Abstract
Dendritic cells (DCs) are key mediators of immune function through robust and tightly regulated presentation of antigen in the context of the MHC Class II. MHC Class II expression is controlled by the transactivator CIITA. CIITA expression in conventional DCs is uniquely dependent on an uncharacterized myeloid cell-specific promoter, CIITApI. We now identify in vivo the promoter structure and factors regulating CIITApI. In immature DCs transcription requires binding of PU.1, IRF8, NFκB, and Sp1 to the promoter. PU.1 binds independently at one site and in a required heterodimer with IRF8 at a composite element. DCs from IRF8-null mice have an unoccupied CIITApI promoter that can be rescued by reconstitution with IRF8 in vitro. Furthermore, mutation of either PU.1 site or the IFR8 site inhibits transcriptional activation. In vivo footprinting and chromatin immunoprecipitation reveals that DC maturation induces complete disassociation of the bound activators paralleled by recruitment of PRDM1/Blimp-1 to the promoter. PRDM1 is a transcriptional repressor with essential roles in B cells, T cells, NK cells, and DCs. We show that PRDM1 co-repressors, G9a and HDAC2, are recruited to CIITApI, leading to a loss of histone acetylation and acquisition of histone H3K9 dimethylation and heterochromatin protein 1γ (HP1γ). PRDM1 binding also blocks IRF8-mediated activation dependent on the PU.1/IRF composite element. Together these findings reveal the mechanisms regulating CIITA and, thus, antigen presentation in DCs, demonstrating that PRDM1 and IRF8/PU.1 counter-regulate expression. The activity of PRDM1 in silencing all three cell type-specific CIITA promoters places it as a central regulator of antigen presentation.
Keywords: Chromatin Histone Modification, Gene Regulation, Gene Transcription, Immunology, MHC, Blimp-1, CIITA, IRF8, PRDM1, PU.1
Introduction
Dendritic cells (DC)2 are primary sentinels of the immune system, recognizing pathogens, responding to inflammation, and providing activation signals to immune effector cells (1). DCs reside in peripheral tissues such as the skin as well as in the spleen and lymph nodes where they survey the environment for pathogens. DCs can also migrate from peripheral tissues to the lymph node upon encounter with pathogens to facilitate antigen presentation to T cells. Recent studies have revealed that DCs comprise multiple subsets with both specialized and overlapping functions and tissue distribution (2). Plasmacytoid DCs (pDC) are unique in the ability to secrete large amounts of Type-I interferon and have lymphoid characteristics. Conventional DCs (cDC) encompass the other subtypes including monocyte-derived DCs and are the predominant type detected in the periphery. Encounter with pathogen or inflammatory stimuli induce cDCs to undergo maturation and increase antigen presentation. This is reflected in increased cell surface expression of major histocompatibility complex class II (MHC-II) and co-stimulatory molecules as well as secretion of TH1- or TH2-inducing cytokines (3, 4).
MHC-II expression is tightly regulated during DC development and maturation (3, 5–7). Immature DCs have highly active antigen processing machinery; however, antigen presentation is limited by the low level of antigen-bound MHC-II located on the cell surface. The expressed MHC-II molecules accumulate in peptide loading compartments within the immature DCs, and those that migrate to the cell surface are ubiquitinated by the E3-ligase MARCH1, which promotes endocytosis and turnover (8, 9). Immature DCs upon receiving a maturation signal such as Toll-like receptor engagement with pathogen-associated molecules switch from antigen processing to antigen presentation facilitated in part by the down-regulation of MARCH1 (10). This is paralleled by a cessation of new MHC-II synthesis to fix the presentation of antigen captured at the time of pathogen recognition. Thus, although MHC-II expression is increased on the cell surface upon maturation, the transcription of MHC-II is silenced.
CIITA is a transcriptional co-activator that acts as a master regulator of MHC-II gene expression (11, 12). CIITA is required for MHC-II expression in DCs and B cells as well as cytokine-induced expression in other cell types (13–16). Loss of CIITA either in cells derived from bare lymphocyte syndrome patients or in CIITA knock-out mice results in a near complete loss of MHC-II (15, 17–19). The CIITA gene is tightly controlled at the level of transcription and utilizes three distinct promoters each transcribing a unique first exon (20). Transcription of CIITA from promoter I (CIITApI) is restricted to cells of the myeloid linage including cDCs and macrophages. CIITA promoter III (CIITApIII) is active primarily in cells of lymphoid lineage including B and T cells, although expression can also be detected in DCs. CIITApIV is responsive to IFNγ in non-hematopoietic cells. Mice with targeted deletion of either CIITApIV or both CIITApIV and CIITApIII confirm that CIITApI is functionally sufficient for CIITA expression in myeloid-derived DCs and macrophages, whereas CIITApIII is required for expression in B cells (6, 21). pDC utilize CIITApIII, not CIITApI, consistent with their lymphoid phenotype (6).
The CIITApIII and CIITApIV promoters have been well characterized. CIITApIII is regulated in B cells through five elements that recruit NF1, Oct1, CREB/ATF, AML2, IRF4, and PU.1 (22–24). The IRF4 and PU.1 factors bind at site C, which represents an Ets-IRF composite element (EICE). Recently, PU.1 binding at a distal enhancer located 11 kb upstream of CIITApIII was also shown to be required for CIITApIII activity in B cells (25). CIITApIV transcription depends on IFNγ-induced STAT1 and IRF1 binding at its proximal promoter (26–28). PU.1 also has a role at CIITApIV in mast cells (29).
Silencing of CIITA and, thus, MHC-II is also tightly regulated. CIITA silencing during B cell to plasma cell differentiation has been directly linked to the transcriptional repressor PRDM1 (Blimp-1) (30, 31). PRDM1 is required for B cell to plasma cell differentiation and influences T cell homeostasis and differentiation into effector T cells (32). In both cases PRDM1 is part of a negative feedback loop with BCL6 (33). In addition, we have recently shown that PRDM1 is expressed in natural killer cells, where it attenuates NK activation (34). PRDM1 can also be induced by proteasome inhibitor treatment in mantle cell lymphoma and contributes to the apoptotic response (35). PRDM1 functions by binding to DNA and serves as a scaffold to recruit chromatin-modifying enzymes, specifically the histone deacetylase HDAC2, the histone H3 lysine 9 dimethyltransferase G9a, and the histone H3 lysine 4 demethylase LSD1 (36–38). Each of these chromatin-modifying enzymes mediates changes associated with inhibition of gene transcription. The PRDM1 DNA binding site consensus has overlapping homology with the IRF consensus (39). At the CIITApIV promoter, PRDM1 can compete with IRF1, whereas at CIITApIII, PRDM1 binds the EICE element at site C (30, 40). Thus, PRDM1 can suppress transcription of its target promoters though displacement of a required IRF factor as well as inducing multiple chromatin structure changes. In addition to its role in suppressing CIITApIII during plasma cell differentiation, PRDM1 also suppresses IFNγ-mediated activation of CIITApIII and CIITApIV in B cells (40, 41).
The factors activating or repressing CIITApI remain unknown. Ablation of IRF8 in mice inhibits CD8+ DC development and prevents expression from CIITApI, although whether this is a direct or indirect effect was not known (42). Landmann et al. (5) revealed loss of CIITA transcription during cDC maturation induced by multiple stimuli. The mechanism was not investigated, but a global loss of histone acetylation across all of the CIITA promoters was observed. LPS suppression of CIITA in cDC requires an intact MyD88-dependent pathway utilizing ERK and p38 MAPK signaling (43). Recently, conditional knock-out of PRDM1 in hematopoietic and endothelial cells was shown to disrupt DC development (44). PRDM1 expression increased in murine bone marrow-derived cDC upon receiving maturation signals. This induction required p38 MAPK and NFκB and directly affected transcription of IL-6 and MCP-1 (Ccl2). This presents the possibility that PRDM1 may also affect CIITA regulation in cDCs.
In this report we define the transcription factors required for CIITA transcription in cDC and show that PU.1 and IRF8 synergize to promote promoter assembly and activate transcription. We also now link the mechanism of CIITA silencing in cDC to direct PRDM1 recruitment at the promoter followed by chromatin remodeling and disassembly of the promoter.
EXPERIMENTAL PROCEDURES
Human DC Isolation
Leukocyte buffy coats were obtained from normal donors (Southwest Florida Blood Bank). Peripheral blood monocytic cells were isolated by sedimentation in Ficoll-Paque (Amersham Biosciences) followed by adhesion for 1 h at 37 °C. Non-adherent cells were removed by gentle washing. Purity of monocytes was greater than 90% as assessed by fluorescence-activated cell sorting (FACS) analysis for CD14 (eBioscience). Differentiation into DCs was initiated by the addition of granulocyte-macrophage colony-stimulating factor (1000 units/ml, Roche Applied Science) and IL-4 (5 ng/ml, Roche Applied Science) in RPMI supplemented with 10% heat-inactivated fetal bovine serum (HyClone). Cytokines were replenished every other day (days 2, 4, and 6) by removing half of the medium and adding back fresh medium with 2× cytokines. On day 3 or 7, non-adherent cells were collected by moderately vigorous aspiration and analyzed. Maturation was induced by the addition of either LPS (10 ng/ml, Sigma) or macrophage-conditioned medium at a final concentration of 50% v/v on day 7 as described (45). The human monocytic cell line THP-1 was cultured in RPMI containing 10% heat-inactivated fetal bovine serum and 100 IU/ml streptomycin and penicillin.
Mouse DC Isolation
Mice DCs were obtained from 6–10-week-old homozygous Irf8−/− and IRF8+/+ mice with a C57BL/6 background as previously described (42) using recombinant human Flt3L (10 ng/ml, Pepro Tech) culture system. After 9 days in culture, non-adherent cells were harvested by gentle aspiration and separated on MACS (Miltenyi Biotec) with CD11c antibody-conjugated beads.
Flow Cytometry
DCs phenotype was monitored by cell surface-staining. Cells at the time period indicated in supplemental Fig. 1a were harvested, counted, and stained with FITC- or phosphatidylethanolamine-conjugated antibodies for CD14, CD1a, CD11C, CD83, CD86, and HLA-DR for human and CD8α, CD40, CD80, and Flt3 for mice (all from eBioscience). Samples were collected on a FACSCalibur (BD Biosciences) and analyzed by FlowJo software (Tree Star). Live cell gates were applied on the basis of fluorescence with propidium iodide and/or light scatter properties.
RNA Isolation and Quantitative Real-time RT-PCR
Total cellular RNA was isolated from human DCs with TRIzol reagent (Invitrogen) according to the manufacturer's instruction. One μg of RNA was DNase-treated using RQ1 DNase (Promega) followed by first-strand cDNA synthesis using the iScript cDNA synthesis kit (Bio-Rad). One-twentieth of the final cDNA reaction volume was used in each PCR reaction. Quantitative real-time PCR analysis was performed using iScript SYBR Green Master Mix and analyzed using a MyIQ real-time PCR detection system (Bio-Rad). Primer quality was determined by single peak on melt curve and efficiencies between 90 and 100%. Primer sequences are provided in supplemental Table 1.
In Vivo Genomic Footprinting
In vivo methylation of human and mouse DCs with dimethyl sulfate and DNA preparation were as described previously (22, 46). Genomic DNA was digested with 100 units of HindIII (New England Biolabs). Ligation-mediated PCR was performed to amplify human and mouse CIITApI promoter. The sequences of the primers used for amplification are shown in supplemental Table 1.
DNA Constructs and Transient Transfection
CIITApI promoter region was cloned by PCR and subcloned into pGL3basic using the EcoRI site at position −612 relative to the transcription start site and includes 97 bp of exon I. Site-directed mutagenesis was done by PCR cloning the mutated sequence into the CIITApI p709 construct. Mutations are the same as used in the electrophoretic mobility shift assays (EMSAs) below and are shown in supplemental Table 1. Plasmids and mutations were confirmed by sequencing. Expression plasmids IRF8, PRDM1, and dominant negative G9a have been described previously (36, 47). THP-1 cells (1 × 106) were transfected by Superfect reagent (Qiagen) according to the manufacturer's instructions. The CIITApI promoter activity was measured after 48 h per the Dual-Luciferase Reporter Assay System protocol (Promega). Luciferase readings were done using the 20/20n luminometer (Turner Biosystems). Firefly luciferase activity was normalized to Renilla luciferase activity in all experiments.
Electrophoretic Mobility Shift Assay
Human immature DC nuclear extracts were prepared according to Dignam et al. (48). Oligonucleotide sequences of probes and competitors are shown in supplemental Table 1. Gel shift probes were end-labeled using T4 polynucleotide kinase (New England Biolabs). Five picomoles of radioactive probe were end-labeled with [γ-32P]ATP and purified using mini Quick Spin DNA columns (Roche Applied Science). 50 pmol of cold competitor probe was synthesized using the same conditions as above. All binding reactions included 3 μl of nuclear extract, 0.5 mm DTT, and 1 μg of poly(dI:dC). Where indicated, 0.2 μg of specific antibody was preincubated with the nuclear extract for 2 h on ice. All oligonucleotide competitions were done at 50-fold molar excess. The antibodies specific to PU.1, IRF4, IRF-1, all NFκB subunits, and Sp1 were from Santa Cruz. Antibody to IRF8 was generated in the Ozato laboratory.
Immunostaining and Confocal Microscopy Analysis
Mature DCs were harvested, seeded on slides, and fixed with 4% paraformaldehyde. Cells were permeabilized in 1% Triton X and methanol (Fisher) to allow intracellular staining. Primary antibody incubation was carried out for 1 h at 4 °C followed by three washes with phosphate-buffered saline (PBS) and incubation with secondary antibodies (Alexa 488 and Alexa 564, Molecular Probes) for 30 min at room temperature. The antibodies and dilutions used were mouse anti-HLA-DRA (L243, 1:100), goat anti-PRDM1 (Abcam, 1:100), and rabbit anti-G9a (Upstate, 1:150). After washing in PBS, slides were mounted in Vector shields with DAPI (Vector Laboratories). Cells were imaged using a Zeiss LSM 510 confocal microscope and analyzed by Zeiss LSM software.
Chromatin Immunoprecipitation Analysis
Monocyte-derived DCs were initially treated with 1% formaldehyde for 10 min to ensure cross-linkage followed by cell and nuclear lysis (50 mm Tris, pH 8.1, 10 mm EDTA, 1% SDS, 0.5 mm PMSF) and shearing. For each experiment, chromatin was pooled from cultured primary DCs derived from four individual donors during differentiation. Immunoprecipitated chromatin was collected and washed sequentially with TSE buffer (20 mm Tris, pH 8.1, 50 mm NaCl, 2 mm EDTA, 0.1% SDS, 1.0% Triton X-100) and LiCl buffer (100 mm Tris, pH 8.1, 50 mm LiCl, 1% Nonidet P-40, 1% sodium deoxycholic acid, 1 mm EDTA). DNA was then eluted with 50 mm NaHCO3 containing 1% SDS from the protein A/G beads (Santa Cruz) and reverse-cross-linked at 65 °C overnight followed by proteinase K treatment. DNA was then purified via phenol/chloroform extraction and ethanol precipitation. For each amplification, 3 μl of DNA was analyzed by quantitative PCR. The amplification primers used are shown in supplemental Table 1.
RESULTS
In Vivo Genomic Footprinting Analysis Detects Multiple Protein/DNA Interactions over the CIITApI Promoter in DCs
MHC-II expression is critical for the antigen presentation function of DCs, and the cell surface MHC-II levels are significantly increased during DC maturation. Somewhat paradoxically, mRNA levels for both MHC-II molecules and the master regulator CIITA are markedly down-regulated during maturation (5). To investigate the mechanism of CIITA activation and suppression in DCs, we generated immature DCs from adherent monocytes isolated from healthy donor peripheral blood and cultured for 7 days in the presence of granulocyte-macrophage colony-stimulating factor and IL-4. Subsequent maturation was induced via LPS or macrophage-conditioned media. Consistent with previous reports, we observed typical DC morphology and surface phenotype as assayed by flow cytometry and down-regulation of CIITA mRNA isoforms using a variety of maturation stimuli (supplemental Fig. 1).
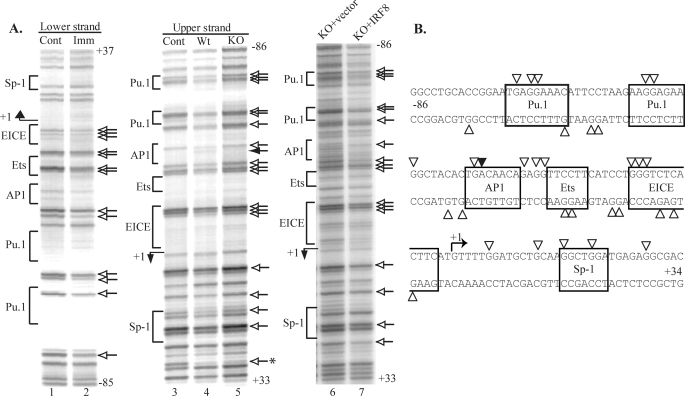
In vivo genomic footprinting analysis has been invaluable in providing unbiased detection of the protein/DNA interactions occurring in the intact cells with nucleotide resolution (46). We used this technique to analyze the occupation status of the CIITApI promoter within both primary human and mouse DCs over the region spanning −134 to +115 in human and −86 to +37 in mouse relative to the transcription start site (Fig. 1 and 2). Close association of transcription factors with the DNA can block or enhance dimethyl sulfate methylation of guanine residues in vivo. The resulting pattern of guanine methylation is then compared with that obtained from genomic DNA purified before dimethyl sulfate treatment. Human immature monocyte-derived DCs display multiple protein/DNA contacts across the region on both upper and lower strands, which is consistent with the strong transcriptional activity in these cells (Fig. 1A, lane 2 versus 1 and lane 5 versus 4). These contacts are summarized in Fig. 1B and can be clustered into five regions. Strikingly, many of the observed contacts lie downstream of the previously reported transcription start site (20). At position +7 to +18 are five contacts on a region of high homology to the consensus EICE. EICE sites cooperatively bind PU.1 and either IRF4 or IRF8 (49). In addition, 10 in vivo contacts are detected from position +35 to +55. This region contains closely adjacent sequence homologies to Sp1 and NFκB binding consensuses. Upstream of the transcription start site five contacts spanning −53 to −33 encompass two putative PU.1 binding sites of which the more distal sequence has very high homology to the consensus PU.1 binding element. At position −85 to −59, eight contacts span a putative Sp1 element and a site labeled Site 1, which has homology to both Ikaros and Oct1 consensus binding elements. Last, a single contact is observed at position −10. A previous analysis of interactions, which focused only upstream of the CIITApI transcription start site, reported five contacts on the upper strand (5). Two contacts (−76 and −50) are also observed in our analysis but are flanked by additional contacts. The three other contacts (−33, −8, and −7) are adjacent to contacts observed in our analysis. Thus, our findings are supportive of previous observations and resolve at high resolution multiple upstream contact points as well as identifying significant regions of contact downstream of the transcription start site.
FIGURE 1.
In vivo footprint analysis of the human CIITApI promoter in DCs. Immature DCs display multiple protein/DNA interactions that are lost upon maturation. A, the lower strand of the promoter is shown in lanes 1–3, and the upper strand is shown in lanes 4–9. Lanes marked cont show the complete guanine sequencing ladder from in vitro methylated DNA. All other lanes are in vivo methylated DNA samples. Imm represents immature monocyte-derived DCs cultured with granulocyte-macrophage colony-stimulating factor and IL-4 for 7 days. Mat and LPS represent mature DC cultured with macrophage-conditioned media or LPS (10 ng/ml) for 48 h, respectively. The transcription start site and direction is indicated by a bent arrow and +1, whereas the sequence position relative to the start site is indicated at the top and bottom of each panel. Residues in immature DCs that are protected or enhanced are indicated by the open and solid arrows, respectively. Elements are indicated on the left and given a putative factor name as discussed under “Results.” In addition, the E2A and Ap1 elements previously reported by Landmann et al. (5) are indicated. B, shown is a schematic of in vivo protein/DNA interactions at the human CIITApI promoter. Protected and enhanced residues are indicated by the open and solid arrowheads, respectively. Putative transcription factor homologies are indicated with boxes. A comparison of the human and mouse promoters is shown in supplemental Fig. 3.
Maturation-induced silencing of CIITApI by exposure to primary macrophage-conditioned media is accompanied by a complete loss of protein/DNA interactions in vivo after 48 h (Fig. 1A, lanes 3 and 6). Previous studies utilizing LPS for 24 h as the maturation signal did not observe any changes in in vivo protein/DNA interactions (5). To determine whether primary macrophage-conditioned media and LPS induce a different response at the CIITApI promoter, we treated human DCs for 24 and 48 h with LPS and profiled the in vivo protein/DNA interactions (Fig. 1A, lanes 7–9). After 48 h of LPS treatment all of the contacts are lost, similar to what we observed in conditioned media. After only 24 h of LPS treatment, the contacts were present but weaker in intensity (data not shown), suggesting that the promoter was in the beginning stages of disassembly. This is consistent with the previous report done at 24 h but now reveals that the silencing of CIITApI involves both an initial inactivation (for example, loss or displacement of a transactivator) followed by a permanent disassembly of the promoter, as the transcription of CIITA p1 gene has already ceased at 24 h as shown in supplemental Fig. 1B. The MHC-II gene HLA-DRA also disassembles upon DC maturation in accordance with the loss of CIITA and HLA-DRA transcriptional inactivation (supplemental Fig. 2).
We next determined whether the CIITApI promoter regulatory elements are conserved between human and mouse. Primary murine bone marrow-derived DCs were subjected to in vivo footprinting across the promoter from −86 to +37 relative to the transcription start site (Fig. 2). Multiple contacts were observed across the entire promoter on both upper and lower strands. Sequence alignment with the human promoter region indicates that all of the putative factor binding elements except for the Site 1 element are conserved between mouse and human (supplemental Fig. 3). Furthermore, the in vivo protein/DNA interactions in the murine DCs cluster at the same regions of homology as seen in human DCs.
FIGURE 2.
In vivo footprint analysis of the mouse CIITApI promoter in wild type and IRF8-null DCs. Immature (Imm) bone marrow-derived DCs display multiple in vivo protein/DNA interactions across the proximal promoter, whereas DCs derived from IRF8-null mice have an unoccupied promoter which can be rescued by IRF8 reconstitution. A, the lower strand of the promoter is shown in lanes 1 and 2, whereas the upper strand is shown in lanes 3–7. Lanes are marked as in Fig. 1A. Lane 5 represents DC generated in vitro from Irf8−/− mice and is compared with wild type mouse DCs generated in parallel (lane 4). Lanes 6 and 7 represent in vivo interactions in Irf8−/− DC reconstituted with either an empty retroviral vector or a retrovirus expressing IRF8, respectively. Occupancy across the entire proximal promoter requires IRF8 and can be rescued by IRF8. B, shown is schematic of in vivo protein/DNA interactions at the mouse CIITApI promoter. Protected and enhanced residues are indicated by the open and solid arrowheads, respectively. Putative transcription factor homologies are indicated with boxes. A comparison of the human and mouse promoters are shown in supplemental Fig. 3.
The EICE element, which is occupied in both human and murine DCs, recruits IRF8 or IRF4 in a complex with PU.1. Ozato and co-workers (42) have previously reported that ablation of IRF8 in mice inhibits CD8+ DC development and prevents expression from CIITApI. In vivo footprint analysis of DCs generated from Irf8−/− mice reveals a nearly complete loss of occupancy across the CIITApI promoter (Fig. 2, lane 5) compared with that of the wild type. To determine whether the loss of promoter occupancy is a direct result of the absence of IRF8 at the promoter or a secondary effect resulting from IRF8 absence during the development of DCs, we reconstituted Irf8−/− DCs with an IRF8 expression vector and then profiled the in vivo protein/DNA interactions. As shown in lanes 6 and 7, reconstitution of IRF8 restored occupancy across the entire CIITApI promoter. Concomitantly, transcription of the CIITApI gene and surface expression of MHC-II is up-regulated in the reconstituted cells (supplemental Fig. 4 and Ref. 42). This indicates that IRF8 is a key factor in CIITApI transcription and is necessary for assembly of the entire CIITApI promoter.
Identification of Multiple Regulatory Motifs Necessary for CIITApI Activation
To identify functional regulatory elements in CIITApI, we cloned an 809-base pair region spanning from −709 to +100 of the human CIITApI promoter into a luciferase reporter construct. Site-directed mutagenesis was done to disrupt the in vivo indentified elements including the NFκB, the conserved PU.1 site at −53, and both the IRF and PU.1 sites in the EICE composite element. Because transfection of primary immature monocyte-derived DCs promotes maturation and precludes analysis, the human monocytic cell line THP-1 was used as a model cell line. The wild type promoter was transcriptionally active, whereas mutation of any of the four sites significantly reduced promoter activity (Fig. 3A). The results demonstrate that these DNA elements are functionally important for the transcriptional activation of CIITApI. To directly address the contribution of factors binding at the EICE as well as possible cooperation between PU.1 and IRFs, we transiently co-transfected CIITApI reporter plasmids with expression plasmids encoding either IRF8 or IRF4. THP-1 cells constitutively express PU.1 but have very low levels of IRF8 and nearly undetectable levels of IRF4 (50). As shown in Fig. 3B, overexpression of IRF8 or IRF4 significantly enhances CIITApI transcriptional activity in THP-1 cells. The dramatic induction in promoter activity is mediated through cooperative interaction with the EICE site as mutation of either the PU.1 or IRF half of the EICE site ablated enhancement by IRF8 and IRF4.
FIGURE 3.
Mutational analysis of human CIITApI promoter function demonstrates that PU.1, EICE, and NFκB elements are critical for transcriptional activity. A, shown is transient transfection of THP-1 cells with the CIITApI-p709 construct and its mutants. Constructs are shown diagrammatically and numbered: 1, wild type; 2, mutation in the distal (−53) PU.1 site; 3, mutation of the IRF homology within the EICE element; 4, mutation of the PU.1 homology within the EICE element; 5, is mutation of the NFκB site. The X in the promoter diagrams indicates the site of the mutation. Data were normalized with Renilla luciferase activity and represent the average of three experiments with the S.D. shown. Significant estimates for each mutant construct are compared with the wild type was p < 0.005. B, overexpression of IRF8 and IRF4 enhances CIITApI promoter activity dependent on the EICE element. CIITApI promoter constructs as indicated on the x axis were co-transfected with expression constructs of IRF8, IRF4, or a control empty vector into THP-1 cells. IRF8 significantly elevated promoter activity (p = 0.029) by at least 7-fold compared with the vector control, whereas IRF4 induced a lesser activation (p = 0.125). Mutation (mut) of either the PU.1 or IRF homologies within the EICE element ablated induction by IRF8 and IRF4, demonstrating a dependence on an intact EICE. Data were normalized with Renilla luciferase activity and represent the average of three experiments with the S.D. shown.
PU.1, IRF8, NFκB, and Sp1 Associate with the CIITApI in Vitro
To examine whether the candidate transcription factors implicated by the in vivo footprinting directly interact with their specific binding motif in vitro, we performed EMSAs using nuclear extracts prepared from primary cultured human immature monocyte-derived DCs. A probe spanning the tandem PU.1 sites at −53 forms two fast migrating complexes that are both diminished by an antibody to PU.1 but not to Sp1 (Fig. 4A). Binding site competition with either a consensus PU.1 element or an unlabeled probe inhibited complex formation, whereas related consensus sites for C/EBPβ and NFAT (nuclear factor of activated T cells) had no effect. Complex formation is also prevented when the probe carries a mutation in the distal half of the tandem PU.1 homology (lane 8), whereas a mutation of the proximal half did not significantly prevent complex formation (lane 9). This suggests that the distal PU.1 site is the dominant binding site required for PU.1 binding in vitro. This is consistent with its higher homology to the PU.1 consensus and its significant effect on transcriptional activity seen in Fig. 3. In addition, PU.1 is present in both complexes, and the upper band may consist of a PU.1 dimer bound at the tandem PU.1 sites.
FIGURE 4.
Identification of PU.1, IRF8, Sp1, and NFκB factors binding at the human CIITApI promoter in vitro. EMSAs using nuclear extracts from immature human DCs and oligonucleotides spanning the tandem PU.1 homologies at −53 base pairs (A), the EICE element (B), and Sp1 and NFκB elements (C) are shown. The relevant region, binding site homologies, and position of mutations are indicated at the bottom of each panel. Reactions in which antibodies were included are indicated above the lane by an α- followed by the antibody specificity. Competitor oligonucleotides were added as indicated above the lanes at 50–200 molar excess. In panel A, lanes 8–10, and panel B, lanes 6–9, the probe used in the binding reaction was created from the mutant sequence indicated at the top of the lanes and illustrated at the bottom. ns indicates nonspecific complex. Complete sequence of each oligonucleotide is shown in supplemental Table 1.
A similar EMSA analysis of the region spanning the EICE site is shown in Fig. 4B. Antibody reactivity indicates that the two fastest migrating complexes contain PU.1. The upper of the two PU.1-containing complexes also is diminished by the IRF8 antibody, consistent with it representing the EICE complex. A slightly slower-migrating complex containing IRF8 is also detected and may represent a complex with another yet to be identified Ets family member. The antibody to IRF4 induces a faint but consistent supershifted band, although none of the complexes is diminished. This indicates that IRF8 and potentially IRF4 can bind to this element. The slowest migrating complex is nonspecific. Mutation at the PU.1 site prevents formation of all three specific complexes when used as the probe, whereas mutation of the IRF site has minimal effect. This indicates that IRF8 does not bind to the DNA in the absence of PU.1.
The region spanning the NFκB and Sp1 sites was also examined by EMSAs (Fig. 4C). Oligonucleotide competition and antibodies against Sp1 as well as individual subunits of NFκB including p50, p52, p65, c-rel, and Rel-B were used to identify the specific protein-DNA complexes. The slowest migrating complex contains Sp1 in association with multiple NFκB subunits with the exception of p52. The fastest migrating complex is nonspecific. The middle complex is not affected by any of the mutations or antibodies and, thus, may also be nonspecific or represent binding outside of the Sp1 and NFκB motifs.
In Vivo Binding of PU.1, IRF8, NFκB, and Sp1 in Immature DCs and Loss of Association upon Maturation
To further investigate if these factors are associated with regulatory regions of CIITA in vivo, we performed chromatin immunoprecipitation (ChIP) experiments using primary human monocyte-derived DC cultures. Using primers specific for the CIITApI, CIITApIII, and HLA-DR promoters, we assessed binding at various times during the maturation. We observed occupancy of PU.1, IRF4, IRF8, p65, and Sp1 in immature DCs at the CIITApI promoter (Fig. 5A). An antibody to NF-Y that is not predicted to associate with CIITApI did not immunoprecipitate the promoter. We also examined the CIITApIII promoter as mRNA from this promoter was detected in monocyte-derived DC, and previous in vivo footprinting studies showed that it was occupied by transcription factors (5). A similar pattern of occupancy by PU.1, IRF8, IRF4, and Sp1 was detected at the CIITApIII promoter with the notable absence of p65, consistent with the lack of potential NFκB binding sites within this region (Fig. 5B).
FIGURE 5.
Chromatin immunoprecipitation analysis of the key transcription activators at the CIITApI promoter. ChIP analysis of CIITApI (A), CIITApIII (B), and HLA-DRA was done in monocyte-derived DCs (C) generated from 12 healthy donors and analyzed as three pools of four donors each. Immature DCs were grown in granulocyte-macrophage colony-stimulating factor/IL-4 for the indicated number of days followed by maturation with macrophage-conditioned media for 24 and 48 h. To account for variability among the three chromatin pools, percent input was calculated then normalized such that each immature day 3 H4-acetyl sample was set to 1 as shown in Fig. 7. Error bars represent S.D.. Immunoprecipitating antibodies are indicated on the x axis; IgG was used as control antibody. Due to the close physical proximity of the PU.1 elements, the signal obtained with the PU.1 antibody represents the collective binding to each of the three PU.1 elements.
A dramatic loss of factor binding was observed at both CIITA promoters upon 24 and 48 h of maturation. The specificity of these protein-DNA interactions was confirmed by examination of the HLA-DRA promoter (Fig. 5C). Consistent with previous findings in other cell types, the HLA-DRA promoter effectively bound NF-Y but not PU.1, IRF8, IRF4, p65, or Sp1. Binding of NF-Y decreased with the time of maturation in parallel with the loss of transcriptional activity. Thus, maturation-induced loss of CIITA mRNA expression is mediated through disassociation of all transcription activators at the promoter consistent with the in vivo footprinting results in Fig. 1A. These findings also reveal that the CIITApI and CIITApIII promoters form highly similar protein-DNA complexes in DCs. Furthermore, the CIITApIII structure in DCs is analogous to that previously detected in B cells. To determine whether the loss of binding at CIITApI is due to a loss of expression of these key transcription factors, we measured protein and mRNA levels of these factors during maturation (supplemental Fig. 5). Only PU.1 was partially diminished in mature cells, whereas expression of the other factors remained the same or increased. Thus, the factors are present in the cell but no longer able to effectively associate with the promoter.
PRDM1 Expression Is Induced upon DC Maturation and Coordinated with the Silencing of CIITA
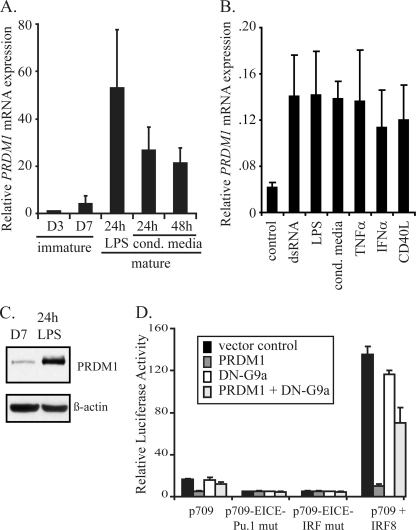
PRDM1 is a potent transcriptional repressor and has recently been shown to be required for optimal DC maturation (44). Furthermore, the PRDM1 recognition sequence is homologous to the core EICE element, which we have shown to be critical for activation of CIITApI. Thus, we hypothesized that PRDM1 contributed to the maturation-induced silencing of CIITA expression within the DC lineage. We fist measured PRDM1 mRNA levels at distinct stages of DC maturation using quantitative real-time-PCR. In immature human monocyte-derived DCs, PRDM1 mRNA is nearly undetectable at day 3 and minimal after 7 days in culture (Fig. 6A). Maturation with either LPS or macrophage-conditioned media results in a significant increase of PRDM1 mRNA. We tested several maturation stimuli, and all are capable of inducing PRDM1 (Fig. 6B). Furthermore, immunoblot analysis confirms that PRDM1 protein levels are also increased during maturation (Fig. 6C). Importantly, the kinetics of PRDM1 expression inversely correlate with CIITA expression, consistent with a negative regulatory role. Thus, we hypothesized that PRDM1 may mediate silencing of CIITApI.
FIGURE 6.
PRDM1 is induced during DC maturation and abrogates IRF8-mediated activation of CIITApI. A, shown is quantitative real-time-PCR analysis of PRDM1 mRNA induction during maturation of monocyte-derived DCs, expressed as -fold induction, relative to day 3. Data represent three independent experiments with S.D. shown. Immature DC were analyzed at day 3 (D3) and day 7 (D7). Maturation was induced by LPS or macrophage-conditioned medium as indicated. B, shown is quantitative real-time-PCR analysis of PRDM1 mRNA induction after 24 h of stimulation using multiple maturation stimuli as indicated on the x axis. Data are expressed relative to GAPDH and represent three independent experiments with S.D. shown. C, immunoblot analysis for PRDM1 in immature and LPS-stimulated monocyte-derived DCs is shown. D, luciferase reporter assays of CIITApI promoter co-transfected with PRDM1 and/or dominant negative G9a (DN-G9a) expression constructs is shown. THP-1 cells were transiently transfected, and luciferase assays were conducted 48 h post-transfection. Data shown represent luciferase activity normalized to co-transfected Renilla expression and are the average of three independent experiments with S.D. shown.
PRDM1 Competes for EICE Sites and Nucleates Chromatin-mediated Silencing of CIITA in Vivo
To assess the ability of PRDM1 to repress activity of the CIITApI promoter, we utilized gene reporter assays in THP-1 cells. Overexpression of PRDM1 is sufficient to repress basal expression of luciferase under control of the wild type CIITApI-p709 promoter by nearly 70% (Fig. 6D). Mutations introduced into either the PU.1 or IRF portion of the EICE element abrogated basal activity with no further repression mediated via PRDM1. Overexpression of IRF8 greatly enhances luciferase activity but is nearly abolished in the presence of PRDM1. These data demonstrate that a functional EICE element is required for PRDM1-mediated repression and that competition between PRDM1 and IRF8 controls CIITApI promoter activity. Importantly, co-transfection of a dominant negative form of the histone methyltransferase G9a abrogated the repressive potential of PRDM1, consistent with our previous report showing a requirement for G9a catalytic activity for PRDM1-mediated silencing of IFNβ (36). Further support of this functional interaction is provided by the observation that both PRDM1 and G9a localize to the nucleus and display significant overlap only in mature DCs (supplemental Fig. 6). The upstream PU.1 element, which has weak homology to the PRDM1 consensus binding motif, is not required for PRDM1-mediated repression (supplemental Fig. 7).
To assess the contribution of PRDM1 to maturation-induced silencing of CIITA, we performed additional ChIP analysis in human monocyte-derived DCs. Upon maturation PRDM1, G9a and HDAC2 accumulate at the CIITApI promoter (Fig. 7A). Progressive loss of acetylation of both histone H3 and H4 was also observed, consistent with loss of CIITA expression during maturation (Fig. 7B). Concomitantly, the repressive marks dimethylated histone H3 lysine 9 and HP1γ, both increased upon maturation. As in previous ChIP experiments, we observed a similar pattern of occupancy at CIITApIII, although at a lesser extent (Fig. 7, C and D). As expected, no binding of PRDM1 or G9a was detected at the HLA-DRA promoter, but a loss of histone acetylation and an increase in dimethylated histone H3 lysine 9 and HP1γ was observed, consistent with MHC-II silencing. When combined with the previous ChIP experiments (Fig. 6), these data suggest a mechanism whereby CIITA silencing is reinforced epigenetically by alterations in histone modifications, which are nucleated via the binding of PRDM1.
FIGURE 7.
Maturation-dependent accumulation of PRDM1, histone modifying enzymes, and repressive chromatin marks at the CIITA promoters. Chromatin immunoprecipitation analysis of CIITApI (A and B), CIITApIII (C and D), and HLA-DRA (E and F) promoters are shown. The upper panels display the changes in repressor binding, whereas the lower panels display changes in histone modifications. Analysis by quantitative PCR is as described in Fig. 5. The antibodies used for immunoprecipitation are indicated on the x axis. G9a and Suv39h1 are both histone H3 lysine 9 methyltransferases that predominantly di- or trimethylate, respectively. Data represents 12 healthy donors and are analyzed as three pools of four donors each and normalized such that each immature day 3 H4-acetyl sample was set to 1. Error bars represent S.D.
DISCUSSION
In this report we have provided evidence for counter regulation of CIITA by IRF8/PU.1 and PRDM1 during the maturation of human monocyte-derived DCs. We have identified functional components of the transcriptionally competent CIITApI in immature DC and demonstrate its disassembly upon maturation. Furthermore, PRDM1 mediates a transition from an active to a repressed chromatin state, resulting in stable silencing of CIITA in mature DCs. This work complements the previous report by Chan et al. (44) and identifies CIITApI as a novel site of PRDM1-mediated regulation. Importantly, PRDM1 is now shown to compete with IRF8 for EICE sites and to regulate all three functional CIITA promoters, suggesting a coordinate mechanism to control MHC-II expression in multiple immune cell lineages.
Numerous transcription factors have been identified that modulate phenotypic changes associated with DC maturation. IRF4 and IRF8 are closely related transcription factors that physically interact with PU.1 to regulate gene expression via recognition of interferon-responsive sequence element and EICE sequence elements (49). Both factors function in monocyte differentiation into DC in vitro and regulate both a common set of genes as well as unique targets (51). Our results suggest that both IRF8 and IRF4 are present in DCs and contribute to CIITA activation during DC differentiation. In our system IRF8 appears to be functionally dominant, as indicated by increased luciferase activity and higher levels of binding both in vivo and in vitro relative to IRF4. Furthermore, a functional EICE element is required for activation. Accordingly, IRF8 transduction into Irf8−/− mice rescues both DC maturation and expression of MHC-II in a DNA binding-dependent manner (42).
Our data indicate that the p65 NFκB subunit and the ubiquitous transcriptional activator Sp1 are components of the active complex at CIITApI in immature DCs. Contacts were detected at the putative binding sites for these factors in footprinting experiments that were lost upon maturation, and site-directed mutagenesis of the potential NFκB binding residues abrogated basal CIITApI activity in luciferase reporter assays. Sp1 has been shown to be required for CD11c expression and may be an important general activator during DC maturation (52). However, p65 itself is likely not sufficient for CIITA expression in DCs as DC maturation proceeds normally, and no defects in MHC-II expression are observed when p65−/− hematopoietic progenitors are adoptively transferred into lethally irradiated hosts (53). However, DC development is severely impaired when both p65 and p50 are simultaneously knocked out, suggesting that cross-talk and compensatory mechanisms exists among the multiple NFκB subunits. Our EMSA experiments confirm this, as binding to a probe spanning the Sp1/NFκB region was markedly diminished in the presence of antibodies to p50, p65, Rel-B, and Sp-1.
Our results also implicate PU.1 in the regulation of CIITApI. We identified two functionally important PU.1 sites within human CIITApI; that is, one upstream of the transcriptional start site and another immediately downstream of the start site and within the context of an EICE element. Both sites displayed maturation-dependent changes by in vivo footprinting, bound a PU.1 site-containing probe in EMSA experiments, and conferred basal activity to CIITApI in luciferase assays. As an interacting partner with IRF4/8, PU.1 is known to be required for the generation of DC. Indeed, ablation of PU.1 diminishes the ability of bone marrow precursors to develop into either conventional or plasmacytoid DCs (54, 55). Several studies have implicated PU.1 in the regulation of CIITA. B cell-specific CIITA expression is conferred through PU.1 binding to pIII in conjunction with E47 and IRF4. In B cells, enforced expression of both PU.1 and IRF8 is not sufficient to drive CIITApIII expression, suggesting important lineage-specific differences between activation requirements of promoter I and promoter III (23). Recently, PU.1 was shown to bind to a distal regulatory element 11 kb upstream of promoter III, which was required for B cell CIITA expression (25). PU.1 also regulates inducible CIITA expression through binding to promoter IV in mast cells in response to IFNγ (29). In the latter two reports, PU.1 binding was also detected at promoter I, albeit at lower levels. Importantly, in DCs PU.1 appears to function both alone and within the context of its interacting partner IRF8 by binding to both PU.1 and EICE sites present within CIITApI.
In response to a Mycobacterium tuberculosis-derived lipoprotein, C/EBPβ binds to PU.1 sites at pI and pIV to prevent MHC-II expression in macrophages (56), and lung DCs infected with Mycobacterium bovis exhibit decreased MHC-II expression relative to uninfected DCs (57). These observations suggest that competition for PU.1 sites regulates CIITA expression. In our EMSA experiments we did not detect binding of C/EBPβ to the PU.1 site under non-pathological conditions. Collectively, these data implicate PU.1 as a critical regulator of CIITA in a variety of contexts and suggest that competition for PU.1 sites may have important implications for proper antigen presentation function during infection.
Given that CIITA mRNA levels decrease upon maturation, we were interested in determining the factors involved in mediating this silencing. PRDM1 has been shown to associate with DC maturation and lead to transcriptional silencing of Il6 and Ccl2 (44). IL-6 is a negative regulator of DC maturation and Il6−/− mice have increased numbers of DCs (58). Here we provide further support for a role of PRDM1 in DC maturation, identifying CIITApI as a novel direct target of binding. PRDM1 has previously been shown to silence both pIII and pIV in B cells (30, 31, 40, 59). PRDM1 is well known to control terminal differentiation of mature B-cells into antibody-producing plasma cells, whereas more recent studies have demonstrated that PRDM1 controls effector functions in both CD4+ and CD8+ T cells (60–65). Furthermore, our group has demonstrated that PRDM1 is involved in the negative regulation of effector cytokine production in human NK cells (34). Thus, regulation of final effector function appears to be a common modality across numerous immune lineages.
PRDM1 mediates silencing of CIITA in DCs through two distinct mechanisms. First, PRDM1 can compete with IRF8 for binding to the promoter. Indeed, we observed reciprocal occupancy of IRF8 and PRDM1 at the CIITApI promoter with kinetics consistent with silencing. Furthermore, IRF8-mediated activation of luciferase under the control of CIITApI is completely abrogated in the presence of PRDM1. PRDM1 has been previously reported to compete with IRF1/2 for DNA binding in vitro, but little competition was observed with IRF8 (39). However, a recent ChIP-on-chip analysis defined AANNGAAA as the dominant motif among PRDM1-bound target genes in the U266 myeloma cell line (66). This suggests that in vivo PRDM1 binding site sequences are less restricted and could encompass many IRF binding factors. Importantly, this motif is present in CIITApI and bound by PRDM1 in mature monocyte-derived DCs. Our in vivo footprinting and chromatin immunoprecipitation analysis reveals that 48-h DC maturation results in a complete loss of the activating factors bound to DNA and large chromatin changes associated with silencing of CIITApI. However, at 24 h of maturation the promoter appears to have an intermediate structure where the factors are mostly bound but histone acetylation is decreasing (Ref. 5 and data not shown). This likely explains the larger number of protein-DNA contacts identified in our study. Expression levels for all of the activating factors except PU.1 are either unchanged or increased upon maturation. Whether this decrease in cellular PU.1 levels contributes to recruitment of PRDM1 to the EICE element remains to be determined. It is possible that low levels of PU.1 destabilizes the PU.1/IRF8 complex and favors PRDM1 binding over IRF8 at the EICE site.
Silencing of CIITA is further reinforced by a second mechanism; that is, the recruitment of chromatin-modifying enzymes by PRDM1. Previous reports have demonstrated the loss of histone acetylation across the CIITApI region during DC maturation (5). We now show that CIITApI in DCs is silenced via recruitment of G9a and HDAC2 by PRDM1 analogous to post-induction silencing of IFNβ upon polyI:C stimulation (36). G9a catalyzes the dimethylation of histone 3 lysine 9, a histone modification known to be associated with silencing of euchromatic genes (67). Our results demonstrate that catalytic activity of G9a is required for silencing as a dominant negative form of G9a, nearly eliminated PRDM1-mediated suppression of CIITApI in reporter assays. Furthermore, HP1γ, which binds methylated histone H3 lysine 9, accumulates to further condense the chromatin in this region. Thus, PRDM1 both competes for sites required for activation and recruits histone modifiers to nucleate a transition from accessible to inaccessible chromatin.
Several aspects of PRDM1 function in DC await elucidation. It will be important to identify other targets of PRDM1 regulation within the DC lineage. Both Il6 and Ccl2 have been previously identified as PRDM1 targets, whereas we show here that PRDM1 binds both CIITApI and pIII. Thus, PRDM1 regulates two essential functions of DCs; that is, cytokine production and antigen presentation. Presumably, PRDM1 will regulate both unique DC-specific and overlapping sets of target genes. Additionally, PRDM1 is reciprocally expressed with the transcriptional repressor Bcl6 and forms a negative feedback loop where each represses the transcription of the other (33). Several examples exist within B and T cells whereby Bcl6 is expressed before PRDM1 and is silenced concomitantly with PRDM1 induction. This antagonistic expression pattern contributes to differentiation of both plasma cells and T follicular helper cells. Bcl6 has been shown to be down-regulated upon maturation of monocyte-derived DC (68); however, the functional interplay between these two transcriptional repressors in DCs has not been characterized. Finally, pDCs differ markedly from cDCs as they continually transcribe MHC-II genes even after antigen encounter (69, 70). CIITA expression in this subset is driven primarily by CIITApIII. It will be important to profile the expression and activity of PRDM1 in pDCs to determine whether these cells fail to induce PRDM1 or utilize an unknown mechanism to alter PRDM1 function at CIITA. The findings in this report now provide detailed characterization of the factors regulating CIITA in cDCs. Through the dedicated function of CIITA in controlling MHC-II expression, this reveals the mechanisms behind antigen presentation in cDCs.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants CA114504 and CA080990 (to K. L. W.) and AI029564.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–7.
- DC
- dendritic cell
- pDC
- plasmacytoid DC
- cDC
- conventional DC
- EICE
- Ets-IRF composite element
- MHC-II
- major histocompatibility complex class II
- PRDM1
- positive regulatory domain I
- CIITA
- counter-regulate MHC Class II transactivator.
REFERENCES
- 1. Liu K., Nussenzweig M. C. (2010) Immunol. Rev. 234, 45–54 [DOI] [PubMed] [Google Scholar]
- 2. Heath W. R., Carbone F. R. (2009) Nat. Immunol. 10, 1237–1244 [DOI] [PubMed] [Google Scholar]
- 3. Pierre P., Turley S. J., Gatti E., Hull M., Meltzer J., Mirza A., Inaba K., Steinman R. M., Mellman I. (1997) Nature 388, 787–792 [DOI] [PubMed] [Google Scholar]
- 4. Sallusto F., Lanzavecchia A. (2000) Immunol. Rev. 177, 134–140 [DOI] [PubMed] [Google Scholar]
- 5. Landmann S., Mühlethaler-Mottet A., Bernasconi L., Suter T., Waldburger J. M., Masternak K., Arrighi J. F., Hauser C., Fontana A., Reith W. (2001) J. Exp. Med. 194, 379–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. LeibundGut-Landmann S., Waldburger J. M., Reis e Sousa C., Acha-Orbea H., Reith W. (2004) Nat. Immunol. 5, 899–908 [DOI] [PubMed] [Google Scholar]
- 7. Santambrogio L., Sato A. K., Fischer F. R., Dorf M. E., Stern L. J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 15050–15055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shin J. S., Ebersold M., Pypaert M., Delamarre L., Hartley A., Mellman I. (2006) Nature 444, 115–118 [DOI] [PubMed] [Google Scholar]
- 9. van Niel G., Wubbolts R., Ten Broeke T., Buschow S. I., Ossendorp F. A., Melief C. J., Raposo G., van Balkom B. W., Stoorvogel W. (2006) Immunity 25, 885–894 [DOI] [PubMed] [Google Scholar]
- 10. De Gassart A., Camosseto V., Thibodeau J., Ceppi M., Catalan N., Pierre P., Gatti E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3491–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Drozina G., Kohoutek J., Jabrane-Ferrat N., Peterlin B. M. (2005) Curr. Top. Microbiol. Immunol. 290, 147–170 [DOI] [PubMed] [Google Scholar]
- 12. Wright K. L., Ting J. P. (2006) Trends Immunol. 27, 405–412 [DOI] [PubMed] [Google Scholar]
- 13. Chin K. C., Mao C., Skinner C., Riley J. L., Wright K. L., Moreno C. S., Stark G. R., Boss J. M., Ting J. P. (1994) Immunity 1, 687–697 [DOI] [PubMed] [Google Scholar]
- 14. Steimle V., Siegrist C. A., Mottet A., Lisowska-Grospierre B., Mach B. (1994) Science 265, 106–109 [DOI] [PubMed] [Google Scholar]
- 15. Steimle V., Otten L. A., Zufferey M., Mach B. (1993) Cell 75, 135–146 [PubMed] [Google Scholar]
- 16. Chang C. H., Fontes J. D., Peterlin M., Flavell R. A. (1994) J. Exp. Med. 180, 1367–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Williams G. S., Malin M., Vremec D., Chang C. H., Boyd R., Benoist C., Mathis D. (1998) Int. Immunol. 10, 1957–1967 [DOI] [PubMed] [Google Scholar]
- 18. Itoh-Lindstrom Y., Piskurich J. F., Felix N. J., Wang Y., Brickey W. J., Platt J. L., Koller B. H., Ting J. P. (1999) J. Immunol. 163, 2425–2431 [PubMed] [Google Scholar]
- 19. Chang C. H., Guerder S., Hong S. C., van Ewijk W., Flavell R. A. (1996) Immunity 4, 167–178 [DOI] [PubMed] [Google Scholar]
- 20. Muhlethaler-Mottet A., Otten L. A., Steimle V., Mach B. (1997) EMBO J. 16, 2851–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Waldburger J. M., Suter T., Fontana A., Acha-Orbea H., Reith W. (2001) J. Exp. Med. 194, 393–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ghosh N., Piskurich J. F., Wright G., Hassani K., Ting J. P., Wright K. L. (1999) J. Biol. Chem. 274, 32342–32350 [DOI] [PubMed] [Google Scholar]
- 23. van der Stoep N., Quinten E., Marcondes Rezende M., van den Elsen P. J. (2004) Blood 104, 2849–2857 [DOI] [PubMed] [Google Scholar]
- 24. van der Stoep N., Quinten E., van den Elsen P. J. (2002) J. Immunol. 169, 5061–5071 [DOI] [PubMed] [Google Scholar]
- 25. Yoon H., Boss J. M. (2010) J. Immunol. 184, 5018–5028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morris A. C., Beresford G. W., Mooney M. R., Boss J. M. (2002) Mol. Cell. Biol. 22, 4781–4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Muhlethaler-Mottet A., Di Berardino W., Otten L. A., Mach B. (1998) Immunity 8, 157–166 [DOI] [PubMed] [Google Scholar]
- 28. Piskurich J. F., Linhoff M. W., Wang Y., Ting J. P. (1999) Mol. Cell. Biol. 19, 431–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ito T., Nishiyama C., Nakano N., Nishiyama M., Usui Y., Takeda K., Kanada S., Fukuyama K., Akiba H., Tokura T., Hara M., Tsuboi R., Ogawa H., Okumura K. (2009) Int. Immunol. 21, 803–816 [DOI] [PubMed] [Google Scholar]
- 30. Ghosh N., Gyory I., Wright G., Wood J., Wright K. L. (2001) J. Biol. Chem. 276, 15264–15268 [DOI] [PubMed] [Google Scholar]
- 31. Piskurich J. F., Lin K. I., Lin Y., Wang Y., Ting J. P., Calame K. (2000) Nat. Immunol. 1, 526–532 [DOI] [PubMed] [Google Scholar]
- 32. Martins G., Calame K. (2008) Annu. Rev. Immunol. 26, 133–169 [DOI] [PubMed] [Google Scholar]
- 33. Crotty S., Johnston R. J., Schoenberger S. P. (2010) Nat. Immunol. 11, 114–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smith M. A., Maurin M., Cho H. I., Becknell B., Freud A. G., Yu J., Wei S., Djeu J., Celis E., Caligiuri M. A., Wright K. L. (2010) J. Immunol. 185, 6058–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Desai S., Maurin M., Smith M. A., Bolick S. C., Dessureault S., Tao J., Sotomayor E., Wright K. L. (2010) Mol. Cancer Res. 8, 907–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gyory I., Wu J., Fejér G., Seto E., Wright K. L. (2004) Nat. Immunol. 5, 299–308 [DOI] [PubMed] [Google Scholar]
- 37. Su S. T., Ying H. Y., Chiu Y. K., Lin F. R., Chen M. Y., Lin K. I. (2009) Mol. Cell. Biol. 29, 1421–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yu J., Angelin-Duclos C., Greenwood J., Liao J., Calame K. (2000) Mol. Cell. Biol. 20, 2592–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kuo T. C., Calame K. L. (2004) J. Immunol. 173, 5556–5563 [DOI] [PubMed] [Google Scholar]
- 40. Tooze R. M., Stephenson S., Doody G. M. (2006) J. Immunol. 177, 4584–4593 [DOI] [PubMed] [Google Scholar]
- 41. Chen H., Gilbert C. A., Hudson J. A., Bolick S. C., Wright K. L., Piskurich J. F. (2007) Mol. Immunol. 44, 1461–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tsujimura H., Tamura T., Gongora C., Aliberti J., Reis e Sousa C., Sher A., Ozato K. (2003) Blood 101, 961–969 [DOI] [PubMed] [Google Scholar]
- 43. Yao Y., Xu Q., Kwon M. J., Matta R., Liu Y., Hong S. C., Chang C. H. (2006) J. Immunol. 177, 70–76 [DOI] [PubMed] [Google Scholar]
- 44. Chan Y. H., Chiang M. F., Tsai Y. C., Su S. T., Chen M. H., Hou M. S., Lin K. I. (2009) J. Immunol. 183, 7039–7046 [DOI] [PubMed] [Google Scholar]
- 45. Romani N., Reider D., Heuer M., Ebner S., Kämpgen E., Eibl B., Niederwieser D., Schuler G. (1996) J. Immunol. Methods 196, 137–151 [DOI] [PubMed] [Google Scholar]
- 46. Mueller P. R., Wold B. (1989) Science 246, 780–786 [DOI] [PubMed] [Google Scholar]
- 47. Weisz A., Marx P., Sharf R., Appella E., Driggers P. H., Ozato K., Levi B. Z. (1992) J. Biol. Chem. 267, 25589–25596 [PubMed] [Google Scholar]
- 48. Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Nucleic. Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Marecki S., Fenton M. J. (2000) Cell Biochem. Biophys. 33, 127–148 [DOI] [PubMed] [Google Scholar]
- 50. O'Reilly D., Quinn C. M., El-Shanawany T., Gordon S., Greaves D. R. (2003) J. Biol. Chem. 278, 21909–21919 [DOI] [PubMed] [Google Scholar]
- 51. Tamura T., Tailor P., Yamaoka K., Kong H. J., Tsujimura H., O'Shea J. J., Singh H., Ozato K. (2005) J. Immunol. 174, 2573–2581 [DOI] [PubMed] [Google Scholar]
- 52. Lopez-Rodriquez C., Hui-Min C., Daniel G. T., Angel L. C. (1995) Eur. J. Immunol. 25, 3496–3503 [DOI] [PubMed] [Google Scholar]
- 53. Ouaaz F., Arron J., Zheng Y., Choi Y., Beg A. A. (2002) Immunity 16, 257–270 [DOI] [PubMed] [Google Scholar]
- 54. Guerriero A., Langmuir P. B., Spain L. M., Scott E. W. (2000) Blood 95, 879–885 [PubMed] [Google Scholar]
- 55. Carotta S., Dakic A., D'Amico A., Pang S. H., Greig K. T., Nutt S. L., Wu L. (2010) Immunity 32, 628–641 [DOI] [PubMed] [Google Scholar]
- 56. Pennini M. E., Liu Y., Yang J., Croniger C. M., Boom W. H., Harding C. V. (2007) J. Immunol. 179, 6910–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pecora N. D., Fulton S. A., Reba S. M., Drage M. G., Simmons D. P., Urankar-Nagy N. J., Boom W. H., Harding C. V. (2009) Cell. Immunol. 254, 94–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Park S. J., Nakagawa T., Kitamura H., Atsumi T., Kamon H., Sawa S., Kamimura D., Ueda N., Iwakura Y., Ishihara K., Murakami M., Hirano T. (2004) J. Immunol. 173, 3844–3854 [DOI] [PubMed] [Google Scholar]
- 59. Piskurich J. F., Gilbert C. A., Ashley B. D., Zhao M., Chen H., Wu J., Bolick S. C., Wright K. L. (2006) Mol. Immunol. 43, 519–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gong D., Malek T. R. (2007) J. Immunol. 178, 242–252 [DOI] [PubMed] [Google Scholar]
- 61. Kallies A., Hawkins E. D., Belz G. T., Metcalf D., Hommel M., Corcoran L. M., Hodgkin P. D., Nutt S. L. (2006) Nat. Immunol. 7, 466–474 [DOI] [PubMed] [Google Scholar]
- 62. Kallies A., Xin A., Belz G. T., Nutt S. L. (2009) Immunity 31, 283–295 [DOI] [PubMed] [Google Scholar]
- 63. Martins G. A., Cimmino L., Shapiro-Shelef M., Szabolcs M., Herron A., Magnusdottir E., Calame K. (2006) Nat. Immunol. 7, 457–465 [DOI] [PubMed] [Google Scholar]
- 64. Rutishauser R. L., Martins G. A., Kalachikov S., Chandele A., Parish I. A., Meffre E., Jacob J., Calame K., Kaech S. M. (2009) Immunity 31, 296–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shin H., Blackburn S. D., Intlekofer A. M., Kao C., Angelosanto J. M., Reiner S. L., Wherry E. J. (2009) Immunity 31, 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Doody G. M., Care M. A., Burgoyne N. J., Bradford J. R., Bota M., Bonifer C., Westhead D. R., Tooze R. M. (2010) Nucleic Acids Res. 38, 5336–5350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tachibana M., Sugimoto K., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., Shinkai Y. (2002) Genes Dev. 16, 1779–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pantano S., Jarrossay D., Saccani S., Bosisio D., Natoli G. (2006) Exp. Cell Res. 312, 1312–1322 [DOI] [PubMed] [Google Scholar]
- 69. Sadaka C., Marloie-Provost M. A., Soumelis V., Benaroch P. (2009) Blood 113, 2127–2135 [DOI] [PubMed] [Google Scholar]
- 70. Young L. J., Wilson N. S., Schnorrer P., Proietto A., ten Broeke T., Matsuki Y., Mount A. M., Belz G. T., O'Keeffe M., Ohmura-Hoshino M., Ishido S., Stoorvogel W., Heath W. R., Shortman K., Villadangos J. A. (2008) Nat. Immunol. 9, 1244–1252 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.