation defects. This could help to explain why different level of Chk1 degradation was observed among various studies, and why some cell lines have difficulty in degrading Chk1 even they express Fbx6 well, for instance, HeLa cell. More work is needed to understand the mechanisms whereby certain tumors acquire a loss of Fbx6 function or bear Fbx6-Chk1 mutations. Together, these studies suggest a model of Darwinian micro-evolution in the tumor tissue, in which cells that acquire phenotypic alterations conferring a survival advantage, for instance, increased or stabilized Chk1 protein expression due to reduced expression or abnormal function of Fbx6 or Cul4A/DDB1, over their neighbors, are better-equipped to handle replicative stress, and resume the S-phase cell cycle progression after the damage is removed. Importantly, those cells will eventually assume dominance within the tumor, and are considered to be a breeding ground for increasingly malignant, drug-resistant cancer cells—the same cells that give rise to tumor recurrence as well as metastatic disease (Fig. 2).
Figure 2.
Roles of the Fbx6-Chk1 axis in cellular sensitivity to anticancer therapy. During a transient treatment or a physiological interruption to the replication movement, the level of Chk1 will maintain constant due to the balance between transcription and degradation. In this situation, this coupled activation-degradation mechanism keeps cell viable. In contrast, during prolonged treatment, continuous phosphorylation of Chk1 will eventually lead to significant reduction in the level of Chk1, causing loss of cell viability. However, when cancer cells acquire Chk1 degradation defects, for instance, due to reduced expression of Fbx6, the protein level of Chk1 will be kept constant or increased, and cells will survive after treatment, which translates into therapy resistance in the clinical setting.
The fact that not all cell lines exhibit an inverse correlation between Chk1 and Fbx6 indicates that Fbx6 is not the sole determinant of Chk1 expression.32 This is nothing more than the reality of human cancer cell biology; in fact, we believe that our cell line panel provides the reader with a perspective on the importance of the Fbx6-Chk1 relationship that cannot be gleaned from the many reductionist studies that focus on one or two cell lines. In addition, we also note that the breast cancer Hs578T cell line, which is also defined as a CPT resistant cell line in the NCI 60 cell line panel, showed Chk1 downregulation, reinforcing the idea that multiple pathways control CPT sensitivity and Chk1 protein level, and Fbx6 expression level is an important but not the sole parameter that governs Chk1 turnover during replication stress. Nonetheless, the heterogeneous expression of Chk1 and Fbx6 proteins in both cancer cell lines and tumor tissues lays the groundwork for more comprehensive analyses of the impact of expression of the Fbx6-Chk1 axis on patient outcome, or as a predictive biomarker for tumor responsiveness to CPT and other replication stress-inducing agents.
Acknowledgements
Y.W.Z. was a recipient of the NCI K99 Career Development Award (5 K99 CA126173), and currently funded by the NCI R00 Career Development Award (4 R00 CA126173) and Case Comprehensive Cancer Center Startup funding.
References
- 1.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–96. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 2.Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem Sci. 2006;31:402–10. doi: 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Pacek M, Walter JC. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 2004;23:3667–76. doi: 10.1038/sj.emboj.7600369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–52. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 6.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–14. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–8. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 8.Kim SM, Kumagai A, Lee J, Dunphy WG. Phosphorylation of Chk1 by ATM- and Rad3-related (ATR) in Xenopus egg extracts requires binding of ATRIP to ATR but not the stable DNA-binding or coiled-coil domains of ATRIP. J Biol Chem. 2005;280:38355–64. doi: 10.1074/jbc.M508673200. [DOI] [PubMed] [Google Scholar]
- 9.Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao H, Piwnica-Worms H. ATR-mediated check-point pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–39. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–59. [PMC free article] [PubMed] [Google Scholar]
- 12.Kumagai A, Dunphy WG. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol Cell. 2000;6:839–49. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- 13.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–55. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Zou L, Lu T, Bao S, Hurov KE, Hittelman WN, et al. Rad17 phosphorylation is required for claspin recruitment and Chk1 activation in response to replication stress. Mol Cell. 2006;23:331–41. doi: 10.1016/j.molcel.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 15.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–7. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takai H, Tominaga K, Motoyama N, Minamishima YA, Nagahama H, Tsukiyama T, et al. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(-/-) mice. Genes Dev. 2000;14:1439–47. [PMC free article] [PubMed] [Google Scholar]
- 17.Kramer A, Mailand N, Lukas C, Syljuasen RG, Wilkinson CJ, Nigg EA, et al. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat Cell Biol. 2004;6:884–91. doi: 10.1038/ncb1165. [DOI] [PubMed] [Google Scholar]
- 18.Zhang YW, Otterness DM, Chiang GG, Xie W, Liu YC, Mercurio F, et al. Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Mol Cell. 2005;19:607–18. doi: 10.1016/j.molcel.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 19.Smits VA, Reaper PM, Jackson SP. Rapid PIKK-dependent release of Chk1 from chromatin promotes the DNA-damage checkpoint response. Curr Biol. 2006;16:150–9. doi: 10.1016/j.cub.2005.11.066. [DOI] [PubMed] [Google Scholar]
- 20.Shimada M, Niida H, Zineldeen DH, Tagami H, Tanaka M, Saito H, et al. Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell. 2008;132:221–32. doi: 10.1016/j.cell.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 21.Niida H, Katsuno Y, Banerjee B, Hande MP, Nakanishi M. Specific role of Chk1 phosphorylations in cell survival and checkpoint activation. Mol Cell Biol. 2007;27:2572–81. doi: 10.1128/MCB.01611-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilsker D, Petermann E, Helleday T, Bunz F. Essential function of Chk1 can be uncoupled from DNA damage checkpoint and replication control. Proc Natl Acad Sci USA. 2008;105:20752–7. doi: 10.1073/pnas.0806917106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker M, Black EJ, Oehler V, Gillespie DA, Scott MT. Chk1 C-terminal regulatory phosphorylation mediates checkpoint activation by de-repression of Chk1 catalytic activity. Oncogene. 2009;28:2314–23. doi: 10.1038/onc.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oe T, Nakajo N, Katsuragi Y, Okazaki K, Sagata N. Cytoplasmic occurrence of the Chk1/Cdc25 pathway and regulation of Chk1 in Xenopus oocytes. Dev Biol. 2001;229:250–61. doi: 10.1006/dbio.2000.9968. [DOI] [PubMed] [Google Scholar]
- 25.Katsuragi Y, Sagata N. Regulation of Chk1 kinase by autoinhibition and ATR-mediated phosphorylation. Mol Biol Cell. 2004;15:1680–9. doi: 10.1091/mbc.E03-12-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen P, Luo C, Deng Y, Ryan K, Register J, Margosiak S, et al. The 1.7 A crystal structure of human cell cycle checkpoint kinase Chk1: implications for Chk1 regulation. Cell. 2000;100:681–92. doi: 10.1016/s0092-8674(00)80704-7. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y, Caldwell JM, Pereira E, Baker RW, Sanchez Y. ATRMec1 Phosphorylation-independent Activation of Chk1 in Vivo. J Biol Chem. 2009;284:182–90. doi: 10.1074/jbc.M806530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kosoy A, O'Connell MJ. Regulation of Chk1 by its C-terminal domain. Mol Biol Cell. 2008;19:4546–53. doi: 10.1091/mbc.E08-04-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collis SJ, Barber LJ, Clark AJ, Martin JS, Ward JD, Boulton SJ. HCLK2 is essential for the mammalian S-phase checkpoint and impacts on Chk1 stability. Nat Cell Biol. 2007;9:391–401. doi: 10.1038/ncb1555. [DOI] [PubMed] [Google Scholar]
- 30.Feng JM, Zhu H, Zhang XW, Ding J, Miao ZH. Proteasome-dependent degradation of Chk1 kinase induced by the topoisomerase II inhibitor R16 contributes to its anticancer activity. Cancer Biol Ther. 2008;7:1726–31. doi: 10.4161/cbt.7.11.6728. [DOI] [PubMed] [Google Scholar]
- 31.Jurvansuu J, Fragkos M, Ingemarsdotter C, Beard P. Chk1 Instability Is Coupled to Mitotic Cell Death of p53-deficient Cells in Response to Virus-induced DNA Damage Signaling. J Mol Biol. 2007;372:397–406. doi: 10.1016/j.jmb.2007.06.077. [DOI] [PubMed] [Google Scholar]
- 32.Leung-Pineda V, Huh J, Piwnica-Worms H. DDB1 targets Chk1 to the Cul4 E3 ligase complex in normal cycling cells and in cells experiencing replication stress. Cancer Res. 2009;69:2630–7. doi: 10.1158/0008-5472.CAN-08-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang YW, Brognard J, Coughlin C, You Z, Dolled-Filhart M, Aslanian A, et al. The F box protein Fbx6 regulates Chk1 stability and cellular sensitivity to replication stress. Mol Cell. 2009;35:442–53. doi: 10.1016/j.molcel.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 2005;19:1162–74. doi: 10.1101/gad.1291305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leung-Pineda V, Ryan CE, Piwnica-Worms H. Phosphorylation of Chk1 by ATR is antagonized by a Chk1-regulated protein phosphatase 2A circuit. Mol Cell Biol. 2006;26:7529–38. doi: 10.1128/MCB.00447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.den Elzen NR, O'Connell MJ. Recovery from DNA damage checkpoint arrest by PP1-mediated inhibition of Chk1. EMBO J. 2004;23:908–18. doi: 10.1038/sj.emboj.7600105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukas C, Bartkova J, Latella L, Falck J, Mailand N, Schroeder T, et al. DNA damage-activated kinase Chk2 is independent of proliferation or differentiation yet correlates with tissue biology. Cancer Res. 2001;61:4990–3. [PubMed] [Google Scholar]
- 38.Hu B, Zhou XY, Wang X, Zeng ZC, Iliakis G, Wang Y. The radioresistance to killing of A1-5 cells derives from activation of the Chk1 pathway. J Biol Chem. 2001;276:17693–8. doi: 10.1074/jbc.M009340200. [DOI] [PubMed] [Google Scholar]
- 39.Perego P, Gatti L, Righetti SC, Beretta GL, Carenini N, Corna E, et al. Development of resistance to a trinuclear platinum complex in ovarian carcinoma cells. Int J Cancer. 2003;105:617–24. doi: 10.1002/ijc.11140. [DOI] [PubMed] [Google Scholar]
- 40.Ng CP, Lee HC, Ho CW, Arooz T, Siu WY, Lau A, et al. Differential mode of regulation of the checkpoint kinases CHK1 and CHK2 by their regulatory domains. J Biol Chem. 2004;279:8808–19. doi: 10.1074/jbc.M312215200. [DOI] [PubMed] [Google Scholar]
- 41.Jin ZH, Kurosu T, Yamaguchi M, Arai A, Miura O. Hematopoietic cytokines enhance Chk1-dependent G2/M checkpoint activation by etoposide through the Akt/GSK3 pathway to inhibit apoptosis. Oncogene. 2005;24:1973–81. doi: 10.1038/sj.onc.1208408. [DOI] [PubMed] [Google Scholar]
- 42.Maude SL, Enders GH. Cdk inhibition in human cells compromises chk1 function and activates a DNA damage response. Cancer Res. 2005;65:780–6. [PubMed] [Google Scholar]
- 43.Matthew EM, Yen TJ, Dicker DT, Dorsey JF, Yang W, Navaraj A, et al. Replication stress, defective S-phase checkpoint and increased death in Plk2-deficient human cancer cells. Cell Cycle. 2007;6:2571–8. doi: 10.4161/cc.6.20.5079. [DOI] [PubMed] [Google Scholar]
- 44.Lam MH, Liu Q, Elledge SJ, Rosen JM. Chk1 is haploinsufficient for multiple functions critical to tumor suppression. Cancer Cell. 2004;6:45–59. doi: 10.1016/j.ccr.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 45.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 46.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–9. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 47.Cho SH, Toouli CD, Fujii GH, Crain C, Parry D. Chk1 is essential for tumor cell viability following activation of the replication checkpoint. Cell Cycle. 2005;4:131–9. doi: 10.4161/cc.4.1.1299. [DOI] [PubMed] [Google Scholar]
- 48.Madoz-Gurpide J, Canamero M, Sanchez L, Solano J, Alfonso P, Casal JI. A proteomic analysis of cell signaling alterations in colorectal cancer. Mol Cell Proteomics. 2007;6:2150–64. doi: 10.1074/mcp.M700006-MCP200. [DOI] [PubMed] [Google Scholar]
- 49.Verlinden L, Vanden Bempt I, Eelen G, Drijkoningen M, Verlinden I, Marchal K, et al. The E2F-regulated gene Chk1 is highly expressed in triple-negative estrogen receptor/progesterone receptor/HER-2 breast carcinomas. Cancer Res. 2007;67:6574–81. doi: 10.1158/0008-5472.CAN-06-3545. [DOI] [PubMed] [Google Scholar]
- 50.Lundgren K, Holm K, Nordenskjold B, Borg A, Landberg G. Gene products of chromosome 11q and their association with CCND1 gene amplification and tamoxifen resistance in premenopausal breast cancer. Breast Cancer Res. 2008;10:81. doi: 10.1186/bcr2150. [DOI] [PMC free article] [PubMed] [Google Scholar]