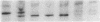Abstract
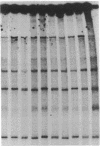
Single base pair differences between otherwise identical DNA molecules can result in altered melting behavior detectable by denaturing gradient gel electrophoresis. We have developed a simplified procedure for using denaturing gradient gel electrophoresis to detect base pair changes in genomic DNA. Genomic DNA is digested with restriction enzymes and hybridized in solution to labeled single-stranded probe DNA. The excess probe is then hybridized to complementary phage M13 template DNA, and the reaction mixture is electrophoresed on a denaturing gradient gel. Only the genomic DNA probe hybrids migrate into the gel. Differences in hybrid mobility on the gel indicate base pair changes in the genomic DNA. We have used this technique to identify two polymorphic sites within a 1.2-kilobase region of human chromosome 20. This approach should greatly facilitate the identification of DNA polymorphisms useful for gene linkage studies and the diagnosis of genetic diseases.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aldridge J., Kunkel L., Bruns G., Tantravahi U., Lalande M., Brewster T., Moreau E., Wilson M., Bromley W., Roderick T. A strategy to reveal high-frequency RFLPs along the human X chromosome. Am J Hum Genet. 1984 May;36(3):546–564. [PMC free article] [PubMed] [Google Scholar]
- Barker D., Schafer M., White R. Restriction sites containing CpG show a higher frequency of polymorphism in human DNA. Cell. 1984 Jan;36(1):131–138. doi: 10.1016/0092-8674(84)90081-3. [DOI] [PubMed] [Google Scholar]
- Fischer S. G., Lerman L. S. Two-dimensional electrophoretic separation of restriction enzyme fragments of DNA. Methods Enzymol. 1979;68:183–191. doi: 10.1016/0076-6879(79)68013-8. [DOI] [PubMed] [Google Scholar]
- Gitschier J., Wood W. I., Goralka T. M., Wion K. L., Chen E. Y., Eaton D. H., Vehar G. A., Capon D. J., Lawn R. M. Characterization of the human factor VIII gene. Nature. 1984 Nov 22;312(5992):326–330. doi: 10.1038/312326a0. [DOI] [PubMed] [Google Scholar]
- Lerman L. S., Fischer S. G., Hurley I., Silverstein K., Lumelsky N. Sequence-determined DNA separations. Annu Rev Biophys Bioeng. 1984;13:399–423. doi: 10.1146/annurev.bb.13.060184.002151. [DOI] [PubMed] [Google Scholar]
- Lerman L. S., Silverstein K., Grinfeld E. Searching for gene defects by denaturing gradient gel electrophoresis. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):285–297. doi: 10.1101/sqb.1986.051.01.034. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Lumelsky N., Lerman L. S., Maniatis T. Detection of single base substitutions in total genomic DNA. Nature. 1985 Feb 7;313(6002):495–498. doi: 10.1038/313495a0. [DOI] [PubMed] [Google Scholar]
- Toole J. J., Knopf J. L., Wozney J. M., Sultzman L. A., Buecker J. L., Pittman D. D., Kaufman R. J., Brown E., Shoemaker C., Orr E. C. Molecular cloning of a cDNA encoding human antihaemophilic factor. Nature. 1984 Nov 22;312(5992):342–347. doi: 10.1038/312342a0. [DOI] [PubMed] [Google Scholar]
- de Martinville B., Schäfer M., White R., Francke U. Chromosomal assignments of three random RFLP loci defined by base-pair changes in MspI sites. Mol Biol Med. 1983 Nov;1(4):415–424. [PubMed] [Google Scholar]