Abstract
Nitric oxide (NO) is a potent activator of the p53 tumor suppressor protein, thereby inducing cell cycle arrest and apoptosis. However, little is known about the regulation of the two other p53-family members, p63 and p73, by nitrogen oxides. We report here an up-regulation of p73 by NO in p53-null K-562 leukemia cells. Chemical NO prodrugs or macrophage iNOS activity induced an accumulation of the TAp73α isoform in these cells, whereas macrophages from iNOS−/− mice did not. NO also up-regulated TAp73 mRNA expression, suggesting a transcriptional regulation. The checkpoint kinases Chk1 and Chk2 can regulate TAp73 induction after DNA damage. We show that these kinases were rapidly phosphorylated upon NO treatment. Genetic silencing or pharmacological inhibition of Chk1 impaired NO-mediated accumulation of TAp73α. Because NO is known to block DNA synthesis through ribonucleotide reductase inhibition, the up-regulation of TAp73α might be caused by DNA damage induced by an arrest of DNA replication forks. In support of this hypothesis, DNA replication inhibitors such as hydroxyurea and aphidicolin similarly enhanced TAp73α expression and Chk1 phosphorylation. Moreover, inhibition of Chk1 also prevented TAp73α accumulation in response to replication inhibitors. The knockdown of TAp73 with siRNA sensitized K-562 cells to apoptosis induced by a nitrosative (NO) or oxidative (H2O2) injury. Therefore, TAp73α has an unusual cytoprotective role in K-562 cells, contrasting with its pro-apoptotic functions in many other cell models. In conclusion, NO up-regulates several p53 family members displaying pro- and anti-apoptotic effects, suggesting a complex network of interactions and cross-regulations between NO production and p53-related proteins.
Keywords: Apoptosis, Checkpoint Control, Gene Regulation, Leukemia, Nitric Oxide, p73, Tumor Suppressor
Introduction
NO is a free radical implicated in numerous physiological functions. In cancer biology, both positive and negative actions of NO have been reported. For example, NO was found to promote tumor growth angiogenesis, and metastasis on the one hand and to induce apoptotic cell death or tumor cytostasis on the other hand (1–4). These opposite responses are linked to the chemistry of nitric oxide in a cellular context. NO not only reacts directly on target molecules but also exerts its effects through derived species with a higher degree of oxidation and generated from NO interaction with O2 (e.g. N2O3, •NO2) or superoxide anion (ONOO−). Those reactive nitrogen species can act as potent nitrosating, nitrating, or oxidizing agents. Given this complex chemistry, the biological outcome of NO is directed by its concentration, the cellular redox environment, and different target susceptibilities. High fluxes of NO are generated by inducible NO synthase (iNOS),3 transcriptionally induced in immune cells such as macrophages by cytokines and bacterial products (e.g. IFN-γ and lipopolysaccharide). Elevated concentrations of NO cause DNA damage, mutation, and apoptotic cell death (5, 6). The tumor suppressor protein p53 is a key player in the DNA damage response and the onset of apoptosis. NO has been shown to activate p53 at high concentrations, which also promote genotoxic and pro-apoptotic effects (3, 6, 7). Because p53 is a transcription factor regulating the expression of several genes involved in DNA repair, cell cycle arrest, and apoptosis, activation of p53 by NO can be considered as a regulatory mechanism preventing the emergence of NO-induced DNA mutations and, hence, tumorigenesis. Augmentation of mutation rates in p53-deficient cells exposed to high NO doses and repression of iNOS gene transcription by p53 in a negative feedback loop are occurrences that strongly support a major role for p53 in the control of NO-induced genotoxicity (7–9).
Two p53-related genes, p63 and p73, have been identified more than 15 years after the discovery of p53 (10, 11). The p53 family members have a similar structural organization comprising an NH2-terminal transactivation domain (TA), a central DNA binding domain, and a COOH-terminal oligomerization domain (12–15). In p63 and p73, an additional sterile-α motif and a transcription inhibitory domain may follow the oligomerization domain. Sequence homology among the three members is limited to 25–40% identity between TA and oligomerization domain regions but increases to 60–80% amino acid identity within the DNA binding domain. These structural similarities enable physical and functional interactions between the p53-family proteins. All three p53-family members are expressed as a variety of isoforms, resulting from the usage of two distinct promoters (P1 and P2) and mRNA differential splicing at the 5′- and 3′-ends. There is now no doubt that p73 and p63 are, like p53, involved in tumor suppression (16–18). However, the p73 and p63 homologues have additional important functions. For instance, in mice p73 and p63 are crucial to proper neural and epithelial development, respectively, whereas the embryonic development of p53-null mice is almost normal (19–21).
The human p73 gene generates two groups of isoforms, one with a complete TA domain comprising nine COOH-terminal splice variants (named TAp73α, β, γ, ..) and the other encompassing at least six other proteins exhibiting a truncated TA domain, produced either from the P1 promoter and NH2-terminal splicing (e.g. ΔEx2p73, ΔEx2/3p73) or from the P2 promoter within intron 3 (ΔNp73). COOH-terminal splice variants such as ΔNp73α and ΔNp73β also exist within the group lacking an entire TA domain. TAp73 isoforms can induce the transcription of an overlapping subset of p53 target genes such as CDKN1A, GADD45, Bax, PUMA, and SFN, but specific targets have been also identified (22, 23). Growth suppression or induction of apoptosis can be accomplished by TAp73 isoforms in cooperation with p53 (24, 25) but also via p53-independent pathways (10, 25, 26). Several studies have shown that p73 is required for apoptosis induction in response to DNA damage by chemotherapeutic drugs, and these findings can explain the relative effectiveness of anti-cancer agents in p53-defective tumor cells (27). Indeed, mutations in the p73 gene are rare in cancer patients. On the contrary, up-regulation of p73 was frequently observed. In particular, deregulated expression of isoforms containing a truncated TA domain, especially ΔNp73, has been demonstrated in several human cancers (27, 28). p73 isoforms lacking the TA domain are transactivation-deficient and do not induce growth suppression or cell death. They behave as dominant-negative proteins by competing with p53 and TAp73 variants at specific DNA binding sequences on promoters and by forming heteroduplexes with TAp73 and TAp63 proteins. The antiapoptotic role of ΔNp73 is crucial during mouse neural development, but it can also confer survival advantages and chemotherapeutic drug resistance in tumors by several mechanisms (27, 29, 30). The oncogenic properties of NH2-terminal-truncated p73 isoforms may help to understand the absence of mutations and overexpression of p73 in tumors. Because ΔNp73 expression is transcriptionally activated by p53 and TAp73, it is implicated in a feedback mechanism that attenuates p53 and TAp73 transcriptional functions, including the p53-dependent DNA damage response, as demonstrated in a recent study using ΔNp73−/− mice (30).
Molecular mechanisms of TAp73 activation by genotoxic agents and γ-irradiation have been investigated and have led to the identification of a variety of transcriptional and post-translational regulatory pathways (Refs. 13, 14 and references therein). Among others, phosphorylation of TAp73 by the c-Abl tyrosine kinase, the JNK and p38 MAP kinases (the latter via c-Abl-mediated activation), or the Chk1 checkpoint kinase stabilizes the protein and increases its transcriptional activity, especially toward pro-apoptotic target genes. The ATM serine-protein kinase activated by DNA damage lies upstream in the pathways leading to TAp73 activation by c-Abl and Chk1. TAp73 activity is also regulated by protein degradation. Indeed, the E3 ubiquitin ligases Itch and MDM2 have opposite effects on p73 half-life. Itch-dependent ubiquitination of p73 targets it to proteasomal degradation, whereas physical association of p73 with MDM2 enhances its stability but inhibits its transcriptional activity. Contrasting with p53 regulation, augmentation of p73 mRNA levels is an important aspect of p73 activation in response to DNA damage. The E2F1 transcription factor implicated in cell-cycle regulation binds to E2F sites within the P1 promoter of the p73 gene and activates the transcriptional expression of TAp73α and TAp73β isoforms in response to oncogenic stress. Phosphorylation of E2F1 by the Chk1/Chk2 checkpoint kinases can increase E2F-1 levels and, as a consequence, transcription of TAp73 isoforms (31).
Several important players acting in the regulatory network controlling p73 activity have been previously shown to be involved in NO-dependent p53 activation. For instance, ATM/ATR DNA damage sensors, the stress-responsive p38 MAP kinase, and MDM2 have been implicated in p53 regulation by NO (32–34). There is, thus, a possibility that those events that activate p53 could also increase p73 transcriptional activity. A number of studies have accumulated that describe the interplay between p53 and nitrogen oxides. However, more than 10 years after the discovery of the other two p53-related proteins and to the best of our knowledge, the potential regulation of p73 or p63 by NO has not been thoroughly investigated. In this report, we started to address this question by examining the effect of genotoxic, high concentrations of NO on TAp73 expression in a p53-deficient human leukemia cell line.
EXPERIMENTAL PROCEDURES
Nitric Oxide Donors
Two NO-generating compounds were used. The S-nitrosothiol S-nitroso-N-acetyl-d,l-penicillamine (SNAP) was synthesized according to Field et al. (35). Depending on the reductive power of its environment, SNAP decomposition generates either the •NO radical or a nitrosating species. It is a powerful nitrosating agent via transnitrosation reactions. (Z)-1-[N-(2-aminoethyl)-N-(2-ammonioethyl)amino]dia-zen-1-ium-1,2-diolate (DETA-NO) was obtained from Cayman chemicals. DETA-NO decomposition gives rise mostly to the free radical •NO.
Biochemicals and Antibodies
Hydroxyurea, resveratrol, 4-propoxyphenol, aphidicolin, adriamycin® (doxorubicin), caffeine, UCN-01, 3-methyladenine, l-buthionine-[S,R]-sulfoximine (BSO), propidium iodide, lipopolysaccharide (LPS) from Salmonella enteritidis, and paraquat (methyl viologen) were purchased from Sigma. 2′,7′-Dichlorodihydrofluoresceine diacetate (H2DCFDA), S-ethyl-isothiourea, FITC-annexin V, and gemcitabine were obtained, respectively, from Invitrogen/Molecular Probes, Merck/Calbiochem, BD Pharmingen, and Lilly France. 5,5′,6,6′-Tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide (JC-1) was also obtained from Invitrogen/Molecular Probes.
Primary antibodies to p73 (IMG-246, Imgenex, and A300-126A, Bethyl Laboratories), Chk1, NQO1, and Crk-L (sc-8408, sc-32793, and sc-319 respectively, Santa Cruz Biotechnology), phospho-Chk1 and phospho-Chk2 (2348 and 2661 respectively, Cell Signaling), β-actin and α-tubulin (A 5316 and T 9026, Sigma) were used for immunoblotting. Appropriate horseradish peroxidase-conjugated anti-mouse (A 8924, Sigma) or anti-rabbit (sc-2030, Santa Cruz Biotechnology) IgG as well as fluorescent Alexa Fluor 680-conjugated anti-mouse (A 21057, Invitrogen/Molecular Probes) or IRDye 800CW-conjugated anti-rabbit (926–32211, LI-COR Biosciences) IgG were used for detection.
Cell Culture and Treatment
The human leukemia cell line K-562 was maintained in RPMI 1640 medium supplemented with 5% fetal bovine serum, 2 mm l-glutamine, 25 mm HEPES and antibiotics. Cultures were grown in a 95% air, 5% CO2 atmosphere in a humidified incubator at 37 °C. Cells were plated in 6-well culture plates at 0.75 × 106 cells per well in 5 ml of culture medium and allowed to recover for 24 h. They were then treated from 30 min to 48 h with NO prodrugs or DNA synthesis inhibitors and finally harvested for protein or RNA extraction. In some experiments caffeine, 3-methyladenine, UCN-01, or Chir-124 were added 30 min before other treatments. For GSH depletion, tumor cells were incubated for 18 h with 100 μm BSO, washed, and resuspended in fresh medium, as described previously (36). Cells were then processed normally for subsequent treatments.
Coculture Conditions
Peritoneal macrophages elicited by the intraperitoneal injection of thioglycolate broth were obtained from C57Bl/6 mice and stimulated as previously described (37). We also employed macrophages isolated from genetically iNOS-deficient mice (38) kindly given by Dr. Charles-Henry Cottart from the Université Paris Descartes, Paris, France). Briefly, macrophages were seeded into 6-well culture plates at a density of 0.3 to 2 × 106 cells/well. Induction of iNOS expression was performed by stimulation for 24 h with mouse IFN-γ and LPS. After elimination of nonadherent peritoneal cells, K-562 cells were added at a concentration of 2 × 106 per well in 5 ml of fresh culture medium containing LPS to maintain iNOS expression. Cocultures were incubated for up to 48 h. Tumor cells were then gently harvested and processed for immunoblotting. Culture supernatant was recovered for nitrite assay with Griess reagent (37).
Immunoblotting Analysis
Cells were washed twice in ice-cold phosphate-buffered saline (PBS). Crude cell extracts were prepared in a 50 mm Tris.HCl lysis buffer, pH 7.4, supplemented with 150 mm NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 1 mm EDTA, 1 mm DTT, and protease inhibitors including 1 mm Pefabloc (Merck/Calbiochem). The soluble proteins (usually 30 μg) were separated by SDS-PAGE using a 10% polyacrylamide gel and transferred onto nitrocellulose membranes. Blocking of the membrane with 5% skimmed milk, incubation with primary and horseradish peroxidase-conjugated secondary antibodies, washings, and chemiluminescent detection of the antigens were performed as described in Guittet et al. (37). To ameliorate the quantification of the immunoblots, we also used the two-color infrared imaging system from LI-COR Biosciences. In these experiments, membranes were blocked with 2.5% skimmed milk or 5% BSA (A3059, Sigma) in PBS containing 0.05% Tween 20 (PBS/Tween). After overnight incubation at 4 °C with primary antibodies and four washings in PBS/Tween, membranes were incubated for 1 h with fluorescent dye-conjugated secondary antibodies. They were then washed four times in PBS/Tween and twice in PBS. The infrared fluorescent signals at 680 and 800 nm were recorded and quantified with an Odyssey scanner (LI-COR Biosciences). Expression of up to three loading controls (Crk-L, β-actin, and α-tubulin) was also analyzed in each experiment. The signal intensity for each band of interest in a treated sample was first normalized relative to the untreated sample. Then, to take into account variations in protein loading and transfer onto the membrane, the mean of all normalized loading control signals was calculated and used to divide the normalized signal intensity for each antigen.
Real-time PCR
Total RNA was prepared using TRIzol (Invitrogen) according to the manufacturer's instructions. A quantity of 2 μg of RNA was used for cDNA synthesis with the High Capacity cDNA Archive kit (Applied Biosystems). Real-time PCR was performed on a Light Cycler 2.0 apparatus (Roche Applied Science) using the LightCycler® FastStart DNA MasterPLUS SYBR Green I kit (Roche Applied Science). Each PCR cycle consisted of 10 s at 95 °C, 10 s at 67 °C, and 15 s at 72 °C. Results were normalized to β-actin RNA levels. The following pairs of oligonucleotides were used: TAp73, 5′-GGCTGCGACGGCTGCAGAGC-3′/5′-GCTCAGCAGATTGAACTGGGCCATG-3′; β-actin, 5′-ACGAGTCCGGCCCCTCCATC-3′/5′-TGGGGGATGCTCGCTCCAAC-3′; ΔNp73, 5′-CAAACGGCCCGCATGTTCCC-3′/5′-TGGTCCATGGTGCTGCTCAGC-3′; and p21CDKN1A, as described by Guittet et al. (37).
RNA Interference
K-562 cells were transiently transfected with siRNA using a Nucleofector kit V and a Nucleofector II device (all from Lonza) set to program T-016 according to the manufacturer's protocol. We used validated siRNA targeted to p53 and TAp73 mRNA (37) as well as a siRNA targeted to Chk1 (stealth siRNA HSS101855, Invitrogen), a siRNA against p63, and a scrambled siRNA control (both from Dharmacon). Depletion of TAp73 mRNA and inhibition of protein expression were maximal at 24 h and decreased thereafter. Therefore, transfected cells were used 24 h after nucleofection.
Apoptosis Analysis
FITC-Annexin V (BD Pharmingen) was used to detect apoptosis by flow cytometry. Briefly, 1 × 106 K-562 cells were transfected with siRNA as described above. After 24 h of recovery, half of these cells were treated by 0.5 mm DETA-NO for 24 h. Cells were harvested 48 h post-transfection, washed twice in cold PBS, and resuspended at a concentration of 1 × 106 cells/ml in 100 μl of a binding buffer containing 10 mm HEPES, pH 7.4, 140 mm NaCl, and 2.5 mm CaCl2. Cells were then stained with 5 μl of FITC-annexin V and 50 μg/ml propidium iodide for 15 min at room temperature in the dark. Finally, 400 μl of binding buffer were added, and the cells were subjected to FACS analysis within 1 h. At least 10,000 events were acquired by a FACScan flow cytometer (BD Biosciences). Data were analyzed using the CellQuestTM program. The percentage of apoptotic cells was calculated by scoring for annexin V-positive cells.
Mitochondrial Membrane Potential Analysis
Variation of mitochondrial membrane potential (ΔΨm) was measured using the JC-1 lipophilic cationic fluorescent dye. K-562 cells were incubated in 0.5 ml of JC-1 solution (10 μg/ml in PBS) and incubated for 20 min at 37 °C in the dark. Cells were analyzed immediately with a FACScan flow cytometer. The green and red fluorescence signals were detected at 530 nm (FL1 channel) and 575 nm (FL2 channel) for JC-1 monomers and aggregates, respectively. We counted 10,000 cells for each sample in acquisition and analyzed them using CellQuestTM software. ΔΨm variations are indicated by fluctuations in the ratio of red to green fluorescence, which is dependent only on the membrane potential. Mitochondrial depolarization is indicated by a decrease in this ratio.
Measurement of Reactive Oxygen Species (ROS) Production
The lipophilic fluorogen H2DCFDA is retained in viable cells and converted intracellularly to a fluorescent fluorescein derivative after oxidation by ROS. K-562 cells previously transfected with siRNA and treated for 24 h with 0.1 mm H2O2 or 6 μg/ml paraquat were harvested, washed once with PBS, and resuspended in Hanks' balanced salt solution supplemented with 10 mm HEPES containing 10 μm H2DCFDA. After dye loading for 1 h at room temperature, cells were centrifuged, resuspended in prewarmed complete culture medium, and incubated for 30 min in a humidified incubator at 37 °C to allow oxidation of the dye. The green fluorescence intensity of the cells was analyzed immediately by a FACScan flow cytometer.
RESULTS
NO Induces TAp73α Expression in K-562 Cells
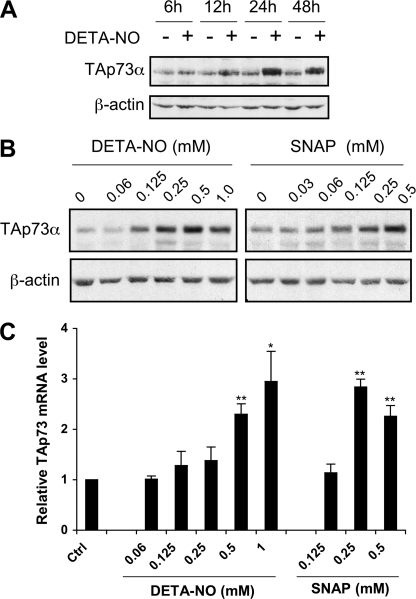
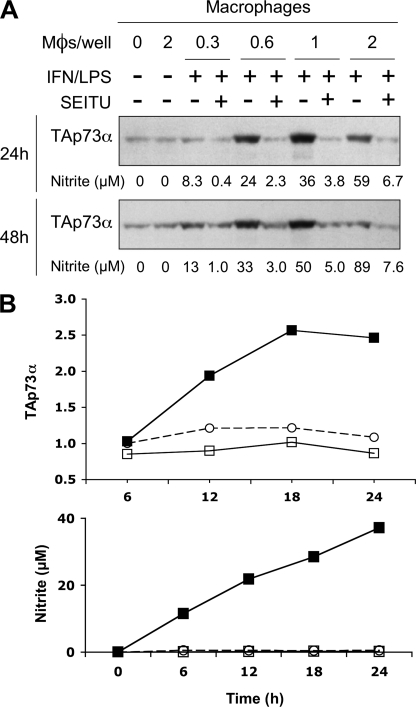
NO has been reported to activate p53 in various cellular models, but much less is known regarding its ability to regulate p53 homologues. The p53-null K-562 leukemia cell line displays moderate but significant levels of the TAp73α isoform (Mr app = 77 kDa) in the absence of any stimulus (Fig. 1A). NO released by the chemical precursor DETA-NO, which mimics macrophage iNOS activity at submillimolar concentrations (36), strongly increased TAp73α expression in these cells. A time-course experiment showed that NO effect was maximal at 24 h and had decreased slightly by 48 h (Fig. 1A). DETA-NO and SNAP, another NO precursor structurally unrelated to DETA-NO, up-regulated TAp73α protein expression in a dose-dependent manner (Fig. 1B). We analyzed p73 regulation by NO further by real-time RT-PCR quantification of TAp73 mRNA levels in K-562 cells before and after treatment with DETA-NO or SNAP (Fig. 1C). Levels of TAp73 mRNA increased up to 3-fold after stimulation by 0.5–1 mm DETA-NO and 0.25–0.5 mm SNAP. Overall, these results are consistent with a transcriptional regulation of TAp73 by NO in K-562 cells. To examine whether TAp73α up-regulation could change the expression of p73 target genes, the relative abundance of p21CDKN1A and ΔNp73 messengers was quantified by RT-quantitative PCR. A more than 5-fold increase in the level of these mRNA was found after treatment with 0.5 mm DETA-NO for 24 h (p21CDKN1A, 5.89 ± 0.67; ΔNp73, 6.01 ± 1.58 versus untreated control, p < 0.05). These data indicate that increased TAp73α expression by NO has functional consequences on gene expression in K-562 cells. The optimal effect of DETA-NO on TAp73α protein expression was reached at a concentration of 0.5 mm (Fig. 1B). Accordingly, DETA-NO was used at this concentration in most experiments described below. To confirm that physiological amounts of NO could up-regulate TAp73α, K-562 cells were cocultured for 24 or 48 h with murine peritoneal macrophages that were first activated with IFN-γ and LPS to induce macrophage iNOS expression. As shown in Fig. 2A, TAp73α levels were strongly increased in cells cocultured for 24 or 48 h with the activated macrophages but not in cells incubated with non-activated macrophages. Induction of TAp73α expression increased with macrophage density, which determines the flux of NO released by the macrophage layer. Overexpression of TAp73α was optimal at 1 × 106 Mφs/well. A higher macrophage density of 2 × 106 Mφs/well was clearly less effective. Thus, NO produced by iNOS activity reproduced a biphasic response already noticed with the chemical NO prodrug DETA-NO (Fig. 1B). We have no explanation for this phenomenon, but it might be a consequence of post-translational modification of TAp73α as it was not observed at the mRNA level (Fig. 1C). The NOS inhibitor S-ethyl-isothiourea, used at a concentration of 50 μm, efficiently inhibited macrophage NOS activity (as assessed by nitrite production) and completely abolished the effect of activated macrophages on TAp73α. To further establish that iNOS activity can up-regulate TAp73, the experiments described above were reproduced using macrophages from iNOS−/− mice. In this case expression levels of TAp73α were measured on immunoblots with a LI-COR infrared Odyssey Imager and Odyssey quantification software. Results from a time-course experiment reported in Fig. 2B, upper panel, show an increase of more than 2-fold of TAp73α protein expression in K-562 cells cocultured for 18 or 24 h with activated macrophages from wild-type mice. As expected, no variations in TAp73α levels were observed in leukemia cells incubated with macrophages obtained from iNOS−/− animals. Lack of iNOS expression in these macrophages was confirmed by the absence of nitrite production in culture supernatants (Fig. 2B, lower panel). These results demonstrate that physiological amounts of NO produced by NO prodrugs or by macrophage iNOS activity induce TAp73α overexpression in K-562 cells.
FIGURE 1.
Induction of TAp73 expression by NO in K-562 cells. A, cells were treated for 6–48 h with 0.5 mm DETA-NO, and TAp73α protein levels were analyzed by immunoblotting using β-actin as a loading control. B, dose-dependent induction of TAp73α protein expression in K-562 cells stimulated for 24 h with the indicated concentrations of DETA-NO and SNAP are shown. C, cells were treated as stated in B, and TAp73 mRNA expression levels were determined by real-time PCR relatively to untreated controls. Significant differences of p < 0.05 (*) or 0.01 (**) versus control cells were calculated by using Student's t test.
FIGURE 2.
Macrophage NO production also induces TAp73α overexpression in K-562 cells. A, K-562 cells (2 × 106) were cocultured with 0.3 to 2 × 106 macrophages (Mφs/well) for 24 or 48 h. These macrophages have been activated (+) or not (−) with IFN-γ and LPS (IFN/LPS) 24 h before addition of tumor cells to induce iNOS expression. The NOS inhibitor S-ethyl-isothiourea (SEITU) was added to some samples to block NO production by activated macrophages. Expression of the TAp73α protein in K-562 cells was evaluated by immunoblotting. Numbers below the immunoblots indicate nitrite concentrations (in μm) measured in the coculture supernatants as an index of iNOS activity. B, upper panel, tumor cells were cultured alone (○) or with 1 × 106 macrophages from either wild-type (■) or iNOS−/− mice (□). Macrophages were activated with IFN-γ and LPS before the addition of K-562 cells as described in A. TAp73α protein amounts in tumor cells were quantified by immunoblotting using fluorescence detection. Results shown are band intensities measured with the Odyssey Imager and expressed in fluorescence intensities normalized to the value measured in the control sample at 6 h. Lower panel, kinetics of nitrite production in culture supernatants are taken as an index of iNOS activity. Results are presented with the same symbols as in the upper panel.
Similar Effects of NO Treatment and DNA Synthesis Inhibition on TAp73 Induction and Checkpoint Kinase Activation
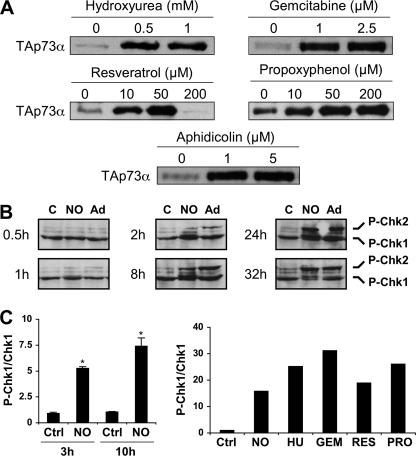
To better understand the molecular mechanisms underlying TAp73 induction by NO, we first tested pharmacological inhibition of several kinases such as c-Abl and p38 MAPK or inhibition of a NQO1-dependent degradation pathway, which have been reported to play important roles in TAp73 regulation (13, 14). None of the inhibitors tested could prevent TAp73 induction by NO (data not shown). We then considered the hypothesis that checkpoint kinases Chk1 and Chk2 might be implicated, as they can induce TAp73 expression (31). The Chk1 and Chk2 kinases are key signal transducers activated by DNA damage and induce cell cycle arrest at specific points in the cell cycle (i.e. G1/S transition, intra-S and G2/M transition). Macrophage iNOS activity blocks DNA synthesis in S phase by inhibiting the supply of dNTPs by ribonucleotide reductase (Rnr) (4). Hence, we considered that blockade of DNA replication forks by NO-mediated dNTP depletion should activate checkpoint kinases, which in turn might induce p73 up-regulation. To test this hypothesis, we examined the effect of several known Rnr inhibitors on TAp73 expression. For this purpose we used hydroxyurea, resveratrol, and 4-propoxyphenol that reduce a tyrosyl free radical present in the Rnr complex and essential for catalysis. We also used gemcitabine, the diphosphate metabolite of which is a mechanism-based inhibitor of the Rnr enzyme. All these molecules strongly increase TAp73α protein levels at concentrations reportedly sufficient to inhibit DNA synthesis (Fig. 3A). Moreover, aphidicolin, a specific DNA polymerase α inhibitor, also increased TAp73α protein amounts in K-562 cells. We, thus, conclude that blockade of DNA replication by inhibition of Rnr or DNA polymerase α activities is sufficient to cause a strong induction of TAp73α expression. To further test our working hypothesis, we investigated whether NO or Rnr inhibitors would activate the cell cycle checkpoint kinases. Activation of Chk1 and Chk2 is mediated by site-specific phosphorylations dependent on the ATM/ATR DNA damage sensors. A time-course of NO-induced phosphorylation of Chk1 and Chk2 at Ser-354 and Thr-68, respectively, is shown on Fig. 3B. Increased phosphorylation of both kinases is already obvious at 2 h and reached a maximum by 8 h and 24 h for Chk1 and Chk2, respectively (Fig. 3B). Phosphorylation of the checkpoint kinases was similar in cells treated by adriamycin, a topoisomerase II inhibitor used as a positive control. Measurements of Chk1 and phospho-Chk1 protein levels by immunoblotting and infrared fluorescence analysis confirm a strong and time-dependent augmentation of the phospho-Chk1/Chk1 ratio after DETA-NO treatment (Fig. 3C, left and right panels). A more than 15-fold increase in this ratio was measured at 24 h. In agreement with our hypothesis, Rnr inhibitors such as hydroxyurea and gemcitabine also induced a great rise in the phospho-Chk1/Chk1 ratio (Fig. 3C, right panel).
FIGURE 3.
NO and DNA synthesis inhibitors increase the phosphorylation of checkpoint kinases. A, K-562 cells were treated for 24 h with the indicated concentrations of inhibitors and processed to analyze TAp73α protein levels by immunoblotting. B, cells were incubated with 0.5 mm DETA-NO (NO) or 0.2 μg/ml adriamycin (Ad) for the indicated times and harvested to measure phospho-Ser-345-Chk1 (P-Chk1) and phospho-Thr68-Chk2 (P-Chk2) by immunoblotting. C, control. C, cells were treated with 0.5 mm DETA-NO (NO), 1 mm hydroxyurea (HU), 1 μm gemcitabine (GEM), 25 μm resveratrol (RES), or 100 μm propoxyphenol (PRO). Phospho-Chk1 (P-Chk1) and total Chk1 amounts were quantified by immunoblotting followed by measurement of band intensities with a LI-COR infrared fluorescence scanner. Results are the ratio between phospho-Chk1 and total Chk1 levels at 3 and 10 h (left panel, mean ± S.E. of 3 experiments) or 24 h (right panel). *, p < 0.05 versus control cells at the same time.
Inhibitors of the Checkpoint Response Compromise TAp73 Regulation by NO Treatment or DNA Synthesis Inhibitors
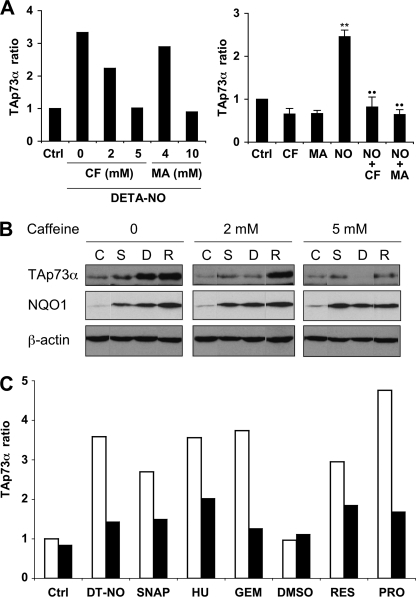
If stalled DNA replication forks activate an intra-S checkpoint that causes an increase in TAp73α expression, then inhibition of the checkpoint response should prevent p73 up-regulation. The PI3K-related protein kinases ATM and ATR are DNA damage sensors that act upstream Chk1/Chk2 in the checkpoint signal transduction pathway. Pharmacological inhibition of ATM/ATR can be achieved by millimolar concentrations of caffeine or 3-methyladenine (39, 40). As shown in the left panel of Fig. 4A, augmentation of TAp73α expression by DETA-NO was inhibited by caffeine and methyladenine in a dose-dependent manner. Complete inhibition was observed at concentrations of 5 mm caffeine and 10 mm methyladenine (Fig. 4A, right panel). These concentrations are consistent with reported doses required for efficient ATM/ATR inhibition. Caffeine was completely ineffective on the induction of NQO1 expression by NO, an effect that is known to depend on the Nrf2 transcription factor (Fig. 4B). In line with our hypothesis, up-regulation of TAp73 by inhibitors of DNA synthesis was also inhibited by 5 mm caffeine (Fig. 4C), strengthening the idea that TAp73 induction was linked to the checkpoint response consecutive to replication arrest.
FIGURE 4.
ATM/ATR inhibition prevents TAp73α up-regulation by NO and DNA synthesis inhibitors. A, K-562 cells were treated for 24 h with 0.5 mm DETA-NO. TAp73α protein levels in cell extracts were measured by immunoblotting using the LI-COR fluorescence scanner and normalized relatively to an untreated control (Ctrl). Caffeine (CF) or 3-methyladenine (MA) were added 30 min before NO treatment at the indicated concentrations (left panel) or at a dose of 5 and 10 mm, respectively (right panel, mean ± S.E. of 4 experiments). Significant differences are p < 0.01 versus control cells (**) or cells treated with DETA-NO (●●). B, accumulation of TAp73α and NQO1 was induced by 250 μm SNAP (S), 0.5 mm DETA-NO (D), or 50 μm resveratrol (R) and measured at 48 h. Only a rise in TAp73α, but not NQO1 protein levels, was prevented by caffeine. β-Actin is shown as a loading marker, and C represents an untreated control. C, TAp73α protein levels in the absence (□) or in the presence of 5 mm caffeine (■) are shown. This checkpoint inhibitor impairs up-regulation of TAp73α expression measured at 24 h after treatment with 0.5 mm DETA-NO (DT-NO), 250 μm SNAP, 1 mm hydroxyurea (HU), 1 μm gemcitabine (GEM), 25 μm resveratrol (RES), or 100 μm propoxyphenol (PRO). Immunoblots of TAp73α were quantified as described in A. Untreated (Ctrl) and DMSO-treated (DMSO) controls are also shown.
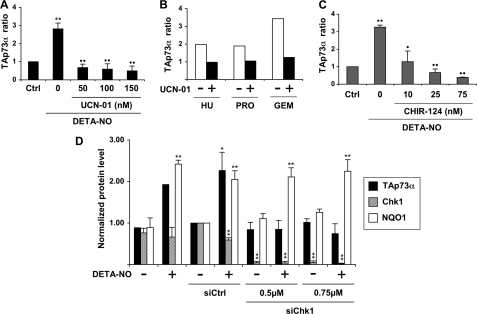
TAp73 Regulation Is Chk1-dependent
The Chk1 and Chk2 kinases control TAp73 mRNA accumulation after DNA damage (31). Our results show that the ATM/ATR-Chk1/Chk2 checkpoint signaling pathway is activated in NO-injured K-562 cells. To determine whether the augmentation of TAp73 levels by NO required Chk1 or Chk2 activities, cells were treated with the radiosensitizing agent UCN-01. This compound is a strong inhibitor of Chk1, although it inhibits Chk2 at higher concentrations (41). Pretreatment of K-562 cells for 30 min with 50–150 nm UCN-01 before the addition of 0.5 mm DETA-NO completely prevented the increase in TAp73α protein levels (Fig. 5A). UCN-01 also inhibited the accumulation of TAp73α after treatment by the Rnr inhibitors hydroxyurea, 4-propoxyphenol, and gemcitabine (Fig. 5B). The strong efficiency of low concentrations of UCN-01 suggested that Chk1 rather than Chk2 could be the main target of UCN-01 in these experiments. To confirm this assumption, the selective Chk1 inhibitor Chir-124 was employed (42). As expected, it prevented NO-induced accumulation of TAp73α as efficiently as UCN-01 (Fig. 5C). Finally, Chk1 expression was knocked down by RNA interference. The Chk1 siRNA we used almost completely depleted the Chk1 protein in K-562 cells (Fig. 5D). Knocking down Chk1 completely prevented accumulation of TAp73α in cells treated with DETA-NO, whereas a control siRNA had no effect. By contrast, induction of NQO1 expression by NO was preserved in the presence of the Chk1 siRNA. A siRNA directed against Chk2 messenger could not impair TAp73α increase (data not shown). These results demonstrate that TAp73 up-regulation by NO requires the Chk1 checkpoint kinase.
FIGURE 5.
Chk1 controls TAp73 induction caused by NO or DNA synthesis inhibitors in K-562 cells. A, immunoblotting of TAp73α was performed and quantified as described under Fig. 4A using lysates of cells treated for 24 h with 0.5 mm DETA-NO. UCN-01 was added at the indicated doses 30 min before NO treatment. Results are the mean ± S.E. of four experiments. ** and ●●, p < 0.01 versus control cells or NO-treated cells, respectively. B, procedures were as in A, except that UCN-01 was used at 50 nm (■). Cells were treated for 24 h with 0.5 mm hydroxyurea (HU), 50 μm propoxyphenol (PRO), or 2.5 μm gemcitabine (GEM). TAp73α levels have been normalized relatively to the amount measured in an untreated control and set to 1 (not shown). C, cells were treated as described in A, except that Chir-124 was used instead of UCN-01. Results are the mean ± S.E. of three determinations. **, p < 0.01 compared with control cells; p < 0.05 (●) or 0.01 (●●) versus cells incubated with DETA-NO. D, K-562 cells were transfected with a control (siCtrl) or a Chk1 (siChk1) siRNA. DETA-NO was added at 0.5 mm 24 h later, and cells were recovered after a further 24-h period for analysis of TAp73α, Chk1, or NQO1 expression by immunoblotting, as described in A. Data are the mean ± S.E. of three determinations. p < 0.05 (*) or 0.01 (**) versus cells transfected with a control siRNA.
Anti-apoptotic Action of TAp73 on K-562 Cells
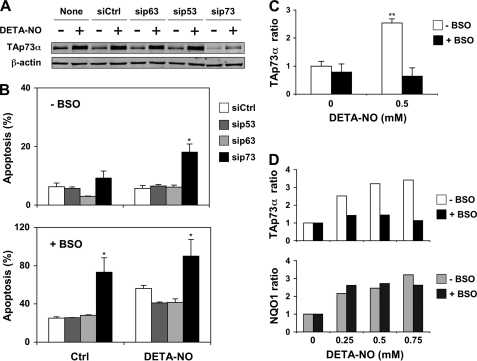
Several TAp73 isoforms have been shown to exert a pro-apoptotic effect in various cellular models, and augmentation of their expression frequently leads to cell death. However, extensive cell death was not apparent in K-562 cell culture after NO treatment despite the reported accumulation of the TAp73α isoform. To further investigate the role of TAp73 in this cell line, TAp73α protein expression was reduced by using a siRNA targeted to TAp73 messengers (Fig. 6A). For comparison, a siRNA control or siRNA against p53 or p63 RNA were also tested. K562 cells do not express p53. Therefore, the siRNA against p53 should be considered as another negative control. As expected, only the siRNA targeting TAp73 reduced TAp73α expression and prevented its accumulation in cells treated by DETA-NO. These siRNA were then used in experiments where K-562 apoptosis was evaluated by annexin V staining. In the absence of additional treatment, there was a low level of apoptosis in cells transfected with any of these siRNA (Fig. 6B, upper panel). Apoptosis markedly increased in cells treated with 0.5 mm DETA-NO but only when TAp73α protein expression was knocked down. This effect was specific, as none of the three other siRNA tested had a similar sensitizing action. In a previous work we showed that GSH depletion increased the sensitivity of K-562 cells to the antiproliferative effect of several NO-donating compounds, including DETA-NO (36). We, therefore, questioned whether GSH depletion would also change the sensitivity of K-562 cells to the pro-apoptotic action of DETA-NO. Pretreatment of cells for 18 h with 100 μm BSO, a specific and irreversible inhibitor of γ-glutamylcysteine synthetase, reduced intracellular glutathione by more than 90% (36). Apoptosis was enhanced in BSO-pretreated cells transfected with a siRNA control compared with cells with a normal GSH content (Fig. 6B, lower panel). Interestingly, knocking down TAp73 considerably increased apoptosis of GSH-depleted cells, whereas apoptosis did not augment in cells transfected with p53 or p63 siRNA. Combining GSH depletion with NO treatment further increased apoptosis in cells transfected with a control siRNA. Apoptosis was higher if TAp73 expression was knocked down, but the incidence of NO treatment could not be evaluated, as apoptosis was already close to 100% in the absence of DETA-NO. It, thus, appeared that either reducing TAp73α levels or depleting GSH in K-562 cells sensitize cells to NO-induced apoptosis. This observation raised the possibility that BSO pretreatment might impair TAp73 expression. Measurements of TAp73α protein levels in GSH-depleted cells confirm this hypothesis (Fig. 6, C and D, upper panel). BSO pretreatment did not significantly change TAp73α levels in control cells but prevented TAp73α accumulation induced by 0.25–0.75 mm DETA-NO. Other responses were not affected, as indicated by a normal up-regulation of NQO1 in GSH-depleted, NO-injured cells (Fig. 6D, lower panel). We concluded from these experiments that the TAp73α isoform was able to protect K-562 cells against NO-induced apoptosis. In cells that were rendered more sensitive to free radical damages by GSH depletion, the protective role of TAp73α was evidenced even in the absence of NO injury, suggesting that the protection mediated by TAp73α might extend to oxygen-derived free radicals.
FIGURE 6.
Knocking down TAp73 sensitizes K-562 cells to NO-induced apoptosis. A, cells were transfected by nucleofection with a 500 nm concentration of a siRNA control (siCtrl) or siRNA targeting p53 (sip53), p63 (sip63), or TAp73 (sip73) transcripts. After 24 h they were treated for a further 24-h period with 0.5 mm DETA-NO. TAp73α expression was analyzed by immunoblotting using β-actin as a loading control. B, cells treated as indicated in A were labeled with FITC-annexin V to quantify apoptosis by flow cytometry. Upper panel, normal cells. Lower panel, GSH depletion amplifies apoptosis evoked by NO. It was induced by an 18-h BSO treatment, which started 6 h after transfection. C and D, GSH depletion inhibits TAp73α up-regulation by NO. Cells were processed as indicated above. TAp73α (C) or TAp73α and NQO1 (D) protein levels were measured by immunoblotting with fluorescence detection as described under Fig. 2B. Results shown in B and C are the mean ± S.E. of three determinations. p < 0.05 (*) and p < 0.01 (**), compared with the indicated control.
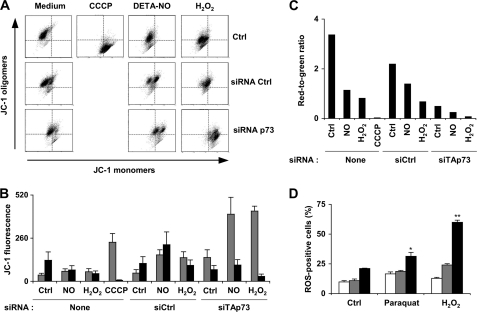
The dissipation of mitochondrial membrane potential (loss of ΔΨm) can be a critical mediator of apoptosis. Quantitative evaluation of mitochondrial membrane depolarization induced by NO treatment was performed by flow cytometry in K-562 cells stained with JC-1 (Fig. 7, A–C). In healthy cells with high ΔΨm, JC-1 accumulates in mitochondria and forms aggregates with intense red fluorescence. In apoptotic cells with low ΔΨm, JC-1 remains in the monomeric form, which shows green fluorescence. Thus, the red-to-green fluorescence ratio is proportional to ΔΨm. The red and green fluorescence signals in cells submitted to various treatments are shown in Fig. 7B, and the calculated red-to-green fluorescence ratio appears in Fig. 7C. This ratio was high in untreated cells and remained elevated in cells transfected with a control siRNA. Loss of ΔΨm was important in p73-knocked down cells and further decreased after exposure to DETA-NO or H2O2. These two chemicals induced a much less important depolarization in cells not transfected with a TAp73 siRNA. Because loss of ΔΨm is also a criterion of apoptosis, these results strengthen our precedent conclusion proposing a protective action of TAp73α against nitrosative (NO) or oxidative stress (H2O2 or GSH depletion) in K-562 cells. The ability of TAp73 to protect cells against oxidative stress was confirmed in another series of experiments. K-562 cells were exposed to H2O2 or to paraquat, a pyridine-based compound that undergoes intracellular redox cycling and generates superoxide anion. ROS production was then measured by intracellular oxidation of H2DCFDA into a green fluorescein derivative. Analysis of the percentage of ROS-positive cells by flow cytometry revealed that knocking down TAp73 expression significantly increased ROS levels in paraquat- and H2O2-treated cells but also, albeit to a lesser extent, in untreated control cells. Transfection of a p53 siRNA, which serves here as a negative control, had no incidence on ROS levels in control and paraquat-treated cells and only slightly increased ROS production in cells exposed to H2O2. These results corroborate our previous findings showing that TAp73 contributes to the regulation of the antioxidant defense system in K-562 cells.
FIGURE 7.
Mitochondrial membrane potential is compromised by knocking down TAp73. A, cells were transfected with 500 nm concentrations of a TAp73 siRNA or a siRNA control (Ctrl). Mitochondrial membrane potential ΔΨm was measured by flow cytometry 48 h later using the JC-1 probe. Some samples were treated by 0.5 mm DETA-NO or 0.1 mm H2O2 during the last 24 h or the last 6 h, respectively. The uncoupling agent carbonyl cyanide p-chlorophenylhydrazone (CCCP) at 50 μm was used as a positive control. Flow cytometry diagrams show the green fluorescent signal of JC-1 monomers in apoptotic cells on the x axis versus the red fluorescent signal of JC-1 oligomers in intact cells on the y axis. B, JC-1 green (gray bars) and red (black bars) fluorescence is shown. siCtrl and siTAp73 refer to a siRNA control and a siRNA against TAp73 mRNA, respectively. Results are the mean ± S.D. of three determinations. C, red-to-green fluorescence ratio was determined from values shown in B. This ratio is proportional to ΔΨm. D, cells were either not transfected (white bars) or transfected with a p53 (gray bars) or a p73 (black bars) siRNA. One day later they were treated with 6 μg/ml paraquat or 0.1 mm H2O2 as indicated. ROS production was analyzed after 24 h by flow cytometry using the H2DCFDA fluorogen. The results shown are the mean ± S.E. of three determinations. p < 0.05 (*) and p < 0.01 (**), compared with untreated and non-transfected cells.
DISCUSSION
NO displays a wide variety of contrasted biological actions as a signaling molecule or in pathophysiological conditions. Attempts to rationalize this complexity have led in particular to the proposal of a series of biological markers indicative of the presence of NO, each characterizing a range of NO concentrations (3). In particular, activation of p53 has been selected as a reliable indicator for high, but still physiological, NO doses. This choice is based on the fact that p53 activation by NO is now regarded as a well established phenomenon of great biological importance. It is most often induced by iNOS activity, because NO fluxes produced by the two other enzymes neuronal NOS (NOS I) and endothelial nitric-oxide synthase (NOS III) do not usually reach the threshold required for p53 activation (3). Two other p53-related proteins, p63 and p73, were identified several years ago, but until now there was a lack of data concerning potential interactions between NO and these homologues. The present work demonstrates that NO levels generated by macrophage iNOS activity or equivalent NO concentrations produced by the decomposition of two NO prodrugs can induce accumulation of a TAp73 isoform in a human cell line. These three sources potentially deliver different nitrogen oxides species (•NO and derived reactive nitrogen species with specific nitrosating and oxidizing properties) that will be referred as NO in the following discussion. We have observed an up-regulation of p53 by similar NO doses (37), and several authors have already described activation of p53 in cells exposed to iNOS activity (6, 7, 34). It, thus, appears that p53 and its homologue, p73, are responsive to comparable high NO levels.
NO and related reactive nitrogen oxides interact with DNA. These interactions can result in base oxidation and nitration and both single and double-strand breaks (43). It has been suggested that these NO-induced DNA damages lead to p53 phosphorylation, stabilization, and transcriptional activation. In agreement with this view, the DNA damage sensors ATM and ATR have been shown to participate in the signaling pathway, leading to p53 activation by genotoxic NO doses (34). However, additional molecular mechanisms have also been described that can explain p53 activation by NO in other experimental models (32, 44). These alternative mechanisms do not appear to be linked to the ATM and ATR kinases or to the DNA-damage checkpoints. In the present study, the NO-induced accumulation of TAp73α in K-562 leukemia cells seems to be dependent on the ATM and ATR damage sensors, as it is abrogated by two inhibitors of these kinases, caffeine and 3-methyladenine (39, 40). ATM and ATR recognize damages on DNA and signal through phosphorylation of downstream components such as the Chk1/Chk2 kinases to initiate cell cycle arrest at specific checkpoints in the cell cycle (45). We have not formally identified which checkpoint is activated in K-562 cells treated by NO. However, we observed that, similar to NO, the Rnr inhibitor hydroxyurea or the DNA polymerase α inhibitor aphidicolin induce an augmentation of TAp73α levels. Both inhibitors cause an arrest of DNA replication forks during the S phase of the cell cycle. Because stalled replication forks are known to trigger the intra-S checkpoint (45), we hypothesized that activation of this checkpoint somehow leads to an augmentation of TAp73α levels in K-562 cells. In agreement with this proposition, other drugs like resveratrol, gemcitabine, and 4-propoxyphenol, that inhibit Rnr but with less selectivity than hydroxyurea (46–48), induce comparable accumulation of TAp73 in K-562 cells. We believe that activation of the intra-S phase checkpoint might also be a relevant mechanism to explain NO effect on p73 expression. Indeed, we have shown previously that macrophage iNOS activity or submillimolar concentrations of DETA-NO inhibit Rnr and induce depletion of dNTPs in K-562 tumor cells (4). Thus, Rnr inhibition by NO might be the primary event leading to activation of a DNA damage checkpoint, probably the intra-S one. However, we cannot formally exclude that direct damages on DNA induced by NO and related nitrogen species might be also involved in the initiation of the DNA damage response, although we previously observed minimal DNA damage induced by 1 mm DETA-NO (37).
The Chk1 and Chk2 signal transducers are phosphorylated by ATM or ATR in several DNA damage checkpoint pathways, including the intra-S checkpoint (45). We effectively observed an increased phosphorylation of Chk1 and Chk2 after NO treatment. Kinetics of Chk1 and Chk2 phosphorylation indicated that activation of the DNA damage checkpoint by NO started after approximately 2 h. Interestingly, depletion of dNTP induced either by DETA-NO or macrophage iNOS activity was evidenced very early, after a similar delay of a few hours (4). We also observed that hydroxyurea and other Rnr inhibitors strongly increase the phosphorylation of Chk1. Again, these findings corroborate our hypothesis, suggesting a central role for Rnr inhibition and S-phase checkpoint activation in the NO-induced DNA damage response. It has been shown that Chk1 and Chk2 control p73 mRNA accumulation after DNA damage (31). Furthermore, reduction of either Chk1 or Chk2 expression impaired p73 protein induction. In this report we also identified Chk1 as an important mediator of TAp73α up-regulation. Knocking-down Chk1 expression or pharmacological inhibition of Chk1 by low concentrations of UCN-01 or Chir-124 was sufficient to abrogate the accumulation of TAp73α induced by NO. Activation of the Chk1 kinase is, therefore, an important step in the signaling pathway connecting the DNA damage response to the augmentation of TAp73α expression mediated by NO. But at odd with Urist et al. (31) report and albeit Chk2 phosphorylation was rapidly induced by NO, participation of this kinase to the up-regulation of TAp73 by NO is unlikely. First, the low concentrations of UCN-01 we employed to inhibit Chk1 activity and TAp73 accumulation are insufficient to inhibit Chk2 activity in K-562 cells (41). Second, the specific Chk1 inhibitor Chir-124 prevented TAp73 induction as efficiently as did UCN-01. Third, a siRNA against Chk2 did not impair the accumulation of TAp73α induced by DETA-NO (data not shown). It is, thus, Chk1, but not Chk2, that transduces the effect of NO on TAp73 expression in K-562 cells. The reason why Chk2 is not required is not clear and requires complete identification of the molecular mechanisms underlying induction of TAp73 by NO. Among the transcription factors known to regulate p73 gene expression, E2F1 occupies a central place (Ref. 14 and references therein). E2F1 is a cell cycle regulator that can also induce apoptosis by p73-independent and p73-dependent pathways. Urist et al. (31) have demonstrated a role for E2F1 stabilization in the transcriptional activation of TAp73 mediated by the Chk1 and Chk2 kinases after DNA damage. We consistently observed an increase in E2F1 protein levels in K-562 cells treated for 24 h with 0.5 mm DETA-NO (data not shown). We, therefore, suspect E2F1 to be a downstream target for Chk1 in the NO-mediated induction of TAp73 expression, but additional investigations are necessary to substantiate this allegation.
Different p73 isoforms have been implicated in cell proliferation, differentiation, and cell death. In particular, extensive studies have explored the pro-apoptotic functions of TAp73 isoforms, which are activated in response to a variety of chemotherapeutic drugs and γ-irradiation (Refs. 14 and 27 and references therein). The ability of distinct TAp73 isoforms, varying in their COOH-terminal region, to transactivate p53-responsive promoters inhibit cell growth or induce apoptosis can differ significantly (49–51). As a general rule, TAp73β is the most potent isoform for transactivation and growth suppression. Both TAp73α and TAp73β can induce apoptosis in different cellular models and experimental conditions (14, 27). Contrasting with these converging data, a few studies highlighted anti-apoptotic or growth-promoting functions of TAp73α or TAp73β (52–54). It is interesting to note that the pro- or anti-apoptotic activity of TAp73α is dependent on the cell type (52). Also, Nyman et al. (52) observed an ambivalent effect of TAp73β which suppressed growth when overexpressed alone but displayed growth-promoting function with the participation of c-Jun. It appears from these studies that increased expression of TAp73 can lead either to growth suppression and cell death on the one hand or to growth progression and cell protection against apoptosis on the other hand. Furthermore, cell fate in response to TAp73 overexpression seems to be determined by the cell type and by functional interplays with other molecular players. In the present study accumulation of TAp73α induced by NO did not cause cell death. This observation is consistent with a previous work showing that K-562 cells were rather resistant to the cytostatic action of NO (36). Unexpectedly, silencing of TAp73 expression compromised cell resistance to NO toxicity and unmasked NO-induced pro-apoptotic effects, as evidenced by exposure of phosphatidylserine on the outer leaflet of the plasma membrane and alteration of mitochondrial membrane potential in cells treated with NO. Loss of ΔΨm was also observed in cells treated by H2O2 when TAp73 expression was knocked down, suggesting a cytoprotective, anti-apoptotic action of TAp73 against oxidative (H2O2) and nitrosative (NO) injuries in K-562 cells. GSH, the most abundant small intracellular thiol, is essential for maintenance of protein thiol and protection against free radical damage, oxidative stress, and NO-induced cytotoxicity (55, 56). Depletion of GSH sensitizes diverse cell types to apoptosis induced by NO or H2O2 treatment (57, 58). In the present study GSH-depleted K-562 cells showed increased susceptibility to NO-induced apoptosis and were also less resistant to apoptosis induced by silencing of TAp73. Because GSH depletion impaired accumulation of TAp73α in cells treated with DETA-NO, TAp73 levels seem to govern the degree of sensitivity to apoptosis of K-562 cells under pro-oxidant (depletion of GSH depletion) or nitrosative (NO treatment) conditions. It has been shown that several anti-oxidant genes are regulated in a p53-dependent manner (59, 60). p53 deficiency increased the production of ROS and the oxidation of molecular components. Because p53 and p73 share common target genes (22), we can imagine that some of the p53-driven antioxidant genes might be also regulated by p73. In support of this assumption, we found enhanced ROS production in K-562 cells with knocked-down TAp73 expression. Analysis of p53-regulated antioxidant genes in further studies might help to understand at a molecular level this interesting cytoprotective and anti-apoptotic function of TAp73 in K-562 cells.
In conclusion, this work identifies p73, besides p53, as another p53-family member susceptible to NO-mediated regulation. Our findings, thus, add a new level of complexicity concerning the regulatory interactions between nitrogen oxides and p53-family members. These proteins (i) share a set of common target genes, (ii) are partly regulated by identical interactors, and (iii) can cooperate in apoptosis and cell cycle control. The fact that similar concentrations of NO can enhance p53 and TAp73 protein expression suggests a possible functional interplay between the two homologues at inflammatory sites or infectious foci where iNOS expression has been demonstrated. In the chronic myeloid leukemia cell line K-562, TAp73α displays unusual cytoprotective effects against oxidative and nitrosative injury. Induction of antioxidant genes by p53 has been already described, and it would be mostly interesting to determine to what extent p53 homologues might substitute to p53 for this function, which contributes to limit DNA damages induced by free radicals.
Acknowledgments
We thank Dr. Charles-Henry Cottart (University Paris-Descartes, Paris, France) for providing iNOS−/− animals, Dr. Pierre-Marie Girard (Institut Curie, Orsay, France) for the generous gift of anti-Chk1 and Chk2 antibodies, and Karine Tuphile for excellent technical assistance. We are grateful to Dr. Roy M. Golsteyn (University of Lethbridge, Lethbridge, Canada) for kindly providing Chir-124.
This work was supported in part by a grant from the Groupement des Entreprises Françaises dans la Lutte contre le Cancer Paris-Ile de France (to M. L.).
- iNOS
- inducible NO synthase
- TA
- transactivation domain
- SNAP
- S-nitrosothiol S-nitroso-N-acetyl-d,l-penicillamine
- DETA-NO
- (Z)-1-[N-(2-aminoethyl)-N-(2-ammonioethyl)amino]dia-zen-1-ium-1,2-diolate
- BSO
- l-buthionine-[S,R]-sulfoximine
- H2DCFDA
- 2′,7′-dichlorodihydrofluoresceine diacetate
- JC-1
- 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide
- ROS
- reactive oxygen species
- Rnr
- ribonucleotide reductase
- ΔΨm
- mitochondrial membrane potential.
REFERENCES
- 1. Jenkins D. C., Charles I. G., Thomsen L. L., Moss D. W., Holmes L. S., Baylis S. A., Rhodes P., Westmore K., Emson P. C., Moncada S. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 4392–4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brüne B., Zhou J. (2007) Cardiovasc. Res. 75, 275–282 [DOI] [PubMed] [Google Scholar]
- 3. Thomas D. D., Ridnour L. A., Isenberg J. S., Flores-Santana W., Switzer C. H., Donzelli S., Hussain P., Vecoli C., Paolocci N., Ambs S., Colton C. A., Harris C. C., Roberts D. D., Wink D. A. (2008) Free Radic. Biol. Med. 45, 18–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roy B., Guittet O., Beuneu C., Lemaire G., Lepoivre M. (2004) Free Radic. Biol. Med. 36, 507–516 [DOI] [PubMed] [Google Scholar]
- 5. Akaike T., Fujii S., Kato A., Yoshitake J., Miyamoto Y., Sawa T., Okamoto S., Suga M., Asakawa M., Nagai Y., Maeda H. (2000) FASEB J. 14, 1447–1454 [DOI] [PubMed] [Google Scholar]
- 6. Messmer U. K., Ankarcrona M., Nicotera P., Brüne B. (1994) FEBS Lett. 355, 23–26 [DOI] [PubMed] [Google Scholar]
- 7. Forrester K., Ambs S., Lupold S. E., Kapust R. B., Spillare E. A., Weinberg W. C., Felley-Bosco E., Wang X. W., Geller D. A., Tzeng E., Billiar T. R., Harris C. C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 2442–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ambs S., Ogunfusika M. O., Merriam W. G., Bennett W. P., Billiar T. R., Harris C. C. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 8823–8828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li C. Q., Trudel L. J., Wogan G. N. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 10364–10369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kaghad M., Bonnet H., Yang A., Creancier L., Biscan J. C., Valent A., Minty A., Chalon P., Lelias J. M., Dumont X., Ferrara P., McKeon F., Caput D. (1997) Cell 90, 809–819 [DOI] [PubMed] [Google Scholar]
- 11. Yang A., Kaghad M., Wang Y., Gillett E., Fleming M. D., Dötsch V., Andrews N. C., Caput D., McKeon F. (1998) Mol. Cell 2, 305–316 [DOI] [PubMed] [Google Scholar]
- 12. Bénard J., Douc-Rasy S., Ahomadegbe J. C. (2003) Hum. Mutat. 21, 182–191 [DOI] [PubMed] [Google Scholar]
- 13. Deyoung M. P., Ellisen L. W. (2007) Oncogene 26, 5169–5183 [DOI] [PubMed] [Google Scholar]
- 14. Vilgelm A., El-Rifai W., Zaika A. (2008) Drug Resist. Updat. 11, 152–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Collavin L., Lunardi A., Del Sal G. (2010) Cell Death Differ. 17, 901–911 [DOI] [PubMed] [Google Scholar]
- 16. Tomasini R., Tsuchihara K., Wilhelm M., Fujitani M., Rufini A., Cheung C. C., Khan F., Itie-Youten A., Wakeham A., Tsao M. S., Iovanna J. L., Squire J., Jurisica I., Kaplan D., Melino G., Jurisicova A., Mak T. W. (2008) Genes and Dev. 22, 2677–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Flores E. R., Sengupta S., Miller J. B., Newman J. J., Bronson R., Crowley D., Yang A., McKeon F., Jacks T. (2005) Cancer Cell 7, 363–373 [DOI] [PubMed] [Google Scholar]
- 18. Guo X., Keyes W. M., Papazoglu C., Zuber J., Li W., Lowe S. W., Vogel H., Mills A. A. (2009) Nat. Cell Biol. 11, 1451–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang A., Walker N., Bronson R., Kaghad M., Oosterwegel M., Bonnin J., Vagner C., Bonnet H., Dikkes P., Sharpe A., McKeon F., Caput D. (2000) Nature 404, 99–103 [DOI] [PubMed] [Google Scholar]
- 20. Mills A. A., Zheng B., Wang X. J., Vogel H., Roop D. R., Bradley A. (1999) Nature 398, 708–713 [DOI] [PubMed] [Google Scholar]
- 21. Sah V. P., Attardi L. D., Mulligan G. J., Williams B. O., Bronson R. T., Jacks T. (1995) Nat. Genet. 10, 175–180 [DOI] [PubMed] [Google Scholar]
- 22. Harms K., Nozell S., Chen X. (2004) Cell. Mol. Life Sci. 61, 822–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fontemaggi G., Kela I., Amariglio N., Rechavi G., Krishnamurthy J., Strano S., Sacchi A., Givol D., Blandino G. (2002) J. Biol. Chem. 277, 43359–43368 [DOI] [PubMed] [Google Scholar]
- 24. Flores E. R., Tsai K. Y., Crowley D., Sengupta S., Yang A., McKeon F., Jacks T. (2002) Nature 416, 560–564 [DOI] [PubMed] [Google Scholar]
- 25. Gonzalez S., Perez-Perez M. M., Hernando E., Serrano M., Cordon-Cardo C. (2005) Cancer Res. 65, 2186–2192 [DOI] [PubMed] [Google Scholar]
- 26. Jost C. A., Marin M. C., Kaelin W. G., Jr. (1997) Nature 389, 191–194 [DOI] [PubMed] [Google Scholar]
- 27. Müller M., Schleithoff E. S., Stremmel W., Melino G., Krammer P. H., Schilling T. (2006) Drug Resist. Updat. 9, 288–306 [DOI] [PubMed] [Google Scholar]
- 28. Domínguez G., García J. M., Peña C., Silva J., García V., Martínez L., Maximiano C., Gómez M. E., Rivera J. A., García-Andrade C., Bonilla F. (2006) J. Clin. Oncol. 24, 805–815 [DOI] [PubMed] [Google Scholar]
- 29. Stiewe T., Stanelle J., Theseling C. C., Pollmeier B., Beitzinger M., Pützer B. M. (2003) J. Biol. Chem. 278, 14230–14236 [DOI] [PubMed] [Google Scholar]
- 30. Wilhelm M. T., Rufini A., Wetzel M. K., Tsuchihara K., Inoue S., Tomasini R., Itie-Youten A., Wakeham A., Arsenian-Henriksson M., Melino G., Kaplan D. R., Miller F. D., Mak T. W. (2010) Genes Dev. 24, 549–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Urist M., Tanaka T., Poyurovsky M. V., Prives C. (2004) Genes Dev. 18, 3041–3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang X., Michael D., de Murcia G., Oren M. (2002) J. Biol. Chem. 277, 15697–15702 [DOI] [PubMed] [Google Scholar]
- 33. Kim S. J., Hwang S. G., Shin D. Y., Kang S. S., Chun J. S. (2002) J. Biol. Chem. 277, 33501–33508 [DOI] [PubMed] [Google Scholar]
- 34. Hofseth L. J., Saito S., Hussain S. P., Espey M. G., Miranda K. M., Araki Y., Jhappan C., Higashimoto Y., He P., Linke S. P., Quezado M. M., Zurer I., Rotter V., Wink D. A., Appella E., Harris C. C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 143–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Field L., Dilts R. V., Ravichandran R., Lenhert P. G., Carnahan G. E. (1978) J. Chem. Soc. Chem. Commun. 1157, 249–250 [Google Scholar]
- 36. Petit J. F., Nicaise M., Lepoivre M., Guissani A., Lemaire G. (1996) Biochem. Pharmacol. 52, 205–212 [DOI] [PubMed] [Google Scholar]
- 37. Guittet O., Tebbi A., Cottet M. H., Vésin F., Lepoivre M. (2008) Nitric. Oxide 19, 84–94 [DOI] [PubMed] [Google Scholar]
- 38. MacMicking J. D., Nathan C., Hom G., Chartrain N., Fletcher D. S., Trumbauer M., Stevens K., Xie Q. W., Sokol K., Hutchinson N. (1995) Cell 81, 641–650 [DOI] [PubMed] [Google Scholar]
- 39. Girard P. M., Pozzebon M., Delacôte F., Douki T., Smirnova V., Sage E. (2008) DNA Repair 7, 1500–1516 [DOI] [PubMed] [Google Scholar]
- 40. Sarkaria J. N., Busby E. C., Tibbetts R. S., Roos P., Taya Y., Karnitz L. M., Abraham R. T. (1999) Cancer Res. 59, 4375–4382 [PubMed] [Google Scholar]
- 41. Busby E. C., Leistritz D. F., Abraham R. T., Karnitz L. M., Sarkaria J. N. (2000) Cancer Res. 60, 2108–2112 [PubMed] [Google Scholar]
- 42. Tao Y., Leteur C., Yang C., Zhang P., Castedo M., Pierré A., Golsteyn R. M., Bourhis J., Kroemer G., Deutsch E. (2009) Cell Cycle 8, 1196–1205 [DOI] [PubMed] [Google Scholar]
- 43. Burney S., Caulfield J. L., Niles J. C., Wishnok J. S., Tannenbaum S. R. (1999) Mutat. Res. 424, 37–49 [DOI] [PubMed] [Google Scholar]
- 44. Glockzin S., von Knethen A., Scheffner M., Brüne B. (1999) J. Biol. Chem. 274, 19581–19586 [DOI] [PubMed] [Google Scholar]
- 45. Sancar A., Lindsey-Boltz L. A., Unsal-Kaçmaz K., Linn S. (2004) Annu. Rev. Biochem. 73, 39–85 [DOI] [PubMed] [Google Scholar]
- 46. Pötsch S., Drechsler H., Liermann B., Gräslund A., Lassmann G. (1994) Mol. Pharmacol. 45, 792–796 [PubMed] [Google Scholar]
- 47. Fontecave M., Lepoivre M., Elleingand E., Gerez C., Guittet O. (1998) FEBS Lett. 421, 277–279 [DOI] [PubMed] [Google Scholar]
- 48. Jordheim L. P., Guittet O., Lepoivre M., Galmarini C. M., Dumontet C. (2005) Mol. Cancer Ther. 4, 1268–1276 [DOI] [PubMed] [Google Scholar]
- 49. De Laurenzi V., Costanzo A., Barcaroli D., Terrinoni A., Falco M., Annicchiarico-Petruzzelli M., Levrero M., Melino G. (1998) J. Exp. Med. 188, 1763–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ozaki T., Naka M., Takada N., Tada M., Sakiyama S., Nakagawara A. (1999) Cancer Res. 59, 5902–5907 [PubMed] [Google Scholar]
- 51. Lee C. W., La Thangue N. B. (1999) Oncogene 18, 4171–4181 [DOI] [PubMed] [Google Scholar]
- 52. Nyman U., Sobczak-Pluta A., Vlachos P., Perlmann T., Zhivotovsky B., Joseph B. (2005) J. Biol. Chem. 280, 34159–34169 [DOI] [PubMed] [Google Scholar]
- 53. Lefkimmiatis K., Caratozzolo M. F., Merlo P., D'Erchia A. M., Navarro B., Levrero M., Sbisa' E., Tullo A. (2009) Cancer Res. 69, 8563–8571 [DOI] [PubMed] [Google Scholar]
- 54. Vikhanskaya F., Toh W. H., Dulloo I., Wu Q., Boominathan L., Ng H. H., Vousden K. H., Sabapathy K. (2007) Nat. Cell Biol. 9, 698–705 [DOI] [PubMed] [Google Scholar]
- 55. Pompella A., Visvikis A., Paolicchi A., De Tata V., Casini A. F. (2003) Biochem. Pharmacol. 66, 1499–1503 [DOI] [PubMed] [Google Scholar]
- 56. Berendji D., Kolb-Bachofen V., Meyer K. L., Kröncke K. D. (1999) Free Radic. Biol. Med. 27, 773–780 [DOI] [PubMed] [Google Scholar]
- 57. Banki K., Hutter E., Colombo E., Gonchoroff N. J., Perl A. (1996) J. Biol. Chem. 271, 32994–33001 [DOI] [PubMed] [Google Scholar]
- 58. Jungas T., Motta I., Duffieux F., Fanen P., Stoven V., Ojcius D. M. (2002) J. Biol. Chem. 277, 27912–27918 [DOI] [PubMed] [Google Scholar]
- 59. Cano C. E., Gommeaux J., Pietri S., Culcasi M., Garcia S., Seux M., Barelier S., Vasseur S., Spoto R. P., Pébusque M. J., Dusetti N. J., Iovanna J. L., Carrier A. (2009) Cancer Res. 69, 219–226 [DOI] [PubMed] [Google Scholar]
- 60. Sablina A. A., Budanov A. V., Ilyinskaya G. V., Agapova L. S., Kravchenko J. E., Chumakov P. M. (2005) Nat. Med. 11, 1306–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]