Abstract
Deep sequencing of viral or bacterial nucleic acids monitors the presence and diversity of microbes in select populations and locations. Metagenomic study of mammalian viromes can help trace paths of viral transmissions within or between species. High throughput sequencing of patient and untreated sewage microbiomes showed many sequences with no similarity to genomic sequences of known function or origin. To estimate the distribution of functional RNAs in these microbiomes, we used the hammerhead ribozyme (HHR) motif to search for sequences capable of assuming its three-way junction fold. Although only two of the three possible natural HHR topologies had been known, our analysis revealed highly active ribozymes that terminated in any of the three stems. The most abundant of these are type II HHRs, one of which is the fastest natural cis-acting HHR yet discovered. Altogether, 13 ribozymes were confirmed in vitro, but only one showed sequence similarity to previously described HHRs. Sequences surrounding the ribozymes do not generally show similarity to known genes, except in one case, where a ribozyme is immediately preceded by a bacterial RadC gene. We demonstrate that a structure-based search for a known functional RNA is a powerful tool for analysis of metagenomic datasets, complementing sequence alignments.
Keywords: Catalytic RNA, Nucleic Acid Chemistry, Nucleic Acid Structure, Ribozyme, RNA Processing, RNA Structure, Hammerhead, Microbiome, Self-cleaving Ribozyme
Introduction
RNAs fulfill diverse biological roles, including regulation and catalysis. The discovery of catalytic RNAs over the last 30 years supports the RNA world hypothesis, which proposes that RNA predated proteins as the information carrier and the catalytic macromolecule (1). Biological catalytic RNAs include phosphotransferases and the ribosomal peptidyl transferase (2, 3). Phosphotransferases include six types of naturally occurring nucleolytic ribozymes capable of cleaving phosphodiester linkages: hammerhead (HHR)2 (4), hairpin (5), hepatitis delta virus (HDV) (6, 7), the Neurospora Varkud satellite (8), the bacterial cofactor-dependent GlmS (9), and in some cases, group I intron-like ribozymes (10, 11). Early work identified these self-cleaving RNAs through analysis of single gene transcripts and pathogen genomes (12). In addition, in vitro selection experiments have identified a variety of self-cleaving ribozymes (13), including the hammerhead motif, which was found independently several times (14). In recent years, structure-based searches and in vitro selection from a genomic library have revealed hammerhead and HDV-like ribozymes in many species ranging from bacteria to mammals (15–20).
Hammerhead ribozymes were originally discovered in plant viroids and virusoids, where they function in the processing of rolling circle transcripts (21). HHRs are also known to exist in the satellite transcripts of various newt species (22), cave crickets (23), and the human blood fluke Schistosoma mansoni (24). The recent discoveries of HHRs in many different species, including mammals, suggest multiple biological functions (15, 19, 20).
The HHR structure consists of three helices (stems I, II, and III) anchored in the 11-nt catalytic core. They are capable of both self-scission and ligation, with ligation occurring at a rate about 100 times slower than scission (25). In self-cleaving (cis-acting) HHRs, two of the stems are capped by loops, resulting in a topology resembling a hammerhead. HHRs can be grouped into three types according to the location of the open-ended stem containing the 5′ and 3′ ends of the ribozyme (types I, II, and III). Biochemical and crystallographic studies have revealed a tertiary interaction, which can enhance catalytic rates by up to 500-fold when compared with the minimal ribozyme under physiological conditions (26–28).
Hammerhead ribozymes have simple structural requirements, robust and tunable biochemical activity, and rather low information content, leading to the possibility of several independent appearances (14). The increasing prevalence of natural HHRs may imply multiple biological roles that evade detection through sequence alignment approaches. Considering that functional RNAs are generally conserved in secondary structure and not necessarily in sequence, they can be described as a set of base-paired domains of variable sequence connected by single-stranded regions. The unpaired strands are typically sequence-conserved in active sites, binding pockets, or tertiary contacts and highly variable in connecting regions. A functional RNA, for example a known ribozyme, can thus be described in terms of its secondary structure and conserved single-stranded elements (29, 30). A search for sequences capable of assuming the same secondary structure and possessing all conserved sequences in the prescribed positions can thus reveal new examples of such RNA, even if its sequence is vastly different from previously identified members of the family. One application of this approach is in identification of known ribozymes in new genomes, as has been demonstrated for the HHR and HDV ribozymes (15, 17, 20, 24, 31).
Another powerful application of structure-based searches is in analysis of metagenomic data. These often originate from heterogeneous samples of potentially unknown biological composition. One of the benefits of metagenomic analysis is to monitor the genetic diversity and geographic presence of known or novel microbes. For example, numerous human viral pathogens have been detected in untreated sewage (32).
Metagenomic data consist of a large number of short sequence reads that may lack sufficient coverage for reconstruction of individual genomes. However, the datasets can be used to estimate the species diversity of a sample or to propose the existence of novel species, depending on whether a read can be matched to a known genome. Both absence of genomic context and lack of similarity to known genomic sequences decrease the utility of such sequences, resulting in a dearth of assignable function. To test such sequences for functional significance requires other approaches, one of which is a motif search for known structured, functional RNAs. Here we used structure descriptors of self-cleaving ribozymes to search for these catalytic RNAs in a set of metagenomic sequences from human and non-human samples including: stools from children with non-poliovirus acute flaccid paralysis (33), cerebrospinal fluids from unexplained cases of encephalitis, plasma from unexplained cases of hepatitis, untreated sewage, attenuated viral vaccines (34), and stools from North American bats (35).
Given the strong base pairing and relatively low sequence requirements of self-cleaving ribozymes, we decided to map the HDV and HHR motifs to the metagenomic sequences. Although a search for HDV-like ribozymes did not reveal any sequences capable of folding into the double-pseudoknot fold, we found several sequences that can independently assume the three topologies of HHR. We prepared the minimum hammerhead constructs, flanked by several nucleotides of the surrounding sequences, and measured their in vitro self-cleavage activity. To test whether the incidence is higher or lower than would be predicted by chance, we performed structure-based searches for HHR through randomly generated sequences of the same length (2.2 × 108 nt) and nucleotide content as our metagenomic dataset, which contained about 8.15 × 105 sequences. The results of these searches agree with the predicted low distribution of HHR motifs in random sequences (29, 31), providing a degree of significance to the incidence of HHRs in the metagenomic dataset.
EXPERIMENTAL PROCEDURES
Sample Preparation
Raw samples from various virome studies were collected from the environment or with patient consent in the case of blood and bodily fluid collection. Purification of viral particles, extraction of viral nucleic acid, and synthesis of cDNA for random PCR were performed as described earlier (36).
Structure-based Searches
The RNABOB program (S. Eddy, Janelia Farm, Howard Hughes Medical Institute) was used to search the sequence file for the HDV and hammerhead ribozyme motifs as described previously (30). The descriptor defines the conserved sequence and structural elements of each motif. All hammerhead descriptors included the minimal construct with the 11 conserved nucleotides of the core and a limitation for each stem to be 3–6 bp long. Loop insertions at the end of helices were allowed to be up to 50 nucleotides long. Scheme 1 shows an example of a descriptor for a type II hammerhead ribozyme requiring strict Watson-Crick base pairing in the helical regions. The cleavage site can consist of any nucleotide except G in element s4. In this descriptor, we require 9-nt regions flanking the 5′ and 3′ ends of the ribozyme motif (s1 and s7, respectively).
SCHEME 1.
Nomenclature
The naming of HHRs follows the scheme previously used for HDV-like ribozymes (17). The identity of the ribozyme family is followed by the type of HHR (e.g. hhrII for type II HHRs). The isolate name matches that found in the original metagenomic data and is followed by a number corresponding to the occurrence of the ribozyme found in the particular isolate.
Primer Sequences
Primers were designed according to the sequence identified by the motif search. Forward primers included the T7 RNA polymerase promoter followed by “gggaga” to facilitate transcription, except in the cases in which the 5′ end of the construct was G-rich. The following is the construct designed for one of the type II HHRs shown in Fig. 1: SewS1_01145s-1, ribozyme construct, 5′-gggaga GGA CUU GGU CUU CUA ACG AGU ACG CGA AAC CCU GGU ACG CCC ACC CAG GGU CGC CGG GAA AUC GGC CCG GCC UGA UGA GCG AUA UUC ACU-3′. The other constructs are: AL1096 (inhibitor), 5′-CCG GCG ACC CU-3′; AL1097 (forward), 5′-UAA UAC GAC UCA CUA UAG GGG AGA GGA CUU GGU CUU CUA ACG AGU ACG CGA AAC CCU GGU ACG CCC ACC C-3′; and AL1098 (reverse), 5′-AGU GAA UAU CGC UCA UCA GGC CGG GCC GAU UUC CCG GCG ACC CUG GGU GGG CGU ACC AGG GUU UCG CGU-3′.
FIGURE 1.
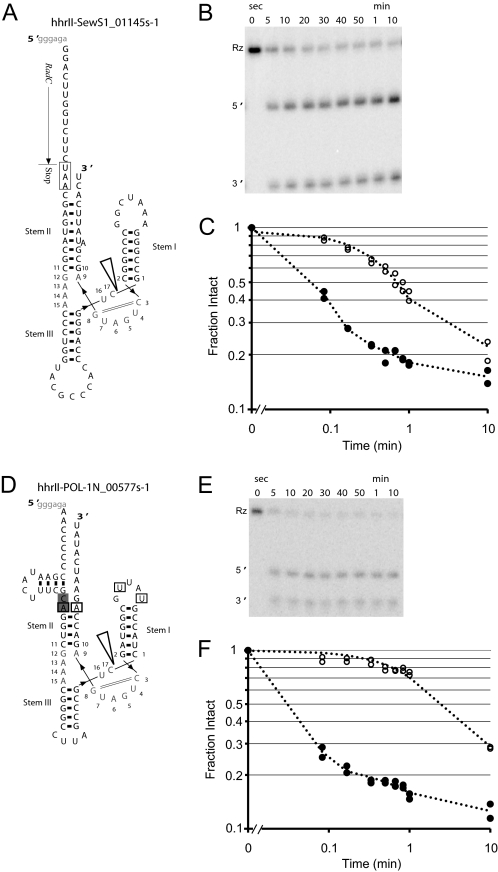
Secondary structure and activity of type II HHRs. A, predicted secondary structure of ribozyme hhII-SewS1_01145s-1. The catalytic core is numbered according to the nomenclature previously proposed (50). The open arrowhead indicates the cleavage site, and the solid arrowheads indicate the direction of the RNA strand. Predicted tertiary contacts between stems I and II are boxed. Conserved CA nts at the end of loop 2 are shaded. Synthetic sequences used to promote transcription are shown in gray lowercase at the beginning of most constructs. B, in vitro self-cleavage activity of the 32P-labeled ribozyme (Rz) incubated in 10 mm MgCl2 at 37 °C. C, log-log graph of ribozyme activities at 10 mm MgCl2 (●) and 1 mm MgCl2 (○), both at 37 °C. D, the predicted secondary structure of ribozyme hhII-POL-1N_00577s-1. E, in vitro self-cleavage activity, measured under the same conditions as B. F, log-log graph of ribozyme activities at 37 °C.
DNA Template Preparation
Constructs were prepared by mutual priming of two synthetic, PAGE-purified oligonucleotides. Primer extensions were carried out in a 100-μl volume containing 0.2 mm of each of the four dNTPs, 5 μm of each primer, and 10 units of Taq DNA polymerase. The primer-extension conditions were 94 °C for 1 min followed by two cycles of 50 °C for 30 s and 72 °C for 2 min as described previously (17).
RNA Transcription
RNA was transcribed at 37 °C for 1 h in a 20-μl volume containing 10 mm DTT; 2.5 mm each GTP, UTP, and CTP; 250 μm ATP; 4.5 μCi of [α-32P]ATP (PerkinElmer Life Sciences); 7.75 mm MgCl2; 20 μm of inhibitor oligonucleotide (specific for each construct); 1 unit of T7 RNA polymerase; and 0.5 pmol of DNA template. Constructs were transcribed in vitro in the presence of limited Mg2+ and cleavage site inhibitor oligonucleotide to prevent co-transcriptional self-scission. Transcripts were purified by denaturing PAGE.
Cleavage Kinetics
In vitro self-scission reactions were performed as described previously (16). Reactions were initiated with the addition of Mg2+ (to a final concentration of 1 or 10 mm) to solutions containing the 32P-labeled ribozyme precursor in 140 mm KCl, 10 mm NaCl, 10 mm Tris buffer, pH 7.4 (final concentrations) at 37 °C and terminated by adding an equal volume of stop buffer containing 20 mm EDTA, 5 mm Tris, pH 7.4, 8 m urea, with xylene cyanol and bromphenol blue loading dyes. The denaturing PAGE gel of self-cleavage products was exposed to phosphorimaging screens and analyzed using a Typhoon PhosphorImager and the ImageQuant software (GE Healthcare).
RESULTS AND DISCUSSION
Motif Searches Identify Type I, II, and III HHRs in Diverse Metagenomic Sequences
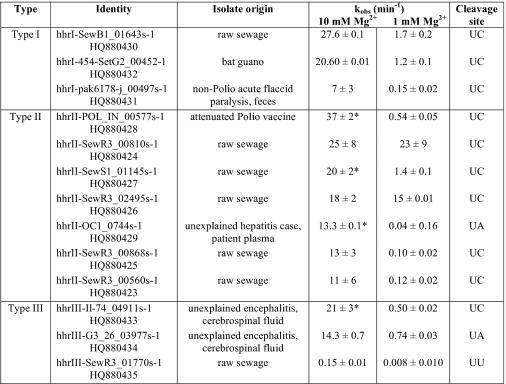
Motif searches revealed a total of 20 candidate ribozymes of all three HHR topologies, of which 13 were confirmed to be active in vitro (Table 1). Our descriptors were similar to those used by Martick et al. (15) to identify the CLEC2 hammerhead ribozyme in rodents, except that we allowed for positions 7 and 17 of the catalytic core to be occupied by any nucleotide. Utilizing the RNABOB algorithm, the permissiveness of a given descriptor can result in a larger output but with a greater incidence of false positives (30). We, therefore, used the RNAfold algorithm of the ViennaRNA package (37) to screen for correctly folding stem-loops. This filtering resulted in 17 candidates showing stable predicted folding. We found two constructs differing by a single point insertion in the flanking single-stranded regions (hhrII-SewR3_00868s-1 and hhrII-SewR3_00560s-1). The motifs reside in the same location within the context of their respective deep sequencing data reads; however, the two reads do not seem to vary in sequence composition enough to rule out differences due to sequencing errors (90% identity). In addition, hhrII-SewR3_02495s-1 and hhrII-SewR3_00810s-1 also reside in the same location; however, there is enough difference between the reads to consider these two HHRs independent finds (73% identity). All 20 original candidates were tested for in vitro catalytic activity.
TABLE 1.
Origin, type, and in vitro activity of microbiome HHRs
All experiments were performed at 140 mm KCl, 10 mm NaCl, 10 mm Tris-HCl, pH 7.4, and incubated at 37 °C at the indicated Mg2+ concentration. The reported kobs values are the average rate constants ± the average deviation of at least two in vitro measurements. GenBank accession numbers for each ribozyme are listed under Identity.
* Fit to biexponential decay and uncleaved residuals model. All other data were fit to a single exponential and uncleaved residuals model.
Self-scission of Microbial Ribozymes
The catalytic activity of 13 novel HHRs identified through structure-based searches of deep-sequencing data from metagenomic sampling was verified through in vitro self-cleavage experiments. The observed cleavage rate constants are given in Table 1. Fig. 1 shows a self-cleavage experiment for two type II HHRs in 10 mm Mg2+ under single-turnover conditions. The ribozymes cleave to about 80%, with the remaining ∼20% representing the lower limit on cleavage yield. Ligation reactions of hammerhead ribozymes have been shown to be efficient in vitro, plateauing at ∼23% in S. mansoni HHR (25). However, self-scission reaction plateaus cannot be directly interpreted as equilibrium with ligation reactions as the kinetics is complicated by the presence of inactive species. Future kinetics experiments will determine ligation rates for these type II HHRs.
All of the discovered ribozymes display robust catalytic activity in vitro. The cleavage kinetics data were fit to a single exponential decay and uncleaved residuals model, giving a rate constant for self-scission, kobs,cleave, reported in Table 1. To examine the ribozyme activity at physiological-like conditions, self-cleavage experiments were also carried out at 1 mm Mg2+.
Comparisons with Known HHRs
Tertiary interactions between peripheral domains have previously been shown to greatly affect the in vitro activity of HHRs. For example, the presence of tertiary interactions in the type I S. mansoni HHR is known to enhance the rate of scission by 50–500-fold under physiological-like conditions (38, 39). To allow for loop-loop contacts, all of our constructs were designed to include at least 9 nucleotides upstream and downstream of the ribozyme motif. It has previously been suggested that ribozymes with different metal dependences experience differences in the rate-limiting step, with tertiary stabilizations placing the molecule farther along the reaction pathway (40). Many of our ribozymes exhibit fast self-scission, suggesting that their catalytic cores are stabilized by tertiary interactions. In addition, although the kinetics experiments were performed at only two Mg2+ concentrations, the 13 HHRs displayed a range of Mg2+ dependences, with a 10-fold difference in Mg2+ concentration resulting in an ∼300× enhanced cleavage in the case of hhrII-OC1_0744s-1 (Table 1). This large cooperativity also suggests that at 10 mm Mg2+, the ribozyme activity is far from maximal. Interestingly, both hhrII-OC1_0744s-1 and hhrII-POL_IN_00577s-1 show high Mg2+ dependence and fast kinetics, whereas neither is predicted by RNAfold (37) to fold into the correct HHR structure within the context of their respective ∼500-nt genomic sequence read. All the remaining ribozymes are predicted to form a stable catalytic core even when embedded within their full sequence reads.
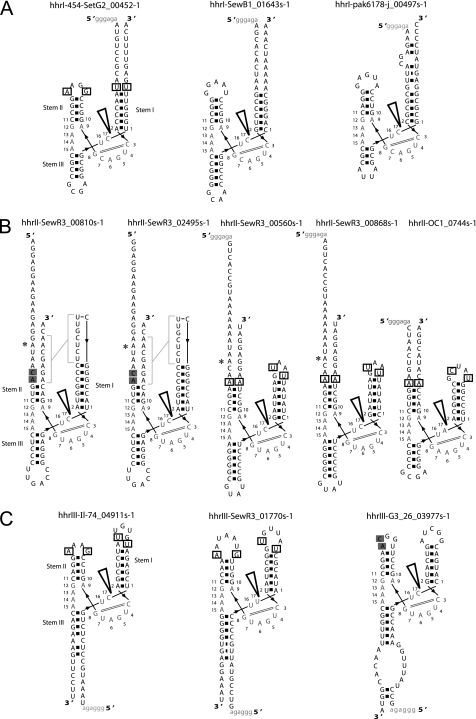
Many naturally occurring HHRs contain conserved motifs in loops 1 and 2, which have been identified as essential features for the loop-loop interactions (41). Base pairing between the 5′-U of loop 1 and the 3′-A of loop 2 as well as the interaction of an extrahelical (penultimate) pyrimidine of loop 1 with a 5′-purine of loop 2 can be predicted for seven of our HHRs (Figs. 1 and 2, boxed). Two others display the latter interaction only. In addition, four of the HHRs predicted to have tertiary interactions contained the last two CA dinucleotides of loop 2, which are conserved in chrysanthemum chlorotic mottle viroid (CChMVd) but also occur in other natural HHRs (41). These two interactions are predicted to be conserved in over 20 naturally occurring HHRs including those of insects, Arabidopsis, CChMVd, and tobacco ringspot virus satellite RNA (41).
FIGURE 2.
Secondary structure of types I (A), II (B), and III (C) HHRs identified in metagenomic sequencing data with in vitro cleavage rate constants reported in Table 1. All labels and symbols are the same as in Fig. 1. Asterisks indicate differences between ribozymes hhrII-SewR3_00810s-1 and hhrII-SewR3_02495s-1 and between ribozymes hhrII-SewR3_00560s-1 and hhrII-SewR3_00868s-1. Gray brackets indicate the putative pseudoknot interactions between loops 1 and 2 in hhrII-SewR3_00810s-1 and hhrII-SewR3_02495s-1.
Several of our ribozymes did not contain conserved loop sequences involved in known tertiary interactions. To predict novel tertiary contacts, we used the DotKnot program (42) to detect potential pseudoknots and kissing loops, such as those known to exist for the S. mansoni HHR (38). DotKnot predicted a pseudoknot between loop 1 and the flanking sequence of stem II in hhrII-SewR3_00810s-1 and hhrII-SewR3_02495s-1 (Fig. 2). This type of tertiary interaction has not previously been proposed in an HHR and points to a potential diversity of tertiary contacts that can stabilize peripheral regions in these ribozymes. These two related ribozymes exhibit the highest rate constants at physiological Mg2+ (1 mm) and vanishing Mg2+ dependence between 1 and 10 mm (Table 1). This result supports the existence of a strong, Mg2+-independent tertiary interaction, which may be achieved by base pairing in a pseudoknot. Overall, there is no apparent trend in the catalytic rates between our HHRs that do and do not display predictable tertiary contacts, further supporting the hypothesis that novel tertiary interactions stabilize the fast-cleaving ribozymes.
Among the 13 ribozymes, 10 cleave a UC sequence, which is identical to the S. mansoni HHR. This sequence is known to have higher activity than other combinations of nucleotides in engineered hammerhead motifs (43). Many other known viral and viroid HHRs, as well as the recently discovered bacterial and eukaryotic HHRs, possess UC or UA at their cleavage sites (18, 19, 44). The sequence conservation of this site suggests that high cleavage activity serves a biologically important function.
Considering that our descriptors allowed large stem-loops to occur, the active HHRs are relatively small, with stem lengths averaging between 3 and 7 bps and loops between 4 and 8 nts (Figs. 1 and 2). Some of the common features include long stem I (6–8 nts) and short stem II (3–4 nts), with the exception of hhrII-SewS1_01145s-1, which has a predicted 8-bp stem II. These lengths follow closely with stem and loop sizes seen in over 20 other naturally occurring HHRs (41, 44).
The remarkable catalytic rates for our fastest HHRs are greater than the rates of other natural HHRs tested in the same format (cis) and similar reaction conditions. One of the most extensively studied natural cis-HHRs is the S. mansoni ribozyme with a kobs = 0.22–0.36 min−1 (30 °C, 10 mm Mg2+, 40 mm Tris, pH 8) (24). To compare cleavage kinetics under identical conditions, we designed a construct (hhrI-Sman) after the S. mansoni ribozyme. Our hhrI-Sman construct displayed kobs = 6.7 ± 0.2 min−1 (37 °C, 10 mm Mg2+, 10 mm Tris, pH 7.4). However, it should be noted that a chimera of Arabis mosaic virus small satellite RNA (satArMV+) in cis-format displayed kobs > 750 min−1 (25 °C, 10 mm Mg2+, 50 mm MES buffer, pH 7.3) (44). Although the diversity of reaction conditions and HHR constructs throughout the literature prevent direct comparison, in the natural cis-format, 12 of our HHRs are among the fastest reported to date.
HHR Incidence in a Randomly Generated Sequence File
To estimate whether the incidence of the HHRs in the metagenomic data is higher or lower than would be expected by chance, we generated a random sequence of identical length and nucleotide content as our metagenomic sequence file. Given the low probability of finding a hammerhead motif (1 per 1013 nt) (24), we would not expect to find stable HHR in this random sequence, and indeed this is the case. Structure-based searches produced four times fewer hits, all of which failed to be confirmed by subsequent secondary structure prediction. By comparison, the HHR motif is over-represented in the 2.2 × 108-nt metagenomic sequences, and the occurrences are unevenly distributed among the sequenced samples. This result implies that HHRs are highly selected for in certain microbiomes and suggests that the samples contain novel HHR-harboring microbes.
The catalytic core of the hammerhead ribozyme can accommodate additional nucleotides between stems I and II. HHRs with this extended core sequence have previously been identified through bioinformatics searches (45). To identify such ribozymes, additional structure-based searches allowing extended core regions were performed with our metagenomic sequences. Three output sequences representing extended core motifs were predicted to have stable secondary structure and were tested for in vitro self-scission; however, none displayed catalytic activity in these assays.
Hammerhead ribozymes have been identified in a multitude of genomes (15, 18–20). Our work shows that HHRs are highly enriched in a diverse set of subject and environmental samples, including human stool and sewage. Identifying structured RNAs is one way to characterize new genomic information and categorize genetic variants or novel enteric viruses. Many of our metagenomic sequences did not align closely with any known genomes. Although the origin of the sample isolates is known, further investigation is needed to identify the viral or bacterial isolate origin of these ribozymes. Two ribozymes were found to bear sequence similarity to previously analyzed HHRs. Of the 13 HHRs, BLASTn (46) revealed only one HHR similar to a genomic ribozyme; hhrIII-Il-74_04911s-1 has a high sequence similarity to the hammerhead ribozymes studied in many viroid-like RNAs and virus satellite RNAs, particularly chicory yellow mottle virus satellite RNA (sChYMV) (E score 3 × 10−4). It has previously been suggested that viroids and satellite RNAs originated from the transcripts of repetitive sequences when the transcripts parasitized viral replication machinery and used viruses as vectors to jump from one organism to another (31). The occurrence of HHRs in repetitive DNA of many species suggests that the catalytic activity is important to the existence of these transcripts. hhrIII-Il-74_04911s-1 may represent another example of such activity, although its flanking sequences do not map to known satellite sequences.
Interestingly, hhrI-454-SetG2_00452-1, isolated from bat guano, bears 89% identity to an artificial RNA construct used to make a three-dimensional model of HHR based on FRET measurements (GenBank accession 1RMN_A) (47, 48). It exhibits fast kinetics (Table 1) and contains both predicted tertiary interactions between stems I and II.
In the case of hhrII-SewS1_011455-1, BLAST searches did not identify another ribozyme with similar sequence, but they did reveal that the upstream sequence is highly similar to the RadC gene (RadC is a DNA repair protein) of the thermophilic photosynthetic bacterium Roseiflexus castenholzii DSM (E value 7 × 10−13). The 5′ end of the HHR construct contains a UAA stop codon, which corresponds to the end of the RadC gene (Fig. 1). The R. castenholzii RadC gene does not have a hammerhead motif at its 3′ end, and a search through its entire genome (GenBank CP000804) did not reveal any HHRs, suggesting that our isolate represents a novel genomic arrangement of RadC and HHR. The close proximity of the ribozyme to the 3′ terminus of the RadC coding region suggests either that the ribozyme is involved in mRNA processing or that it forms a 5′ terminus of a novel genetic element close enough to the RadC gene to form a single transcriptional unit with the mRNA.
It will be interesting to learn whether the viral or bacterial sources of these 13 ribozymes display pathogenicity or are merely commensal microbes in the human microbiome. The presence of the ribozymes identified on the RNA strands of yet uncharacterized viral genomes will require further studies. The sequence reads containing these ribozymes will be extended by overlapping with other unclassifiable reads, potentially forming contigs that can be analyzed for signatures of viral genomes. Deeper sequencing of those microbiomes harboring our ribozymes is needed to assemble new microbial genomes. Given that metagenomic sampling can be used to monitor genetic variation over time and location, functional HHRs may be evolutionarily important to these viral or bacterial genomes. In general, although the biological functions of all HHRs have not been identified, the association with satellites and RT ORFs suggests a role in retrotransposition (18), which has recently been suggested for HDV ribozymes as well (17, 49).
In this study, structure-based searches revealed the fastest known natural HHRs, the first natural type II HHRs, and a putative novel pseudoknot stabilizing the most robust type II ribozymes. Because the sources of these ribozymes remain to be identified, their detection will motivate a new search for HHR-containing microbes. In the case of metagenomic sequences, identifying the presence of catalytic motifs can be used to evaluate the distribution of functional RNAs in a sample and possibly assist in the identification of novel viruses. The expansion of libraries of coding and non-coding RNAs will ultimately help characterize new genomes. The discovery of new ribozymes will further help us understand their distribution in nature and their biological importance.
Supplementary Material
Acknowledgments
We thank the Lupták group, B. Brejová, T. Vinař, and the Lathrop group for fruitful discussions. We also thank Drs. C. Wang and J. Victoria.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM094929 (to A. L.) and R01 HL083254 (to E. D.). This work was also supported by the University of California, Irvine, startup funds and the Pew Charitable Funds (to A. L.).
This article was selected as a Paper of the Week.
- HHR
- hammerhead ribozyme
- HDV
- hepatitis delta virus
- nt
- nucleotide
- contig
- group of overlapping clones
- Rz
- ribozyme.
REFERENCES
- 1. Gesteland R. F., Cech T., Atkins J. F. (2006) The RNA world: The Nature of Modern RNA Suggests a Prebiotic RNA World, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 2. Scott W. G., Martick M., Chi Y. I. (2009) Biochim. Biophys. Acta 1789, 634–641 [DOI] [PubMed] [Google Scholar]
- 3. Fedor M. J., Williamson J. R. (2005) Nat. Rev. Mol. Cell Biol. 6, 399–412 [DOI] [PubMed] [Google Scholar]
- 4. Forster A. C., Symons R. H. (1987) Cell 50, 9–16 [DOI] [PubMed] [Google Scholar]
- 5. Hampel A., Tritz R. (1989) Biochemistry 28, 4929–4933 [DOI] [PubMed] [Google Scholar]
- 6. Sharmeen L., Kuo M. Y., Dinter-Gottlieb G., Taylor J. (1988) J. Virol. 62, 2674–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu H. N., Lin Y. J., Lin F. P., Makino S., Chang M. F., Lai M. M. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 1831–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Saville B. J., Collins R. A. (1990) Cell 61, 685–696 [DOI] [PubMed] [Google Scholar]
- 9. Winkler W. C., Nahvi A., Roth A., Collins J. A., Breaker R. R. (2004) Nature 428, 281–286 [DOI] [PubMed] [Google Scholar]
- 10. Nielsen H., Johansen S. D. (2009) RNA Biol. 6, 375–383 [DOI] [PubMed] [Google Scholar]
- 11. Nielsen H., Westhof E., Johansen S. (2005) Science 309, 1584–1587 [DOI] [PubMed] [Google Scholar]
- 12. Cech T. R. (2002) Biochem. Soc. Trans. 30, 1162–1166 [DOI] [PubMed] [Google Scholar]
- 13. Tang J., Breaker R. R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5784–5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Salehi-Ashtiani K., Szostak J. W. (2001) Nature 414, 82–84 [DOI] [PubMed] [Google Scholar]
- 15. Martick M., Horan L. H., Noller H. F., Scott W. G. (2008) Nature 454, 899–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Salehi-Ashtiani K., Lupták A., Litovchick A., Szostak J. W. (2006) Science 313, 1788–1792 [DOI] [PubMed] [Google Scholar]
- 17. Webb C. H., Riccitelli N. J., Ruminski D. J., Lupták A. (2009) Science 326, 953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de la Peña M., García-Robles I. (2010) RNA 16, 1943–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de la Peña M., García-Robles I. (2010) EMBO Rep. 11, 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seehafer C., Kalweit A., Steger G., Gräf S., Hammann C. (2011) RNA 17, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Prody G. A., Bakos J. T., Buzayan J. M., Schneider I. R., Bruening G. (1986) Science 231, 1577–1580 [DOI] [PubMed] [Google Scholar]
- 22. Epstein L. M., Gall J. G. (1987) Cold Spring Harb. Symp. Quant. Biol. 52, 261–265 [DOI] [PubMed] [Google Scholar]
- 23. Rojas A. A., Vazquez-Tello A., Ferbeyre G., Venanzetti F., Bachmann L., Paquin B., Sbordoni V., Cedergren R. (2000) Nucleic Acids Res. 28, 4037–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ferbeyre G., Smith J. M., Cedergren R. (1998) Mol. Cell Biol. 18, 3880–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Canny M. D., Jucker F. M., Pardi A. (2007) Biochemistry 46, 3826–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Khvorova A., Lescoute A., Westhof E., Jayasena S. D. (2003) Nat. Struct. Biol. 10, 708–712 [DOI] [PubMed] [Google Scholar]
- 27. De la Peña M., Gago S., Flores R. (2003) EMBO J. 22, 5561–5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martick M., Scott W. G. (2006) Cell 126, 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bourdeau V., Ferbeyre G., Pageau M., Paquin B., Cedergren R. (1999) Nucleic Acids Res. 27, 4457–4467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Riccitelli N. J., Lupták A. (2010) Methods 52, 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ferbeyre G., Bourdeau V., Pageau M., Miramontes P., Cedergren R. (2000) Genome. Res. 10, 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blinkova O., Rosario K., Li L., Kapoor A., Slikas B., Bernardin F., Breitbart M., Delwart E. (2009) J. Clin. Microbiol. 47, 3507–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Victoria J. G., Kapoor A., Li L., Blinkova O., Slikas B., Wang C., Naeem A., Zaidi S., Delwart E. (2009) J. Virol. 83, 4642–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Victoria J. G., Wang C., Jones M. S., Jaing C., McLoughlin K., Gardner S., Delwart E. L. (2010) J. Virol. 84, 6033–6040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li L., Victoria J. G., Wang C., Jones M., Fellers G. M., Kunz T. H., Delwart E. (2010) J. Virol. 84, 6955–6965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Symonds E. M., Griffin D. W., Breitbart M. (2009) Appl. Environ. Microbiol. 75, 1402–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hofacker I. L. (2003) Nucleic Acids Res. 31, 3429–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Canny M. D., Jucker F. M., Kellogg E., Khvorova A., Jayasena S. D., Pardi A. (2004) J. Am. Chem. Soc. 126, 10848–10849 [DOI] [PubMed] [Google Scholar]
- 39. Nelson J. A., Uhlenbeck O. C. (2006) Molecular Cell 23, 447–450 [DOI] [PubMed] [Google Scholar]
- 40. Roychowdhury-Saha M., Burke D. H. (2006) RNA 12, 1846–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dufour D., de la Peña M., Gago S., Flores R., Gallego J. (2009) Nucleic Acids Res. 37, 368–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sperschneider J., Datta A. (2010) Nucleic Acids Research 38, e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kore A. R., Vaish N. K., Kutzke U., Eckstein F. (1998) Nucleic Acids Res. 26, 4116–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shepotinovskaya I. V., Uhlenbeck O. C. (2008) Biochemistry 47, 7034–7042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. De la Peña M., Flores R. (2001) J. Biol. Chem. 276, 34586–34593 [DOI] [PubMed] [Google Scholar]
- 46. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Menger M., Eckstein F., Porschke D. (2000) Nucleic Acids Res. 28, 4428–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tuschl T., Gohlke C., Jovin T. M., Westhof E., Eckstein F. (1994) Science 266, 785–789 [DOI] [PubMed] [Google Scholar]
- 49. Eickbush D. G., Eickbush T. H. (2010) Mol. Cell Biol. 30, 3142–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hertel K. J., Pardi A., Uhlenbeck O. C., Koizumi M., Ohtsuka E., Uesugi S., Cedergren R., Eckstein F., Gerlach W. L., Hodgson R., et al. (1992) Nucleic Acids Res. 20, 3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.