Abstract
Utilization of gene silencing effectors, such as microRNA (miRNA) and small hairpin RNA (shRNA), provides a powerful new strategy for human skin care in vivo, particularly for hyperpigmentation treatment and aging prevention. In this study, tyrosinase (Tyr), the rate-limiting enzyme of melanin (black pigment) biosynthesis, was served as a target for treatment of hyperpigmentation in mouse and human skins. There are over 54 native microRNA capable of silencing human tyrosinase for skin whitening and lightening. To this, we have designed a mir-434-5p homologue and used it to successfully demonstrate the feasibility of miRNA-mediated skin whitening and lightening in vitro and in vivo. Under the same experimental condition in the trials, Pol-II-directed intronic mir-434-5p expression did not cause any detectable sign of cytotoxicity, whereas siRNAs targeting the same sequence often induced certain nonspecific mRNA degradation as previously reported. Because the intronic miRNA-mediated gene silencing pathway is tightly regulated by multiple intracellular surveillance systems, including Pol-II transcription, RNA splicing, exosomal digestion and nonsense-mediated RNA decay (NMD), the current findings underscore the fact that intronic miRNA agents, such as manually re-designed mir-434-5p homologues, are effective, target-specific and safe to be used for skin whitening without any detectable cytotoxic effect. Given that the human skins also express a variety of other native miRNAs, we may re-design these miRNAs based on their individual functions for skin care, which may provide significant insights into areas of opportunity for new cosmetic and/or therapeutical applications.
Keywords: microRNA, miRNA, mir-434, intron, gene silencing, RNAi, tyrosinase, melanin, cosmetics, pigmentation, skin whitening
Introduction
Prevention of hyperpigmentation (ie, sun-burn) and aging is the key means for having healthy skins. However, many of the skin pigmentation and aging processes are associated with personal gene activities. For example, tyrosinase (Tyr), a melanocytic membrane-bound glycoprotein, is the rate-limiting enzyme critical for melanin (black pigment) biosynthesis in skins and hairs, while hyaluronidase (Hyal) often causes skin wrinkle by degrading subcutaneous hyaluronan (HA), the major antiaging extracellular matrix in skins. Therefore, a good skin care can be achieved by suppressing the activities of these enzymes.
Among a variety of currently available skin whitening and lightening products, many chemical and naturally extracted agents have been applied to inhibit tyrosinase function, using materials such as hormone-derived inhibitory oligopeptides, hydroxytetronic acid derivatives, benzoyl compounds, hydroquinone compositions, alcohol diol and triol analogues, kojic acid derivatives, ascomycete-derived enzymes, and plant extracts. Although these cosmetic agents work well in vitro, only a few of them, such as hydroquinone and its derivatives, are able to induce decent hypopigmenting effects in clinical trials (Solano et al 2006). Nevertheless, all hydroquinone derivatives leading to a reactive quinone are putative cytotoxic agents. Thus, the gap between in vitro and in vivo studies suggests that innovative strategies are needed for validating their safety and efficacy.
With the advance of recent RNA interference (RNAi) technologies, novel small RNA agents have been found to mediate more potent effects in targeted gene suppression. These include double-stranded short interfering RNA (eg, dsRNA/siRNA) and doxyribonucleotidylated-RNA interfering molecules (eg, D-RNAi) (Fire et al 1998; Elbashir et al 2001; Lin and Ying 2001). Conceivably, these small RNA agents may be used to develop new cosmetic designs and products for skin care. In principle, the RNAi mechanism elicits a post-transcriptional gene silencing (PTGS) phenomenon capable of inhibiting specific gene function with high potency at a few nanomolar dosage, which has been proven to be effective longer and much less toxic than conventional gene-knockout methods using antisense oligonucleotides or small molecule chemical inhibitors (Lin and Ying 2001). As reported in many previous studies, the siRNA-induced gene silencing effects may last over one week, while the D-RNAi effects can even sustain up to one month after one treatment (Grant 1999; Elbashir et al 2001; Lin and Ying 2001; Lin and Ying 2004). These siRNA/D-RNAi agents evoke a series of intracellular sequence-specific mRNA degradation and/or translational suppression processes, affecting all highly homologous gene transcripts, namely co-suppression. It has been observed that such co-suppression results from the generation of small RNA products (21–25 nucleotide bases) by the enzymatic activities of RNaseIII endoribonucleases (Dicer) and/or RNA-directed RNA polymerases (RdRp) on aberrant RNA templates, which are usually the derivatives of foreign transgenes or viral genomes (Grant 1999; Elbashir et al 2001; Lin and Ying 2001).
Limitations of siRNA/shRNA-based gene silencing agents
Although the modern RNAi technologies may offer a new avenue for suppressing unwanted gene function in skins, the applications thereof have not been demonstrated to work consistently and safely in higher vertebrates, including fish, avian, mammal and human. For example, almost all of the current siRNA agents are based on a double-stranded RNA (dsRNA) conformation, which has been shown to cause interferon-mediated nonspecific RNA degradation in vertebrates (Stark et al 1998; Elbashir et al 2001). Such an interferon-mediated cytotoxic response reduces the target specificity of siRNA-induced gene silencing effects and often results in global RNA degradation in vertebrate cells. Particularly in mammalian cells, it has been noted that the RNAi effects are disturbed when the siRNA/dsRNA size is longer than 25 basepairs (bp) (Elbashir et al 2001). Transfection of siRNA or small hairpin RNA (shRNA) sized less than 25 bp may not completely overcome such a problem, because both Sledz and colleagues (2003) and Lin and Ying (2004) have reported that the high dosage of siRNAs and shRNAs (such as >250 nM in human T cells) is able to cause strong cytotoxic effects similar to those of long dsRNAs. This toxicity is due to their double-stranded RNA conformation, which activates the interferon-mediated nonspecific RNA degradation and programmed cell death through the activation of cellular PKR and 2-5A signaling pathways. It has been well established that interferon-activated protein kinase PKR can trigger cell apoptosis, while the activation of interferon-induced 2′,5′-oligoadenylate synthetase (2-5A) system leads to extensive cleavage of single-stranded RNAs, such as mRNAs (Stark 1998). Both PKR and 2-5A systems contain dsRNA-binding motifs, which possess high affinity to the double-stranded RNA conformation. Further, the most difficult problem is that these small siRNA/shRNA agents are not stable enough to be maintained at an optimal dose in vivo due to the abundant RNase activities in higher vertebrates (Brantl 2002).
As the RNAi effects are naturally caused by the production of small RNA products (21–25 nucleotide bases) from a transcriptional template derived from foreign transgenes or viral genomes (Grant 1999; Lin and Ying 2001), the recent utilization of Pol-III-directed siRNA/shRNA expression vectors has been shown to offer relatively stable RNAi effects in vivo (Tuschl 2002). Although previous studies using such a vector-based siRNA approach have demonstrated constant gene silencing effects (Lee et al 2002; Miyagishi and Tira 2002; Paul et al 2002), their strategies failed to focus the RNAi effects on a targeted cell or tissue population because of the ubiquitous existence of type III RNA polymerase (Pol-III) activity. Pol-III promoters, such as U6 and H1, are activated in almost all cell types, making tissue-specific gene silencing impossible. Moreover, because the leaky read-through activity of Pol-III transcription often occurs on a short DNA template in the absence of proper termination, large RNA products longer than desired 25 bps can be synthesized and cause unexpected interferon-mediated cytotoxicity (Gunnery et al 1999; Schramm 2002). Such a problem can also result from the competitive conflict between the Pol-III promoter and another vector promoter (ie, LTR and CMV promoters). Furthermore, it is recently noted that high siRNA/shRNA concentrations generated by the Pol-III-directed RNAi systems can over-saturate the cellular native microRNA (miRNA) pathway and thus cause global miRNA inhibition and cell death (Grimm et al 2003). These disadvantages discourage the use of Pol-III-based RNAi vector systems in health care. In order to improve the delivery stability, targeting specificity and safety aspects of modern RNAi technologies for healthy skin care, a better transduction and maintenance strategy is highly desired.
Intronic microRNA-mediated RNAi mechanism
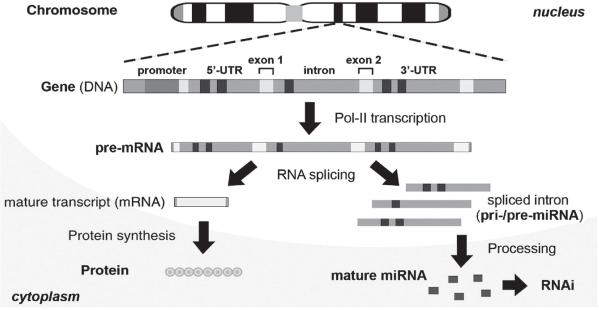
Research based on gene transcript (eg, mRNA), an assembly of protein-coding exons, is fully described throughout the literature, taking the fate of spliced noncoding introns to be completely digested for granted (Nott et al 2003). Is it true that the intronic portion of a gene is destined to be a genetic waste without function or is there an yet undiscovered function for it? Recently, this misconception was corrected by the discovery of intronic microRNA (miRNA) (Lin et al 2003; Ying and Lin 2004; Ying and Lin 2005). Intronic miRNA is a new class of small single-stranded regulatory RNAs derived from the introns, which are spliced out of the precursor messenger RNA (pre-mRNA) of the encoding gene and further processed into small mature miRNAs (Lin et al 2003). A miRNA is usually about 18–27 nucleotides (nt) in length and is capable of either directly degrading its messenger RNA (mRNA) target or suppressing the protein translation of its targeted mRNA, depending on the complementarity between the miRNA and its target. In this way, the intronic miRNA is functionally similar to previously described siRNA/shRNA, but differs from them in the requirement of intracellular type II RNA polymerase (Pol-II) transcription and RNA splicing processes for its biogenesis (Lin et al 2003). In addition, since introns naturally contain multiple translational stop codons for recognition by the intracellular nonsense-mediated decay (NMD) system (Zhang et al 1994; Danin-Kreiselman et al 2003), most of the unstructured intron sequences can be quickly degraded after RNA splicing to prevent excessive accumulation, which is toxic to the cells. It has been determined that approximately 10%–30% of a spliced intron is preserved after the exosome and NMD digestion in cytoplasm with a relatively long half-life, indicating the cellular origin of native intronic miRNAs (Clement et al 1999).
Natural intronic miRNA biogenesis relies on the coupled interaction between nascent Pol-II-mediated pre-mRNA transcription and intron splicing/excision (Figure 1), which take place within certain nuclear regions proximal to genomic perichromatin (Ghosh and Garcia-Blanco 2000; Lin and Ying 2004). In eukaryotic cells, gene transcription is mediated by Pol-II, and the resulting precursor mRNA (pre-mRNA) consists of four major components: 5′-untranslated region (UTR), protein-coding exon, noncoding intron and 3′-UTR. Broadly speaking, both 5′- and 3′-UTR can be seen as a kind of intron extension. Introns occupy the largest proportion of the noncoding sequences in the pre-mRNA. Each intron can range up to 30 kilobases and is excised out of the pre-mRNA during RNA splicing executed by intracellular spliceosomes. Subsequently, some of the intron-derived RNA fragments are further processed to form microRNA (miRNA) derivative molecules, which can effectively silence their respective targeted genes through an RNA interference (RNAi)-like mechanism. Exons, on the other hand, are ligated together to form a mature mRNA for protein synthesis.
Figure 1.
Biogenesis of native intronic microRNA (miRNA). Intronic miRNA is co-transcribed within a precursor messenger RNA (pre-mRNA) by eukaryotic type-II RNA polymerases (Pol-II) and cleaved out of the pre-mRNA by RNA splicing. While the exons are ligated to form a mature messenger RNA (mRNA) for protein synthesis, the spliced intron(s) with a high secondary structure (ie, hairpin and/or stem-loop) is further processed into mature miRNA capable of triggering post-transcriptional gene silencing (PTGS) effects, such as RNA interference (RNAi).
Differences between miRNA and siRNA biogenesis pathways
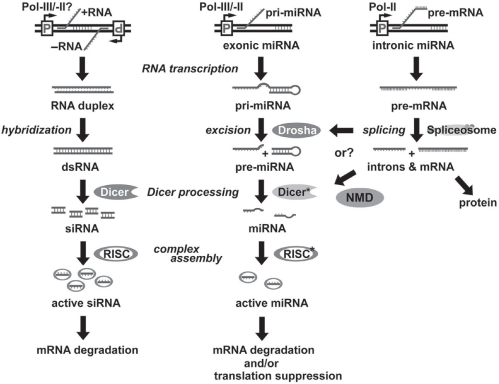
We have demonstrated that effective mature miRNAs can be generated from the introns of vertebrate genes, of which the biogenetic process is different from those of siRNA and intergenic miRNA (Lin et al 2003; Lin et al 2005). Figure 2 compares the native biogenesis and RNAi mechanisms among siRNA, intergenic (exonic) miRNA and intronic miRNA. Presumably, an siRNA is formed by two perfectly complementary RNAs transcribed by two reversely positioned promoters from one DNA template; these complementary RNAs are then hybridized and further processed into 20–25-bp duplexes by RNaseIII endoribonucleases, namely Dicer. Different from this siRNA model, the biogenesis of intergenic miRNA, eg, lin-4 and let-7, involves a long noncoding precursor RNA transcript (pri-miRNA), which is directly transcribed from a Pol-II or Pol-III RNA promoter, whereas intronic miRNA is co-transcribed with its encoding gene by only Pol-II and released after RNA splicing as a spliced intron. The spliced intron then serves as a pri-miRNA for processing into an intronic precursor miRNA (pre-miRNA) or a multi-pre-miRNA cluster. In the cell nucleus, the pri-miRNA is further excised by either Drosha-like RNases (for intergenic miRNA) or spliceosomal components (for intronic miRNA) to form a hairpin-like stem-loop precursor or a cluster of multiple stem-loop structures, termed pre-miRNA, and then exported to cytoplasm for final processing into mature miRNA by a miRNA-associated Dicer (Dicer*) (Lee et al 2003). Subsequently, all three small regulatory RNAs are incorporated into a RNA-induced silencing complex (RISC), which contains either strand of siRNA or the mature strand of miRNA. The Dicers and RISCs for siRNA and miRNA pathways are known to be different (Tang 2005). For example, some enzymes of the nonsense-mediated decay (NMD) system may play the role of Dicer* in the mechanism of intronic miRNA maturation. As a result, the effect of miRNA is generally more specific and less adverse than that of siRNA because only one strand is involved. On the other hand, siRNAs primarily trigger mRNA degradation, whereas miRNAs can induce either mRNA degradation or suppression of protein synthesis, or both, depending on the sequence complementarity to their targeted gene transcripts. Because the intronic miRNA pathway is tightly regulated by multiple intracellular surveillance systems, such as Pol-II transcription, RNA splicing, exosomal digestion and NMD processing, the gene silencing effects of intronic miRNA are considered to be more effective, specific and safer than those of siRNA and intergenic miRNA (Lin et al 2008).
Figure 2.
Comparison of various biogenesis and PTGS mechanisms among siRNA, intergenic (exonic) miRNA and intronic miRNA. SiRNA is formed by annealing of two perfectly complementary short RNAs transcribed from two opposite promoters (remains to be determined), and then further processed into 19–22 bp double-stranded duplexes by RNaseIII-familial endonucleases, Dicer. Intergenic (exonic) miRNA, eg, lin-4 and let-7, is directly transcribed as a long single-strand noncoding RNA precursor (pri-miRNA), probably by either a Pol-II or a Pol-III RNA promoter, whereas intronic miRNA is mainly transcribed by the Pol-II promoter of its encoded gene and co-existed within the intron region of the encoded gene transcript (pre-mRNA). After pre-mRNA splicing, the spliced intron functions as a pri-miRNA to form intronic miRNA. In the nucleus, the pri-miRNA is excised by either a Drosha RNase (intergenic miRNA) or spliceosomal/NMD components (intronic miRNA) to form hairpin-like miRNA precursors (pre-miRNA) and then exported to cytoplasm for further processing by Dicer* to form mature miRNAs. The Dicers for siRNA and miRNA pathways may be different. For instance, some spliceosomal/NMD components are likely involved in the maturation of intronic miRNA but not siRNA. All three small regulatory RNAs are finally incorporated into a RNA-induced silencing complex (RISC), which contains either strand of siRNA or the single-strand of miRNA. The effect of miRNA is considered to be more specific and less adverse than that of siRNA because only one strand is involved. On the other hand, siRNAs primarily trigger mRNA degradation, whereas miRNAs can induce either mRNA degradation or translational suppression, depending on the sequence complementarity to their targeted gene transcripts.
Development of miRNA-based gene silencing agents
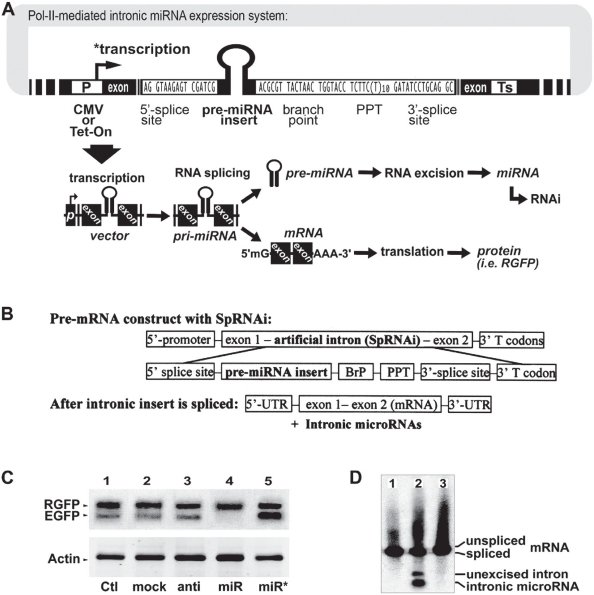
Based on the intronic RNA splicing and processing mechanisms (Figures 3A and 3B), we have designed and developed a Pol-II-mediated recombinant gene expression system containing at least a splicing-competent intron, namely SpRNAi, capable of inhibiting the EGFP gene with high complementarity to the intron sequence. The SpRNAi is co-transcribed with the precursor mRNA (pre-mRNA) of the recombinant gene by Pol-II RNA polymerases (P) and cleaved out of the pre-mRNA by RNA splicing. Subsequently, the spliced SpRNAi is further processed into mature gene silencing agents, such as shRNA and miRNA, capable of triggering RNAi-related gene silencing. After intron removal, the exons of the recombinant gene transcript are linked together to form a mature mRNA molecule for translational synthesis of a marker or functional protein.
Figure 3.
Structural composition of the SpRNAi-incorporated recombinant RGFP gene (SpRNAi-RGFP) in an expression-competent vector (A), and the strategy (B) of using this composition to generate manmade microRNA (miRNA), mimicking the biogenesis mechanism of natural intronic miRNAs. In vivo tests of the SpRNAi-RGFP gene composition against a targeted green EGFP gene in Tg(actin-GAL4:UAS-gfp) zebrafish showed a significant knockdown of over 85% of the EGFP gene expression, as determined by western blot analyses (C). The intron-derived anti-EGFP miRNA and its spliced precursor were clearly observed on a 1% formaldehyde agarose gel electrophoresis after northern blotting (D).
As shown in Figure 3A, the essential components of the SpRNAi intron include several consensus nucleotide elements, consisting of a 5′-splice site, a branch-point motif (BrP), a poly-pyrimidine tract (PPT), and a 3′-splice site. In addition, a hairpin RNA-like pre-miRNA sequence is inserted inside the SpRNAi intron located between the 5′-splice site and the branch-point motif (BrP). This portion of the intron normally forms a lariat structure during RNA splicing and processing. We have observed that spliceosomal U2 and U6 snRNPs, both helicases, are involved in the unwinding and excision of the lariat RNA fragment into pre-miRNA; however, the detailed processing remains to be elucidated. Further, the 3′-end of the SpRNAi construct contains a multiple translational stop codon region (T codon) in order to increase the accuracy of intronic RNA splicing and NMD processing. When presented in a cytoplasmic mRNA, this T codon will signal the activation of the nonsense-mediated decay (NMD) pathway to degrade any unstructured RNA accumulation in the cell. However, the highly secondary structured hairpin RNA and pre-miRNA insert will be preserved for further Dicer cleavage, so as to form mature siRNA and miRNA, respectively. Moreover, for intracellular expression, we manually incorporate the SpRNAi construct in the DraII restriction site of a red fluorescent protein (RGFP) gene isolated from mutated chromoproteins of the coral reef Heteractis crispa, so as to form a recombinant SpRNAi-RGFP gene. The cleavage of RGFP at its 208th nucleotide site by the restriction enzyme DraII generates an AG–GN nucleotide break with three recessing nucleotides in each end, which will form 5′- and 3′-splice sites respectively after the SpRNAi insertion. Because this intronic insertion disrupts the structure of a functional RGFP protein, which can be recovered by intron splicing, we can determine the release of intronic shRNA/miRNA and RGFP-mRNA maturation through the appearance of red RGFP around the affected cells. The RGFP gene also provides multiple exonic splicing enhancers (ESEs) to increase RNA splicing accuracy and efficiency.
In this intronic miRNA expression system (Figure 3B), we provide a genetic engineering method for using synthetic RNA splicing and processing elements, such as 5′-splice site, branch-point motif (BrP), poly-pyrimidine tract (PPT), and 3′-splice site, to form an artificial SpRNAi intron containing at least a desired RNA insert for antisense RNA, small hairpin RNA (shRNA) and/or microRNA (miRNA) production. A DNA synthesizer can chemically produce and link these elements. Alternatively, the linkage of these elements can be achieved by enzymatic restriction and ligation. The intron so obtained can be used directly for transfection into cells of interest or further incorporated into a cellular gene for co-expression along with the gene transcript (ie, pre-mRNA) by Pol-II. During RNA splicing and mRNA maturation, the desired RNA insert will be excised and released by intracellular spliceosome, exosome and NMD mechanisms and then triggers a desired gene silencing effect on specific gene transcripts with high complementarity to the inserted RNA sequence, while the exons of the recombinant gene transcript are linked together to form mature mRNA for expression of a desirable gene function, such as translation of a reporter or marker protein selected from the group of red/green fluorescent protein (RGFP/EGFP), luciferase, lac-Z, and their derivative homologues. The presence of the reporter/marker protein is useful for signaling the production of the inserted shRNA/miRNA molecules in treated cells, thus facilitating the identification of the desired gene silencing/RNAi effects.
In accordance with the biogenesis of intronic miRNA, mature mRNA formed by the linkage of exons can also be useful in conventional gene therapy to replace impaired or missing gene function, or to increase specific gene expression. Alternatively, this method provides novel compositions and means for inducing cellular production of gene silencing molecules through intronic RNA splicing and processing mechanisms to elicit either antisense-mediated gene knockout or RNA interference (RNAi) effects, which are useful for inhibiting targeted gene function. The intron-derived gene silencing molecules so obtained may include antisense RNA, ribozyme, short temporary RNA (stRNA), double-stranded RNA (dsRNA), small interfering RNA (siRNA), tiny noncoding RNA (tncRNA), short hairpin RNA (shRNA), microRNA (miRNA), and RNAi-associated precursor RNA constructs (pri-/pre-miRNA). The use of these intronic RNA-derived gene silencing agents is a powerful tool for targeting and silencing unwanted genes selected from the group consisting of pathogenic transgenes, viral genes, mutant genes, oncogenes, disease-related small RNA genes and any other types of protein-coding as well as noncoding genes.
Using this novel Pol-II-mediated SpRNAi-RGFP expression system, we have successfully generated mature shRNA and miRNA molecules with full gene silencing capacity in vitro in human prostate cancer (LNCaP), human cervical cancer (HeLa) and rat neuronal stem (HCN-A94-2) cells as well as in vivo in zebrafish, chicken and mouse (Lin and Ying 2006; Lin et al 2006). We have tested different pre-miRNA insert constructs targeting against green EGFP and other cellular gene expression in zebrafish and various human cell lines, and have learned that effective gene silencing miRNAs are derived from the 5′-proximity of the intron sequence between the 5′-splice site and the branching point. As shown in Figure 3C, a strong gene silencing effect was observed only in the transfection of anti-EGFP pre-miRNA insert (lane 4), whereas no effect could be detected in those of other inserts indicated by lanes from left to right: 1, blank vector control (Ctl); 2, pre-miRNA insert targeting HIV-p24 (mock); 3, antisense EGFP insert without the hairpin loop structure (anti); and 5, reverse pre-miRNA sequence which was completely complementary to the anti-EGFP pre-miRNA (miR*). No effect was detected on off-target genes, such as marker RGFP and housekeeping β-actin, suggesting that such an intronic miRNA-mediated gene silencing effect is highly target-specific. To confirm the role of RNA splicing in this intronic RNAi effect, we further tested three different SpRNAi-RGFP expression systems as shown in Figure 3D: 1, vector expressing intron-free RGFP without any pre-miRNA insert; 2, vector expressing RGFP with an intronic anti-EGFP pre-miRNA insert; and 3, vector similar to the 2 construct but with a defective 5′-splice site in the SpRNAi intron. As a result, northern blotting revealed that mature miRNAs were released only from the spliced intron of the vector 2 construct, which was exactly identical to the SpRNAi vector construct with the anti-EGFP pre-miRNA insert in Figure 3C. Thus, RNA splicing is required for intronic miRNA biogenesis.
Evaluation of natural antityrosinase miRNA agents
We have adopted the proof-of-principle design of the SpRNAi-RGFP expression system and use it for developing novel cosmetic products for skin care. In this new approach, we apply skins a nonnaturally occurring intron capable of being processed into hairpin-like precursor microRNA (pre-miRNA) molecules by the skin cells and thus inducing specific gene silencing effects on epidermal pigment-related genes and/or aging-causing genes. In this case, the RNA splicing- and processing-generated gene silencing molecule is the hairpin-like pre-miRNA insert located within the intron area of the recombinant gene and was capable of silencing a targeted gene, such as tyrosinase (Tyr), hyaluronidase (Hyal), hyaluronan receptors CD44 and CD168, and other pigmentation-related and/or aging-related genes and oncogenes. Alternatively, such a pre-miRNA insert can also be artificially incorporated into the intron region of a cellular gene in the skin. In general, this kind of intronic insertion technology includes plasmid-like transgene transfection, homologous recombination, transposon delivery, jumping gene integration and retroviral infection.
In the present design, the recombinant SpRNAi-RGFP gene expresses an intronic insert construct reminiscent of a hairpin-like pre-mRNA structure. The recombinant gene consists of an exon and an intron. The exon is ligated after RNA splicing to form a functional mRNA and protein for identification of the intronic RNA release, while the intron is spliced out of the recombinant gene transcript and further processed into the desired intronic RNA molecule that functions as a gene silencing effector. These desired intronic RNA molecules may comprise a hairpin-like stem-loop structure containing a sequence motif homologous to 5′-GCTAAGCCAG GC-3′ or 5′-GCCTGGCTTA GC-3′, which facilitates not only accurate excision of the desired RNA molecule out of the intron but also nuclear exportation of the desired RNA molecule to the cytoplasm. Also, the stem-arms of these intron-derived RNA molecules contain homology or complementarity, or both, to a targeted gene or a coding sequence of the targeted gene transcript. The homologous or complementary sequences of the desired RNA molecules are sized from approximately 18 to 27 nucleotide bases. The homology and/or complementarity rate of the desired intronic RNA molecule to the targeted gene sequence is ranged from approximately 30%∼100%, more preferably 35%∼49%, for a desired hairpin-like intronic RNA and 90%∼100% for a linear intronic RNA molecule.
In addition, the 5′-end of the nonnaturally occurring intron contains a donor splice site homologous to 5′-GTAAGAGK-3′ motifs, while its 3′-end is an acceptor splice site that is homologous to 5′-GWKSCYRCAG-3′ motifs. Moreover, a branch point sequence is located between the 5′- and 3′-splice sites, containing homology to 5′-TACTWAY-3′ motifs. The adenosine “A” nucleotide of the branch-point sequence forms a part of (2′–5′)-linked lariat intron RNA by cellular (2′–5′)-oligoadenylate synthetases and spliceosomes in almost all spliceosomal introns. Furthermore, a poly-pyrimidine tract is closely located between the branch-point and 3′-splice site, containing a high T or C content oligonucleotide sequence homologous to either 5′-(TY)m(C/−)(T)nS(C/−)-3′ or 5′-(TC)nNCTAG(G/−)-3′ motifs. The symbols of “m” and “n” indicate multiple repeats ≥1; most preferably, the m number is equal to 1∼3 and the n number is equal to 7∼12. The symbol “−” refers to an empty nucleotide in the sequence. There are also some linker nucleotide sequences for the connection of all these intron components. By definition, the symbol W refers to an adenine (A) or thymine (T)/uracil (U), the symbol K refers to a guanine (G) or thymine (T)/uracil (U), the symbol S refers to a cytosine (C) or guanine (G), the symbol Y refers to a cytosine (C) or thymine (T)/uracil (U), the symbol R refers to an adenine (A) or guanine (G), and the symbol N refers to an adenine (A), cytosine (C), guanine (G) or thymine (T)/uracil (U).”
Based on the above design, we have tested an optimized SpRNAi-RGFP gene construct expressing either anti-Tyr or anti-Hyal pre-miRNA directed against the pigmentation-related gene Tyr or aging-related gene Hyal in mouse skins. These pre-miRNAs target a highly conserved region (>98% homology) in both human Tyr and mouse Hyal genes. In nature, there are 54 native miRNAs capable of targeting human tyrosinase (Tyr; 2082 bp) for pigmentation gene silencing, including mir-1, mir-15a, mir-16, mir-31, mir-101, mir-129, mir-137, mir-143, mir-154, mir-194, mir-195, mir-196b, mir-200b, mir-200c, mir-206, mir-208, mir-214, mir-221, mir-222, mir-292-3p, mir-299-3p, mir-326, mir-328, mir-381, mir-409-5p, mir-434-5p, mir-450, mir-451, mir-452, mir-464, mir-466, mir-488, mir-490, mir-501, mir-522, mir-552, mir-553, mir-570, mir-571, mir-582, mir-600, mir-619, mir-624, mir-625, mir-633, mir-634, mir-690, mir-697, mir-704, mir-714, mir-759, mir-761, mir-768-5p, and mir-804. According to the miRNA-target database of the miRBase::Sequences program (http://microrna.sanger.ac.uk), all these anti-Tyr miRNAs are directed against a region within the first 300 nucleotides of the Tyr gene transcript (NCBI accession number NM000372). On the other hand, there are 9 native miRNAs capable of targeting hyaluronidase (Hyal; 2518 bp; NCBI accession number NM007312) for aging gene silencing, including mir-197, mir-349, mir-434-5p, mir-549, mir-605, mir-618, mir-647, mir-680, mir-702, and mir-763. Among these native miRNAs, mir-434-5p is the only one that targets both Tyr and Hyal genes in human and it is also one of the most efficient miRNAs targeting the least off-target genes other than Tyr and Hyal. However, because almost all native miRNAs target several to over fifty cellular genes and they tend to bind with some of the target genes more strongly than others, the use of these native miRNAs is likely not specific and safe enough for the purpose of skin care.
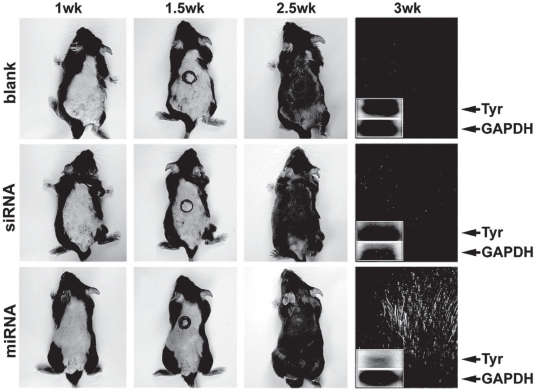
To test the feasibility of miRNA-mediated skin whitening, we utilized an SpRNAi-RGFP expression vector to express native pre-mir-434-5p in mouse skin. As shown in Figure 4, patched albino (white) skins of melanin-knockdown mice (W-9 black) were observed after a daily intra-cutaneous (i.c.) injection of the pre-mir-434-5p expression vector (50 μg) for four times (total 200 μg). Tyr, a type-I membrane protein and copper-containing enzyme, catalyzes the critical and rate-limiting step of tyrosine hydroxylation in the biosynthesis of melanin (black pigment) in skins and hairs; thus, the silencing of Tyr expression will result in a great loss of skin/hair pigments. Approximately two weeks after the first i.c. injection, we could clearly observe that the skin and hair pigments were significantly lost only in mice transfected with the pre-mir-434-5p. In contrast, the blank control and the transfection of a Pol-III (U6)-transcribed siRNA agent against the same Tyr sequence presented no significant effects. Northern blot analyses using mRNAs isolated from the hair follicles of the pre-mir-434-5p-transfected mice showed a markedly reduction of the Tyr expression (76.1% ± 5.3%) two days post-transfection, whereas mild, nonspecific degradation of random gene transcripts was detected in the siRNA-transfected skins (seen from the smearing patterns of both house-keeping control GAPDH and targeted Tyr mRNAs). Since Grimm et al have reported that high concentrated siRNA/shRNA can over-saturate the cellular miRNA pathway and cause global mRNA degradation (Grimm et al 2006), our results indicate that the siRNA pathway is incompatible with the native miRNA pathway in skin cells. Thus, the use of miRNA likely will provide a more effective, compatible and safe means for skin care. However, because the native mir-434-5p is able to silence five other cellular genes, including TRPS1, PITX1, LCOR, LYPLA2 and Hyal, its off-target gene silencing effects remain to be determined.
Figure 4.
Effects of miRNA-mediated tyrosinase (Tyr) gene silencing on the depigmentation of mouse skins and hairs. Targeted gene silencing in epidermal tissues was shown to be feasible using the ectopic transfection of a recombinant SpRNAi-RGFP gene vector expressing the native mir-434 pre-miRNA. Subcutaneous transfection of mir-434-5p induced a strong and specific gene silencing effect on Tyr but not house-keeping GAPDH expression, whereas transfection of a U6 promoter-driven siRNA against the same Tyr sequence resulted in nonspecific RNA degradation of both Tyr and GAPDH gene transcripts. Because Tyr plays an essential role in melanin (black pigment) production, a successful Tyr gene silencing effect can be manifested by the significant loss of black color in mouse skins and hairs. The circles indicate the location of i.c. injections. Small windows show the northern blotting of Tyr mRNA levels in local hair follicles, confirming the gene silencing effect of mir-434-5p on Tyr expression.
Re-design of mir-434-5p for skin whitening use in human
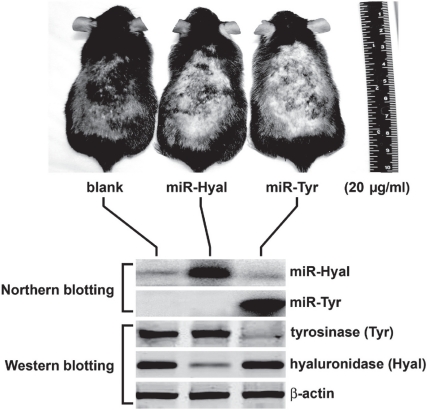
In order to improve the target-specificity and safety of anti-Tyr miRNA agents, we have re-designed the seed sequence of the mir-434-5p to form a highly matched region binding to either Tyr nucleotides 3–25 (miR-Tyr) or Hyal nucleotides 459–482 (miR-Hyal). The pre-miRNA insert sequence for pre-miR-Tyr is 5′-GTCCGATCGT CGCCCTACTC TATTGCCTAA GCCGCTAAGC CAGGCGGCTT AGGCAATAGA GTAGGGCCGA CGCGTCAT-3′, which forms a hairpin-like RNA after splicing and can be further processed into a mature miR-Tyr microRNA (miRNA) sequence containing or homologous to 5′-GCCCTACTCT ATTGCCTAAG CC-3′. Alternatively, the sequence for pre-miR-Hyal is 5′-GTCCGATCGT CAGCTAGACA GTCAGGGTTT GAAGCTAAGC CAGGCTTCAA ACCCTGACTG TCTAGCTCGA CGCGTCAT-3′, which forms a different kind of hairpin-like RNA after splicing and is further processed into a mature miR-Hyal miRNA sequence containing or homologous to 5′-AGCTAGACAG TCAGGGTTTG AA-3′. Although both pre-miR-Tyr and pre-miR-Hyal constructs were re-designed based on the same mir-434-5p backbone and mir-302 stem-loop, the mature miR-Tyr and miR-Hyal were shown to be totally different from each other. As shown in Figure 5, the transfective expressions of miR-Tyr and miR-Hyal in mouse skins specifically knocked down the targeted Tyr (reduction >90%) and Hyal genes (reduction >67%), respectively, without any crossover off-target effect. The expression levels of mature miR-Tyr and miR-Hyal microRNAs were directly measured by northern blot analyses, while the knockdown rates of the targeted Tyr and Hyal proteins were assessed by western blot analyses.
Figure 5.
Effects of Tyr and Hyal gene silencing on skin pigmentation in mice. The specificity of tyrosinase (Tyr) gene silencing was markedly improved, using a man-made anti-Tyr pre-miRNA (miR-Tyr) expressed by the recombinant SpRNAi-RGFP gene vector in mouse skins, showing a much higher target-specific and less off-target silencing effect on the targeted Tyr gene expression. Neither off-target (hyaluronidase) nor housekeeping (ß-actin) genes were affected by miR-Tyr treatment.
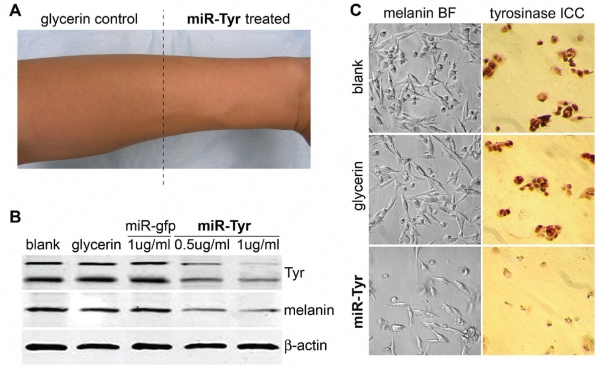
After understanding the optimized gene silencing effects of the re-designed miR-Tyr and miR-Hyal miRNAs, we continued to test their efficacy, target specificity and safety in human skins. For efficient vector transfection into the human epidermal cell layers, a 1 μg/ml SpRNAi-RGFP vector solution was prepared by mixing 100 μg of the purified SpRNAi-RGFP vector in 1 ml of autoclaved ddH2O with 99 ml of 100% DNase-free glycerin (or glycerol). DNase-free glycerin was used to encapsulate miR-Tyr for deep skin delivery and cell membrane penetration. This formed the major ingredient base for skin whitening and lightening products. Based on this, more other cosmetic ingredients could be added to increase the color, fragrance, effectiveness and/or stability of the final cosmetic products. As shown in Figure 6A, Asian male arm skins were treated with 2 ml of this major ingredient base expressing either miR-Tyr (right) or empty SpRNAi-RGFP vector alone (glycerin control, left). The whitening effect of the miR-Tyr treatment on human skins (loss of the black pigment–melanin) was clearly observed within three days after two treatments per day.
Figure 6.
Effects of Tyr gene silencing on skin pigmentation in human skins. Human trials showed that over 50% of the Tyr expression was suppressed by the re-designed miR-Tyr, confirming the results of Figure 5 but in the human arm skins (A) and primary skin cell cultures (B and C), as determined by western blotting and immunocytochemical (ICC) staining analyses.
Next, a primary skin cell culture was obtained by trypsin-dissociated skin explants from the tested donor with personal consent. The SpRNAi-RGFP vector transfection (final 6 μg/ml) in the primary skin culture was performed using FuGene liposomal reagent (Roche Biochemicals, Indianapolis, IN), as described previously (Lin et al 2003; Lin et al 2006). Figure 6B shows that western blot analyses of the loss of the targeted tyrosinase proteins and its substrate melanin were statistically significant (p > 0.001). The reduction levels of Tyr and its substrate melanin in skins were proportional to the treated concentrations of the miR-Tyr expression vector, suggesting a positive correlation between the increase of the miR-Tyr treatment and the loss of the targeted Tyr proteins and its substrate melanin. No effect was observed in other treatments, such as SpRNAi-RGFP vector alone (glycerin control) and an SpRNAi-RGFP vector expressing an anti-EGFP pre-miRNA (miR-gfp). At 1 μg/ml of the miR-Tyr expression vector transfection, the optimal gene silencing rates were approximately 55%–60% for Tyr and 30%–45% for melanin, while the expression of nontarget house-keeping control ß-actin was not affected by miR-Tyr treatment. Taken together, these results demonstrated the high target-specificity of our re-designed manmade miRNA agents.
Figure 6C further showed that the skin melanin levels were significantly reduced as shown in bright-field (BF) photographs of the primary skin cell culture (upper panels), while melanin (black dots around the cell nuclei) is highly expressed in normal skin cells without miR-Tyr treatment (ie, blank and glycerin only). The miR-Tyr-treated skin cells showed very limited melanin accumulation, demonstrating an effective skin-whitening effect in vivo. In regard to this loss of skin melanin, the targeted tyrosinase expression was concurrently reduced in the miR-Tyr-treated skin cells, as determined by immunocytochemical (ICC) staining analysis (Figure 6C, lower panels). According to these results, the re-designed miR-Tyr microRNA effectively knocked down the expression of Tyr proteins and successfully blocked melanin production in human skins in vivo.
Microarray analyses of target specificity and safety
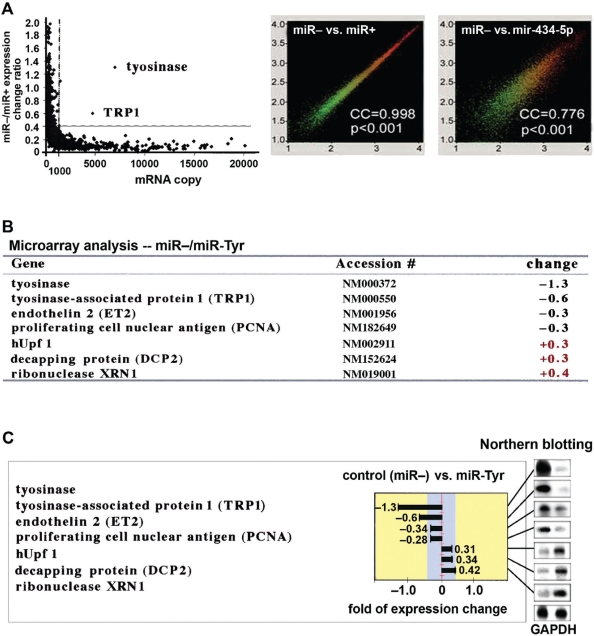
After the efficacy of miR-Tyr-mediated gene silencing was established in human skins, we applied human genome microarray analyses (Human GeneChip U133A&B arrays, Affymetrix, Santa Clara, CA) to assess the changes of approximately 32,668 human gene expression patterns in miR-Tyr-transfected and control primary skin cell cultures. As shown in Figure 7A (left), microarray analyses in nontreated (miR−) versus miR-Tyr-transfected (miR+) primary skin cell cultures showed that only two cellular genes, Tyr and its associated TRPI, whose expression levels were altered more than 1.5-fold (>50% change of gene expression; Figure 7B), indicating that the miR-Tyr-mediated gene silencing effect is highly specific to the targeted Tyr. Furthermore, no gene related to either cytotoxicity or interferon-mediated PKR/2-5A pathway was affected, suggesting that this gene silencing effect is safe for skin care treatments. We have also used northern blot analyses to compare and assess the gene expression levels of the microarray-identified genes (Figure 7C), confirming the results of Figures 7A and 7B. Furthermore, the correlation coefficiency (CC) rate clearly indicated that a high 99.8% population of the 32,668 tested human genes remained unchanged in the miR-Tyr-transfected (miR+) cells, compared to a significantly lower 77.6% CC rate in the mir-434-5p-transfected cells (Figure 7A, right). This indicates that the expression patterns of as few as 65 cellular genes were altered in the cells transfected with the re-designed miR-Tyr. On the other hand, over 7,317 genes might be changed in the cells transfected with the native mir-434-5p. Since it is known that almost all native miRNAs target multiple cellular genes due to their mismatched stem-arms, our present study demonstrates that the re-design of these miRNA stem-arm regions is necessary for increasing the safety and efficiency of these miRNAs used in cosmetic applications.
Figure 7.
Human genome microarray analyses (Affymetrix human GeneChip U133A&B, CA) of altered gene expression patterns in the human primary skin cell cultures with or without miR-Tyr transfection. This procedure was performed to demonstrate the higher target-specificity and lower off-target effect of the miR-Tyr compared to those of native microRNAs, such as mir-434-5p.
Materials and methods
Construction of SpRNAi-containing recombinant transgene (SpRNAi-RGFP)
Synthetic nucleic acid sequences used for generation of three different SpRNAi introns containing either sense-, antisense- or hairpin-EGFP insert were listed as follows: N1-sense, 5′-pGTAAGAGGAT CCGATCGCAG GAGCGCACCA TCTTCTTCAA GA-3′; N1-antisense, 5′-pCGCGTCTTGA AGAAGATGGT GCGCTCCTGC GATCGGATCC TCTTAC-3′; N2-sense, 5′-pGTAAGAGGAT CCGATCGCTT GAAGAAGATG GTGCGCTCCT GA-3′; N2-antisense, 5′-pCGCGTCAGGA GCGCACCATC TTCTTCAAGC GATCGGATCC TCTTAC-3′; N3-sense, 5′-pGTAAGAGGAT CCGATCGCAG GAGCGCACCA TCTTCTTCAA GTTAACTTGA AGAAGATGGT GCGCTCCTGA-3′; N3-antisense, 5′-pCGCGTCAGGA GCGCACCATC TTCTTCAAGT TAACTTGAAG AAGATGGTGC GCTCCTGCGA TCGGATCCTC TTAC-3′; N4-sense, 5′-pCGCGTTACTA ACTGGTACCT CTTCTTTTTT TTTTTGATAT CCTGCAG-3′; N4-antisense, 5′-pGTCCTGCAGG ATATCAAAAA AAAAAGAAGA GGTACCAGTT AGTAA-3′. In addition, two exon fragments were generated by DraII restriction enzyme cleavage of a red fluorescent RGFP gene at its 208th nucleotide (nt) site and the 5′ fragment was further blunt-ended by T4 DNA polymerase. RGFP refers to a new red-shifted fluorescent chromoprotein gene generated by insertion of an additional aspartate at the 69th amino acid (a.a.) site of HcRed1 chromoproteins from Heteractis crispa. (BD Biosciences, CA), developing less aggregate and almost twice intense far-red fluorescent emission at 570-nm. We also cleaved a pHcRed1-N1/1 plasmid (BD Biosciences, CA) with XhoI and XbaI restriction enzymes and purified a full 769-bp RGFP gene fragment and a 3,934-bp empty plasmid separately isolated from 2% agarose gel electrophoresis.
Hybridization of N1-sense to N1-antisense, N2-sense to N2-antisense, N3-sense to N3-antisense and N4-sense to N4-antisense was separately performed by heating each complementary mixture of sense and antisense (1:1) sequences to 94 °C for 2 min and then 70 °C for 10 min in 1x PCR buffer (eg, 50 mM Tris-HCl, pH 9.2 at 25 °C, 16 mM (NH4)2SO4, 1.75 mM MgCl2). Continuously, sequential ligation of either N1, N2 or N3 hybrid to the N4 hybrid was performed by gradually cooling the mixture of N1–N4, N2–N4 or N3–N4 (1:1) hybrids respectively from 50 °C to 10 °C over a period of 1 hr, and then T4 ligase and relative buffer (Roche, IN) were mixed with the mixture for 12 hr at 12 °C, so as to obtain introns for insertion into exons with proper ends. After the RGFP exon fragments were added into the reaction (1:1:1), T4 ligase and buffer were adjusted accordingly to reiterate ligation for another 12 hr at 12 °C. To clone the properly sized recombinant RGFP gene, 10 ng of the ligated nucleotide sequences were amplified by PCR with a pair of RGFP-specific primers 5′-CTCGAGCATG GTGAGCGGCC TGCTGAA-3′ and 5′-TCTAGAAGTT GGCCTTCTCG GGCAGGT-3′ at 94 °C, 1 min, 52 °C, 1 min and then 68 °C, 2 min for 30 cycles. The resulting PCR products were fractionated on a 2% agarose gel, and a ∼900-bp nucleotide sequences was extracted and purified by a Gel Extraction kit (Qiagen, CA). The composition of this ∼900 bp SpRNAi-containing RGFP gene was further confirmed by sequencing.
Since the recombinant SpRNAi-RGFP gene possesses an XhoI and an XbaI restriction site at its 5′- and 3′-end, respectively, it can be easily cloned into a vector with cohesive ends to the XhoI and XbaI cloning sites. The vector must be a skin-compatible, expressing-competent organism or suborganism selected from the group consisted of plasmids, cosmids, jumping genes, transposons, adeno-associated viral and retroviral vectors. Moreover, since the insert within the intron is also flanked with a PvuI and an MluI restriction site at its 5′- and 3′-end, respectively, we can remove and replace the insert with another different insert sequence possessing cohesive ends to the PvuI and MluI cloning sites. The inserted sequence is preferably a hairpin-like gene silencing effector containing high complementarity to a gene target selected from the group consisted of fluorescent protein (GFP) genes, luciferase genes, lac-Z genes, viral genes, bacterial genes, plant genes, animal genes and human genes. The complementarity and/or homology rate between the gene-silencing effector insert and its targeted gene is ranged from about 30%–100%, more preferably 35%–49% for a hairpin-shRNA insert and 90%–100% for a sense-stRNA or antisense-siRNA insert.
Cloning of the SpRNAi-containing transgene into an expression-competent vector
We mixed the SpRNAi-RGFP gene with the linearized 3,934-bp empty pHcRed1-N1/1 plasmid at 1:16 (w/w) ratio, cooled the mixture from 65 °C to 15 °C over a period of 50 min, and then added T4 ligase and relative buffer accordingly into the mixture for ligation at 12 °C for 12 hrs. This forms the SpRNAi-RGFP expression vector, which can be propagated in an E.coli DH5α LB culture containing 50 μg/ml kanamycin (Sigma Chemical, St. Louis, MO). A positive clone was confirmed by PCR with the RGFP-specific primers at 94 °C, 1 min and then 68 °C, 2 min for 30 cycles, and for further sequencing. For cloning into viral vectors, the same ligation procedure was repeated except using an XhoI/XbaI-linearized pLNCX2 retroviral vector (BD). Since the insert within the SpRNAi intron was flanked with a PvuI and a MluI restriction site at its 5′- and 3′-end, respectively, we could remove and replace the anti-EGFP shRNA insert with the miR-Tyr or miR-Hyal insert sequences possessing cohensive ends to the PvuI and MluI cloning sites.
Synthetic nucleic acid sequences used for generating various SpRNAi introns containing either miR-Tyr or miR-Hyal insert were listed as follows: miR-Tyr-sense, 5′-GTCCGATCGT CGCCCTACTC TATTGCCTAA GCCGCTAAGC CAGGCGGCTT AGGCAATAGA GTAGGGCCGA CGCGTCAT-3′; miR-Tyr-antisense, 5′-ATGACGCGTC GGCCCTACTC TATTGCCTAA GCCGCCTGGC TTAGCGGCTT AGGCAATAGA GTAGGGCGAC GATCGGAC-3′; and miR-Hyal-sense, 5′-GTCCGATCGT CAGCTAGACA GTCAGGGTTT GAAGCTAAGC CAGGCTTCAA ACCCTGACTG TCTAGCTCGA CGCGTCAT-3′; miR-Hyal-antisense, 5′-ATGACGCGTC GAGCTAGACA GTCAGGGTTT GAAGCCTGGC TTAGCTTCAA ACCCTGACTG TCTAGCTGAC GATCGGAC-3′. These inserts were formed via hybridization of miR-Tyr-sense to miR-Tyr-antisense and miR-Hyal-sense to miR-Hyal-antisense, respectively. These miR-Tyr- and miR-Hyal-expressing vectors could be propagated in E.coli DH5α LB-culture containing either 50 μg/ml kanamycin (for pHcRed1-N1/1 plasmid-based vector) or 100 μg/ml ampicillin (for pLNCX2 viral vector). The propagated SpRNAi-RGFP vectors could be isolated and purified using a Mini-prep or Maxi-prep Plasmid Extraction kit (Qiagen, CA), following the manufacturer’s suggestion. For pLNCX2 vectors, we also used a packaging cell line PT67 (BD) for producing infectious but replication-incompetent virus. The transfected PT67 cells were grown in 1x DMEM medium supplemented with 10% fetal bovine serum with 4 mM L-glutamine, 1 mM sodium pyruvate, 100 μg/ml streptomycin sulfate and 50 μg/ml neomycin (Sigma Chemical, MO) at 37 °C under 5% CO2. The titer of virus produced by PT67 cells was determined to be at least 106 cfu/ml before transfection.
Vector transfection
For vector transfection into cultivated cells or mouse skins, we first mixed the SpRNAi-RGFP expression plasmid vectors containing either anti-EGFP, miR-Tyr or miR-Hyal pre-miRNA insert using FuGene reagent (Roche, IN), following the manufacturer’s suggestion. Thereafter, the mixtures were directly applied to the cultivated cells (ie, primary human skin culture) or mouse skins, respectively. Vectors containing an insert-free RGFP gene and an SpRNAi-RGFP gene with a pre-miRNA insert against the HIV gag-p24 gene were used as negative controls. Tissue or cell morphology and fluorescence imaging was photographed at 0-, 24-, 48-, and 72-hr time points after the first transfection. For transfection to the human skins in vivo, a pre-made SpRNAi-RGFP vector solution was formed by mixing certain amounts (ie, 1–1000 μg) of the purified SpRNAi-RGFP vector with or without either anti-EGFP, miR-Tyr or miR-Hyal pre-miRNA insert in 1 ml of autoclaved ddH2O with 99 ml of 100% DNase-free glycerin (or glycerol). This solution was then directly applied to the skins with gentle massage for 3 min.
Northern blot analysis
RNA (20 μg total RNA or 2 μg poly[A+] RNA) was fractionated on 1% formaldehyde-agarose gels and transferred onto nylon membranes (Schleicher and Schuell, Keene, NH). Synthetic probes complementary to either the 75-bp junction sequence flanking between the RGFP 5′-exon and the anti-EGFP pre-miRNA insert or miR-Tyr, or miR-Hyal, were labeled with the Prime-It II kit (Stratagene, La Jolla, CA) by random primer extension in the presence of [32P]-dATP (>3000 Ci/mM, Amersham International, Arlington Heights, IL), and purified with 10 bp-cutoff Micro Bio-Spin chromatography columns (Bio-Rad, Hercules, CA). Hybridization was carried out in the mixture of 50% freshly deionized formamide (pH 7.0), 5x Denhardt’s solution, 0.5% SDS, 4x SSPE and 250 mg/mL denatured salmon sperm DNA fragments (18 hr, 42 °C). Membranes were sequentially washed twice in 2x SSC, 0.1% SDS (15 min, 25 °C), and once in 0.2x SSC, 0.1% SDS (45 min, 37 °C) before autoradiography.
SDS-PAGE electrophoresis and Western blot analysis
For detection of targeted proteins, isolated cells were rinsed with ice cold PBS after growth medium was removed, and then treated with the CelLytic-M lysis/extraction reagent (Sigma, MO) supplemented with protease inhibitors, Leupeptin, TLCK, TAME and PMSF, following manufacture’s recommendations. The lysates were incubated at room temperature on a shaker for 15 min, scraped into microtubes, and centrifuged for 5 min at 12,000 × g to pellet the cell debris. Supernatants that contain lysate proteins were collected and stored at −70 °C until use. Protein determinations were measured with SOFTmax software package on an E-max microplate reader (Molecular Devices, Sunnyvale, CA). Each 30 μg cell lysate was added into SDS-PAGE sample buffer either with (reduced) or without (unreduced) 50 mM DTT, and boiled for 3 min before being loaded onto 8% polyacylamide gels, while the reference lane was loaded with 2∼3 μl molecular weight markers (Bio-Rad). SDS-polyacrylamide gel electrophoresis was performed according to the standard protocols (Molecular Cloning, 3rd ED). Protein fractionations were electroblotted onto a nitrocellulose membrane, blocked with Odyssey blocking reagent (Li-Cor Biosciences, Lincoln, NB) for 1∼2 hr at the room temperature. Protein expression was subsequently detected using primary antibodies directed against EGFP (1:5,000; JL-8, BD), RGFP (1:10,000; BD), Tyr (1:2,000; Santa Crutz), or Hyal (1:2,000; Santa Crutz) incubated overnight at 4 °C. The protein blots were then rinsed 3 times with TBS-T and incubated with goat antimouse secondary antibody conjugated with Alexa Fluor 680 reactive dye (1:2,000; Molecular Probes) for 1 hr at room temperature. After three more TBS-T rinses, scanning and image analysis were completed with Li-Cor Odyssey Infrared Imager and Odyssey Software v.10 (Li-Cor).
Intronic mir-434-5p and miR-Tyr-mediated gene silencing in mouse skins in vivo
Patched albino (white) skins of melanin-knockdown mice (W-9 black strain) were created either by a succession of intra-cutaneous (i.c.) injection of an isolated SpRNAi-RGFP gene expression vector with the native mir-434-5p pre-miRNA insert for 4 days (total 200 μg) or by a direct skin infusion of a liposome-encapsulated SpRNAi-RGFP gene expression vector with the designed miR-Tyr pre-miRNA insert for two times per day for six days (total 240 μg). To generate the SpRNAi-RGFP gene expression vector with the native mir-434-5p pre-miRNA insert, we followed the same procedure as aforementioned, except using a synthetic mir-434-5p pre-miRNA for intronic insertion (ie, 5′-GTCCGATCGT CUCGACUCUG GGUUUGAACC AAAGCUCGAC UCAUGGUUUG AACCAUUACU UAAUUCGUGG UUUGAACCAU CACUCGACUC CUGGUUCGAA CCAUCCGACG CGTCAT-3′). For efficient delivery into target tissues, the construct was mixed with a FuGene liposomal transfection reagent (Roche, IN).
Intronic miR-Tyr-mediated gene silencing in human skins
For efficient vector transfection into the human multiple skin cell layers, a 1 μg/ml SpRNAi-RGFP vector solution is made by mixing 100 μg of the purified SpRNAi-RGFP vector in 1 ml of autoclaved ddH2O with 99 ml of 100% DNase-free glycerin (or called glycerol). DNase-free glycerin is used to encapsulate miR-Tyr for deep skin delivery and cell membrane penetration. This forms the major ingredient base for our present skin whitening and lightening inventions. Thereafter, one (left arm) of the author’s arm skins was directly treated with one 2 ml of the major ingredient base solution containing SpRNAi-RGFP vector with (right side) or without (left side) miR-Tyr. The effect of miR-Tyr treatment on skin whitening (loss of the black pigment–melanin) was assessed three days after two single treatments per day.
Immunocytochemical (ICC) staining assay
An immunochemical staining kit was obtained from Imgenex (San Diego, CA) and used according to the manufacturer’s suggestion. Specimens were first rinsed in PBS three times and incubated with Zeller’s solution (10 mM Tris, 100 mM MgCl2, 5% fetal calf serum, 1% BSA and 0.5% Tween-20, pH 7.4) for 30 min. They were then incubated with primary antibody (diluted in Zeller’s solution) overnight in a humidified chamber at 4 °C. Thereafter, the specimens were washed with TBST three times and incubated with secondary antibody for 2 hr, using biotinylated goat antirabbit or horse antimouse antibody as the secondary antibody (Chemicon, Temecula, CA). Specimens were then washed once with TBST and incubated with streptavidin-HRP as the tertiary antibody for 2 hrs and followed by PBT washings. The bound antibody was subsequently detected with DAB substrates and observed under a 100x microscope with whole field scanning and recorded at 100x and 400x magnification (TE2000 inverted microscopic quantitation system).
Microarray analysis
To prepare labeled probes for microarray hybridization, total RNAs (2 μg) were converted into double-stranded cDNAs, using a Superscript Choice system kit (Gibco/BRL, Gaithersburg, MD) with a modified oligo(dT)24-T7 promoter primer, such as 5′-GGCCAGTGAA TTGTAATACG ACTCACTATA GGGAGGCGG-(dT)24-3′, following the manufacturer’s protocol. Double-stranded cDNAs were purified by phenol/chloroform extractions, precipitated with ethanol, and resuspended at a concentration of 0.5 μg/μl in diethyl pyrocarbonate (DEPC)-treated ddH2O. Phase-Lock Gel (5′Prime →3′Prime, Inc., Boulder, CO) was used for all organic extractions to increase recovery. In-vitro transcription was performed with T7 RNA polymerase and with 1 μg of cDNA, 7.5 mM unlabeled ATP and GTP, 5 mM unlabeled UTP and CTP, and 2 mM biotin-labeled CTP and UTP (biotin-11-CTP, biotin-16-UTP, Enzo Diagnostics). Reactions were carried out for 4 hr at 37 °C, and cRNA was purified by RNeasy spin columns (Qiagen, CA). A sample was separated on a 1% agarose gel to check the size range, and then μg of cRNA was fragmented randomly to an average size of 50 bases by heating at 94 °C for 35 min in 40 mM Tris-acetate, pH 8.0, 100 mM KOAc/30 mM MgOAc.
Sets of four oligonucleotide microarrays (GeneChip U133A&B arrays, Affymetrix, Santa Clara, CA) containing total 32,668 genes were used for hybridization. Hybridizations were completed in 200 μl of AFFY buffer (Affymetrix) at 40 °C for 16 hr with constant mixing. After hybridization, arrays were rinsed three times with 200 μl of 6x SSPE-T buffer (1 × 0.25 M sodium chloride/15 mM sodium phosphate, pH 7.6/1 mM EDTA/0.005% Triton) and then washed with 200 μl of 6x SSPE-T for 1 hr at 50 °C. The arrays were rinsed twice with 0.5x SSPE-T and washed with 0.5x SSPE-T at 50 °C for 15 min. Staining was done with 2 μg/ml streptavidin-phycoerythrin (Molecular Probes) and 1 mg/ml acetylated BSA (Sigma) in 6x SSPE-T (pH 7.6). The arrays were read at 7.5 μm with a confocal scanner (Molecular Dynamics) and analyzed with Affymetrix Microarray Suite version 4.0 software. The samples were normalized by using the total average difference between perfectly matched probe and the mismatched probe. The differential signals that were induced greater than 2-fold are collected.
Statistical analysis
Results were presented as mean ±SE. Statistical analysis of data was performed by one-way ANOVA. When main effects were significant, the Dunnett’s post-hoc test was used to identify the groups that differed significantly from the controls. For pairwise comparison between two treatment groups, the two-tailed student t test was used. For experiments involving more than two treatment groups, ANOVA was performed followed by a post-hoc multiple range test. Probability values of p < 0.05 were considered significant. All p values were determined from two-tailed tests.
Conclusion
In summary, the utilization of intronic hairpin-like miRNA expression provides a potentially powerful and innovative strategy for human skin care, particularly for hyperpigmentation and aging prevention. Under the same treatment in animal trials, Pol-II-directed intronic miRNA expression did not cause any detectable cytotoxicity, whereas Pol-III-directed siRNAs induced nonspecific mRNA degradation as previously reported (Elbashir 2001; Lin and Ying 2001; Sledz et al 2003; Lin and Ying 2004). This underscores the fact that the intronic miRNA agent is effective, target-specific and safe for use in skin care. Because the intronic miRNA pathway is regulated by multiple intracellular surveillance systems, such as Pol-II transcription, RNA splicing, exosomal digestion and NMD processing, the gene silencing effect of intronic miRNA is considered to be most effective, specific and safest among all three currently known RNAi pathways. By utilizing the intronic miRNA expression strategy, many cosmetic applications can be designed and developed for skin care, offering more long-term effectiveness, better target-specificity and higher safety in skin gene manipulation, which prevents the unspecific off-target cytotoxicity as commonly seen in the conventional siRNA methods.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- Brantl S. Antisense-RNA regulation and RNA interference. Biochimica et Biophysica Acta. 2002;1575:15–25. doi: 10.1016/s0167-4781(02)00280-4. [DOI] [PubMed] [Google Scholar]
- Clement JQ, Qian L, Kaplinsky N, et al. The stability and fate of a spliced intron from vertebrate cells. RNA. 1999;5:206–20. doi: 10.1017/s1355838299981190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danin-Kreiselman M, Lee CY, Chanfreau G. RNAse III-mediated degradation of unspliced pre-mRNA and lariat introns. Mol Cell. 2003;11:1279–89. doi: 10.1016/s1097-2765(03)00137-0. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–8. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Garcia-Blanco MA. Coupled in vitro synthesis and splicing of RNA polymerase II transcripts. RNA. 2000;6:1325–34. doi: 10.1017/s1355838200992537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SR. Dissecting the mechanisms of posttranscriptional gene silencing: divide and conquer. Cell. 1999;96:303–6. doi: 10.1016/s0092-8674(00)80541-3. [DOI] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–41. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Gunnery S, Ma Y, Mathews MB. Termination sequence requirements vary among genes transcribed by RNA polymerase III. J Mol Biol. 1999;286:745–57. doi: 10.1006/jmbi.1998.2518. [DOI] [PubMed] [Google Scholar]
- Lee NS, Dohjima T, Bauer G, et al. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat Biotechnol. 2002;20:500–5. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lin SL, Chang D, Wu DY, et al. A novel RNA splicing-mediated gene silencing mechanism potential for genome evolution. Biochem Biophys Res Commun. 2003;310:754–60. doi: 10.1016/j.bbrc.2003.09.070. [DOI] [PubMed] [Google Scholar]
- Lin SL, Chang D, Ying SY. Asymmetry of intronic pre-microRNA structures in functional RISC assembly. Gene. 2005;356:32–8. doi: 10.1016/j.gene.2005.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SL, Chang SJE, Ying SY. Transgene-like animal model using intronic microRNAs. Methods Mol Biol. 2006;342:321–34. doi: 10.1385/1-59745-123-1:321. [DOI] [PubMed] [Google Scholar]
- Lin SL, Kim H, Ying SY. Intron-mediated RNA interference and microRNA (miRNA) Front Biosci. 2008;13:2216–30. doi: 10.2741/2836. 2008. [DOI] [PubMed] [Google Scholar]
- Lin SL, Ying SY. D-RNAi (messenger RNA-antisense DNA interference) as a novel defense system against cancer and viral infections. Current Cancer Drug Targets. 2001;1:241–7. doi: 10.2174/1568009013334151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SL, Ying SY. Combinational therapy for HIV-1 eradication and vaccination. Int J Oncol. 2004;24:81–8. [PubMed] [Google Scholar]
- Lin SL, Ying SY. Novel RNAi therapy – Intron-derived microRNA drugs. Drug Design Reviews. 2004;1:247–55. [Google Scholar]
- Lin SL, Ying SY. Gene silencing in vitro and in vivo using intronic microRNAs. Methods Mol Biol. 2006;342:295–312. doi: 10.1385/1-59745-123-1:295. [DOI] [PubMed] [Google Scholar]
- Miyagishi M, Taira K. U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat Biotechnol. 2002;20:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- Nott A, Meislin SH, Moore MJ. A quantitative analysis of intron effects on mammalian gene expression. RNA. 2003;9:607–17. doi: 10.1261/rna.5250403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul CP, Good PD, Winer I, et al. Effective expression of small interfering RNA in human cells. Nat Biotechnol. 2002;20:505–8. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- Sledz CA, Holko M, de Veer MJ, et al. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5:834–9. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- Solano F, Briganti S, Picardo M, et al. Hypopigmenting agents: an updated review on biological, chemical and clinical aspects. Pigment Cell Res. 2006;19:550–71. doi: 10.1111/j.1600-0749.2006.00334.x. [DOI] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BR, et al. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–64. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Tang G. siRNA and miRNA: an insight into RISCs. Trends Biochem Sci. 2005;30:106–14. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Tuschl T. Expanding small RNA interference. Nat Biotechnol. 2002;20:446–8. doi: 10.1038/nbt0502-446. [DOI] [PubMed] [Google Scholar]
- Ying SY, Lin SL. Intronic microRNAs. Biochem Biophys Res Commun. 2005;326:515–20. doi: 10.1016/j.bbrc.2004.10.215. [DOI] [PubMed] [Google Scholar]
- Ying SY, Lin SL. Intron-derived microRNAs–fine tuning of gene functions. Gene. 2004;342:25–8. doi: 10.1016/j.gene.2004.07.025. [DOI] [PubMed] [Google Scholar]
- Zhang G, Taneja KL, Singer RH, et al. Localization of pre-mRNA splicing in mammalian nuclei. Nature. 1994;372:809–12. doi: 10.1038/372809a0. [DOI] [PubMed] [Google Scholar]