Abstract
RNA 2′,3′-cyclic phosphate ends play important roles in RNA metabolism as substrates for RNA ligases during tRNA restriction-repair and tRNA splicing. Diverse bacteria from multiple phyla encode a two-component RNA repair cassette, comprising Pnkp (polynucleotide kinase-phosphatase-ligase) and Hen1 (RNA 3′-terminal ribose 2′-O-methyltransferase), that heals and then seals broken tRNAs with 2′,3′-cyclic phosphate and 5′-OH ends. The Pnkp-Hen1 repair operon is absent in the majority of bacterial species, thereby raising the prospect that other RNA repair systems might be extant. A candidate component is RNA 3′-phosphate cyclase, a widely distributed enzyme that transforms RNA 3′-monophosphate termini into 2′,3′-cyclic phosphates but cannot seal the ends it produces. Escherichia coli RNA cyclase (RtcA) is encoded in a σ54-regulated operon with RtcB, a protein of unknown function. Taking a cue from Pnkp-Hen1, we purified E. coli RtcB and tested it for RNA ligase activity. We report that RtcB per se seals broken tRNA-like stem-loop structures with 2′,3′-cyclic phosphate and 5′-OH ends to form a splice junction with a 2′-OH, 3′,5′-phosphodiester. We speculate that: (i) RtcB might afford bacteria a means to recover from stress-induced RNA damage; and (ii) RtcB homologs might catalyze tRNA repair or splicing reactions in archaea and eukarya.
Keywords: Enzyme Catalysis; Metals; Nucleic Acid Enzymology; RNA Processing; Transfer RNA (tRNA); 2′,3′-Cyclic Phosphodiester; tRNA Repair; tRNA Splicing
Introduction
Programmed breakage of the tRNA anticodon loop by site-specific endoribonucleases (ribotoxins) is an ancient mechanism by which microbes either respond to virus infection or distinguish self from non-self (1–5). tRNA ribotoxins employ a transesterification mechanism that generates 5′-OH and 2′,3′-cyclic phosphate termini. Analogous site-specific transesterifications are catalyzed by archaeal and eukaryal endonucleases that initiate pre-tRNA splicing by incising the intron-exon borders (6). tRNA splicing and viral evasion of tRNA restriction demand that the broken tRNA ends be resealed faithfully by dedicated cellular or viral RNA repair enzymes. In fungal and plant tRNA splicing and in bacteriophage T4 tRNA restriction-repair, this entails sequential enzymatic end-healing and end-sealing steps (1, 7–11). In the healing phase, the 2′,3′-cyclic phosphate end is hydrolyzed to a 3′-OH, and the 5′-OH end is phosphorylated by an NTP-dependent polynucleotide kinase to yield a 5′-monophosphate. The healed 3′-OH and 5′-PO4 termini are then suitable substrates for sealing by a “classical” ATP-dependent RNA ligase that restores the 3′,5′-phosphodiester backbone. An alternative mechanism of RNA sealing, via direct attack of a 5′-OH end on a 2,3′-cyclic phosphate to yield a 3′,5′-phosphodiester, is thought to comprise the major pathway of tRNA splicing in archaea and animals (12–15).
Ribotoxin-encoding genes are widespread in prokarya, where their expression or activity is induced in response to cellular stress (16–18). This raises the question of whether prokarya might have RNA repair systems to recover from stress-induced RNA damage. Candidate RNA repair enzymes have been identified and characterized in diverse bacterial and archaeal taxa (19–26). The best case for a bona fide bacterial RNA repair pathway can be made for the many species that encode the enzymes Pnkp2 (polynucleotide 5′-kinase/3′-phosphatase) and Hen1 in an operon-like gene cassette (27, 28). Bacterial Pnkp is a multifunctional end-healing and -sealing enzyme composed of three catalytic domains: N-terminal kinase (5′-end-healing), central phosphoesterase (3′-end-healing), and C-terminal adenylyltransferase/ligase (sealing) (21, 29–32). The adenylyltransferase domain is homologous to classical ATP-dependent RNA ligases and reacts with ATP to form a covalent enzyme-AMP adduct (21), but it cannot catalyze RNA sealing per se. Hen1 serves two functions: (i) it enables the end-sealing function of the Pnkp ligase module; and (ii) it is a manganese-dependent 3′-terminal ribose 2′-O-methyltransferase that installs a methyl “mark” at the 3′-healed tRNA end prior to the sealing step and thereby immunizes the repair junction against further rounds of damage (27, 28).
Most bacteria, including Escherichia coli, do not have the Pnkp-Hen1 cassette. In considering alternative RNA repair strategies, our attention has focused on E. coli RNA 3′-phosphate cyclase (RtcA) as a candidate RNA repair enzyme. RNA cyclase transforms RNA 3′-monophosphate termini (which are not substrates for strand sealing) into potentially ligatable 2′,3′-cyclic phosphate ends. Cyclization entails a series of three nucleotidyl transfer reactions (33, 34). In the first step, Rtc reacts with ATP to form a covalent RtcA-AMP intermediate and liberate PPi. Adenylate is linked via a phosphoamide bond to a histidine Nϵ atom (35, 36). In the second step, the adenylate is transferred from RtcA-AMP to the RNA 3′-phosphate terminus to form an activated phosphoanhydride intermediate, RNA(3′)pp(5′)A. In the third step, the terminal ribose 2′-OH attacks the 3′-phosphate of RNA(3′)pp(5′)A to generate an RNA 2′,3′-cyclic phosphate product and release AMP. The cyclase pathway is reminiscent of the three nucleotidyl transfer steps catalyzed by classical RNA/DNA ligases, also via enzyme-AMP and polynucleotide-adenylate intermediates, notwithstanding that the tertiary structure and active site of RNA cyclase (Fig. 1) have nothing in common with polynucleotide ligases (36–38). Nonetheless, recent studies revealed that the catalytic repertoire of RNA cyclase overlaps that of RNA/DNA ligases; to wit, E. coli RtcA catalyzes adenylylation of 5′-phosphate ends of DNA or RNA strands to form AppDNA and AppRNA products (39). However, RtcA is unable to convert a polynucleotide-adenylate into a sealed phosphodiester. Thus, if RtcA is involved directly in an RNA-sealing pathway, it either requires a protein partner (à la Pnkp-Hen1) or plays a supportive role in RNA repair by providing a 2′,3′-cyclic phosphate substrate for a separate bacterial RNA ligase enzyme.
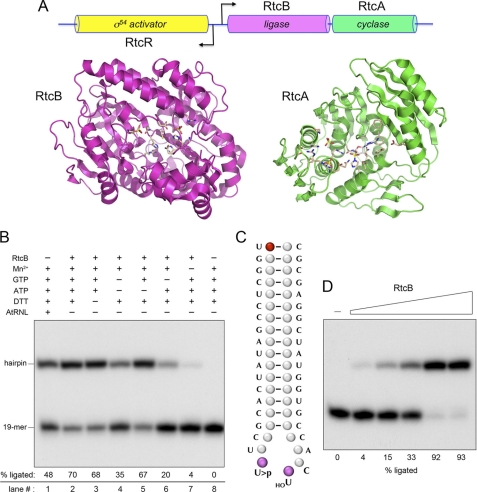
FIGURE 1.
The RtcB protein encoded by the E. coli rtcBA operon is an RNA ligase. A, genetic organization of the E. coli rtc operon. The tightly clustered rtcB and rtcA ORFs encoding the 408-aa RtcB and 338-aa RtcA polypeptides are transcribed rightward from a σ54 promoter under the control of the transcriptional regulator encoded by the oppositely oriented rtcR gene (40). RtcA is an RNA 3′-phosphatase cyclase; the structure of the E. coli RtcA-adenylate intermediate is rendered in green (36). The structure of Pyrococcus RtcB is shown in magenta (41). A primary structure alignment of E. coli and Pyrococcus RtcBs is provided in supplemental Fig. S3. Here we show that E. coli RtcB is an RNA ligase. B, ligase reaction mixtures (10 μl) containing 50 mm Tris-HCl (pH 7.4), 0.2 μm 5′-32P-labeled broken stem-loop RNA (panel C), and (where indicated by + above the lanes) 10 mm MnCl2, 2 mm DTT, 100 μm GTP, 100 μm ATP, and either 1 μm E. coli RtcB or 0.1 μm AtRNL were incubated at 37 °C for 30 min. The reactions were quenched with EDTA/formamide, and the products were analyzed by electrophoresis through a 20% polyacrylamide gel containing 7 m urea in 90 mm Tris borate, 2.5 mm EDTA. An autoradiograph of the gel is shown. The positions of the radiolabeled 19-mer substrate strand and the ligated hairpin product are indicated on the left. The extents of ligation were quantified by scanning the gel and are specified below the lanes. C, the broken tRNA-like anticodon stem-loop substrate shown was generated by K. lactis γ-toxin incision at the wobble U of the UUC anticodon. The 5′-32P-labeled 19-mer strand has a uridine 2′,3′-cyclic phosphate at the break. The distal 20-mer strand is unlabeled and has a 5′-OH uridine terminus at the break. D, reaction mixtures (10 μl) containing 50 mm Tris-HCl (pH 7.4), 0.2 μm 5′-32P-labeled broken stem-loop RNA, 2 mm MnCl2, 100 μm GTP, and either no RtcB (lane −) or 0.05, 0.1, 0.2, 0.5, or 1 μm RtcB (proceeding from left to right) were incubated at 37 °C for 30 min. The products were analyzed by PAGE and visualized by autoradiography. The extents of ligation are specified below the lanes.
Taking a cue from Pnkp-Hen1, we queried whether RtcB, the protein encoded by the flanking gene in the E. coli rtcBA operon (40) (Fig. 1), might be the missing ligase per se or in combination with RtcA. E. coli RtcB is a 408-aa polypeptide of heretofore unknown function. RtcB homologs are widely prevalent in bacteria, archaea, protozoa, and metazoa (40), although not in fungi and plants. Crystal structures of Pyrococcus horikoshii and Thermus thermophilus RtcB homologs have been solved (41) (PDB ID 2EPG)3 and have virtually identical folds, exemplified by the Pyrococcus RtcB shown in Fig. 1. This view of RtcB highlights a deep and wide hydrophilic pocket lined by conserved histidines (suggestive of an enzyme active site) and the nearby binding of two sulfate anions (which are potential mimetics of nucleic acid or nucleotide phosphates). The E. coli rtcBA operon is regulated by RtcR, a σ54-specific activator encoded by the divergently transcribed neighboring rtcR gene (Fig. 1). The operon is repressed under standard laboratory conditions of bacterial growth (40) and is presumably activated in response to an as yet unknown cellular stress.
Here we transiently expressed a plasmid-borne copy of rtcB in E. coli under the control of a T7 RNA polymerase promoter. The protein was produced as a His10-Smt3 fusion and isolated from a soluble bacterial extract by nickel-agarose chromatography (38) (see supplemental material for detailed methods). The tag was then removed by the Smt3-specific protease Ulp1, and the tag-free RtcB was separated from His10-Smt3 by a second nickel affinity step. This material was purified further by gel filtration, which resolved a single major protein component from high molecular weight aggregates recovered in the void volume (supplemental Fig. S1). SDS-PAGE analysis of the peak fractions revealed a single 45-kDa polypeptide (supplemental Fig. S1), consistent with the predicted size of recombinant RtcB. The gel filtration profile of RtcB, when compared with standards used to calibrate the column, was consistent with it being a monomer in solution.
To test whether RtcB might have RNA ligase activity, we prepared a substrate that mimicked the broken tRNAGlu(UUC) anticodon stem-loop generated by Kluyveromyces lactis γ-toxin (Fig. 1C). In brief, we 5′-32P-labeled a 39-nucleotide synthetic RNA hairpin that included the exact nucleotide sequence of the anticodon stem-loop of yeast tRNAGlu(UUC). The labeled hairpin was reacted in vitro with purified recombinant K. lactis γ-toxin (42), which breaks the RNA at a single phosphodiester 3′ of the wobble uridine, leaving 2′,3′-cyclic phosphate and 5′-OH ends (4, 43) (Fig. 1C). The broken 19-mer and 20-mer strands were copurified and separated from residual uncut hairpin by preparative gel electrophoresis.
As a positive control for RNA repair, the broken stem-loop substrate was reacted with recombinant Arabidopsis thaliana tRNA ligase (AtRNL) (11) in a mixture containing a divalent cation and ATP and GTP substrates (100 μm each) for the ligase and 5′-kinase components of the trifunctional plant enzyme. AtRNL sealed the broken ends, as evinced by the conversion of the input 32P-labeled 19-mer strand (Fig. 1B, lane 8) into a 32P-labeled ligated hairpin product (Fig. 1B, lane 1).
The salient finding was that recombinant RtcB also repaired the broken stem-loop under the same reaction conditions (Fig. 1B, lane 2). The requirements for the RtcB ligase reaction were gauged by systematic omission of reaction components (Fig. 1B), which showed that activity was nearly abolished by omitting manganese (lane 7) but was unaffected by omission of either DTT (lane 3) or ATP (lane 5). Withdrawal of GTP reduced RtcB ligase activity (lanes 4 and 6). The extent of sealing of the broken stem-loop increased with increasing RtcB concentration and was nearly quantitative at saturating enzyme (Fig. 1D). Inclusion of recombinant E. coli RtcA in the reaction mixtures (at the same concentration as RtcB) had no apparent effect on the efficiency of RNA sealing (not shown). Thus, we surmise that RtcB is a novel stand-alone RNA ligase.
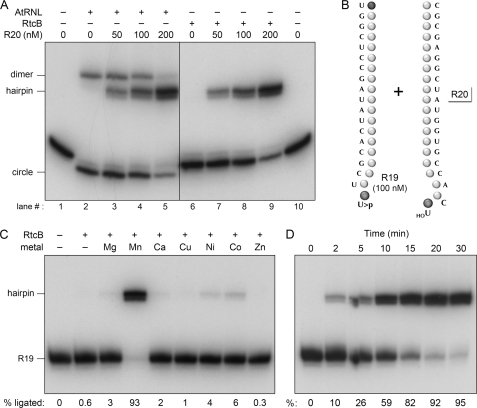
To better gauge the substrate specificity of the RtcB ligase reaction and to compare it with that of AtRNL, we presented the enzymes with a 5′-32P-labeled single-stranded RNA (R19) comprising just the 5′ half of the broken stem-loop (Fig. 2B). To prepare this RNA, we end-labeled a synthetic RNA comprising the 5′ 30 nucleotides of the RNA hairpin in Fig. 1C. Cleavage of this RNA by purified γ-toxin yielded the 32P-labeled R19 strand plus an unlabeled 11-mer that was separated from R19 during preparative gel electrophoresis. Reaction of AtRNL with the R19 strand resulted in its conversion to two novel products (Fig. 2A, compare lanes 1 and 2). The predominant product, migrating slightly faster than the substrate strand during denaturing PAGE, corresponds to a 19-mer RNA circle (11), and the minority product is a dimer of the 19-mer strand (presumably a dimer circle). It is well established that AtRNL is adept at intramolecular circularization of RNA single strands with 2′,3′-cyclic phosphate ends, both in vitro and in vivo (11, 44). By contrast, RtcB was unreactive with the R19 strand (Fig. 2A, lane 6). We surmise that the 5′-monophosphate end of the R19 substrate, which is essential for the ligation step of the plant tRNA ligase pathway (whether preformed or generated in situ by the AtRNL kinase), is actually inimical to an RtcB intramolecular RNA circularization reaction.
FIGURE 2.
RtcB substrate and cofactor specificity. A, AtRNL and RtcB have distinct substrate specificities. Reaction mixtures (10 μl) containing 50 mm Tris-HCl (pH 7.4), 2 mm MnCl2, 100 μm GTP, 100 nm 5′-32P-labeled R19 strand substrate plus 0, 50, 100, or 200 nm cold R20 strand, and 0.1 μm AtRNL or 1 μm RtcB were incubated at 37 °C for 30 min. The reaction mixtures were analyzed by denaturing PAGE. The products corresponding to R19 circle, R19 dimer, and R19-R20 hairpin are indicated on the left. B, the 5′-32P-labeled R19 single strand with a 2′,3′-cyclic phosphate end is shown. The unlabeled complementary R20 oligonucleotide was added to the ligation reactions as specified. C, metal cofactor requirement. Reaction mixtures (10 μl) containing 50 mm Tris-HCl (pH 7.4), 0.1 μm 5′-32P-labeled R19 strand, 1 μm cold R20 strand, 100 μm GTP, 1 μm RtcB, and 2 mm MgCl2, MnCl2, CaCl2, CuCl2, NiCl2, CoCl2, or ZnCl2 were incubated at 37 °C for 30 min. Divalent cation and RtcB were omitted from control reactions (−). The extents of ligation are specified below the lanes. D, time course. A reaction mixture (100 μl) containing 50 mm Tris-HCl (pH 7.4), 2 mm MnCl2, 0.1 μm 5′-32P-labeled R19 strand, 1 μm cold R20 strand, 100 μm GTP, and 1 μm RtcB was incubated at 37 °C. Aliquots (10 μl) were withdrawn at the times specified and quenched immediately with EDTA/formamide. The time 0 sample was taken prior to the addition of RtcB. The extents of ligation are specified below the lanes.
The fates of the R19 strand in the respective plant and E. coli ligase reactions were altered by inclusion of increasing amounts of an unlabeled 5′-OH RNA oligonucleotide (R20) complementary to R19, which can anneal to it and reconstitute the broken tRNA-like stem-loop (Fig. 2B). In the AtRNL reactions, the R20 strand suppressed intramolecular circularization of R19 and promoted the joining of 32P-labeled R19 to R20 to yield a hairpin product (Fig. 2A, lanes 3–5). In the RtcB mixtures, inclusion of R20 triggered its ligation to the previously inert 32P-labeled R19 strand to form the stem-loop hairpin product (Fig. 2A, lanes 7–9). (The hairpin product migrated as a doublet, reflecting the presence of a minority fraction of n − 1 length RNA in the synthetic oligonucleotide preparation.) These results are consistent with a 5′-OH terminus being the immediate substrate for the RtcB ligase.
Further characterization of the RtcB sealing reaction was performed with mixtures containing 100 nm 32P-labeled R19 strand plus 1 μm cold R20 strand. Formation of the hairpin product required a divalent cation, and this requirement was satisfied specifically by 2 mm manganese (Fig. 2C). Other divalent cations at 2 mm concentration were very weakly active (nickel, cobalt, magnesium) or virtually inactive (calcium, copper, zinc) (Fig. 2C). The selectivity for manganese might reflect the spatial clustering of a constellation of conserved histidines and a cysteine in the presumptive active site pocket of RtcB (Fig. 1A). These residues have been suggested to comprise a potential metal-binding site (40, 41) that, being dominated by “soft” metal contacts to nitrogens and sulfur, could account for the preference for manganese over magnesium. Additional insights to the metal specificity of RtcB were provided by mixing experiments, in which reactions containing 2 mm manganese were supplemented with 2 mm of another divalent cation. Copper and zinc abolished ligase activity in the presence of manganese, whereas nickel and cobalt were strongly inhibitory (supplemental Fig.S2), suggesting that each of these four soft metals might out-compete manganese for a putative metal-binding site on the enzyme, wherein engaged they are unable to support reaction chemistry. By contrast, magnesium and calcium had no such deleterious effect in combination with manganese (supplemental Fig. S2), implying that these “hard” metals do not bind effectively to the RtcB active site. The kinetic profile of the RtcB ligation reaction is shown in Fig. 2D; the hairpin ligation product accumulated progressively with reaction time and was virtually complete in 20–30 min.
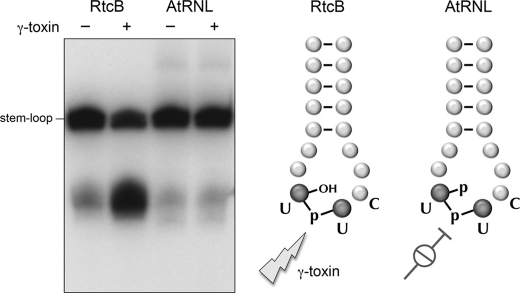
The end product of the AtRNL end-healing and -sealing reactions is a 2′-PO4, 3′,5′-phosphodiester splice junction (10) (Fig. 3). Our hypothesis is that RtcB performs direct ligation without end-healing, in which case the end product should be a standard 2′-OH, 3′,5′-phosphodiester splice junction (Fig. 3). A key prediction is that the repaired tRNAGlu(UUC)-like product of the AtRNL reaction should be refractory to recleavage by K. lactis γ-toxin, which requires a 2′-OH nucleophile for transesterification (42), whereas the product of the RtcB-sealing reaction should be susceptible to γ-toxin. Indeed, we found that this was the case (Fig. 3), fortifying our inference that RtcB catalyzes direct ligation. It is noteworthy that of the two product species comprising the hairpin doublet, only the slower migrating “full-length” repaired tRNAGlu(UUC) anticodon loop was susceptible to recleavage by γ-toxin, whereas the shorter species, resulting from sealing to an n − 1 contaminant of the 20 strand, was refractory (Fig. 3, and other data not shown). This is entirely consistent with the stringent requirement of γ-toxin for a 5′-UUC anticodon triplet (43).
FIGURE 3.
RtcB forms a cleavable 3′,5′-phosphodiester at the repair junction. Ligation reactions containing 50 mm Tris-HCl (pH 7.4), 100 nm 5′-32P-labeled R19 strand substrate, 1 μm cold R20 strand, and either (i) 10 mm MgCl2, 2 mm DTT, 100 μm ATP, 100 μm GTP, and 0.1 μm AtRNL or (ii) 2 mm MnCl2, 100 μm GTP, and 1 μm RtcB were incubated at 37 °C for 30 min. The RNA products were isolated by phenol extraction and ethanol precipitation. Aliquots of the resuspended RNAs were incubated for 60 min at 4 °C with (+) or without (−) 1.8 μm K. lactis γ-toxin (lanes +) in reaction mixtures (10 μl) containing 20 mm Tris-HCl (pH 7.5), 2 mm DTT, and 2 m trimethylamine oxide. The treated and mock-treated RNAs were analyzed by denaturing PAGE and visualized by autoradiography. The predicted RtcB and AtRNL repair junctions, showing just the tRNAGlu(UUC) anticodon stem-loop, and their susceptibility or resistance to cleavage by γ-toxin are illustrated at the right.
Here we identified E. coli RtcB as the exemplar of a new RNA ligase family. RtcB is unrelated structurally to classical ATP-dependent polynucleotide ligases; indeed, RtcB is indifferent to the presence of ATP as a reaction component. RtcB activity is stimulated by GTP, via an as yet unclear mechanism. We suspect an allosteric effect rather than the participation of GTP in the RtcB ligation chemistry, which appears to entail direct ligation, analogous to tRNA splicing in extracts of archaeal and animal cells. The sealing reaction of a partially purified human RNA ligase depended on ATP or dATP (not GTP) (45); the role of ATP in the human RNA ligase reaction was not determined.
The genetic organization of RtcA and RtcB in a two-gene operon in E. coli suggests that they provide the healing and sealing functions, respectively, in an RNA repair pathway. In this scenario, the healing refers to the restoration of ligatable 2′,3′-cyclic phosphate ends in the event that the inciting RNA damage directly generates RNA 3′-monophosphates or that the 2′,3′-cyclic phosphate products of RNA transesterification are further processed to a 3′-monophosphate by a bacterial 2′,3′-cyclic phosphodiesterase. The fact that the rtcBA operon in E. coli is regulated by σ54 suggests that the RNA repair functions are induced in response to cellular stress. In this vein, it is worth noting that E. coli has several ribotoxin systems that are turned on in stress situations (18, 46).
Many bacterial taxa encode RtcB and RtcA homologs, although it is not always the case that they are genetically linked on the respective bacterial chromosomes. RtcB and RtcA homologs are found in many archaeal proteomes, but they are not grouped together in operons. Among eukarya, RtcB homologs are present in metazoa and protozoa but missing in most fungi and plants. The phylogenetic distribution of RtcB points to its candidacy as a tRNA splicing enzyme in animals and archaea.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM46330 (to S. S.).

The on-line version of this article (available at http://www.jbc.org) contains detailed methods and supplemental Figs. S1–S3.
S. Sekine, Y. Bessho, and S. Yokoyama, unpublished results.
- Pnkp
- polynucleotide 5′-kinase/3′-phosphatase
- aa
- amino acid
- AtRNL
- Arabidopsis thaliana tRNA ligase.
REFERENCES
- 1. Amitsur M., Levitz R., Kaufmann G. (1987) EMBO J. 6, 2499–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ogawa T., Tomita K., Ueda T., Watanabe K., Uozumi T., Masaki H. (1999) Science 283, 2097–2100 [DOI] [PubMed] [Google Scholar]
- 3. Tomita K., Ogawa T., Uozumi T., Watanabe K., Masaki H. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8278–8283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lu J., Huang B., Esberg A., Johansson M. J., Byström A. S. (2005) RNA 11, 1648–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Klassen R., Paluszynski J. P., Wemhoff S., Pfeiffer A., Fricke J., Meinhardt F. (2008) Mol. Microbiol. 69, 681–697 [DOI] [PubMed] [Google Scholar]
- 6. Xue S., Calvin K., Li H. (2006) Science 312, 906–910 [DOI] [PubMed] [Google Scholar]
- 7. Konarska M., Filipowicz W., Gross H. J. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 1474–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Greer C. L., Peebles C. L., Gegenheimer P., Abelson J. (1983) Cell 32, 537–546 [DOI] [PubMed] [Google Scholar]
- 9. Schwer B., Sawaya R., Ho C. K., Shuman S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2788–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Englert M., Beier H. (2005) Nucleic Acids Res. 33, 388–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nandakumar J., Schwer B., Schaffrath R., Shuman S. (2008) Mol. Cell 31, 278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Filipowicz W., Shatkin A. J. (1983) Cell 32, 547–557 [DOI] [PubMed] [Google Scholar]
- 13. Filipowicz W., Konarska M., Gross H. J., Shatkin A. J. (1983) Nucleic Acids Res. 11, 1405–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Laski F. A., Fire A. Z., RajBhandary U. L., Sharp P. A. (1983) J. Biol. Chem. 258, 11974–11980 [PubMed] [Google Scholar]
- 15. Zofallova L., Guo Y., Gupta R. (2000) RNA 6, 1019–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang Y., Zhang J., Hara H., Kato I., Inouye M. (2005) J. Biol. Chem. 280, 3143–3150 [DOI] [PubMed] [Google Scholar]
- 17. Nariya H., Inouye M. (2008) Cell 132, 55–66 [DOI] [PubMed] [Google Scholar]
- 18. Christensen-Dalsgaard M., Jørgensen M. G., Gerdes K. (2010) Mol. Microbiol. 75, 333–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ho C. K., Shuman S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12709–12714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martins A., Shuman S. (2004) J. Biol. Chem. 279, 50654–50661 [DOI] [PubMed] [Google Scholar]
- 21. Martins A., Shuman S. (2005) RNA 11, 1271–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nandakumar J., Shuman S., Lima C. D. (2006) Cell 127, 71–84 [DOI] [PubMed] [Google Scholar]
- 23. Raymond A., Shuman S. (2007) Nucleic Acids Res. 35, 839–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brooks M. A., Meslet-Cladiére L., Graille M., Kuhn J., Blondeau K., Myllykallio H., van Tilbeurgh H. (2008) Protein Sci. 17, 1336–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Torchia C., Takagi Y., Ho C. K. (2008) Nucleic Acids Res. 36, 6218–6227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jain R., Shuman S. (2009) RNA 15, 923–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chan C. M., Zhou C., Huang R. H. (2009) Science 326, 247. [DOI] [PubMed] [Google Scholar]
- 28. Jain R., Shuman S. (2010) RNA 16, 316–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Keppetipola N., Shuman S. (2006) RNA 12, 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Keppetipola N., Shuman S. (2006) J. Biol. Chem. 281, 19251–19259 [DOI] [PubMed] [Google Scholar]
- 31. Keppetipola N., Shuman S. (2007) Nucleic Acids Res. 35, 7721–7732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Keppetipola N., Nandakumar J., Shuman S. (2007) Nucleic Acids Res. 35, 3624–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Filipowicz W., Strugala K., Konarska M., Shatkin A. J. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 1316–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reinberg D., Arenas J., Hurwitz J. (1985) J. Biol. Chem. 260, 6088–6097 [PubMed] [Google Scholar]
- 35. Billy E., Hess D., Hofsteenge J., Filipowicz W. (1999) J. Biol. Chem. 274, 34955–34960 [DOI] [PubMed] [Google Scholar]
- 36. Tanaka N., Smith P., Shuman S. (2010) Structure 18, 449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Palm G. J., Billy E., Filipowicz W., Wlodawer A. (2000) Structure 8, 13–23 [DOI] [PubMed] [Google Scholar]
- 38. Tanaka N., Shuman S. (2009) RNA 15, 1865–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chakravarty A. K., Shuman S. (2011) J. Biol. Chem. 286, 4117–4122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Genschik P., Drabikowski K., Filipowicz W. (1998) J. Biol. Chem. 273, 25516–25526 [DOI] [PubMed] [Google Scholar]
- 41. Okada C., Maegawa Y., Yao M., Tanaka I. (2006) Proteins 63, 1119–1122 [DOI] [PubMed] [Google Scholar]
- 42. Keppetipola N., Jain R., Meineke B., Diver M., Shuman S. (2009) RNA 15, 1036–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lu J., Esberg A., Huang B., Byström A. S. (2008) Nucleic Acids Res. 36, 1072–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mori T., Ogasawara C., Inada T., Englert M., Beier H., Takezawa M., Endo T., Yoshihisa T. (2010) Mol. Biol. Cell 21, 3722–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Perkins K. K., Furneaux H., Hurwitz J. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 684–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Blanga-Kanfi S., Amitsur M., Azem A., Kaufmann G. (2006) Nucleic Acids Res. 34, 3209–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.