Abstract
Using an ensemble approach, we demonstrate that an oligomeric RecA species is required for the extension phase of RecA filament formation. The RecA K72R mutant protein can bind but not hydrolyze ATP or dATP. When mixed with other RecA variants, RecA K72R causes a drop in the rate of ATP hydrolysis and has been used to study disassembly of hydrolysis-proficient RecA protein filaments. RecA K72R filaments do not form in the presence of ATP but do so when dATP is provided. We demonstrate that in the presence of ATP, RecA K72R is defective for extension of RecA filaments on DNA. This defect is partially rescued when the mutant protein is mixed with sufficient levels of wild type RecA protein. Functional extension complexes form most readily when wild type RecA is in excess of RecA K72R. Thus, RecA K72R inhibits hydrolysis-proficient RecA proteins by interacting with them in solution and preventing the extension phase of filament assembly.
Keywords: ATPases, DNA, DNA Recombination, DNA Repair, Protein-DNA Interaction, RecA
Introduction
The bacterial RecA protein catalyzes recombinational DNA repair. Homologs of RecA are present in all forms of life including the RadA protein of Archaea and the Rad51 and Dmc1 proteins of eukaryotes (1–6). The conservation of RecA proteins highlights their importance to genome integrity. In Escherichia coli, the 352-amino acid 37,842-Da RecA protein also plays a role inducing the expression of DNA repair proteins in response to DNA damage through a regulatory network known as the SOS response.
To carry out recombinational DNA repair or to stimulate the SOS response, RecA protein must first bind to DNA. RecA can bind both single-stranded and double-stranded DNA (ssDNA and dsDNA, respectively), but it binds preferentially to ssDNA (7). When binding either type of DNA, RecA forms extended, oligomeric filaments along the length of the DNA deoxyribose-phosphate backbone. The RecA/DNA nucleoprotein filament is a right-handed helix with the DNA on the inside of the filament groove and the RecA along the outside. There are six RecA monomers per turn of the filament and three nucleotides (or base pairs) per monomer, although each monomer makes contacts with the nucleotide triplet of the adjacent monomer (8).
RecA nucleoprotein filaments can assume a variety of conformational states depending on the presence and identity of the nucleotide cofactor and the DNA substrate (for review, see Ref. 7). RecA can bind to ssDNA in the absence of a nucleotide cofactor or in the presence of ADP and form a compact, “collapsed” filament with a pitch of 64 Å that is not active for RecA-related activities (9, 10). This filament state is also referred to as the O state (11). In the presence of ATP, dATP, ATPγS,3 or ADP-AlF4−, RecA filaments are more extended, with a pitch of 95 Å (10). These filaments are said to be in the “active” or A state (11). RecA filaments bound to dsDNA or that actively catalyze exchange of homologous DNA strands exist in the P or “pairing enhanced” state (12).
The formation of RecA filaments on DNA is a dynamic process. The first and rate-limiting step of filament formation is nucleation (13–15). RecA protein nucleates onto DNA as oligomers of 4–6 monomers (16, 17). Nucleation is inhibited by the single-stranded DNA-binding (SSB) protein (18–20). Inhibition of nucleation by SSB can be overcome with the help of the RecF, RecO, and RecR proteins (for review, see Ref. 21). The rate of nucleation onto SSB-bound DNA can also be enhanced using dATP as a nucleotide cofactor (22) by deleting the C-terminal 17 amino acid residues of RecA (23) or by mutating residue Glu-38 to Lys (RecA E38K or RecA730) (24, 25).
After nucleation, the RecA filament rapidly extends in the 5′ to 3′ direction with respect to the bound DNA (26, 27). Extension of the filament can occur at rates greater than 1000 monomers/min at 37 °C (28). Information on the RecA species involved in the extension phase of filament formation is limited. Two single-molecule studies have addressed the issue but come to different conclusions with either monomers (20) or a larger oligomer (17) being the relevant species. Secondary structure in long ssDNA substrates can inhibit RecA filament extension (18). In RecA assays in vitro, SSB is often added after nucleation takes place to melt out secondary structure in the DNA. SSB does not pose a barrier to RecA filament extension in vitro (18, 20).
RecA filament disassembly occurs from the 5′-proximal end of the filament and also proceeds in the 5′ to 3′ direction with respect to the bound DNA (27, 29). The rate of RecA filament disassembly is slower than the rate of filament extension, occurring at 60–70 monomers/min/filament end (30). Although some RecA protein can add to the 5′-proximal end of the filament and some disassembly can occur from the interior of the filament or at the 3′-proximal end of the filament, these events are limited in rate and in scope (20). Thus, filament assembly and disassembly are generally end-dependent processes occurring at opposite filament ends, especially when ATP is being hydrolyzed.
Once RecA protein is bound to DNA, it can hydrolyze ATP. All subunits throughout the filament hydrolyze ATP with no enhancement of hydrolysis at the filament ends (31). RecA can hydrolyze several other (d)NTPs (for review, see Ref. 28), although the most thoroughly studied alternative cofactor used in RecA assays is dATP. When bound to ssDNA, RecA hydrolyzes ATP with a monomer kcat of 30 min−1 (7, 31–33). The kcat for RecA bound to dsDNA is 20 min−1 (for review, see Ref. 7). Binding and hydrolysis of the nucleotide cofactor occur at the monomer-monomer interfaces of the RecA filament. RecA contains canonical Walker A and Walker B motifs (residues 66–73 and 140–144, respectively) for ATP binding (Ref. 34, and for review, see Ref. 7). These motifs are present on the 3′-proximal face of a RecA monomer within a filament (8). Residues from the 5′-proximal face of the adjacent monomer, including Lys-248 and Lys-250, also make contact with the nucleotide cofactor (8, 35). Amino acids on both sides of the interface participate in hydrolysis of the nucleotide cofactor including Glu-96, Lys-72, and Lys-248 (35–38). Several RecA-mediated activities depend on ATP hydrolysis (for review, see Ref. 11). RecA filament disassembly requires ATP hydrolysis. Exchange of homologous DNA strands that are more than 1000 bp long, exchange past sites of DNA damage or heterology, and exchange between two homologous duplex DNA molecules also require ATP hydrolysis.
The importance of ATP hydrolysis to RecA function has been addressed by mutation of the conserved Lys-72 of the Walker A motif to arginine (K72R) (38). Mutation of the equivalent lysine residue in the Walker A motifs of other nucleotide-binding proteins had been made previously and resulted in nonfunctional mutant proteins (39–43). In RecA, the conservative K72R mutant protein binds to DNA and nucleotide cofactors but catalyzes negligible amounts of ATP hydrolysis (38). Additional experiments with the RecA K72R mutant protein reinforced the finding that ATP binding, but not hydrolysis, was necessary for pairing homologous strands of DNA (38, 44, 45).
After its discovery, RecA K72R became a useful tool in studying the filament dynamics of RecA proteins that are hydrolysis-proficient. It was used to demonstrate that very little disassembly of RecA filaments occurs from the interior of the filament (46), that RecA filaments on dsDNA are more dynamic than those bound to ssDNA (46), that RecA filaments disassemble over the course of DNA strand exchange (47), and that RecA filament disassembly is necessary to complete exchange of long homologous DNA substrates (48). If differing ratios of the wild type and the mutant RecA proteins are premixed before incubating them with circular single-stranded DNA (cssDNA), the decrease in ATP hydrolysis rate is greater than that expected if the hydrolysis-deficient RecA K72R were simply occluding binding sites on the DNA (46). This result provides evidence for interaction between the two RecA proteins and formation of mixed filaments on DNA. The addition of RecA K72R to a wild type RecA filament on cssDNA in the presence of ATP leads to a decrease in the rate of ATP hydrolysis (46). A decrease in wild type RecA-mediated ATP hydrolysis suggests that the hydrolysis-deficient RecA K72R can interact with a wild type filament even if the mutant protein cannot form its own filaments on cssDNA. Lysine 72 is present at the RecA monomer-monomer interface. Mutation of lysine 72 to arginine may distort the interface or may alter the position of the nucleotide cofactor such that additional RecA protein cannot join the growing end of the filament. This filament extension defect in RecA K72R would effectively cap the growing end of the filament and eventually result in net disassembly of the wild type RecA filament. The E. coli RecX protein is also thought to inhibit RecA by capping the growing end of a filament (49).
Use of the RecA K72R mutant in vivo initially suggested that ATP hydrolysis is necessary for RecA to trigger the SOS response (50). A later study by the same group showed that a lack of filament formation by the RecA K72R protein in vivo was what had inhibited induction of the SOS response (51). Introducing a Glu-38 to Lys mutation in addition to K72R restores RecA filament formation in the presence of ATP in vitro. ATP is still bound but not hydrolyzed by the double mutant protein, which also supports SOS induction in vivo (51).
The RecA E38K/K72R provides a better platform for studying some aspects of the function of RecA-mediated ATP hydrolysis than does RecA K72R. However, this double mutant protein does not allow investigation of RecA filament dynamics, as we will show. A continued need for RecA K72R as a tool to investigate the filament dynamics of other RecA proteins led us to better characterize the filament formation defect of this mutant protein in the presence of ATP. A lack of RecA filament formation could be attributed to defects in either the nucleation or the extension phases of this process or both. We find that RecA K72R is defective in the extension phase of filament formation. In particular, our results help to resolve the literature controversy about the RecA species involved in filament extension. Using ensemble rather than single molecule methods, we provide evidence that RecA protein adds to the 3′-proximal end of a filament as oligomers during the extension phase of RecA filament formation. This further implicates an oligomeric RecA species in both phases of RecA filament formation.
EXPERIMENTAL PROCEDURES
DNA Substrates
Circular single-stranded DNA was derived from M13mp18 bacteriophage and was purified by CsCl banding as previously described (12). Concentrations of M13mp18 are reported in terms in of μm nucleotides (μm nt) using a conversion factor of 108 μm nt A260 nm−1. A 50-nt, 3′ fluorescein-labeled oligomer with the sequence 5′-GGC CTC GCG GTA GCT GAG CTC GGA GCG CAC GAT TCG CAC TGC TGA TGT TC was ordered from Integrated DNA Technologies. The concentration of fluorescent oligonucleotide is reported in terms of μm molecules.
Proteins
The wild type E. coli RecA, RecA K72R, RecA E38K/K72R, and SSB proteins were purified as previously described (45, 48, 51–53), and their concentrations were determined using native extinction coefficients: ϵ280 = 2.23 × 104 m−1 cm−1 for the RecA proteins (54), and ϵ280 = 2.38 × 104 m−1 cm−1 for SSB protein.
Fluorescence Polarization (FP) Experiments
Fluorescence polarization was used to measure direct binding of RecA K72R or wild type RecA to ssDNA using either ATP, dATP, or ATPγS as a nucleotide cofactor. The fluorescent entity in these experiments was the 3′ fluorescein-labeled 50-nt oligomer described above. FP experiments had a reaction solution of 25 mm Tris acetate (pH 7.8, 80% cation), 10 mm magnesium acetate, 3 mm potassium glutamate, 5% w/v glycerol, 1 mm dithiothreitol (DTT), and 3 mm ATP, dATP, or ATPγS. RecA was serially diluted in the storage buffer. Fluorescent oligonucleotide at a final concentration of 10 nm molecule was added to the diluted RecA and incubated in the dark for 20 min at 37 °C. Each dilution was done in triplicate. The polarization of each sample was measured in triplicate on a Beacon 2000 fluorescence polarization system (Invitrogen) at 37 °C. The data were plotted and fit to a modified form of the Hill equation, y = (Bmin + (Bmax − Bmin))(xn/(Kdn + xn)) (55), and apparent Kd values were determined using the Prism software.
Electron Microscopy (EM) Experiments
A modified Alcian method was used to visualize RecA filaments on cssDNA. Activated carbon grids were prepared as described previously (56). All EM assays contained a reaction solution of 25 mm Tris acetate (pH 7.5, 88% cation), 10 mm magnesium acetate, 3 mm potassium glutamate, 5% w/v glycerol, 12 mm phosphocreatine, and 10 units/ml creatine phosphokinase to regenerate ATP, and 4 μm M13mp18 cssDNA. All reactions contained 3 mm ATP and 0.4 μm SSB, although the timing of their addition varies among experiments.
In the EM experiment with a substoichiometric amount of wild type RecA to form nucleation points for RecA K72R, the reaction solution was incubated with ATP and SSB for 10 min at 37 °C before the addition of wild type RecA to a final concentration of 0.133 μm. The reactions were incubated for 20 additional min, and then a sample was removed for spreading before RecA K72R, or additional wild type RecA was added to achieve a final concentration of 2 μm, with the total RecA concentration being 2.133 μm. Subsequent samples were removed at the indicated times for spreading on activated carbon grids and eventual visualization.
EM experiments in which wild type and RecA K72R proteins were premixed before the addition to M13mp18 cssDNA contained varying ratios of wild type RecA and RecA K72R proteins as listed in the legends to Figs. 3, 5, and 7 and accompanying text. The total RecA concentration was maintained at 2 μm. The RecA proteins were incubated with the reaction solution at 37 °C for 10 min before the addition of ATP and SSB. The complete reaction was incubated at 37 °C for 20 min before spreading on activated carbon grids.
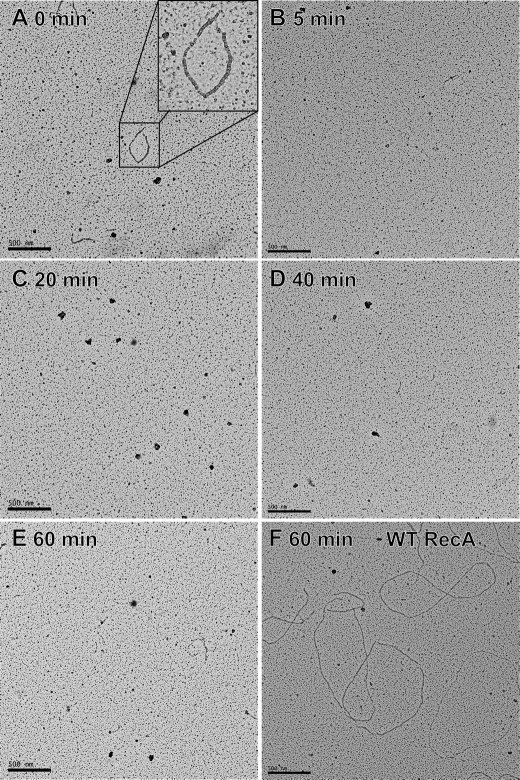
FIGURE 3.
RecA K72R fails to form extended filaments on cssDNA using wild type RecA as a nucleation factor. Images of RecA filaments bound to cssDNA were obtained at 15,000 × magnification by EM as described under “Experimental Procedures.” Wild type RecA formed filaments on SSB-coated cssDNA, and reaction samples were imaged before and 5, 20, 40, and 60 min after RecA K72R addition. The ratio of wild type RecA to RecA K72R was 1:15. Reactions contained 4 μm M13mp18 cssDNA, 3 mm ATP, 0.4 μm SSB, 0.133 μm wild type RecA, and 2 μm RecA K72R (or an additional 2 μm wild type RecA in the 60-min time point only). The small wild type RecA filaments in the 0 time point disassemble within 5 min of RecA K72R addition and do not reassemble in the presence of RecA K72R. For comparison, panel F shows a 60-min time point for a reaction in which the wild type RecA protein, instead of the mutant protein, was added to the wild type RecA nuclei.
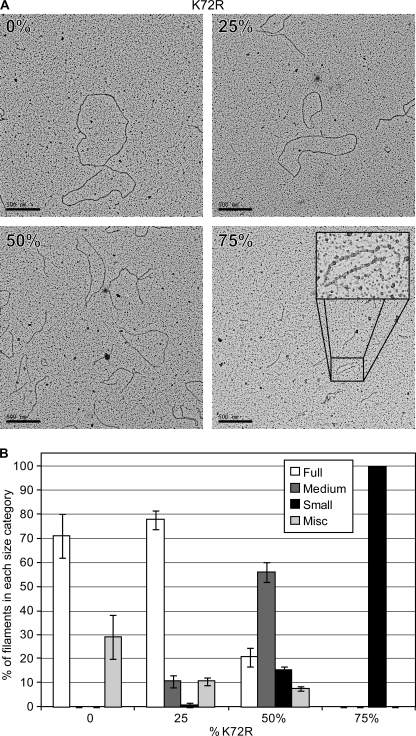
FIGURE 5.
Wild type RecA and RecA K72R from shortened filaments on cssDNA when premixed in solution. Wild type RecA and RecA K72R were mixed in the same ratios as in Fig. 4 before the addition to cssDNA, and images of the resulting RecA filaments were obtained by EM as described under “Experimental Procedures.” The concentrations of DNA, RecA, ATP, and SSB are the same as in the legend to Fig. 4. A, images of RecA filaments at 0, 25, 50, and 75% RecA K72R were obtained at 15,000× magnification. B, more than 200 filaments at 0, 25, 50, and 75% RecA K72R were visually inspected and grouped into Full, Medium, Small, or Miscellaneous (Misc) length categories as described under “Experimental Procedures.” Bars represent the average percentage of RecA filaments in each length category as counted from four randomly selected squares on carbon grids containing spread reaction samples with 0, 25, 50, or 75% RecA K72R. Error bars represent 1 S.D. from the mean.
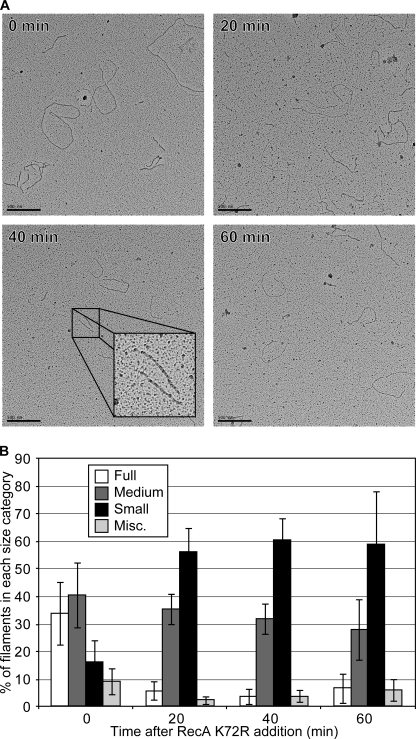
FIGURE 7.
RecA filaments decrease in length after wild type RecA bound to cssDNA is challenged with RecA K72R. Wild type RecA filaments formed on cssDNA were challenged with varying concentrations of RecA K72R as in Fig. 6, and the resultant filaments were imaged by EM as described under “Experimental Procedures.” The concentrations of DNA, RecA, ATP, and SSB are the same as in the legend to Fig. 6B. A, images of RecA filaments before, 20, 40, and 60 min after the addition of RecA K72R were obtained at 15,000× magnification. B, more than 400 filaments at 0, 20, 40, and 60 min after RecA K72R addition were visually inspected and grouped into the same length categories as Fig. 5B and as described under “Experimental Procedures.” Bars represent the average percentage of RecA filaments in each length category as counted from four randomly selected squares on carbon grids containing spread reaction samples from each time point. Error bars represent 1 S.D. from the mean.
The EM experiment in which RecA K72R is added as a challenge to established wild type RecA filaments contained wild type RecA protein at 1.3 μm that was incubated with the reaction solution for 10 min at 37 °C before the addition of ATP and SSB. A sample was removed for spreading after an additional 20 min of incubation at 37 °C, and then RecA K72R was added to achieve a final concentration of 2.6 μm. Subsequent samples were removed at the indicated times for spreading on activated carbon grids.
Samples from the reaction mixtures described above were diluted 3.25-fold with 200 mm ammonium acetate, 10 mm HEPES, and 10% (w/v) glycerol, pH adjusted to 7.5, and adsorbed to an activated carbon grid for 3 min. The grid was then touched to a drop of the above buffer followed by floating on a drop of the same buffer for 1 min. The sample was then stained by touching to a drop of 5% uranyl acetate followed by floating on a fresh drop of the same solution for 30 s. Finally, the grid was washed by touching to a drop of double-distilled water followed by immersion in two, 10-ml beakers of double distilled water. After the sample was dried, it was rotary-shadowed with platinum. This protocol is designed for visualization of complete reaction mixtures, and no attempt was made to remove proteins not bound to DNA, which can lead to a high background of unbound proteins. Prepared samples were observed, and images were obtained using a TECNAI G2 12 Twin Electron Microscope (FEI Co.) equipped with a GATAN 890 CCD camera. Pictures were taken at 15,000 × magnification.
To determine how RecA filament length changed when RecA K72R was present, at least 200 filaments per time point or RecA K72R/wild type RecA ratio from at least four squares on the carbon grids were counted at an identical magnification. The observed filaments were grouped into one of three broad categories based on the extent of RecA filament coverage of the cssDNA. “Full” filaments completely encompassed the circular DNA molecule or had small discontinuities in the regular striated pattern of the filament but were indistinguishable in length from filaments without discontinuities. “Medium” filaments were distinctly smaller in length than full filaments and may have had gaps large enough to observe SSB bound to the DNA. “Small” filaments encompassed the least amount of cssDNA, appearing as short fragments of RecA filament on a cssDNA molecule that was otherwise coated with SSB. Filaments that did not fall into one of the above categories, like filaments that formed on linear ssDNA molecules, aggregated in solution or twisted in on themselves such that filament length was unknown were classified as “miscellaneous.”
ATP Hydrolysis (ATPase) Assays
A coupled enzyme, spectrophotomeric assay (57, 58) was used to measure RecA-mediated ATP hydrolysis. The ADP generated by hydrolysis was converted back to ATP by a regeneration system of pyruvate kinase and phosphoenolpyruvate. The resultant pyruvate was converted to lactate by lactate dehydrogenase using NADH as a reducing agent. The conversion of NADH to NAD+ was monitored as a decrease in absorbance at 380 nm. The amount of ATP hydrolyzed over time was calculated using the NADH extinction coefficient ϵ380 = 1.21 mm−1cm−1. The assays were carried out in a Varian Cary 300 dual beam spectrometer, with a temperature controller and a 12-position cell changer. The path length was 0.5 or 1 cm, and the band pass was 2 nm. All ATPase assays contained a reaction solution of 25 mm Tris acetate (pH 7.5, 88% cation), 10 mm magnesium acetate, 3 mm potassium glutamate, 5% w/v glycerol, 1 mm DTT, 3 mm phosphoenolpyruvate, 10 units/ml pyruvate kinase, 10 units/ml lactate dehydrogenase, 4.5 mm NADH, and 4 μm M13mp18 cssDNA.
For the ATPase assays in which wild type and RecA K72R were premixed, varying ratios of wild type and RecA K72R proteins were mixed together and then added to the reaction solution. The total RecA concentration was maintained at 2 μm. The ratios can be found in the legend to Fig. 4 and accompanying text. The RecA proteins were incubated with the reaction solution at 37 °C for 10 min before the addition of 0.4 μm SSB protein and 3 mm ATP to start RecA-mediated hydrolysis.
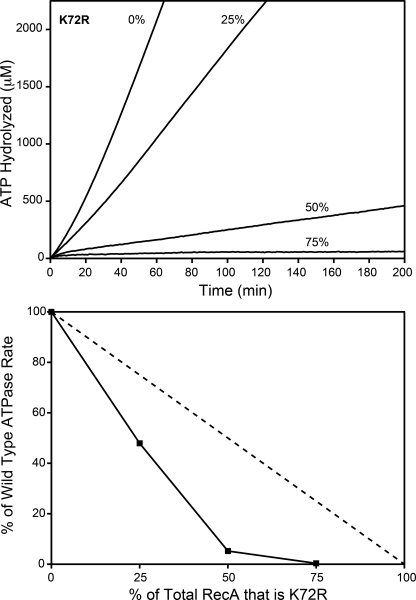
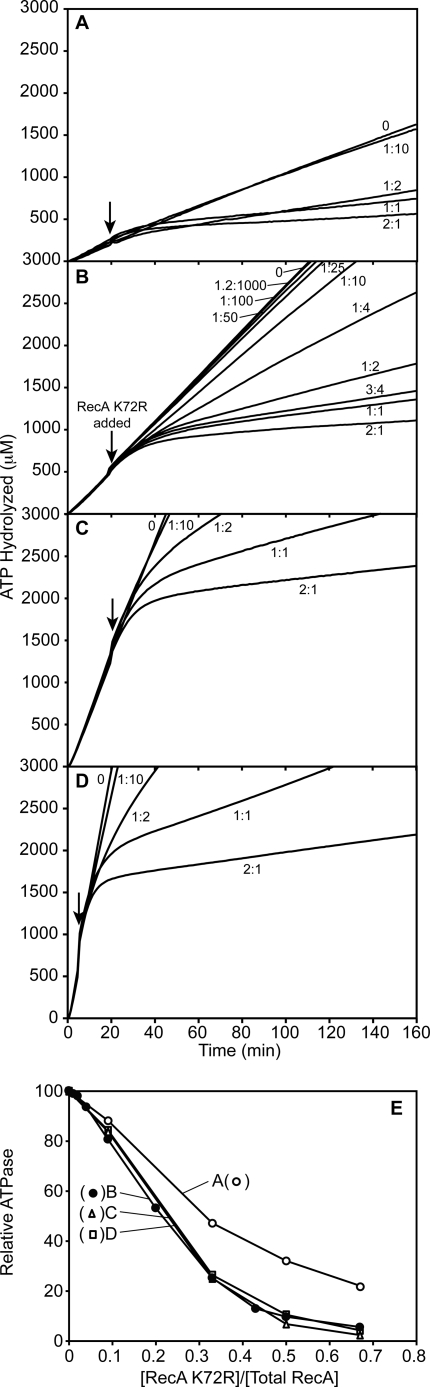
FIGURE 4.
The rate of wild type RecA-catalyzed ATP hydrolysis decreases disproportionately with increasing RecA K72R concentrations. Wild type RecA and RecA K72R were mixed in varying ratios before the addition to cssDNA. The resulting rate of wild type RecA-catalyzed ATP hydrolysis in the presence of RecA K72R was measured as described under “Experimental Procedures.” Reactions contained 4 μm M13mp18 cssDNA, 2 μm total RecA, 3 mm ATP, and 0.4 μm SSB. RecA K72R made up 0, 25, 50, or 75% of the total RecA in each reaction. Upper panel, the concentration of hydrolyzed ATP in μm is plotted versus time in minutes at 0, 25, 50, or 75% RecA K72R. Lower panel, the rate of ATP hydrolysis is plotted as a percentage of the hydrolysis rate at 0% RecA K72R versus the RecA K72R percentage of total RecA. The dashed line represents the percentage of wild type RecA-mediated ATP hydrolysis expected if the rate decreased proportionally to the RecA K72R concentration.
ATPase assays comparing the extent of wild type RecA inhibition by RecA K72R or by RecA E38K/K72R when either mutant RecA protein was added as a challenge to wild type RecA filaments contained wild type RecA protein at 2 μm, which was incubated with the reaction solution for 10 min at 37 °C before the addition of 0.4 μm SSB protein and 3 mm ATP as described above. ATP hydrolysis by wild type RecA was monitored for 50 min before RecA E38K/K72R or RecA K72R was added to achieve a final concentration of 4 μm. Compensating RecA K72R storage buffer was also added to these reactions to keep the volume change of each reaction the same. The absorbance of the reaction was monitored at 380 nm until the ATP regeneration system was exhausted.
For the ATPase assays in which only RecA K72R was added as a challenge to wild type RecA filaments (in Fig. 6), wild type RecA protein at the concentration indicated in the legend was incubated with the reaction solution for 10 min at 37 °C before the addition of SSB protein and 3 mm ATP as described above. ATP hydrolysis by wild type RecA was monitored for 20 min (or 5 min as indicated) before RecA K72R was added to achieve a final concentration indicated in the legend to Fig. 6 as a proportion of the wild type RecA concentration. Compensating RecA K72R storage buffer was also added to these reactions to keep the volume change and solution conditions of each reaction the same. The absorbance of the reaction was monitored at 380 nm until the ATP regeneration system was exhausted.
FIGURE 6.
A significant proportion of RecA K72R is needed to fully inhibit ATP hydrolysis by established wild type RecA filaments on cssDNA. Wild type RecA filaments formed on cssDNA were challenged with varying concentrations of RecA K72R, and the resultant decrease in the rate of wild type RecA-catalyzed ATP hydrolysis was monitored over time. ATPase assays were carried out as described under “Experimental Procedures.” The reactions all contained 3 mm ATP. Other conditions for panels A, B, C, and D, respectively, were: M13mp18 cssDNA, 2, 4, 8, and 16 μm; wild type RecA, 0.67, 1.33, 2.66, and 5.32 μm; SSB, 0.2, 0.4, 0.8, and 1.6 μm. In each experiment the RecA K72R was varied in concentration from 0 μm to twice the concentration of wild type RecA. RecA K72R concentrations are presented as a ratio of RecA K72R to wild type RecA. Wild type RecA filaments were allowed to hydrolyze ATP for 20 min before RecA K72R addition in panels A, B, and C and 5 min in panel D. In panel E, the wild type RecA ATPase in the unchallenged reaction is taken as 100%, and the proportional decline in the ATPase rate in the new steady state is plotted as a function of the mutant to wild type protein ratio after the addition of RecA K72R. The new steady-state rates were defined beginning 30 min after the mutant protein challenge in panels A and B and beginning at 20 min for most reactions (1:2, 1:1, and 2:1) in panels C and D. In the latter two panels, the new rate after a 1:10 challenge was determined during the 10 min immediately before the exhaustion of the ATP regeneration system.
RESULTS
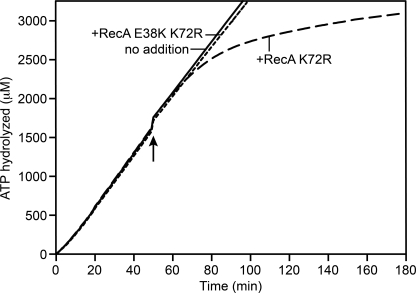
RecA K72R does not form filaments on cssDNA as observed by EM (51). A combination of the K72R mutation with a mutation producing E38K generates a double mutant that does bind to ssDNA and form extended filaments in the presence of ATP (51). The double mutant, thus, has advantages for studies of the function of RecA-mediated ATP hydrolysis. However, this same double mutant is unable to reveal wild type RecA protein filament dynamics when added as a challenge to an ATPase reaction with wild type RecA bound to ssDNA. When added to wild type RecA filaments bound to circular ssDNA and hydrolyzing ATP, the K72R mutant leads to a decline in ATPase activity, whereas the double mutant has no effect (Fig. 1). The clear inhibition imposed by the single K72R mutation here and in previous studies (46) triggered the present effort to better characterize this mutant protein so as to understand its effects on RecA reactions and refine our interpretations of earlier work. We demonstrate that in the presence of ATP, RecA K72R has a defect in the extension phase of RecA filament formation and that this process involves an oligomeric species of RecA.
FIGURE 1.
A RecA K72R challenge of wild type RecA protein filaments reveals RecA filament dynamics, whereas a challenge with RecA E38K/K72R does not. Wild type RecA filaments formed on cssDNA were challenged with RecA K72R or RecA E38K/K72R, and the rate of wild type RecA-catalyzed ATP hydrolysis was monitored over time. ATPase assays were carried out as described under “Experimental Procedures.” Each reaction contained 4 μm M13mp18 cssDNA, 2 μm wild type RecA, 3 mm ATP, 0.4 μm SSB, and 4 μm RecA K72R or 4 μm E38K/K72R or an equivalent volume of RecA storage buffer. The challenge was made after 50 min of wild type RecA and cssDNA incubation.
RecA K72R Has a Defect in the Extension Phase of RecA Filament Assembly
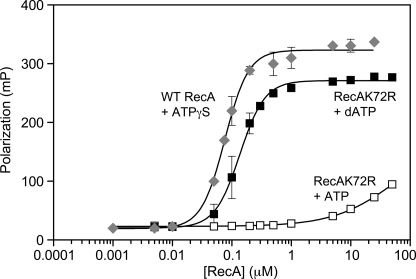
Direct association of RecA K72R with ssDNA was measured by FP. Varying concentrations of RecA K72R or wild type RecA were incubated with 10 nm concentrations of a fluorescein-labeled 50-nt oligonucleotide. RecA K72R binding to DNA was measured in the presence of ATP or dATP. The non-hydrolysable ATPγS nucleotide was used with the wild type RecA-DNA complexes to provide an appropriate comparison for the hydrolysis-deficient RecA K72R protein. ATP-hydrolysis-mediated dissociation is, thus, not an issue for either protein. The binding curves of Fig. 2 demonstrate that RecA K72R is defective in DNA binding compared with wild type RecA. The steep rise in polarization as the concentration of RecA K72R was increased shows that RecA K72R associates with ssDNA in the presence of dATP and is consistent with the facile formation of RecA K72R filaments under this condition observed by Gruenig et al. (51). However, the 0.14 μm measured Kd for RecA K72R-ssDNA binding in the presence of dATP is still somewhat higher than the 0.08 μm measured Kd of wild type RecA bound to ssDNA. The best-fit binding curves produce Hill coefficients of 2.19 and 2.07 for the wild type and K72R mutant protein (with dATP), respectively. The Hill coefficients can be interpreted to mean that there is cooperativity in DNA binding involving at least two RecA binding units (monomers or oligomers). We note that on this 50-mer oligonucleotide, there is room for only 16 RecA subunits or 2–3 RecA oligomers if they each involve 4–6 RecA subunits.
FIGURE 2.
RecA K72R has a defect in ssDNA binding as measured by FP. Direct binding to a fluorescein-labeled 50-nt oligonucleotide by wild type RecA protein in the presence of ATPγS (diamonds) or RecA K72R in the presence of ATP (open squares) or dATP (filled squares) was measured via FP as described under “Experimental Procedures.” The reactions contained 10 nm oligonucleotide, 3 mm ATP, dATP, or ATPγS, and 0.005–50 μm RecA K72R or 0.001–25 μm wild type RecA. Polarization (mP) is plotted versus log scale RecA concentration (μm). Data points represent the mean of three independent trials, and error bars represent 1 S.D. from the mean. The data are fit to a modified form of the Hill Equation as described under “Experimental Procedures.”
RecA K72R binds DNA with much lower affinity in the presence of ATP. Because saturation of polarization was not achieved even at the highest concentrations of RecA K72R, we can only estimate that the apparent Kd is greater than 50 μm. Although binding is clearly compromised, it is not clear to what extent nucleation of RecA K72R is inhibited relative to wild type RecA because the wild type RecA-DNA binding curve presumably is a result of both nucleation and the subsequent short extension of a filament on the 50-nt oligonucleotide. Extension could thus play a significant role in the curves of Fig. 2.
To isolate the extension phase, we tested whether filament extension by RecA K72R could be observed by EM if a substoichiometric amount of wild type RecA were used to provide nucleation points for RecA K72R. Enough wild type RecA protein to bind 10% of the available DNA binding sites was incubated with 4 μm SSB-coated cssDNA. After 20 min of incubation, 2 μm concentrations of either wild type RecA or RecA K72R was added to the reaction. When the mutant was added, this resulted in a 1:15 ratio of wild type RecA to RecA K72R. Samples were removed for EM imaging before and 5, 20, 40, and 60 min after RecA K72R addition (Fig. 3). The small wild type RecA filaments visible in the 0-min image were not extended upon the addition of RecA K72R. In fact, the small wild type RecA filaments disassembled within 5 min of RecA K72R addition and failed to reassemble even 60 min after RecA K72R addition. In contrast, the addition of more wild type protein generated numerous extended filaments that were evident by EM imaging in samples removed at 5 (not shown) or 60 min (Fig. 3F) after the addition. The lack of filament extension by RecA K72R demonstrates that this mutant protein is deficient for the extension phase of RecA filament assembly. Because RecA is a DNA-dependent ATPase, the disassembly of the wild type RecA filaments after RecA K72R addition is consistent with the ability of RecA K72R to inhibit wild type RecA-mediated ATP hydrolysis.
Models for Wild Type RecA Inhibition by RecA K72R
The inability of RecA K72R to form filaments on cssDNA in the presence of ATP appears to reflect, in large measure at least, a defect in the extension phase of RecA filament formation. However, RecA K72R inhibits ATP hydrolysis by wild type RecA (46, 47). It had been assumed that the hydrolysis-deficient RecA K72R replaced the wild type RecA, as it hydrolyzed ATP and disassembled from the DNA, thereby blocking rebinding of the wild type protein. The lack of filament formation by RecA K72R in Fig. 3 clearly demonstrates that this is not the case.
RecA K72R does not inhibit wild type RecA by replacing it on the DNA, yet the wild type RecA protein is unable to form or to maintain filaments when RecA K72R is present. The mutant protein must somehow affect the normal filament formation properties of the wild type protein, and this must involve protein-protein interactions. The wild type RecA protein and the RecA K72R mutant can form mixed filaments in the presence of dATP (46). Interactions between the two RecA variants in the presence of ATP could allow RecA K72R to inhibit wild type RecA by conferring its filament assembly defect on wild type RecA. To explore this alternative possibility, we needed to understand the molecular form of the RecA species involved in the filament assembly processes that are deficient in RecA K72R.
The nucleation phase of RecA filament formation uses oligomers of 4–6 RecA subunits (16, 17, 20). If RecA K72R and wild type RecA form mixed oligomers in solution, then a nucleation defect in RecA K72R may limit the ability of the oligomers to nucleate onto ssDNA.
The molecular form of the RecA species involved in filament extension is more contentious, with published biochemical data supporting both the addition of RecA monomers (20) or oligomers (17) to growing filament ends. Depending on which species is relevant, two issues arise with respect to RecA K72R inhibition of wild type RecA. If monomers are the unit of filament extension, then it is necessary to know whether RecA K72R can add to a filament and, if so, whether more monomers of RecA can be added after it. If oligomers are the extending species, then it is necessary to know if RecA K72R oligomers or mixed wild type and mutant oligomers are active (if they form at all). The considerations listed above along with the results in Figs. 2 and 3 led to the following models for RecA K72R inhibition of wild type RecA.
Nucleation Mechanism: RecA K72R Inhibits Nucleation of Mixed Wild Type and RecA K72R Oligomers
RecA K72R inhibits wild type RecA-mediated ATP hydrolysis if the two RecA variants are mixed in solution before binding to ssDNA (46). Because the nucleating species is an oligomer, pre-mixing the two RecA proteins might allow a RecA K72R nucleation defect to contribute to its inhibition of wild type RecA. After a challenge by the mutant protein, a nucleation defect could in principle block the replacement of RecA that dissociates from preformed wild type filaments. The results of Fig. 3 argue that a nucleation defect cannot fully explain the inhibition afforded by a RecA K72R challenge. If nucleation alone were affected and extension could continue, the short wild type filaments used in that experiment should grow when the mutant protein is added. We, therefore, focused on the defect in extension.
Extension Mechanisms
The results of Fig. 3 imply that the addition of RecA K72R somehow blocks further extension of the filament. There are at least three possible mechanisms by which this could occur.
Extension Mechanism 1: RecA Filament Extension Involves the Addition of Monomers to Growing Filaments, and the Addition of RecA K72R Prevents Further Extension
This model is analogous to the RecA filament-capping mechanism proposed for the E. coli RecX protein (49) in which a protein bound at the 3′ assembling end of a RecA filament prevents extension but allows dissociation from the 5′-proximal end of the filament to proceed. We note that the Lys-72 residue of RecA protein is in the ATP hydrolysis active site and faces the 3′-proximal end of a filament. If RecA K72R added to the filament as a monomer, the interface between a wild type filament and the first RecA K72R monomer to be added would in principle be entirely normal. If overall folding of RecA K72R is unaffected by the mutation (as suggested by the extended filaments made by the mutant protein in the presence of dATP), the arginine substitution for Lys-72 would only affect the interface between that first monomer and the second monomer to be added.
Thus, if RecA extension proceeded by the addition of monomers, one RecA K72R monomer could be added more or less normally to the 3′ assembling end. In that circumstance, the filament capping model makes two testable predictions. First, if the binding of that first RecA K72R monomer is sufficiently tight, then very little RecA K72R with respect to the wild type RecA might be necessary to eventually bring about full inhibition of hydrolysis. Second, full inhibition of hydrolysis would occur through complete disassembly of wild type RecA filaments, which could be observed by EM. The filament capping mechanism was tested by challenging established wild type RecA filaments with RecA K72R. In this scheme, the nucleation aspect of RecA K72R inhibition of wild type RecA can be neglected.
Extension Mechanism 2: Weak Filament Capping Reflecting the Addition of RecA K72R Monomers That Bind Only Weakly to the 3′ Assembling End
If filament extension involves the addition of monomers and if the K72R substitution has long range effects on conformation that increase the Kd for interaction of that first monomer with a wild type filament, then higher amounts of RecA K72R may be needed to effect inhibition. The inhibition will reflect Kd, and the degree of inhibition at any particular RecA K72R concentration will depend upon the concentration of available filament ends.
Extension Mechanism 3: Extension Involves the Addition of Oligomers to Growing Filaments, and Mixtures of Wild Type and RecA K72R Protein Could Render the Extension Oligomer Inactive, Prevent Oligomer Formation Altogether, or Form an Altered Oligomeric Cap on the Filament to Which Additional Oligomers Cannot Be Added
In this case in the EM assay of Fig. 3, the formation of competent extension complexes in the presence of so much RecA K72R could not occur. If oligomers form normally but are inactive when they include some threshold of the K72R mutant protein, the degree of inhibition will reflect the ratio of free wild type and K72R mutant protein. The results below are focused on distinguishing between the use of monomeric or oligomeric species in the RecA filament extension process.
Wild Type RecA and RecA K72R Form Shortened Filaments on cssDNA When Premixed in Solution
When wild type RecA and RecA K72R were mixed in solution before cssDNA binding, Shan and Cox (46) observed a decrease in the rate of ATP hydrolysis that was disproportionate to the amount of RecA K72R in the reaction. They concluded that the mechanisms of RecA K72R inhibition of wild type RecA involved more than just the binding of the mutant protein so as to block DNA binding sites. We confirmed this finding in ATPase assays with conditions that were conducive to EM imaging. In Fig. 4, wild type RecA and RecA K72R were mixed together in varying ratios before being added to cssDNA. Ten minutes later, ATP and SSB were added to begin ATP hydrolysis and to melt out secondary structure in the DNA, respectively. As expected, the rate of ATP hydrolysis declined as the concentration of RecA K72R in solution increased (Fig. 4, upper panel), and the decline was disproportionate to the amount of RecA K72R in the reaction solution (Fig. 4, lower panel).
To investigate the state of the RecA filaments formed when RecA K72R and wild type RecA were premixed, we obtained EM images of samples from each ratio of the two RecA variants after 20 min of incubation with the DNA, the ATP, and the SSB protein (Fig. 5). As the proportion of RecA K72R increased, the length of the filaments visibly decreased (Fig. 5A). We counted more than 200 randomly selected RecA filaments from each ratio of the RecA variants and grouped them according to length judgments as described under “Experimental Procedures” (Fig. 5B). There is little difference in RecA filament length distribution between the 0% RecA K72R and the 25% RecA K72R conditions with most of the filaments falling into the full category. When wild type RecA and RecA K72R are present in equal amounts (50% RecA K72R), the filament length distribution changes considerably, with the majority of the filaments falling into the medium category. When RecA K72R is in excess of wild type RecA (75% RecA K72R), only short RecA filaments are observed by EM.
The decrease in RecA filament length is consistent with the filament extension defect of RecA K72R preventing full filament formation on the DNA. The results generally eliminate extension inhibition Mechanism 1. If filament extension involved the addition of tight binding monomers, the chance addition of the mutant protein would limit filament extension severely even when the mutant was only 25% that of the RecA protein present. Instead, the rather abrupt collapse in filament formation when the mutant protein represents 50% or more of the RecA protein present suggests that either monomers are added and the interaction is weak (relatively high Kd) or that oligomers are formed, and they are inactive for extension when the content of RecA K72R exceeds a certain threshold, perhaps 50% of the oligomer subunits.
Oligomers Are the RecA Filament Extension Species
Distinguishing between Mechanisms 2 and 3 requires a determination of whether the degree of inhibition reflects a particular Kd or a particular stoichiometry of mutant to wild type protein. Using ATP hydrolysis as an indirect measure of RecA binding, wild type RecA filaments were challenged with RecA K72R set up as described previously (46) but with a more thorough titration of RecA K72R concentrations and under conditions that were amenable to EM assays being done in parallel. Wild type RecA was incubated with cssDNA in the presence of ATP and SSB, and ATP hydrolysis was monitored for at least either 5 or 20 min before varying concentrations of RecA K72R were added to the reaction solution (Fig. 6). To determine whether the effects of RecA K72R reflected the wild type RecA/RecA K72R stoichiometry or instead reflected the binding constant of RecA K72R for filament ends, the challenge reactions were carried out at a series of DNA and RecA concentrations, keeping all protein to DNA ratios constant (Fig. 6, panels A–D). DNA, wild type RecA, and SSB concentrations were varied over an 8-fold range. In Fig. 6, panel B uses the RecA concentrations comparable with Figs. 4 and 5.
The degree of inhibition realized in the mutant protein challenge depended upon the ratio of mutant protein to wild type protein. Regardless of the starting concentration of wild type RecA protein, there was very little drop in the rate of ATP hydrolysis when the ratio of RecA K72R to wild type RecA was less than 1:10. When the ratio was at least 1:10, the rate of ATP hydrolysis decreased over time and then settled into a slower steady-state rate of hydrolysis. The greatest declines in ATPase rate were seen when the K72R mutant concentration was equal to or in excess of the concentration of the wild type RecA protein. These ratios are similar to the levels of RecA K72R that produced the calamitous decline in RecA filament lengths as seen in the electron microscope in Fig. 5. The extent to which RecA K72R inhibited wild type RecA-mediated ATP hydrolysis reflected the ratio of the two proteins, as seen in Fig. 6E. The effects of the mutant RecA protein as a proportion of the total RecA protein were identical in panels B–D (in which the starting wild type protein and DNA concentrations are varied over a 4-fold range). In panel A, where the overall RecA protein concentrations are below 1 μm (where RecA-promoted DNA strand exchange is suboptimal and RecA filament formation is less reliable (61)), the overall effects are similar, although the extent of inhibition is reduced.
EM images of RecA filaments on cssDNA were taken before and 20, 40, and 60 min after RecA K72R was added under the conditions of Fig. 6B to achieve a 2:1 ratio of mutant to wild type RecA protein (Fig. 7). A tight filament capping mechanism for RecA K72R inhibition of wild type RecA (mechanism 1) predicts full disassembly of RecA filaments, but this was not observed. RecA filaments on ssDNA persisted even 60 min after the challenge with RecA K72R (Fig. 7A). However, the filaments that did remain had decreased in length. To get an idea of how the distribution of RecA filament lengths changed over time after RecA K72R addition, we counted RecA filaments from each time point as described under “Experimental Procedures” (Fig. 7B). Before RecA K72R addition, most of the filaments fell into the full-to-medium-length categories. After RecA K72R addition, the distribution shifted to mostly medium and small filaments, with small filaments being the most numerous. Because RecA K72R was added to established wild type RecA filaments on cssDNA, a nucleation defect of RecA K72R could not contribute to the inhibition of wild type RecA. Thus, the filament extension defect of RecA K72R must lead to the decrease in filament length. The decrease in filament length resulted in less wild type RecA being associated with the DNA, which explains the drop in the rate of ATP hydrolysis. The overall results are most easily explained if the species involved in extension is an oligomer. Because the extent of wild type RecA inhibition in Fig. 6 (especially panels B–D) is dependent on the ratio of RecA K72R to wild type RecA protein, we conclude that there must be a certain proportion of wild type RecA present in the extension oligomer for it to bind DNA. This is consistent with the shortened filament lengths we see in Fig. 7 and also with the progressively shorter filaments we see in Fig. 5 as the RecA K72R concentration increases.
DISCUSSION
The main finding of this study is that the extension unit for RecA filament formation is an oligomer of RecA protein. In the presence of ATP, the extension phase of RecA K72R nucleoprotein filament formation is essentially absent. This deficiency can be partially overcome in vitro by including wild type RecA protein in the reaction. Formation of mixed oligomers blocks filament extension when RecA K72R is present in amounts that are similar to or in excess of the wild type RecA protein.
Using FP, we observed that RecA K72R has a greatly diminished capacity to bind a short ssDNA oligonucleotide in the presence of ATP (Fig. 2). At least some ability of RecA K72R to nucleate onto ssDNA is retained, although a nucleation defect in RecA K72R could have masked a filament extension defect. We found that RecA K72R failed to form extended filaments even when wild type RecA was used to create nucleation points for those filaments (Fig. 2); thus, the extension phase of RecA filament assembly is severely constrained in the K72R mutant protein.
The conclusion that oligomers are used as the species for RecA filament extension is based on our detailed observations of filaments formed in the presence of different ratios of mutant and wild type protein. In experiments where mutant and wild type protein were premixed (Figs. 4 and 5), the distribution of filament lengths was almost indistinguishable from 100% wild type RecA, when 25% of the total RecA protein was mutant. A substantial collapse in extended filament formation was seen as the mutant content increased to 50 and 75% that of the total. As determined by direct EM observation, the decline in ATP hydrolysis reflected the absence of RecA filaments on the DNA.
In Fig. 6 we document that the inhibitory effects of RecA K72R in a challenge of preformed filaments of wild type RecA protein reflect the mutant-to-wild type stoichiometry, as opposed to the binding affinity of the mutant protein for either RecA filaments or RecA subunits in solution. As seen in Fig. 6E, the proportional effects of a challenge by the mutant protein are quite consistent when the starting wild type RecA concentration is above a 1 μm threshold and varied over a 4-fold range. This result is inconsistent with models relying on extension by the addition of monomeric subunits, where the effects would reflect the Kd for monomer binding to the filament end.
Even though RecA K72R does not inhibit wild type RecA by blocking DNA binding sites as expected, RecA K72R can still be used to monitor filament disassembly by hydrolysis-proficient RecA proteins. RecA K72R inhibits filament extension through interaction with other RecA proteins in solution, so RecA K72R-mediated inhibition of ATP hydrolysis catalyzed by an established filament depends on disassembly of the hydrolysis-proficient RecA. When wild type RecA filaments are challenged by sufficient concentrations of RecA K72R, any wild type RecA protein that dissociates would be sequestered in a pool of inactive oligomers that includes RecA K72R.
The lack of filament formation by RecA K72R in the presence of ATP described previously (51) and examined in more detail in this study seems to be at odds with the original paper that characterized RecA K72R and its binding to DNA (38). In the original paper describing RecA K72R, the authors measured appreciable binding of RecA K72R to ssDNA in the presence of ATP. The difference between their findings and more current studies of RecA K72R may lie in their choice of DNA substrate. To measure association of RecA K72R and DNA, Rehrauer and Kowalczykowski (38) used a modified fluorescent DNA substrate called etheno-DNA (62). It is known that affinity of RecA protein for ssDNA increases with the extent of etheno-modification (38, 62). Rehrauer and Kowalczykowski (38) did find that the state of RecA K72R filaments on etheno-DNA in the presence of ATP more closely resembled the compact, inactive, O state of RecA filaments usually found in the absence of nucleotide cofactor or in the presence of ADP. They also observed that dATP and ATPγS nucleotide cofactors strengthened the interaction of RecA K72R with DNA, which is consistent with visible filament formation in the EM using these cofactors (51).
The altered state of RecA K72R filaments may explain how wild type RecA is able to partially overcome the filament formation defects of RecA K72R. RecA filament formation is a cooperative process (63–65). RecA-mediated ATP hydrolysis takes place in trans across the subunit-subunit interface. ATP hydrolysis occurs in coordinated waves along the filament (60). All of these observations require RecA subunits in a filament to communicate with their neighbors. One way that this communication could be achieved is by altering the conformational state of an adjacent RecA. In the work of Haruta et al. (12) that described the pairing enhanced or “P” conformational state of RecA filaments, there is evidence that RecA monomers bound to dsDNA at a ds-ssDNA junction can induce neighboring RecA monomers to assume the P-state even though they are bound to ssDNA. Perhaps wild type RecA partially overcomes the filament formation defect of RecA K72R by inducing it to adopt a conformation competent to bind DNA. This idea seems especially reasonable in light of the fact that RecA K72R itself can bind DNA if given the right nucleotide cofactor, and the identity of the nucleotide cofactor has long been known to affect the conformational state of RecA filaments (for review, see Ref. 11).
Finally, the results of this study explain in more detail the general defects of RecA K72R in vivo (51), where it behaves like a null mutation. Studies with a RecA protein double mutant, RecA E38K/K72R, which can more readily form filaments with ATP but still does not hydrolyze ATP, demonstrated that extended filament formation and not ATP hydrolysis was necessary to induce the SOS response. RecA K72R, which cannot support SOS induction, is unable to form filaments on DNA. Because ATP is in vast excess over dATP in the cell (59), the presence of dATP cannot rescue any RecA K72R phenotype. Even in the presence of RecOR in vivo, the filament extension defect of RecA K72R demonstrated in this study would prevent filament formation. In general, we recommend the use of the RecA E38K/K72R double mutant for any study that seeks to isolate the role of ATP hydrolysis in a RecA function in vitro or a phenotype in vivo.
However, the E38K/K72R double mutant is not suitable for studies of RecA filament dynamics, as it does not affect the ATPase activity or the filaments formed by the wild type protein when it is added as a challenge. We speculate that the E38K mutation may prevent an interaction with wild type RecA protein so that it cannot form either mixed oligomers or mixed filaments.
Acknowledgments
We thank Elizabeth Wood for cloning and providing for the expression of RecA K72R. We thank the Keck laboratory for use of their fluorescence polarimeter. We also thank Dennis Harris for purification of RecA K72R and Marielle Gruenig and David Taggart for helpful discussions of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM32335.
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- SSB protein
- ssDNA-binding protein
- cssDNA
- circular ssDNA
- FP
- fluorescence polarization
- nt
- nucleotide(s).
REFERENCES
- 1. Brendel V., Brocchieri L., Sandler S. J., Clark A. J., Karlin S. (1997) J. Mol. Evol. 44, 528–541 [DOI] [PubMed] [Google Scholar]
- 2. Bishop D. K., Park D., Xu L., Kleckner N. (1992) Cell 69, 439–456 [DOI] [PubMed] [Google Scholar]
- 3. Shinohara A., Gasior S., Ogawa T., Kleckner N., Bishop D. K. (1997) Genes to Cells 2, 615–629 [DOI] [PubMed] [Google Scholar]
- 4. Seitz E. M., Brockman J. P., Sandler S. J., Clark A. J., Kowalczykowski S. C. (1998) Genes Dev. 12, 1248–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ogawa T., Yu X., Shinohara A., Egelman E. H. (1993) Science 259, 1896–1899 [DOI] [PubMed] [Google Scholar]
- 6. Sung P. (1994) Science 265, 1241–1243 [DOI] [PubMed] [Google Scholar]
- 7. Lusetti S. L., Cox M. M. (2002) Ann. Rev. Biochem. 71, 71–100 [DOI] [PubMed] [Google Scholar]
- 8. Chen Z., Yang H., Pavletich N. P. (2008) Nature 453, 489–494 [DOI] [PubMed] [Google Scholar]
- 9. Heuser J., Griffith J. (1989) J. Mol. Biol. 210, 473–484 [DOI] [PubMed] [Google Scholar]
- 10. Egelman E. H., Stasiak A. (1986) J. Mol. Biol. 191, 677–697 [DOI] [PubMed] [Google Scholar]
- 11. Cox M. M. (2003) Annu. Rev. Microbiol. 57, 551–577 [DOI] [PubMed] [Google Scholar]
- 12. Haruta N., Yu X., Yang S., Egelman E. H., Cox M. M. (2003) J. Biol. Chem. 278, 52710–52723 [DOI] [PubMed] [Google Scholar]
- 13. Kowalczykowski S. C., Clow J., Krupp R. A. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 3127–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pugh B. F., Cox M. M. (1987) J. Biol. Chem. 262, 1326–1336 [PubMed] [Google Scholar]
- 15. Pugh B. F., Cox M. M. (1988) J. Mol. Biol. 203, 479–493 [DOI] [PubMed] [Google Scholar]
- 16. Galletto R., Amitani I., Baskin R. J., Kowalczykowski S. C. (2006) Nature 443, 875–878 [DOI] [PubMed] [Google Scholar]
- 17. van Loenhout M. T., van der Heijden T., Kanaar R., Wyman C., Dekker C. (2009) Nucleic Acids Res. 37, 4089–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kowalczykowski S. C., Krupp R. A. (1987) J. Mol. Biol. 193, 97–113 [DOI] [PubMed] [Google Scholar]
- 19. Thresher R. J., Christiansen G., Griffith J. D. (1988) J. Mol. Biol. 201, 101–113 [DOI] [PubMed] [Google Scholar]
- 20. Joo C., McKinney S. A., Nakamura M., Rasnik I., Myong S., Ha T. (2006) Cell 126, 515–527 [DOI] [PubMed] [Google Scholar]
- 21. Cox M. M. (2007) Crit. Rev. Biochem. Mol. Biol. 42, 41–63 [DOI] [PubMed] [Google Scholar]
- 22. Menetski J. P., Kowalczykowski S. C. (1989) Biochemistry 28, 5871–5881 [DOI] [PubMed] [Google Scholar]
- 23. Eggler A. L., Lusetti S. L., Cox M. M. (2003) J. Biol. Chem. 278, 16389–16396 [DOI] [PubMed] [Google Scholar]
- 24. Witkin E. M., McCall J. O., Volkert M. R., Wermundsen I. E. (1982) Mol. Gen. Genet. 185, 43–50 [DOI] [PubMed] [Google Scholar]
- 25. Lavery P. E., Kowalczykowski S. C. (1992) J. Biol. Chem. 267, 20648–20658 [PubMed] [Google Scholar]
- 26. Register J. C., 3rd, Griffith J. (1985) J. Biol. Chem. 260, 12308–12312 [PubMed] [Google Scholar]
- 27. Shan Q., Bork J. M., Webb B. L., Inman R. B., Cox M. M. (1997) J. Mol. Biol. 265, 519–540 [DOI] [PubMed] [Google Scholar]
- 28. Roca A. I., Cox M. M. (1997) Prog. Nucleic Acid Res. Mol. Biol. 56, 129–223 [DOI] [PubMed] [Google Scholar]
- 29. Bork J. M., Cox M. M., Inman R. B. (2001) J. Biol. Chem. 276, 45740–45743 [DOI] [PubMed] [Google Scholar]
- 30. Arenson T. A., Tsodikov O. V., Cox M. M. (1999) J. Mol. Biol. 288, 391–401 [DOI] [PubMed] [Google Scholar]
- 31. Brenner S. L., Mitchell R. S., Morrical S. W., Neuendorf S. K., Schutte B. C., Cox M. M. (1987) J. Biol. Chem. 262, 4011–4016 [PubMed] [Google Scholar]
- 32. Cox M. M. (2004) in The Bacterial Chromosome (Higgins N. P. ed) pp. 369–388, American Society for Microbiology, Washington, D. C. [Google Scholar]
- 33. Cox M. M. (2007) Nat. Rev. Mol. Cell Biol. 8, 127–138 [DOI] [PubMed] [Google Scholar]
- 34. Walker J. E., Saraste M., Runswick M. J., Gay N. J. (1982) EMBO J. 1, 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cox J. M., Abbott S. N., Chitteni-Pattu S., Inman R. B., Cox M. M. (2006) J. Biol. Chem. 281, 12968–12975 [DOI] [PubMed] [Google Scholar]
- 36. Campbell M. J., Davis R. W. (1999) J. Mol. Biol. 286, 417–435 [DOI] [PubMed] [Google Scholar]
- 37. Campbell M. J., Davis R. W. (1999) J. Mol. Biol. 286, 437–445 [DOI] [PubMed] [Google Scholar]
- 38. Rehrauer W. M., Kowalczykowski S. C. (1993) J. Biol. Chem. 268, 1292–1297 [PubMed] [Google Scholar]
- 39. Sung P., Higgins D., Prakash L., Prakash S. (1988) EMBO J. 7, 3263–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dombroski A. J., Brennan C. A., Spear P., Platt T. (1988) J. Biol. Chem. 263, 18802–18809 [PubMed] [Google Scholar]
- 41. Myles G. M., Hearst J. E., Sancar A. (1991) Biochemistry 30, 3824–3834 [DOI] [PubMed] [Google Scholar]
- 42. Seeley T. W., Grossman L. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 6577–6581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Seeley T. W., Grossman L. (1990) J. Biol. Chem. 265, 7158–7165 [PubMed] [Google Scholar]
- 44. Cox M. M., Lehman I. R. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 3433–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shan Q., Cox M. M., Inman R. B. (1996) J. Biol. Chem. 271, 5712–5724 [DOI] [PubMed] [Google Scholar]
- 46. Shan Q., Cox M. M. (1996) J. Mol. Biol. 257, 756–774 [DOI] [PubMed] [Google Scholar]
- 47. Shan Q., Cox M. M. (1997) J. Biol. Chem. 272, 11063–11073 [DOI] [PubMed] [Google Scholar]
- 48. Britt R. L., Haruta N., Lusetti S. L., Chitteni-Pattu S., Inman R. B., Cox M. M. (2010) J. Biol. Chem. 285, 3211–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Drees J. C., Lusetti S. L., Chitteni-Pattu S., Inman R. B., Cox M. M. (2004) Mol. Cell 15, 789–798 [DOI] [PubMed] [Google Scholar]
- 50. Renzette N., Sandler S. J. (2008) Mol. Microbiol. 67, 1347–1359 [DOI] [PubMed] [Google Scholar]
- 51. Gruenig M. C., Renzette N., Long E., Chitteni-Pattu S., Inman R. B., Cox M. M., Sandler S. J. (2008) Mol. Microbiol. 69, 1165–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Petrova V., Chitteni-Pattu S., Drees J. C., Inman R. B., Cox M. M. (2009) Mol. Cell 36, 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lohman T. M., Green J. M., Beyer R. S. (1986) Biochemistry 25, 21–25 [DOI] [PubMed] [Google Scholar]
- 54. Craig N. L., Roberts J. W. (1981) J. Biol. Chem. 256, 8039–8044 [PubMed] [Google Scholar]
- 55. Bindslev N. (2008) Drug-Acceptor Interactions: Modeling Theoretical Tools to Test and Evaluate Experimental Equilibrium Effects, Co-Action Publishing, Stockholm, Sweden [Google Scholar]
- 56. Lusetti S. L., Wood E. A., Fleming C. D., Modica M. J., Korth J., Abbott L., Dwyer D. W., Roca A. I., Inman R. B., Cox M. M. (2003) J. Biol. Chem. 278, 16372–16380 [DOI] [PubMed] [Google Scholar]
- 57. Morrical S. W., Lee J., Cox M. M. (1986) Biochemistry 25, 1482–1494 [DOI] [PubMed] [Google Scholar]
- 58. Lindsley J. E., Cox M. M. (1990) J. Biol. Chem. 265, 9043–9054 [PubMed] [Google Scholar]
- 59. Bochner B. R., Ames B. N. (1982) J. Biol. Chem. 257, 9759–9769 [PubMed] [Google Scholar]
- 60. Cox J. M., Tsodikov O. V., Cox M. M. (2005) PLoS Biol. 3, 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cox M. M., Lehman I. R. (1982) J. Biol. Chem. 257, 8523–8532 [PubMed] [Google Scholar]
- 62. Menetski J. P., Kowalczykowski S. C. (1987) J. Biol. Chem. 262, 2093–2100 [PubMed] [Google Scholar]
- 63. Takahashi M. (1989) J. Biol. Chem. 264, 288–295 [PubMed] [Google Scholar]
- 64. Stasiak A., Di Capua E., Koller T. (1981) J. Mol. Biol. 151, 557–564 [DOI] [PubMed] [Google Scholar]
- 65. Menetski J. P., Kowalczykowski S. C. (1985) J. Mol. Biol. 181, 281–295 [DOI] [PubMed] [Google Scholar]