Abstract
Educational immune tolerance to self-antigens is induced primarily in the thymus where tissue-restricted antigens (TRAs) are presented to T lymphocytes by cells of the thymic stroma – a process known as central tolerance. The expression of these TRAs is controlled in part by a transcription factor encoded by the autoimmune regulatory (Aire) gene. Patients with a mutation of this gene develop a condition known as autoimmune–polyendocrinopathy–candidiasis–ectodermal–dystrophy (APECED), characterized by autoimmune destruction of endocrine organs, fungal infection and dental abnormalities. There is now evidence for TRA expression and for mechanisms of functional tolerance outside the thymus. This has led to a number of studies examining Aire expression and function at these extra-thymic sites. These investigations have been conducted across different animal models using different techniques and have often shown discrepant results. Here we review the studies of extra thymic Aire and discuss the evidence for its expression and function in both human and murine systems.
Keywords: Aire, diabetes, ectopic expression, mTEC, tolerance
Introduction
T cells originate in the bone marrow (BM) but migrate to the thymus to undergo development and maturation [1–3]. This maturation is central to the development of a functional immune system and relies initially on the selection of high-quality T cell receptor (TCR) clones. TCR generation in the thymus is achieved through random somatic gene rearrangements [4], giving rise to T cells with an array of receptors capable of recognizing an immense repertoire of antigens. In the classic model of tolerance, TCRs in the thymus cortex are presented initially in the context of major histocompatibility (MHC)-restricted self-antigens, and only those with sufficient affinity/avidity survive. In contrast, TCRs with very low or no affinity/avidity die by neglect [5]. T cells pass subsequently to the thymic medulla, where they again encounter MHC-restricted self-antigens. Here, receptors that show high affinity/avidity for self-antigens are deleted in a process called negative selection. In principle, this process of positive and negative selection gives rise to a T cell repertoire tolerant to self-antigens, and which is able to recognize non-self-antigens in the context of self-MHC.
The issue of how T cells in the thymus can be exposed to self-antigens, which should be expressed only in a tissue-restricted fashion outside the thymus, was previously unclear. It has become apparent that cells in the thymus can express a selected set of proteins which are otherwise limited to specific tissues within the body. Expression of these tissue-restricted antigens (TRAs) within the thymus has now been demonstrated for a number of antigens in both mice and humans, and at both RNA and protein levels [6–11]. While TRAs are expressed primarily by medullary thymic epithelial cells (mTECs) within the thymus [6–8,10,12–15], cells of haematopoietic origin such as dendritic cells (DCs) also appear capable of expressing these self-antigens [15–20]. Indeed, some TRAs may be expressed by both cell types [6,7,15,16].
Prevention of autoimmune disease is therefore dependent on the correct display of TRAs in the thymus. Indeed, low levels of thymic insulin expression is associated with progression to Type I diabetes, whereas high levels appear to be protective, possibly reflecting more efficient deletion of insulin-reactive T cells [9,11]. The control of thymic TRA expression is clearly an area of considerable interest because it may hold the key to understanding the pathogenesis of autoimmune diseases.
More recently it has become clear that TRAs are also expressed in peripheral lymphoid tissues [21–26] and it is likely that here also they contribute to the process of immune tolerance.
A transcription factor encoded by the autoimmune regulatory gene (AIRE – human, Aire – murine) appears to be involved in thymic TRA expression and this has received enormous attention over the past 10 years [27]. Given the peripheral expression of TRAs, it is possible that Aire also plays a role in peripheral TRA expression.
The aim of this review is to examine the evidence for the expression and function of Aire in peripheral, non-thymic tissue.
Structure and function of Aire in thymus
The human Aire gene was reported independently by two groups in 1997 [28,29]. Three isoforms are described: Aire-1 as the full-length functional protein (Fig. 1), and Aire-2 and -3 are shorter forms generated by alternative splicing [28,30]. Based on its domain structure, the Aire protein is believed to be a transcription factor. It has two zinc fingers of plant homeodomain (PHD)-type motifs [31,32], a Sp100, AIRE-1, NucP41/75 and DEAF-1/suppressin (SAND) domain [33], and at least one functional nuclear localization signal [34], all of which are consistent with this role [35–37]. However, Aire may not act as a classical transcription factor given, for example, the number of genes it influences, its dependence on different co-factors and start sites in the thymus [27,38,39]. Studies indicate that other factors interact with Aire and contribute to its function. A recent study by Abramson et al. identified an Aire complex centred on DNA-PK, which appeared to potentiate TRA transcription, and another complex which regulated pre-mRNA processing of TRAs [38]. A second study showed Aire to act through a gene node, Gucy2d, which linked together the TRAs in mTECs, indicative of a cascade control mechanism for these proteins [40]. In contrast, another study indicated that the interaction between Aire and the DAXX protein inhibits transactivation by Aire [32]. Thus, transactivation by Aire may require interactions with other factors.
Fig. 1.
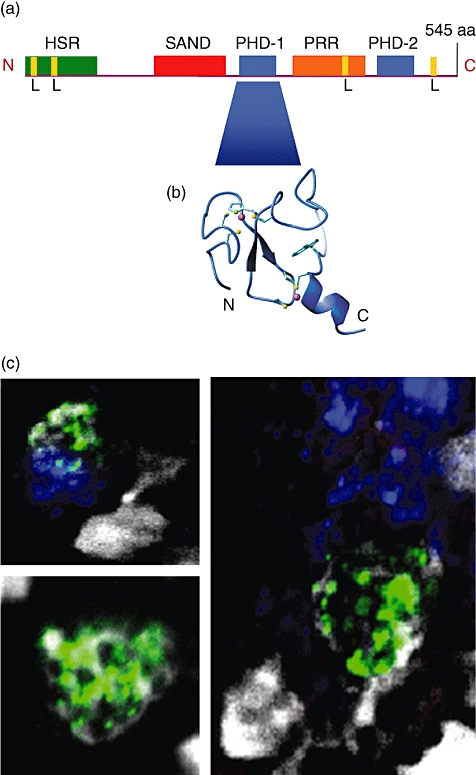
The autoimmune regulatory (Aire) protein. (a) Schematic representation of the human AIRE protein indicating the functional protein domains (adapted from [28,36]). (b) Ribbon representation of the PHD1 domain of AIRE 1 (taken from [35]). (c) AIRE staining (green) [using Santa Cruz, AIRE (D17)] in the thymus has a characteristic punctate nuclear appearance; here, Ulex europaeus agglutinin (UEA) staining (blue) identifies the medulla and 4′-6-diamidino-2-phenylindole (DAPI) staining (white) the nucleus (Eldershaw et al., unpublished data). L: LXXLL nuclear receptor interaction motif; HSR: homogeneously staining region; SAND: Sp100, AIRE, NucP41/75 and DEAF-1; PHD: plant homeodomain zinc finger; PRR: proline-rich region; N: N-terminus; C: C-terminus. Fig.1(b) reproduced by kind permission of Journal of Biological Chemistry. © 2005 The American Society for Biochemistry and Molecular Biology. Bottomley MJ, Stier G, Pennacchini D et al. NMR Structure of the First PHD Finger of Autoimmune Regulator Protein (AIRE1). Insights into autoimmune polyendocrinopathy-candiasis-ectodermal dystrophy (APECED) disease. J Biol Chem 2005; 280:11505–11512.
Mutations in the human AIRE gene result in a condition known as autoimmune–polyendocrinopathy–candiasis–ectodermal–dystrophy (APECED) [41] or autoimmune polyglandular syndrome type 1 (APS-1) [42,43]. The condition is particularly well described in Finnish [41,44], Iranian Jewish [45] and Sardinian [46] populations. The syndrome is characterized by the presence of at least two of the following conditions: autoimmune adrenal failure (Addison's disease), hypoparathyroidism and chronic mucocutaneous candidiasis [42,47,48]. APECED is also associated with a number of other conditions (Table 1), some of which have a clear autoimmune component and others (such as keratoconjunctivitis [48,49] and diarrhoea [48]) which do not.
Table 1.
The diagnostic dyad† and the most common disease components and median age ranges at their appearance in the series of 91 APECED patients studied (modified from [48]).
| Age range (median) at appearance (years) | |
|---|---|
| Diagnostic dyad† | 2·2–35 (7·0) |
| Classic triad | |
| •Candidiasis | 0·2–31 (5·4) |
| •Hypoparathyroidism | 1·6–43 (6·0) |
| •Adrenal failiure | 3·5–41 (10·0) |
| •All three | 3·5–43 (11·3) |
| Other endocrine disorders | |
| •Ovarian failure | −36 |
| •Testicular failure | −37 |
| •Diabetes mellitus | 4·1–58 (23·5) |
| •Hypothyroidism | 4·7–45 (26·5) |
| Skin disorders | |
| •Alopecia | 2·5–30 (10·3) |
| •Vitiligo | 0·7–45 (12·7) |
| •Rash with fever | 0·7–31 (2·8) |
| Gastrointestinal disorders | |
| •Pernicious anaemia | 6·1–48 (17·2) |
| •Severe obstipation | 1·0–31 (13·0) |
| •Chronic diarrhoea | 2·5–27 (6·6) |
| •Hepatitis | 0·7–16 (8·0) |
| Eye disorders | |
| •Keratoconjunctivitis | 1·3–16 (5·4) |
Estimated from the observed incidence rates over the age intervals, assuming that all patients live until the age of 50 years.
At least two of candidiasis, hypoparathyroidism and adrenocortical failure.
Murine Aire shares 71% homology with human AIRE and conservation of all functional protein motifs [50,51]. Aire knock-out mice display some features of human APECED, including circulating autoantibodies to ovary, testis, salivary gland, retina, pancreas, liver, adrenal gland and stomach [8,52]. Lymphocytic infiltrates can be seen within these organs as well as the thyroid and liver and they manifest as infertility, blepharitis, keratoconjunctivitis and thymic, adrenal and ovarian atrophy. In contrast to humans, mice do not appear to develop adrenal failure, hypoparathyroidism or candidiasis, the three prerequisite features of human APECED; neither do they appear to display the gastrointestinal or skin manifestations described in Table 1. These differences aside, murine models have allowed a detailed examination of the contribution of Aire to immune tolerance.
Aire is expressed at highest levels within the thymus in both mice and humans and is detectable at both RNA and protein levels [28,30,53–56]. Expression is most prominent within mTECs [6,55,57], but Aire is also seen at lower levels in myeloid DCs [25,54,56,58]. Transgenic mice studies demonstrate that Aire up-regulates TRA expression in the thymus [59] and that Aire deficiency prevents such expression [8]. Studies to date have been at the RNA level, with few published studies on protein expression. None the less, functional studies indicate that lower TRA expression is associated with inefficient negative selection of high-avidity organ-specific autoreactive T cells, and with the subsequent development of autoimmunity [60–62].
TRA expression by lymphoid tissue outside the thymus
The deletion of self-reactive T cells in the thymus is an incomplete process, and self-reactive T cells are detectable in the peripheral blood of clinically healthy individuals [63,64]. Mechanisms of peripheral tolerance must therefore be in place to control the autoimmune potential of these cells.
Extra-thymic TRA expression has been described in both human and murine tissue, raising the possibility that mechanisms of tolerance seen to act within the thymus could also be at work in the periphery. Expression of pancreatic islet autoantigens including proinsulin has been demonstrated by circulating bone marrow-derived cells in humans [17,19,65] as well as rodents [17,66]. Neural self-antigens have also been detected in bone marrow-derived cells in a number of mouse models [67,68] and a recent study suggests that a range of TRAs, including retinal and pancreatic antigens, are expressed by lymph node stromal cells [22]. A number of transgenic studies in mice further support TRA expression in the periphery [21,23–26].
Aire expression by lymphoid tissue outside the thymus
The identification of extra-thymic TRA expression raises the possibility that here, too, Aire may contribute to transcriptional control of TRAs.
Evidence for extrathymic Aire expression at an RNA level
The expression of Aire in tissues outside the thymus has been recognized since the original studies which cloned the Aire gene [28]. Although the highest expression of AIRE was found within the thymus in these studies of human tissues, RNA transcripts were also identified in lymph node, fetal liver and appendix tissues. The techniques of in-situ hybridization, Northern blotting, polymerase chain reaction (PCR) and real-time PCR have all been used subsequently to study the expression of AIRE in extra-thymic tissue, and these are summarized in Table 2. However, there are significant discrepancies between studies, particularly those in mice. For example, Naval Medical Research Institute (NMRI) mice strongly express Aire in lymph node (LN) and spleen [69], whereas only weak expression has been reported in C57BL/6 mice [23,55,56], with one study reporting stronger expression in the lymph node than the thymus [70]. In human tissue, extra-thymic AIRE mRNA has been detected in lymph nodes, tonsils and gut-associated lymphoid tissues (GALT) [28,54,71], spleen [71], fetal liver [28,71] and peripheral blood mononuclear cells (PBMCs) [54,58,72], but here again, not all studies have been in agreement [28,54,71].
Table 2.
Summary of findings for studies of autoimmune regulatory (Aire) gene expression (measured as RNA) in peripheral tissue.
| Author | Species | Tissue | Technique | Findings |
|---|---|---|---|---|
| Nagamine et al. (1997) [28] | Human | PCR screening of cDNA library | 3 Aire proteins, 1–3. Aire 1 is full length | |
| Thymus, spleen, LN, appendix, PBMC, BM, F liver | Northern blot | Expressed detected in thymus and LN. Weak expression in appendix and F liver but not detected in spleen, PBMC or BM | ||
| Heino et al. (1999) [71] | Human | Thymus, LN, spleen, F liver, adrenal cortex, thyroid glands, gonads, pancreas, kidney, liver, lung, skin, heart, PBMC | In situ hybridization | Expression detected only in thymus, LN, spleen and F liver |
| Blechsmidt et al (1999) [50] | Mouse | Heart, brain, spleen, lung, liver, skeletal muscle, kidney and testis | Northern blot of PCR products spanning exons 1–7 | Failed to detect transcripts |
| Embryonic thymus | In situ hybridization of histological sections | Expression detected from 14·5 days post coitum in developing thymus | ||
| Embryonic stem cells Adult heart, brain, spleen, lung, liver, skeletal muscle, kidney and testis | RT–PCR amplification on normalized first-strand cDNAs followed by sequencing of cloned PCR products | Aire transcripts detected in 11 days post coitum embryos and adult heart, spleen, lung, skeletal muscle and testis only | ||
| Ruan et al. (1999) [30] | Mouse (C57BL/6) | Spleen, thymus, liver, stomach, small intestine, muscle, ovary, lung, testis, kidney, adrenal, thyroid, heart, pancreas | Northern blot | No detectable expression |
| RT–PCR | Expression in thymus, ovary, lung, testis, kidney, adrenal gland and barely detected in thyroid and heart but not the other tissues | |||
| Competitive RT–PCR | Expression strongest in thymus – approximately 10× higher than ovary. No clear signal detected for other tissues | |||
| Zuklys et al. (2000) [55] | Mouse (C57BL/6, 6 weeks) | Thymus, liver, brain, BM, LN, stomach, skin, kidney | Northern blot | Detected in thymus and more weakly in LN and spleen |
| Heino et al. (2000) [74] | Mouse (BALB/c, 6–12 weeks) | Thymus, LN, spleen, PBMC, adrenal, pancreas, testis, muscle, liver | RT–PCR 1st round | Expression detected in thymus, LN, spleen only |
| Southern hybridization with amplification products | Detected in testis in addition to thymus, LN and spleen | |||
| Nested primers and 2 rounds of PCR | Detected in thymus, LN, spleen, adrenal, testis, muscle and liver | |||
| Halonen et al. (2001) [69] | Mouse (NMRI, adult) | Thymus, spleen, LN, BM, ovary | In situ hybridization using cRNA probes | Strongly expressed in cells of thymus, spleen, LN. Moderately expressed in cells of BM and ovary |
| Adamson et al. (2004) [53] | Mouse (CD1 and C57BL/6J) aged 6–8 weeks | Thymus, spleen, testis, kidney, liver, brain, lung, heart, gut, adrenal | In situ hybridization | Detected in thymic medulla |
| RT–PCR expression relative to expression in CD1 mouse thymus | Thymus tissue from C57BL/6J mice Aire expression approx. 4× higher than CD1 thymus control. Low levels detected in spleen, testis, kidney, liver, brain, lung, ovary, heart, gut and barely detectable in adrenal gland | |||
| C57BL/6J at embryonic days (E) 14·5, 16·5 and 18·5 | In situ hybridization RT–PCR relative to adult | Detected in thymus at E14·5 but lower level than seen in adult Detected in spleen at E16·5 but 20-fold less than detected in adult. | ||
| Otherwise undetected | ||||
| Zheng et al. (2004) [26] | Mouse (TRAMP, bone marrow chimeras) | Thymus, spleen, CD11c+ and CD11c– from spleen | Real-time PCR | Expression in thymus, spleen (10-fold less than thymus) and CD11c+ cells from spleen |
| Lee et al. (2007) [23] | Mouse (C57BL/6) | Thymus, spleen, MLN, SKLN, small intestine | RT–PCR | Detected in thymus, MLN, SKLN and weakly in spleen |
| Schaller et al. (2008) [76] | Mouse (HD – C57BL/6 background) (LA – unknown strain contributions) | Thymus, testis, sorted SKLN cells | Quantitative RT–PCR | Detected in thymus and testis, enriched in CD45- sorted SKLN cells |
| Hubert et al. (2008) [56] | Mouse (C57BL/6) | Thymus, spleen and DC subsets from these tissues | Nested PCR | Expression in all DC subsets and mTECs |
| Real-time quantitative PCR of different regions of the Aire mRNA transcript (exons 1–2, exons 7–8, exons 13–14) | Expression in all DC subsets (100-fold less than mTECs but greater than Aire–/–) | |||
| Suzuki et al. (2008) [72] | Human | PBMC | Real-time quantitative PCR | Expression in B cells, low level in T cells, and at low levels in 1 of 4 CD14+ cell samples |
| Pöntynen et al. (2008) [58] | Human | PBMC from buffy coats | Real-time quantitative PCR | Detected at relatively low but almost equivalent levels in monocyte-derived DCs, plasmacytoid DCs and myeloid DCs |
| Kont et al. (2008) [70] | Mouse (C57BL/6) | Thymus and LN | Real-time quantitative PCR | Detected in thymus and LN |
| Gardner et al. (2008) [73] | Mouse (NOD) | Thymus, spleen and LN-cells expressing identified eTAC markers | Real-time quantitative PCR | Detected in thymus and at low but levels in spleen and LN |
| Poliani et al. (2010) [54] | Human | Thymus, LN, tonsil, spleen, GALT | Real-time quantitative PCR | Detected in all tissues except spleen |
| Cohen et al. (2010) [21] | Mouse (C57BL/6) | LN stromal subsets | RT–PCR | Haematopoietic cells and 1 of 4 stromal subsets |
| Fletcher et al. (2010) [22] | Mouse (C57BL/6) | LN stromal subsets | RT–PCR | Detected at varying levels in all 4 stromal subsets |
LN, lymph node; MLN, mesenteric lymph node; SKLN, skin draining lymph node; GALT, gut-associated lymphoid tissue; PBMC, peripheral blood mononuclear cells; BM, bone marrow; F liver, fetal liver; DC, dendritic cell; NOD, non-obese diabetic; RT–PCR, reverse transcription–polymerase chain reaction; NMRI, Naval Medical Research Institute; mTEC, medullary thymic epithelial cells; eTAC, extra-thymic Aire expressing cell; TRAMP, transgenic adenocarcinoma of the mouse prostate.
The lineage of extra-thymic Aire expressing cells have been described as both myeloid [26,54,56,58,72] and epithelial [73]. Zheng et al. 2004 and Hubert et al. 2008 demonstrated Aire mRNA expression in DCs in different strains of mouse, albeit at levels far lower than that detected in the thymus [26,56]. The latter study demonstrated expression across different subsets of DCs and is in agreement with human studies, where Aire transcripts have also been detected in monocyte-derived, myeloid as well as plasmacytoid and interdigitating DCs [54,58]. More recently in mice, CD45– lymph node stromal cells that do not express the folate receptor gp38 have been noted to express Aire concomitantly with a number of TRAs [21,22,73]. Notably, this Aire expression has not yet been confirmed at a protein level, and mRNA was at very low levels needing 40 cycles of PCR for their detection. Lymph node stromal cells positive for gp38 in these mice appear to express a different subset of TRAs and do so independently of Aire [21,22].
Despite discrepancies between studies there is steadily accumulating evidence for low-level Aire RNA expression in extrathymic tissues, and particularly within peripheral lymphoid tissues. There is also evidence to suggest that AIRE is expressed in PBMCs, principally cells of the myeloid lineage such as monocytes and DCs [26,54,56,58,72]
Evidence for extrathymic Aire expression at a protein level
Some, but not all, studies have demonstrated congruency between Aire RNA and protein expression within tissues (Table 3). Halonen et al. previously described Aire RNA expression across peripheral lymphoid organs in the NMRI mouse [69] and were able to reproduce these findings at a protein level using immunohistochemistry. Aire was also detected in bone marrow, peripheral blood cells and ovary. Although detecting a perhaps surprisingly wide distribution of extra-thymic Aire (leading to concerns about antibody specificity), these findings were consistent across the techniques of reverse transcription (RT)–PCR, in-situ hybridization and immunohistochemistry.
Table 3.
Summary of findings for studies of autoimmune regulatory (Aire) protein expression in peripheral tissue.
| Author | Species | Tissue | Technique | Antibody | Findings |
|---|---|---|---|---|---|
| Heino et al. (1999) [71] | Human | Thymus, spleen, LN, F liver, adrenal cortex, thyroid gland, gonad, pancreas, kidney, skin, lung, heart, PBL Adult and fetal | Immunohistochemistry | Mouse polyclonal against human AIRE protein, rabbit polyclonal against a peptide to the 20 C-terminal amino acids of mouse Aire | Staining found in thymus medulla, medulla and paracortical regions in lymph node, in spleen and F liver. No staining was seen in the remaining tissues. Identical results obtained with the mouse and rabbit anti-AIRE |
| Heino et al. (2000) [74] | Mouse (BALB/c and NOD) | Thymus, skeletal muscle, testis, ovaries, kidney, adrenal gland, lung, LN, spleen, liver | Western blotting | Rabbit polyclonal against 11 peptides from the mouse Aire sequence | 60kDa Aire-specific band detected in thymus only. Band was abolished by pre-absorbing antibody with immunizing serum confirming specificity for Aire staining. No staining seen for any of the other tissues |
| Halonen et al. (2001) [69] | Mouse (NMRI) | Thymus, spleen, LN, BM, PBMCs | Immunohistochemistry | Rabbit polyclonal against mouse Aire polypeptide | Staining found predominantly in epithelial cells of thymus as nuclear dots. Some staining of splenic macrophages, lymphocytes and neutrophils. Some staining of lymphocytes, and reticular cells in LN. Some staining of large megakaryocyte subpopulations and possibly lymphoblasts and myeloblasts in BM. Some staining of lymphocytes, polymorphonuclear leukocyte and monocyte nuclei in PBMC smears |
| Ramsey et al. (2002) [52] | Mouse (C57BL/6 Aire+/+ and Aire–/–) | Thymus, liver, brain | Immunohistochemistry | Rabbit polyclonal against a peptide from mouse Aire protein | Thymus, liver and brain in Aire+/+ mice showed immunoreactivity for Aire. No reactivity detected in Aire–/– mouse tissues confirming specificy for Aire staining |
| Adamson et al. (2004) [53] | Mouse (CD1 and C57BL/6J at 6–8 weeks) | Thymus, LN, spleen, lung, kidney, ovary, testis, goblet cells of gut, brain, fallopian tubes, heart, liver, adrenal glands | Immunohistochemistry | Rabbit polyclonal against a peptide from the human AIRE protein | Aire expression detected in all tissues other than heart, liver and adrenal glands in both mouse strains |
| C57BL/6J E14·5 | In contrast to adult tissue described above, Aire staining was seen in liver but decreased towards term. Like the adult, staining was found in lung and kidney. Although staining was seen in the gut at this embryonic stage, it was in different cells to the adult gut | ||||
| Suzuki et al. (2008) [72] | Human | PBMC | FACS | Rabbit polyclonal against a peptide from the human AIRE protein | Barely expressed on T cell, intermediate expression on B cells and CD14+ cells which appeared to be DC/macrophages |
| Hubert et al. (2008) [56] | Mouse | Thymus, spleen, LN | Immunohistochemistry/ immunofluorescence | Rat monoclonal against a peptide from mouse Aire protein | Staining seen in the medullar and cortico-medullar junction of thymus |
| Thymus, spleen | Transgenic mice expressing β-galactosidase under control of Aire promoter | β-galactosidase expression | Staining seen in the medullar and cortico-medullar junction of thymus | ||
| Thymus, spleen and LN DCs CD45–UEA+ cells from spleen and LN | FACS | Rat monoclonal against a peptide from mouse Aire protein | No staining seen in any tissues examined | ||
| Schaller et al. (2008) [76] | Mouse (HD C57BL/6 background) (LA unknown strain contributions) | Thymus, testis | Immunohistochemistry | Rat monoclonal against a peptide from mouse Aire protein – also used by Hubert et al. 2008 [45] | Staining seen in the medullary cells of the thymus, spermatogonia and spermatocytes of the testis |
| Gardner et al. (2008) [73] | Mouse [Aire-driven Igrp-GFP (ADIG)] | Thymus, LN and spleen | Immunofluorescence | Anti-GFP staining | GFP was expressed in mTECs and found in LN and spleen |
| Rat monoclonal against a peptide from mouse Aire protein – also used by Hubert et al. 2008 [45] | Aire staining seen in LN | ||||
| Poliani et al. (2010) [54] | Human | Fetal and adult tissues of the cardiovascular system, endocrine glands, eye and annexa, female genital system, gastrointestinal tract, haemolymphopoietic system, integumentary system, male genital system, mammary gland, nervous system, respiratory system, salivary gland, soft tissues and urinary system | Immunohistochemistry | Mouse monoclonal raised against human Aire protein | In fetal tissue – Aire only detected in thymus |
| In adult tissue – Aire detected in thymus, LN, tonsils and GALT but not in spleen. Aire-positive cells regularly found in LN but at lower numbers than thymus. Aire staining rarer still in tonsils and GALT. All other tissues were negative for AIRE staining |
LN, lymph node; MLN, mesenteric lymph node; SKLN, skin draining lymph node; GALT, gut-associated lymphoid tissue; PBMC, peripheral blood mononuclear cells; BM, bone marrow; F liver, fetal liver; DC, dendritic cell; NOD, non-obese diabetic; FACS, fluorescence activated cell sorting; GFP, green fluorescent protein; PBL, peripheral blood leucocytes; igrp, islet-specific glucose-6-phosphatase-related protein; ADIG, Aire-driven Igrp-GFP.
Immunohistochemical staining studies of both the inbred C57BL/6J and the outbred CD1 mice strains by Adamson et al. detected Aire expression in thymic mTECs, but also sporadic staining of the non-epithelial cells within both the medulla and cortex. Consistent with their RNA findings, Aire staining was detected in spleen, specifically within nuclei of B and T cells. Strong staining was also detected in lymph node, gut, gonadal and brain tissue, and within epithelial cells of the lung, kidney and fallopian tube. For the most part, these immunohistochemical findings support their RNA findings, the exception being for the liver and heart where protein expression could not be confirmed [53]. A recent study by Poliani et al. also confirmed the presence of AIRE mRNA and protein in adult human lymph nodes [54], but were unable to detect Aire at any level in a large number of other peripheral tissue (including the ovaries and fallopian tubes, bone marrow, spleen, testis, lung and kidneys).
In contrast, other studies have been unable to confirm their RNA findings at a protein level. Heino et al., who had previously reported Aire mRNA in thymus, lymph node, spleen and other peripheral tissues in the normal BALB/c mouse, were unable to detect Aire protein using Western blotting or immunohistochemistry in any tissue other than thymus in a number of mouse models [74]. Similarly, and in contrast to their findings at the RNA level, Hubert et al. were unable to detect Aire protein in murine spleen or lymph nodes [56]. They further examined expression using β-galactosidase under the control of the Aire promoter. Although seen in the thymus this signal was not detectable in the spleen, nor in thymic, lymph node or splenic-derived DCs. Notably in human tissue, Heino et al. observed Aire staining in thymus medulla, paracortical regions and medulla of lymph node, spleen and fetal liver [71]. They demonstrated that 10–20% of Aire-expressing cells in lymph node express CD83, which would be consistent with a DC phenotype.
As in the RNA studies, the lineage of Aire protein expressing cells has been examined. First, the study by Poliani et al. demonstrate these cells in human lymph node tissue to be CD45 negative DCs [54]. Further analysis by real-time PCR showed thar these cells selectively express RNA encoding indoleamine 2,3-dioxygenase and interleukin-10. Expression of these molecules are consistent with a role in the maintenance of tolerance [75]. Secondly, Suzuki et al., who reported AIRE mRNA expression in human peripheral blood B and T cells, with less convincing expression in CD14+ myeloid cells (Table 2), were able to confirm their findings at a protein level for all three cell types [72]. From their size and CD65 expression Suzuki et al. took these CD14+ cells to be DC/macrophages. The results of these studies suggest that peripheral Aire-expressing cells are of DC lineage, but subsequent transgenic studies provide some evidence for epithelial cells to also express Aire.
Evidence for extrathymic Aire expression from transgenic studies
Gardner et al. modified the Aire locus to drive expression of green fluorescent protein (GFP) fused to a pancreatic auto-antigen, islet-specific glucose-6-phosphatase-related protein (Igrp) [73]. In this Aire-driven Igrp-GFP (ADIG) mouse, thymic histology demonstrated GFP expression to be restricted to Aire-expressing mTEC cells (although GFP+ cells that did not co-localize with endogenous Aire were also apparent). They also demonstrated GFP staining in the spleen and lymph nodes. The majority of these extra-thymic Aire-expressing cells, termed eTACs, appeared to be stromal in origin and shared many characteristics with thymic mTECs. Perhaps surprisingly, eTACs lacked the expression of CD80 and CD86 co-stimulatory molecules, as well as the mTEC marker Ulex europaeus agglutinin (UEA)-1. They appeared to be characterized most clearly by the expression of MHC II+, epithelial cell adhesion molecule (EpCAM+) but CD45–. Such cells were also subsequently detectable in a non-transgenic murine system, and moderate levels of Aire were described in these cells within the spleen and lymph node by quantitative PCR. These levels of expression were higher than DCs resident in the spleen and lymph node. Aire was detectable at a protein level through immunohistochemical staining, and expression co-localized with GFP. However, as noted previously in non-transgenic systems, Aire expression was much lower than in the thymus, and near the limits of detection.
The studies described above and summarized in Tables 2 and 3 provide significant evidence for extra-thymic Aire expression at both a protein and RNA level. Reports are most consistent for peripheral lymphoid tissues – lymph nodes and spleen. However, the lack of a good antibody for the Aire protein is a clear limiting factor in the reproducibility of these studies. Currently, individual research groups generate many of the antibodies used in their reported studies. Most of the antibodies used are polyclonal and generated against peptides rather than whole protein. Polyclonal antibodies can be useful, given the low levels of Aire expression in the periphery, the existence of splice variants of Aire [28] and for detecting post-translationally modified isoforms. However, polyclonal antibodies are more likely to cross-react with non-specific proteins. Despite the use of appropriate controls to demonstrate specificity, studies remain conflicting. It is inevitable that some of the results will be artefactual, resulting from a combination of low expression levels and suboptimal antibodies.
Function of extra-thymic Aire
What, then, is the function of this low level of peripherally expressed Aire? A direct approach to investigating this would involve selective blocking of extra-thymic Aire expression and observation of the effect on (auto)immunity. Such a study has yet to be reported. However, analogous to the thymus, where transcription of TRA is controlled by Aire, studies have tested the hypothesis that Aire controls the transcription of TRA, and that this expression induces immune tolerance. Perhaps unsurprisingly, results have been conflicting.
Gardner et al. utilized microarray analysis on sorted GFP+ eTAC cells and compared the genes up-regulated by Aire in these cells with those of sorted mTECs. The total number and the fold-change of Aire-regulated genes was lower in eTACS than in mTECs, but the results suggested that there were genes in the periphery under the control of Aire [73]. There was very little similarity between Aire-reglated genes in these two cell types. This same study also looked at the evidence for a tolerogenic function of peripheral Aire-dependent TRA expression. The ability of eTACs to affect tolerance was tested using an IGRP-specific 8·3 TCR transgenic line; 8·3 T cells proliferated in the pancreatic lymph nodes when transferred into wild-type hosts (where Igrp is only expressed in the pancreas), and persisted in non-pancreatic lymph nodes, suggesting that these T cells are not tolerized. Conversely, in ADIG mice (where Igrp expression is driven by Aire in eTACs), the transferred 8·3 T cells underwent rapid proliferation and cell death in all the major secondary lymphoid organs. Such data indicate that Aire-expressing cells are able to present self-antigens and induce T cell tolerance.
In contrast, Schaller et al. examined the testicular tissue of Aire+/− and Aire−/− mice for expression of TRAs known previously to be under Aire control in the thymus [76]. With the possible exception of a salivary protein (and in contrast to the thymus), transcription of these TRAs in the peripheral tissue was not influenced by knock-out of the Aire gene. The study by Zheng et al. described previously (Table 2) reported Aire RNA in splenic CD11c+ myeloid cells [26]. Enrichment of these cells resulted in a clear increase in transcripts for insulin but not for glutamic acid decarboxylase 67 (GAD67) or cytochrome P450, with the latter found previously to be Aire-dependent in the thymus.
An explanation for these discrepancies is that Aire regulates transcriptionally a different panel of TRAs in the periphery to the thymus (Fig. 2). A cartoon illustrating this and a summary of thymic and peripheral Aire-dependent TRAs are presented in Fig. 2. Although not an exhaustive list, it helps to illustrate the complexity of peripheral Aire function in the periphery. It is also worth noting that Aire-controlled TRAs identified in the thymus have often been examined and reproduced across studies [8,59,60,70,76], but those in the periphery have been identified only in individual studies [26,73].
Fig. 2.
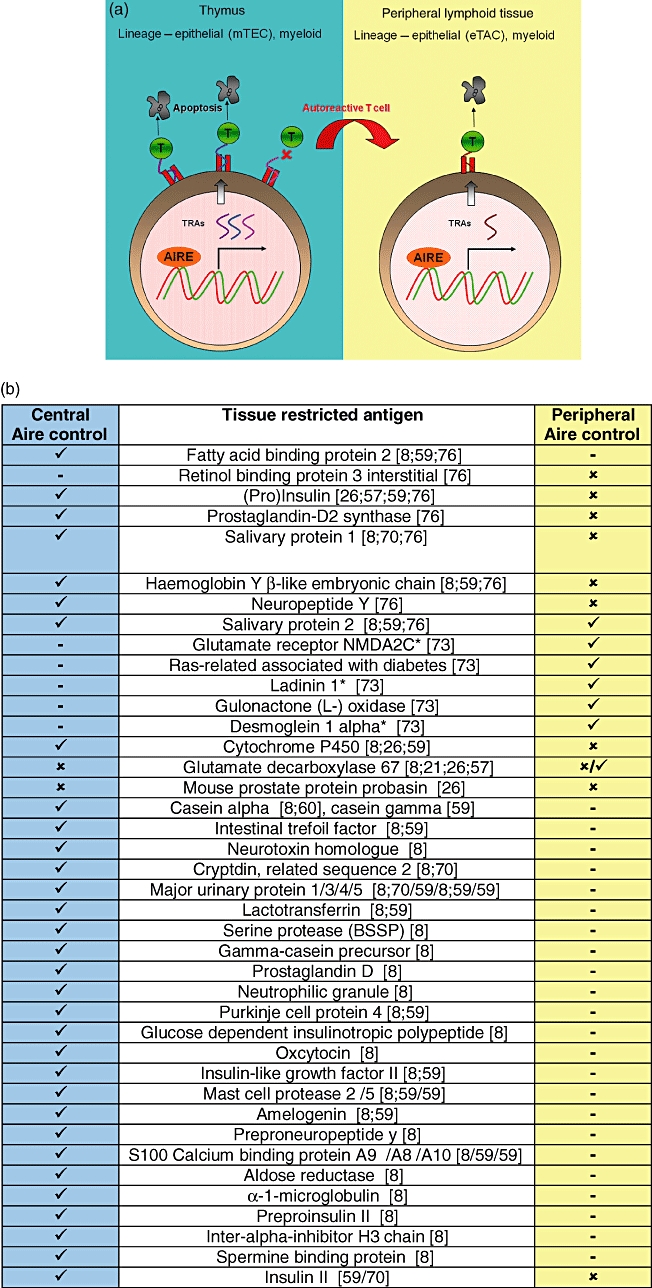
Autoimmune regulatory (Aire) protein and TRAs in the thymus and periphery. (a) Cartoon of Aire expression and function in the thymus and peripheral lymphoid tissue. TRAs are expressed by medullary thymic epithelial cells (mTECs) and myeloid cells. T cells which recognize these TRAs with too high affinity/avidity die by apoptosis. Self-reactive T cells which do not interact with their cognate antigen escape to the periphery and are eliminated following exposure to TRAs displayed in the periphery. Aire expressed in the periphery may control the expression of different TRAs to those under Aire control in the thymus. TRAs: tissue restricted antigens; DCs: dendritic cells; ✓: expressed; ✗: not expressed;  : not done. (b) Comparison of thymic and peripheral Aire-restricted TRAs. *Human homologues of these mouse proteins have been described as autoantigens in human autoimmune diseases.
: not done. (b) Comparison of thymic and peripheral Aire-restricted TRAs. *Human homologues of these mouse proteins have been described as autoantigens in human autoimmune diseases.
Another explanation for discrepancies is differential expression of Aire by subpopulations of cells in the same tissue. For example, Cohen et al. [21] demonstrate that subpopulations of lymph node cells differ in their expression of Aire. Perhaps predictably, Aire knock-out does not affect TRA expression within subpopulations that do not express Aire. For example, GAD67 was found to be expressed in three lymph node cell subpopulations, two of which expressed Aire. Analysis of the Aire−/− mice revealed loss of GAD67 expression only in those cell populations where Aire would normally be expressed. Meanwhile cells from the Aire−/− mice which would not normally express Aire were still able to present antigen to antigen-specific T cells.
Onwards from Aire
There is increasing evidence that the TRAs can be regulated transcriptionally in the absence of Aire [21,22]. Evidence has emerged recently for another transcription, factor Deaf1, in the control of TRA expression in pancreatic lymph nodes [77]. Yip et al. report a link between alternative spice forms of Deaf1 and TRA expression, which associates with risk for islet autoimmunity. In addition Deaf1 expression was present in lymph node cell subsets low in expression of Aire [22].
Conclusion
There is now strong evidence for Aire expression in peripheral tissues. These levels are significantly lower than in thymic stromal cells, and low enough to be difficult to detect reproducibly with current techniques. Lack of good-quality antibodies to examine protein expression further hampers progress, particularly in humans. In addition, reports of Aire expression in myeloid cells both in lymphoid organs and peripheral blood also supports a role for Aire in peripheral tissue. While Aire appears to be involved in transcriptional control of peripheral TRAs, there is mounting evidence to suggest that it is not involved with control of all TRAs. However, those antigens controlled by Aire in the periphery are not identical to those controlled by Aire in the thymus. Furthermore, even within peripheral lymphoid tissue, TRA expression may be influenced by the expressing cell lineage. Understanding the control of peripheral TRA expression is clearly only just beginning and much remains to be unravelled. None the less, this developing field offers the possibility of uncovering new layers of immune tolerance to self-antigens that are likely to be relevant in our understanding of autoimmunity.
Disclosure
None to declare.
References
- 1.Chi AW, Bell JJ, Zlotoff DA, Bhandoola A. Untangling the T branch of the hematopoiesis tree. Curr Opin Immunol. 2009;21:121–6. doi: 10.1016/j.coi.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hiesche KD, Revesz L. Cellular repopulation in irradiated mouse thymus and bone marrow. Beitr Pathol. 1974;151:304–16. doi: 10.1016/s0005-8165(74)80007-7. [DOI] [PubMed] [Google Scholar]
- 3.Zlotoff DA, Schwarz BA, Bhandoola A. The long road to the thymus: the generation, mobilization, and circulation of T-cell progenitors in mouse and man. Semin Immunopathol. 2008;30:371–82. doi: 10.1007/s00281-008-0133-4. [DOI] [PubMed] [Google Scholar]
- 4.Chien YH, Gascoigne NR, Kavaler J, Lee NE, Davis MM. Somatic recombination in a murine T-cell receptor gene. Nature. 1984;309:322–6. doi: 10.1038/309322a0. [DOI] [PubMed] [Google Scholar]
- 5.Klein L, Hinterberger M, Wirnsberger G, Kyewski B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat Rev Immunol. 2009;9:833–44. doi: 10.1038/nri2669. [DOI] [PubMed] [Google Scholar]
- 6.Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol. 2001;2:1032–9. doi: 10.1038/ni723. [DOI] [PubMed] [Google Scholar]
- 7.Smith KM, Olson DC, Hirose R, Hanahan D. Pancreatic gene expression in rare cells of thymic medulla: evidence for functional contribution to T cell tolerance. Int Immunol. 1997;9:1355–65. doi: 10.1093/intimm/9.9.1355. [DOI] [PubMed] [Google Scholar]
- 8.Anderson MS, Venanzi ES, Klein L, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 9.Pugliese A, Zeller M, Fernandez A, Jr, et al. The insulin gene is transcribed in the human thymus and transcription levels correlated with allelic variation at the INS VNTR–IDDM2 susceptibility locus for type 1 diabetes. Nat Genet. 1997;15:293–7. doi: 10.1038/ng0397-293. [DOI] [PubMed] [Google Scholar]
- 10.Sospedra M, Ferrer-Francesch X, Dominguez O, Juan M, Foz-Sala M, Pujol-Borrell R. Transcription of a broad range of self-antigens in human thymus suggests a role for central mechanisms in tolerance toward peripheral antigens. J Immunol. 1998;161:5918–29. [PubMed] [Google Scholar]
- 11.Vafiadis P, Bennett ST, Todd JA, et al. Insulin expression in human thymus is modulated by INS VNTR alleles at the IDDM2 locus. Nat Genet. 1997;15:289–92. doi: 10.1038/ng0397-289. [DOI] [PubMed] [Google Scholar]
- 12.Derbinski J, Gabler J, Brors B, et al. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med. 2005;202:33–45. doi: 10.1084/jem.20050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein L, Klein T, Ruther U, Kyewski B. CD4 T cell tolerance to human C-reactive protein, an inducible serum protein, is mediated by medullary thymic epithelium. J Exp Med. 1998;188:5–16. doi: 10.1084/jem.188.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein L, Roettinger B, Kyewski B. Sampling of complementing self-antigen pools by thymic stromal cells maximizes the scope of central T cell tolerance. Eur J Immunol. 2001;31:2476–86. doi: 10.1002/1521-4141(200108)31:8<2476::aid-immu2476>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 15.Kyewski B, Derbinski J, Gotter J, Klein L. Promiscuous gene expression and central T-cell tolerance: more than meets the eye. Trends Immunol. 2002;23:364–71. doi: 10.1016/s1471-4906(02)02248-2. [DOI] [PubMed] [Google Scholar]
- 16.Gallegos AM, Bevan MJ. Central tolerance to tissue-specific antigens mediated by direct and indirect antigen presentation. J Exp Med. 2004;200:1039–49. doi: 10.1084/jem.20041457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia CA, Prabakar KR, Diez J, et al. Dendritic cells in human thymus and periphery display a proinsulin epitope in a transcription-dependent, capture-independent fashion. J Immunol. 2005;175:2111–22. doi: 10.4049/jimmunol.175.4.2111. [DOI] [PubMed] [Google Scholar]
- 18.Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: learning self-control in the thymus. Nat Rev Immunol. 2005;5:772–82. doi: 10.1038/nri1707. [DOI] [PubMed] [Google Scholar]
- 19.Pugliese A, Brown D, Garza D, et al. Self-antigen-presenting cells expressing diabetes-associated autoantigens exist in both thymus and peripheral lymphoid organs. J Clin Invest. 2001;107:555–64. doi: 10.1172/JCI10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Throsby M, Homo-Delarche F, Chevenne D, Goya R, Dardenne M, Pleau JM. Pancreatic hormone expression in the murine thymus: localization in dendritic cells and macrophages. Endocrinology. 1998;139:2399–406. doi: 10.1210/endo.139.5.5989. [DOI] [PubMed] [Google Scholar]
- 21.Cohen JN, Guidi CJ, Tewalt EF, et al. Lymph node-resident lymphatic endothelial cells mediate peripheral tolerance via Aire-independent direct antigen presentation. J Exp Med. 2010;207:681–8. doi: 10.1084/jem.20092465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fletcher AL, Lukacs-Kornek V, Reynoso ED, et al. Lymph node fibroblastic reticular cells directly present peripheral tissue antigen under steady-state and inflammatory conditions. J Exp Med. 2010;207:689–97. doi: 10.1084/jem.20092642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JW, Epardaud M, Sun J, et al. Peripheral antigen display by lymph node stroma promotes T cell tolerance to intestinal self. Nat Immunol. 2007;8:181–90. doi: 10.1038/ni1427. [DOI] [PubMed] [Google Scholar]
- 24.Magnusson FC, Liblau RS, von Boehmer H, et al. Direct presentation of antigen by lymph node stromal cells protects against CD8 T-cell-mediated intestinal autoimmunity. Gastroenterology. 2008;134:1028–37. doi: 10.1053/j.gastro.2008.01.070. [DOI] [PubMed] [Google Scholar]
- 25.Nichols LA, Chen Y, Colella TA, Bennett CL, Clausen BE, Engelhard VH. Deletional self-tolerance to a melanocyte/melanoma antigen derived from tyrosinase is mediated by a radio-resistant cell in peripheral and mesenteric lymph nodes. J Immunol. 2007;179:993–1003. doi: 10.4049/jimmunol.179.2.993. [DOI] [PubMed] [Google Scholar]
- 26.Zheng X, Yin L, Liu Y, Zheng P. Expression of tissue-specific autoantigens in the hematopoietic cells leads to activation-induced cell death of autoreactive T cells in the secondary lymphoid organs. Eur J Immunol. 2004;34:3126–34. doi: 10.1002/eji.200425177. [DOI] [PubMed] [Google Scholar]
- 27.Mathis D, Benoist C. Aire. Annu Rev Immunol. 2009;27:287–312. doi: 10.1146/annurev.immunol.25.022106.141532. [DOI] [PubMed] [Google Scholar]
- 28.Nagamine K, Peterson P, Scott HS, et al. Positional cloning of the APECED gene. Nat Genet. 1997;17:393–8. doi: 10.1038/ng1297-393. [DOI] [PubMed] [Google Scholar]
- 29.The Finnish–German APECED Consortium. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat Genet. 1997;17:399–403. doi: 10.1038/ng1297-399. [DOI] [PubMed] [Google Scholar]
- 30.Ruan QG, Wang CY, Shi JD, She JX. Expression and alternative splicing of the mouse autoimmune regulator gene (Aire) J Autoimmun. 1999;13:307–13. doi: 10.1006/jaut.1999.0326. [DOI] [PubMed] [Google Scholar]
- 31.Bjorses P, Halonen M, Palvimo JJ, et al. Mutations in the AIRE gene: effects on subcellular location and transactivation function of the autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy protein. Am J Hum Genet. 2000;66:378–92. doi: 10.1086/302765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meloni A, Fiorillo E, Corda D, et al. DAXX is a new AIRE-interacting protein. J Biol Chem. 2010;285:13012–21. doi: 10.1074/jbc.M109.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibson TJ, Ramu C, Gemund C, Aasland R. The APECED polyglandular autoimmune syndrome protein, AIRE-1, contains the SAND domain and is probably a transcription factor. Trends Biochem Sci. 1998;23:242–4. doi: 10.1016/s0968-0004(98)01231-6. [DOI] [PubMed] [Google Scholar]
- 34.Pitkanen J, Vahamurto P, Krohn K, Peterson P. Subcellular localization of the autoimmune regulator protein. Characterization of nuclear targeting and transcriptional activation domain. J Biol Chem. 2001;276:19597–602. doi: 10.1074/jbc.M008322200. [DOI] [PubMed] [Google Scholar]
- 35.Bottomley MJ, Stier G, Pennacchini D, et al. NMR structure of the first PHD finger of autoimmune regulator protein (AIRE1). Insights into autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy (APECED) disease. J Biol Chem. 2005;280:11505–12. doi: 10.1074/jbc.M413959200. [DOI] [PubMed] [Google Scholar]
- 36.Peterson P, Pitkanen J, Sillanpaa N, Krohn K. Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy (APECED): a model disease to study molecular aspects of endocrine autoimmunity. Clin Exp Immunol. 2004;135:348–57. doi: 10.1111/j.1365-2249.2004.02384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pitkanen J, Doucas V, Sternsdorf T, et al. The autoimmune regulator protein has transcriptional transactivating properties and interacts with the common coactivator CREB-binding protein. J Biol Chem. 2000;275:16802–9. doi: 10.1074/jbc.M908944199. [DOI] [PubMed] [Google Scholar]
- 38.Abramson J, Giraud M, Benoist C, Mathis D. Aire's partners in the molecular control of immunological tolerance. Cell. 2010;140:123–35. doi: 10.1016/j.cell.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 39.Villasenor J, Benoist C, Mathis D. AIRE and APECED: molecular insights into an autoimmune disease. Immunol Rev. 2005;204:156–64. doi: 10.1111/j.0105-2896.2005.00246.x. [DOI] [PubMed] [Google Scholar]
- 40.Macedo C, Evangelista AF, Magalhaes DA, et al. Evidence for a network transcriptional control of promiscuous gene expression in medullary thymic epithelial cells. Mol Immunol. 2009;46:3240–4. doi: 10.1016/j.molimm.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 41.Ahonen P. Autoimmune polyendocrinopathy–candidosis–ectodermal dystrophy (APECED): autosomal recessive inheritance. Clin Genet. 1985;27:535–42. doi: 10.1111/j.1399-0004.1985.tb02037.x. [DOI] [PubMed] [Google Scholar]
- 42.Neufeld M, Blizzard RM. Polyglandular autoimmune diseases. In: Pinchera A, Doniach D, Fenzi GF, Baschieri L, editors. Symposium on autoimmune aspects of endocrine disorders. New York: Academic Press; 1980. pp. 357–65. [Google Scholar]
- 43.Neufeld M, Maclaren NK, Blizzard RM. Two types of autoimmune Addison's disease associated with different polyglandular autoimmune (PGA) syndromes. Medicine (Baltimore) 1981;60:355–62. doi: 10.1097/00005792-198109000-00003. [DOI] [PubMed] [Google Scholar]
- 44.Ahonen P, Myllarniemi S, Sipila I, Perheentupa J. Clinical variation of autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy (APECED) in a series of 68 patients. N Engl J Med. 1990;322:1829–36. doi: 10.1056/NEJM199006283222601. [DOI] [PubMed] [Google Scholar]
- 45.Zlotogora J, Shapiro MS. Polyglandular autoimmune syndrome type I among Iranian Jews. J Med Genet. 1992;29:824–6. doi: 10.1136/jmg.29.11.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosatelli MC, Meloni A, Meloni A, et al. A common mutation in Sardinian autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy patients. Hum Genet. 1998;103:428–34. doi: 10.1007/s004390050846. [DOI] [PubMed] [Google Scholar]
- 47.LeBoeuf N, Garg A, Worobec S. The autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy syndrome. Pediatr Dermatol. 2007;24:529–33. doi: 10.1111/j.1525-1470.2007.00510.x. [DOI] [PubMed] [Google Scholar]
- 48.Perheentupa J. Autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy. J Clin Endocrinol Metab. 2006;91:2843–50. doi: 10.1210/jc.2005-2611. [DOI] [PubMed] [Google Scholar]
- 49.Betterle C, Greggio NA, Volpato M. Clinical review 93: autoimmune polyglandular syndrome type 1. J Clin Endocrinol Metab. 1998;83:1049–55. doi: 10.1210/jcem.83.4.4682. [DOI] [PubMed] [Google Scholar]
- 50.Blechschmidt K, Schweiger M, Wertz K, et al. The mouse AIRE gene: comparative genomic sequencing, gene organization, and expression. Genome Res. 1999;9:158–66. [PMC free article] [PubMed] [Google Scholar]
- 51.Wang CY, Shi JD, Davoodi-Semiromi A, She JX. Cloning of Aire, the mouse homologue of the autoimmune regulator (AIRE) gene responsible for autoimmune polyglandular syndrome type 1 (ASP1) Genomics. 1999;55:322–6. doi: 10.1006/geno.1998.5656. [DOI] [PubMed] [Google Scholar]
- 52.Ramsey C, Bukrinsky A, Peltonen L. Systematic mutagenesis of the functional domains of AIRE reveals their role in intracellular targeting. Hum Mol Genet. 2002;11:3299–308. doi: 10.1093/hmg/11.26.3299. [DOI] [PubMed] [Google Scholar]
- 53.Adamson KA, Pearce SHS, Lamb JR, Seckl JR, Howie SEM. A comparative study of mRNA and protein expression of the autoimmune regulator gene (Aire) in embryonic and adult murine tissues. J Pathol. 2004;202:180–7. doi: 10.1002/path.1493. [DOI] [PubMed] [Google Scholar]
- 54.Poliani PL, Kisand K, Marrella V, et al. Human peripheral lymphoid tissues contain autoimmune regulator-expressing dendritic cells. Am J Pathol. 2010;176:1104–12. doi: 10.2353/ajpath.2010.090956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zuklys S, Balciunaite G, Agarwal A, Fasler-Kan E, Palmer E, Hollander GA. Normal thymic architecture and negative selection are associated with Aire expression, the gene defective in the autoimmune-polyendocrinopathy–candidiasis–ectodermal dystrophy (APECED) J Immunol. 2000;165:1976–83. doi: 10.4049/jimmunol.165.4.1976. [DOI] [PubMed] [Google Scholar]
- 56.Hubert FX, Kinkel SA, Webster KE, et al. A specific anti-Aire antibody reveals Aire expression is restricted to medullary thymic epithelial cells and not expressed in periphery. J Immunol. 2008;180:3824–32. doi: 10.4049/jimmunol.180.6.3824. [DOI] [PubMed] [Google Scholar]
- 57.Devoss JJ, Anderson MS. Lessons on immune tolerance from the monogenic disease APS1. Curr Opin Genet Dev. 2007;17:193–200. doi: 10.1016/j.gde.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 58.Pontynen N, Strengell M, Sillanpaa N, et al. Critical immunological pathways are downregulated in APECED patient dendritic cells. J Mol Med. 2008;86:1139–52. doi: 10.1007/s00109-008-0374-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnnidis JB, Venanzi ES, Taxman DJ, Ting JP, Benoist CO, Mathis DJ. Chromosomal clustering of genes controlled by the Aire transcription factor. Proc Natl Acad Sci USA. 2005;102:7233–8. doi: 10.1073/pnas.0502670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anderson MS, Venanzi ES, Chen Z, Berzins SP, Benoist C, Mathis D. The cellular mechanism of Aire control of T cell tolerance. Immunity. 2005;23:227–39. doi: 10.1016/j.immuni.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 61.Liston A, Lesage S, Wilson J, Peltonen L, Goodnow CC. Aire regulates negative selection of organ-specific T cells. Nat Immunol. 2003;4:350–4. doi: 10.1038/ni906. [DOI] [PubMed] [Google Scholar]
- 62.Liston A, Gray DH, Lesage S, et al. Gene dosage – limiting role of Aire in thymic expression, clonal deletion, and organ-specific autoimmunity. J Exp Med. 2004;200:1015–26. doi: 10.1084/jem.20040581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lohmann T, Leslie RD, Londei M. T cell clones to epitopes of glutamic acid decarboxylase 65 raised from normal subjects and patients with insulin-dependent diabetes. J Autoimmun. 1996;9:385–9. doi: 10.1006/jaut.1996.0052. [DOI] [PubMed] [Google Scholar]
- 64.Semana G, Gausling R, Jackson RA, Hafler DA. T cell autoreactivity to proinsulin epitopes in diabetic patients and healthy subjects. J Autoimmun. 1999;12:259–67. doi: 10.1006/jaut.1999.0282. [DOI] [PubMed] [Google Scholar]
- 65.Narendran P, Neale AM, Lee BH, et al. Proinsulin is encoded by an RNA splice variant in human blood myeloid cells. Proc Natl Acad Sci USA. 2006;103:16430–5. doi: 10.1073/pnas.0607380103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kojima H, Fujimiya M, Matsumura K, Nakahara T, Hara M, Chan L. Extrapancreatic insulin-producing cells in multiple organs in diabetes. Proc Natl Acad Sci USA. 2004;101:2458–63. doi: 10.1073/pnas.0308690100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goolsby J, Marty MC, Heletz D, et al. Hematopoietic progenitors express neural genes. Proc Natl Acad Sci USA. 2003;100:14926–31. doi: 10.1073/pnas.2434383100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marty MC, Alliot F, Rutin J, Fritz R, Trisler D, Pessac B. The myelin basic protein gene is expressed in differentiated blood cell lineages and in hemopoietic progenitors. Proc Natl Acad Sci USA. 2002;99:8856–61. doi: 10.1073/pnas.122079599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Halonen M, Pelto-Huikko M, Eskelin P, Peltonen L, Ulmanen I, Kolmer M. Subcellular location and expression pattern of autoimmune regulator (Aire), the mouse orthologue for human gene defective in autoimmune polyendocrinopathy candidiasis ectodermal dystrophy (APECED) J Histochem Cytochem. 2001;49:197–208. doi: 10.1177/002215540104900207. [DOI] [PubMed] [Google Scholar]
- 70.Kont V, Laan M, Kisand K, Merits A, Scott HS, Peterson P. Modulation of Aire regulates the expression of tissue-restricted antigens. Mol Immunol. 2008;45:25–33. doi: 10.1016/j.molimm.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heino M, Peterson P, Kudoh J, et al. Autoimmune regulator is expressed in the cells regulating immune tolerance in thymus medulla. Biochem Biophys Res Commun. 1999;257:821–5. doi: 10.1006/bbrc.1999.0308. [DOI] [PubMed] [Google Scholar]
- 72.Suzuki E, Kobayashi Y, Kawano O, et al. Expression of AIRE in thymocytes and peripheral lymphocytes. Autoimmunity. 2008;41:133–9. doi: 10.1080/08916930701773941. [DOI] [PubMed] [Google Scholar]
- 73.Gardner JM, Devoss JJ, Friedman RS, et al. Deletional tolerance mediated by extrathymic Aire-expressing cells. Science. 2008;321:843–7. doi: 10.1126/science.1159407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heino M, Peterson P, Sillanpaa N, et al. RNA and protein expression of the murine autoimmune regulator gene (Aire) in normal, RelB-deficient and in NOD mouse. Eur J Immunol. 2000;30:1884–93. doi: 10.1002/1521-4141(200007)30:7<1884::AID-IMMU1884>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 75.Mellor AL, Keskin DB, Johnson T, Chandler P, Munn DH. Cells expressing indoleamine 2,3-dioxygenase inhibit T cell responses. J Immunol. 2002;168:3771–6. doi: 10.4049/jimmunol.168.8.3771. [DOI] [PubMed] [Google Scholar]
- 76.Schaller CE, Wang CL, Beck-Engeser G, et al. Expression of Aire and the early wave of apoptosis in spermatogenesis. J Immunol. 2008;180:1338–43. doi: 10.4049/jimmunol.180.3.1338. [DOI] [PubMed] [Google Scholar]
- 77.Yip L, Su L, Sheng D, et al. Deaf1 isoforms control the expression of genes encoding peripheral tissue antigens in the pancreatic lymph nodes during type 1 diabetes. Nat Immunol. 2009;10:1026–33. doi: 10.1038/ni.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]


