Abstract
Endotoxin tolerance reprograms Toll-like receptor 4 responses by impairing LPS-elicited production of pro-inflammatory cytokines without inhibiting expression of anti-inflammatory or anti-microbial mediators. In septic patients, Toll-like receptor tolerance is thought to underlie decreased pro-inflammatory cytokine expression in response to LPS and increased incidence of microbial infections. The impact of endotoxin tolerance on recruitment, post-translational modifications and signalosome assembly of IL-1 receptor-associated kinase (IRAK) 4, IRAK1, TNF receptor-associated factor (TRAF) 6, TGF-β-activated kinase (TAK) 1, and IκB kinase (IKK) γ is largely unknown. We report that endotoxin tolerization of THP1 cells and human monocytes impairs LPS-mediated receptor recruitment and activation of IRAK4, ablates K63-linked polyubiquitination of IRAK1 and TRAF6, compromises assembly of IRAK1-TRAF6 and IRAK1-IKKγ platforms, and inhibits TAK1 activation. Deficiencies in these signaling events in LPS-tolerant cells coincided with increased expression of A20, an essential deubiquitination enzyme, and sustained A20-IRAK1 associations. Overexpression of A20 inhibited LPS-induced activation of NF-κB and ablated NF-κB reporter activation driven by ectopic expression of MyD88, IRAK1, IRAK2, TRAF6, and TAK1/TAB1, while not affecting the responses induced by IKKβ and p65. A20 shRNA knockdown abolished LPS tolerization of THP1 cells, mechanistically linking A20 and endotoxin tolerance. Thus, deficient LPS-induced activation of IRAK4 and TAK1, K63-linked polyubiquitination of IRAK1 and TRAF6, and disrupted IRAK1-TRAF6 and IRAK1-IKKγ assembly associated with increased A20 expression and A20-IRAK1 interactions are new determinants of endotoxin tolerance.
Keywords: Innate Immunity, Lipopolysaccharide (LPS), Macrophage, Signal Transduction, Toll-like Receptors (TLR)
Introduction
Innate immune cells, such as macrophages, neutrophils, and dendritic cells, detect pathogens by virtue of nonclonally expressed pattern recognition receptors, including Toll-like receptors (TLRs)4 (1). TLRs expressed on the cell surface (TLR1, 2, 4, 5, 6, 10, and 11) or in endosomes (TLR3 and 7–9) recognize pathogen-associated molecular patterns (PAMPs), e.g. lipoproteins (TLR2), LPS (TLR4), flagellin (TLR5), and microbial nucleic acids (TLR3 and 7–9) (2–4). All TLRs express an ectodomain involved in PAMP detection, a transmembrane region, and an intracellular tail with a Toll-IL-1 receptor (TIR) domain essential for signal transduction (3). Upon PAMP recognition, TLRs dimerize, which brings together their TIR domains, forming docking platforms to enable recruitment of adapter proteins that associate with TLRs via homotypic TIR-TIR domain interactions (2, 4–6). TLR4 utilizes signaling adapters myeloid differentiation response gene (MyD) 88 and TIR domain containing adapter-inducing IFN-β (TRIF), and “sorting” adapters MyD88 adapter-like (Mal) and TRIF-related adapter molecule that “bridge” signaling adapters to TLR4 (4, 5). MyD88 molecules cluster within TLR4 to recruit IL-1 receptor-associated kinase (IRAK) 4, IRAK1, and IRAK2, initiating autophosphorylation and activation of IRAK4 (7–10). IRAK4 phosphorylates and activates IRAK1 and IRAK2, leading to their interactions with TNF receptor-associated factor (TRAF) 6 (11–13). IRAK1 undergoes K63-linked polyubiquitination that facilitates its interaction with TGF-β-activated kinase (TAK) 1 and TAK1 activation, resulting in activation of MAPKs and IκB kinases (IKK) (11, 13–15). K63-polyubiquitinated IRAK1 also directly interacts with IKKγ, leading to IKK activation (15). IKKs phosphorylate IκB proteins, resulting in their K48-linked ubiquitination and proteasomal degradation, releasing NF-κB to translocate to the nucleus and activate transcription of pro-inflammatory cytokines. Following MyD88 dissociation, TLR4 and TRIF-related adapter molecule translocate to endosomes where TRIF and TRAF3 are recruited, activating TRAF-associated NF-κB activator-binding kinase (TBK) 1 and IKKϵ. TBK1 and IKKϵ phosphorylate interferon regulatory factors 3 and 7 (16, 17) that translocate to the nucleus and activate expression of type I IFN (17). Transcription factors activated via the MyD88- or TRIF-dependent pathways turn on expression of multiple inflammatory genes, MHC, and co-stimulatory and accessory molecules (4).
Sepsis is characterized by the acute pro-inflammatory “cytokine storm,” followed by the “silencing” phase signified by suppressed TLR-inducible expression of pro-inflammatory cytokines (18). This “silencing” phase does not globally deactivate monocytes and macrophages, because expression of anti-inflammatory mediators and antimicrobial effectors is not inhibited and has been termed “TLR reprogramming” (18). Similar TLR reprogramming occurs in endotoxin tolerance that develops after prior exposure to LPS and suppresses expression of pro-inflammatory cytokines in response to subsequent LPS challenge, without inhibiting anti-inflammatory and antimicrobial mediators (19–22). The development of endotoxin tolerance in monocytes of septic patients curtails excessive pro-inflammatory responses but increases the risk of infections (18, 21, 23). Studies in LPS-tolerized human monocytes revealed unaltered TLR4 expression but suppressed TLR4 tyrosine phosphorylation, MyD88-TLR4 and IRAK1-MyD88 interactions, IRAK1 activation and increased expression of negative regulators of TLR4 signaling (20, 24–29). The impact of endotoxin tolerance in human monocytes on LPS-induced recruitment of IRAK4 to TLR4, IRAK4 and TAK1 activation, and polyubiquitination of IRAK1, TRAF6, and IKKγ has not been defined. Little is known about the involvement of deubiquitination enzymes in endotoxin tolerance.
We report herein impaired LPS-mediated recruitment of IRAK4 to TLR4 and IRAK4 activation in endotoxin-tolerant THP1 cells and human monocytes that are associated with ablated K63-linked polyubiquitination of IRAK1 and TRAF6 and disrupted IRAK1-TRAF6 and IRAK1-IKKγ interactions. We found that LPS tolerization increases expression of A20, a key deubiquitination enzyme that removes K63-linked polyubiquitin chains from TRAF6 and IKKγ (30, 31). We showed that A20 gene silencing abolishes induction of endotoxin tolerance in THP1 cells, mechanistically linking increased expression of A20 to endotoxin tolerization.
EXPERIMENTAL PROCEDURES
Reagents and Cell Culture
Antibodies (Abs) against IκB-α, IRAK1, TAK1, TRAF6, IKKγ, A20, ubiquitin (Ub), tubulin, Myc, HA, and anti-HA-HRP conjugate were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-total p38 Ab was from Promega (Madison, WI), anti-phospho (p)-p38, anti-IRAK4, anti-p-IRAK1, anti-p-TAK1, and anti-IKKβ Abs were obtained from Cell Signaling (Danvers, MA). Abs against K48-linked and K63-linked polyUb chains were purchased from Millipore (Billerica, MA). Ultrapure Escherichia coli 0111:B4 LPS free of lipoproteins was obtained from Invivogen (San Diego, CA). THP1 cells were obtained from the ATCC (Manassas, VA) and maintained in RPMI 1640 supplemented with 2 mm l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin (all reagents were from Mediatech Inc., Manassas, VA), 5 × 10−5 m β-mercaptoethanol (Invitrogen), and 10% FBS (Sigma) (complete RPMI). Human monocytes were prepared by counter flow elutriation and cultured in complete RPMI. HEK 293T cells were from ATCC and maintained in DMEM containing 2 mm l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% FBS (complete DMEM). The 293/TLR4/MD2 stable cell line was described previously (26, 32) and maintained in complete DMEM containing 1 mg/ml G418 (Sigma).
Recombinant Plasmids, Transient Transfection, and Reporter Assays
pcDNA3-HA-A20 expression vector was kindly provided by Dr. Antonio Leonardi (University of Naples, Naples, Italy). pcDNA3-AU1-MyD88, pRK5-IRAK1, pcDNA3-Myc-IRAK2, and pcDNA3-FLAG-TRAF6 were described previously (32, 33), and the plasmids pCMV1-FLAG-TAK1, pcDNA3-HA-TAB1, and pcDNA3-FLAG-p65 were provided by Dr. Katherine Fitzgerald (University of Massachusetts Medical School, Worcester, MA). pcDNA3-FLAG-IKKβ was obtained from Addgene (plasmid 23298) and was initially reported by Geleziunas et al. (34). THP1 cells were plated in 150-mm tissue culture dishes (10 × 106 cells/dish) and differentiated for 72 h with 20 ng/ml phorbol myristate acetate (PMA, Sigma) to attain macrophage-like phenotype (35). Thereafter, the cells were transfected for 3 h with pcDNA3 or pcDNA3-HA-A20 (10 μg of total plasmid DNA/dish) using Superfect transfection reagent (Qiagen). After 24 h, the cells were collected, and cell extracts were prepared as reported (32). Transient transfection of HEK293T and 293/TLR4/MD2 cells was carried out as reported previously (20, 26, 32). For NF-κB reporter assays, HEK293T or 293/TLR4/MD2 cells were plated in 24-well plates (105 cells/well), grown for 20 h, and transfected with the indicated amounts of expression vectors along with pELAM-luciferase (500 ng/well) and pTK-Renilla luciferase (50 ng/well), using Superfect transfection reagent (Qiagen) according to the manufacturer's protocol. After recovery and treatments, firefly luciferase and Renilla luciferase activities were measured in cell lysates as described (20, 26, 32).
Isolation of RNA and Real Time RT-PCR Analysis
Total RNA was isolated with RNeasy kits (Qiagen), using in-column digestion of residual genomic DNA, as recommended by the manufacturer. cDNA was prepared from 1 μg of total RNA using a reverse transcription system (Promega) and subjected to real time PCR with gene-specific primers for human hypoxanthine phosphoribosyltransferase (5′-ACCAGTCAACAGGGGACATAAAAG-3′, forward, and 5′-GTCTGCATTGTTTTGCCAGTGTC-3′, reverse), TNF-α (5′-CCCAGGCAGTCAGATCATCTTC-3′, forward, and 5′-GCTTGAGGGTTTGCTACAACATG-3′, reverse), A20 (5′-AACATTTTGCTGCTGCCTC-3′, forward, and 5′-AGGTGCTTTGTGTGGTTCG-3′, reverse) on a MyIQ real time PCR detection system (Bio-Rad). Purified and sequenced PCR products were used as standards, and the data were analyzed by the 2−ΔΔCT method (36).
Co-immunoprecipitation and Immunoblotting
Whole cell lysates (0.5–3 mg of total protein) were precleared with protein G-agarose beads (20 μl/sample; Roche Applied Science) for 4 h at 4 °C upon rotation. Precleared cell extracts were incubated overnight at 4 °C with 1 μg/ml of the respective Ab in lysis buffer containing 20 mm HEPES, pH 7.4, 1% Triton X-100, 150 mm NaCl, 12.5 mm β-glycerophosphate, 50 mm NaF, 1 mm DTT, 1 mm sodium orthovanadate, 2 mm EDTA, 1 mm PMSF, and protease inhibitor mixture (Roche Applied Science). For ubiquitination experiments, SDS was added to cell lysates to 1% concentration to disrupt noncovalent protein-protein interactions, and cell lysates were boiled for 5 min, cooled down, and diluted with lysis buffer to reduce SDS concentration to 0.1%, as published (15). Protein G-agarose beads were added (45 μl/sample), and incubation continued for 4 h. The beads were washed five times with lysis buffer and resuspended in Laemmli sample buffer (50 mm Tris-Cl, pH 6.8, 10% glycerol, 2% SDS, 0.1% bromphenol blue, 5% 2-mercaptoethanol). Immunoprecipitated proteins were separated on 4–20% mini-gels (Invitrogen), electrotransferred to Immobilon-P membranes, blocked, and probed with the respective Abs.
In Vitro Kinase Assays
In vitro kinase assays were performed as described (8, 25, 37, 38) using inactive MAPKK kinase 6 (MKK6, Millipore) and myelin basic protein (MBP, Sigma) as substrates for TAK1 and IRAK4, respectively. Endogenous IRAK4 or TAK1 proteins were immunoprecipitated with the respective Abs/protein G-agarose. Immune complexes were washed twice in a co-immunoprecipitation lysis buffer; washed three times in a kinase buffer containing 20 mm HEPES, pH 7.6, 50 mm NaCl, 10 mm MgCl2, 20 mm β-glycerophosphate, 1 mm sodium orthovanadate, 1 mm ATP; and then incubated for 25 min at 30 °C in a kinase reaction with 1.0 μg of the respective recombinant substrates, 1 mm ATP, and 5 μCi of [32P]ATP (PerkinElmer Life Sciences) made up to a total volume of 20 μl with kinase buffer. The samples were then resolved by SDS-PAGE, gels were dried, exposed to an x-ray film, and visualized by autoradiography.
shRNA Gene Knockdowns
A20 shRNA lentiviral particles, control shRNA lentiviral particles, co-pGFP lentiviral particles, puromycin dihydrochlorate, and polybrene were purchased from Santa Cruz. THP1 cells were incubated with lentiviral particles according to the manufacturer's protocol, followed by selection of A20 shRNA- or control shRNA-expressing cells, as well as copGFP-expressing transfectants (to monitor transfection efficiency) in cRPMI containing 5 μg/ml puromycin. The specificity of shRNA-mediated gene silencing was controlled by immunoprecipitation and immunoblot analyses of cell lysates obtained from A20 shRNA-, control shRNA-expressing, or GFP-expressing THP1 cells with A20 Ab. The effect of A20 and control shRNA gene knockdowns on endotoxin tolerization was studied by pretreatment of A20 shRNA- or control shRNA-transduced THP1 cells for 20 h with medium or 10 ng/ml LPS, followed by washing and addition of medium, 100 ng/ml LPS, or 100 ng/ml PMA. After 24 h, the supernatants were collected and assayed for TNF-α by ELISA.
Determination of Cytokine Production by ELISA
Levels of TNF-α in cell-free supernatants were determined by ELISA in the University of Maryland Baltimore Cytokine Core Laboratory using commercially paired antibodies and recombinant standards obtained from R & D (TNF-α), with the lower detection limit of 3.9 pg/ml.
Statistical Analysis
Statistical analysis was performed using the GraphPad Prism 5 program for Windows (GraphPad Software Inc., San Diego, CA). Statistical differences among experimental groups were evaluated by the Student's t test with the level of significance set at p < 0.05. The values are expressed as the means ± S.D.
RESULTS
Endotoxin Tolerance Impairs Recruitment of IRAK4 to TLR4 and IRAK4 Activation
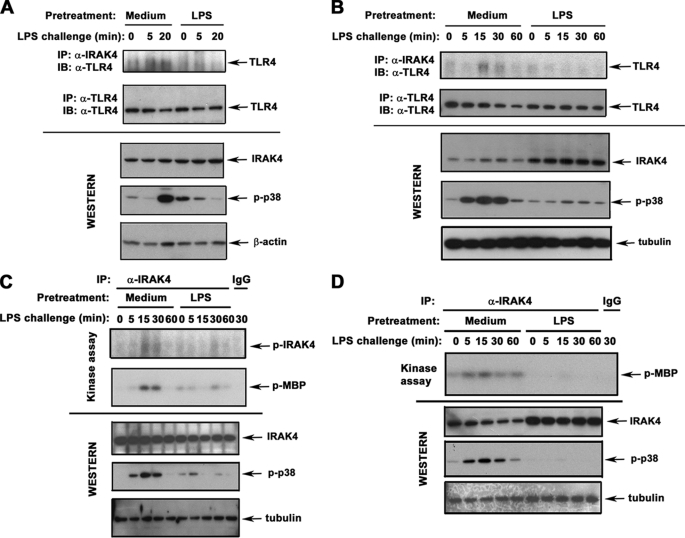
To examine whether endotoxin tolerance affects receptor recruitment, phosphorylation, and activation of IRAK4, we used THP1 cells differentiated with PMA to acquire the macrophage phenotype (35) and primary human monocytes. LPS induced IRAK4-TLR4 association in control THP1 cells and monocytes within 5–20 min, accompanied by p38 phosphorylation, whereas induction of endotoxin tolerance blunted LPS-induced IRAK4 recruitment to TLR4 and p38 activation (Fig. 1, A and B). To determine IRAK4 activation, IRAK4 proteins were immunoprecipitated from control and endotoxin-tolerant cells and subjected to in vitro kinase assays, using MBP as a substrate. Because of differences in molecular masses between IRAK4 (53 kDa) and MBP (19 kDa), it is possible to examine both parameters of IRAK4 activation in a single assay. LPS induced IRAK4 autophosphorylation and kinase activity in control THP1 cells and monocytes, whereas endotoxin-tolerant cells had significantly reduced responses (Fig. 1, C and D, and data not shown). Induction of endotoxin tolerance did not reduce total levels of TLR4 and IRAK4 in THP1 cells and monocytes (Fig. 1), indicating that inhibited LPS-inducible TLR4-IRAK4 associations and IRAK4 activation noted in endotoxin tolerance were not due to lower expression of TLR4 and IRAK4. Thus, endotoxin tolerization of THP1 cells and monocytes inhibits LPS-induced IRAK4 recruitment to TLR4 and IRAK4 activation.
FIGURE 1.
Endotoxin tolerization inhibits LPS-induced IRAK4 recruitment to TLR4 and IRAK4 activation. THP1 cells (A and C) and human monocytes (B and D) were pretreated for 20 h with medium or 10 ng/ml LPS. After washing, the cells were treated for the indicated time points with medium or 100 ng/ml LPS (LPS challenge). A and B, IRAK4 and TLR4 proteins were immunoprecipitated (IP) from whole cell lysates and subjected to immunoblot (IB) analyses with anti-TLR4 Ab, and total expression of IRAK4 proteins was examined by immunoblotting of cell lysates with α-IRAK4 Ab. To assess LPS-inducible cell activation, cell lysates were immunoblotted with anti-p-p38 Ab. β-Actin and tubulin immunoblots were used to control for protein loading. C and D, IRAK4 proteins were immunoprecipitated from cell lysates and subjected to in vitro kinase assays in the absence (IRAK4 autophosphorylation, top panels) or in the presence of MBP (IRAK4 kinase activity, middle panels). Whole cell lysates were immunoblotted with Abs against IRAK4, p-p38, and tubulin to determine total expression levels of IRAK4, LPS inducibility/tolerization, and protein loading, respectively. Shown are the results of a representative (n = 3) experiment.
Endotoxin Tolerization Compromises LPS-inducible K63-linked but Not K48-linked Polyubiquitination of IRAK1
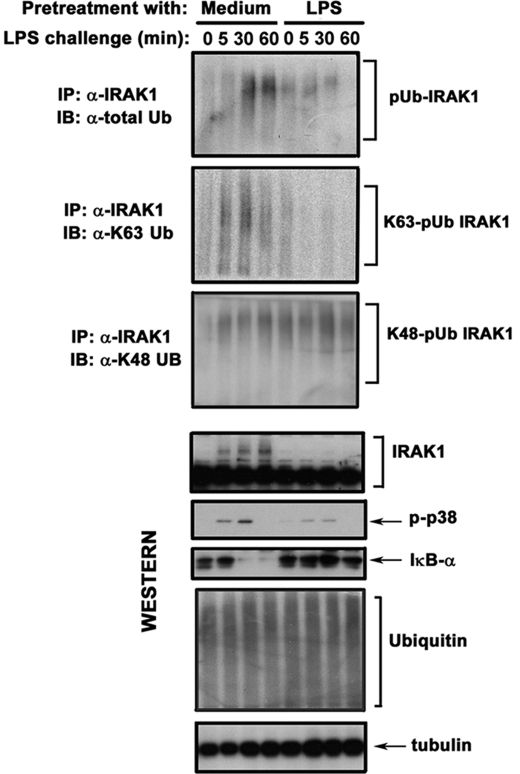
IRAK4 induces phosphorylation, polyubiquitination, and kinase activity of IRAK1 that recruits TRAF6 and IKKγ, leading to activation of TAK1 and IKKs (11–14, 39–42). Although K48-linked polyubiquitination is linked to proteasomal degradation, K63-linked polyubiquitination promotes assembly of signaling platforms and activation of protein functions (6). To determine the impact of endotoxin tolerance on LPS-inducible ubiquitination of IRAK1, we employed immunoblot analyses of immunoprecipitated IRAK1 with α-total Ub Ab or Abs discriminating K48- versus K63-linked polyUb chains. LPS challenge of medium-pretreated THP1 cells induced polyubiquitination of IRAK1 resulting from conjugation of both K48- and K63-linked polyUb chains (Fig. 2). LPS-tolerant THP1 cells (Fig. 2) and monocytes (data not shown) had reduced levels of IRAK1 proteins immunoreactive with Abs against total Ub, failed to respond to LPS by up-regulation of K63-linked polyubiquitination of IRAK1, but maintained its persistent K48-linked polyubiquitination. Comparable levels of total IRAK1 were observed in control and LPS-tolerant THP1 cells and monocytes (Figs. 2 and 3), indicating that differences in K63-linked polyubiquitination of IRAK1 were not due to variations in IRAK1 expression. Thus, endotoxin tolerance mitigates LPS-induced K63-linked polyubiquitination of IRAK1.
FIGURE 2.
Endotoxin-tolerant THP1 cells exhibit impaired LPS-mediated K63-linked polyubiquitination of IRAK1. THP1 cells were pretreated for 20 h with medium or 10 ng/ml LPS, washed, and incubated with medium or 100 ng/ml LPS as indicated. IRAK1 was immunoprecipitated (IP) from cell lysates, followed by immunoblot (IB) analyses with anti-total Ub Ab or Abs specific for K63-linked Ub and for K48-linked Ub chains. For controlling total IRAK1 levels and determining cell activation and endotoxin tolerance induction, IRAK-1 and p38 phosphorylation were examined by immunoblotting of whole cell lysates with the corresponding Abs. Tubulin immunoblot was carried out to control protein loading. The data of a representative experiment (n = 3) are shown.
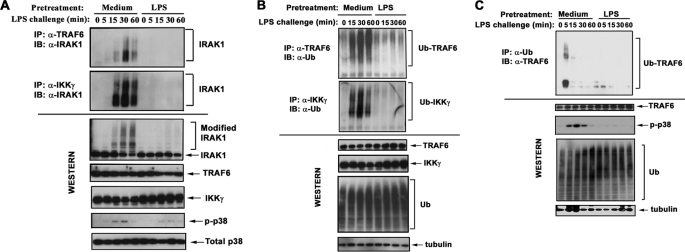
FIGURE 3.
Endotoxin tolerization impairs LPS-induced IRAK1 interactions with TRAF6 and IKKγ and compromises LPS-induced K63-linked polyubiquitination of TRAF6 and IKKγ. After exposure to medium or 10 ng/ml LPS for 20 h, THP1 cells or monocytes were washed and treated with medium or challenged with 100 ng/ml LPS for the indicated time course. A and B, TRAF6 and IKKγ proteins were immunoprecipitated (IP) and subjected to Western blot analyses with α-IRAK1 Ab (A) or α-Ub Ab (B). C, whole cell lysates of human monocytes were immunoprecipitated with α-Ub Ab, and the proportion of TRAF6 was detected by immunoblot (IB) analysis using α-TRAF6 Ab. A–C, whole cell lysates were examined by immunoblotting, using Abs against IRAK1, TRAF6, IKKγ, and Ub to determine their total protein levels and with α-p-p38 Ab to control for LPS inducibility and tolerization. Immunoblotting of total p38 or tubulin was performed to control protein loading. The results of a representative experiment (n = 3) are shown.
Induction of Endotoxin Tolerance Attenuates LPS-elicited IRAK1-TRAF6 and IRAK1-IKK-γ Assemblies and Inhibits K63-linked Polyubiquitination of TRAF6
Because post-translational modifications of IRAK1 are necessary for recruitment and activation of TRAF6 (6) and IKKγ (15), we addressed the impact of endotoxin tolerance on IRAK1 signalosome assembly, using immunoblot analysis of TRAF6 or IKKγ immunoprecipitates with α-IRAK1 Ab. Endotoxin-tolerant THP1 cells had deficient LPS-induced recruitment of TRAF6 and IKKγ to native or modified IRAK1, in contrast to robust recruitment observed in LPS-responsive cells (Fig. 3A). Comparable levels of unmodified IRAK1 and TRAF6 were seen in LPS-tolerant versus control THP1 cells, whereas IKKγ levels were even higher in LPS-tolerant cells (Fig. 3). Thus, deficient interactions of IRAK1 with TRAF6 and IKKγ in endotoxin-tolerant cells were not due to lower expression of the interacting components.
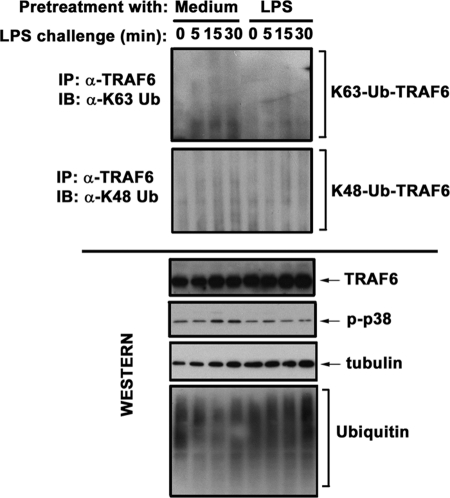
TRAF6 is an E3 Ub ligase activated via K63-linked autoubiquitination (13, 43–45), and conjugation of K63-linked polyUb moieties to IRAK1 promotes recruitment and K63-linked polyubiquitination of IKKγ (15, 46). Therefore, we examined the impact of endotoxin tolerization on the ubiquitination status of TRAF6 and IKKγ. LPS induced polyubiquitination of TRAF6 and IKKγ in medium-pretreated THP1 cells and monocytes, whereas endotoxin-tolerant cells failed to respond to LPS by polyubiquitination of TRAF6 and IKKγ (Fig. 3). To determine what type of ubiquitination is affected by LPS tolerization, we used Abs discriminating K48- versus K63-linked polyUb chains. LPS stimulation of control THP1 cells resulted in the appearance of modified TRAF6 bands immunoreactive with Ab specific for K63-linked polyUb, whereas conjugation of K48-linked Ub moieties was manifested to a significantly lower extent (Fig. 4). Induction of endotoxin tolerance only minimally affected the abundance of K48-linked polyubiquitinated TRAF6 but significantly decreased the abundance of K63-linked polyubiquitinated TRAF6 (Fig. 4). Immunoprecipitation and immunoblot analyses with Abs discriminating K48- and K63-linked polyUb revealed that endotoxin tolerization selectively inhibits K63-linked polyubiquitination of IKKγ without affecting K48-linked polyubiquitination (data not shown). Similar levels of total unmodified TRAF6, IKKγ, and Ub proteins were seen in LPS-responsive and -tolerant THP1 cells or monocytes (Figs. 3 and 4), indicating the suppressive effect of LPS tolerization on ubiquitination of these intermediates was not caused by a decrease in the abundance of total Ub or TRAF6. These data indicate that endotoxin tolerization of THP1 cells and monocytes disrupts LPS-induced IRAK1 interactions with TRAF6 and IKKγ, coinciding with compromised K63-linked polyubiquitination of TRAF6 and IKKγ.
FIGURE 4.
Induction of endotoxin tolerance inhibits K63-linked, but not K48-linked, polyubiquitination of TRAF6. THP1 cells were pretreated for 20 h with medium or 10 ng/ml LPS, washed and treated with medium or 100 ng/ml LPS for the indicated times. TRAF6 proteins were immunoprecipitated (IP) from whole cell lysates with α-TRAF6 Ab and immunoblotted (IB) with Abs discriminating K48- versus K63-linked polyubiquitin chains. Whole cell lysates were analyzed by immunoblotting using α-TRAF6, α-Ub, α-p-p38, and α-tubulin Abs to determine total expression of unmodified TRAF6, ubiquitin, and tubulin and to assess phosphorylation status of p38. Shown are the results of a representative (n = 3) experiment.
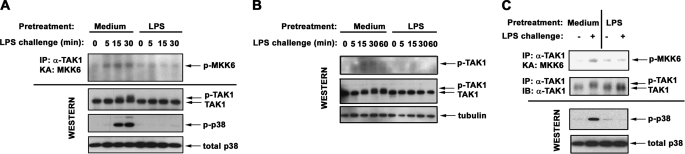
LPS Tolerization Suppresses TLR4-inducible TAK1 Activation
Before this study, it was unknown whether LPS tolerization affects TLR4-mediated activation of TAK1. Hence, we analyzed the impact of endotoxin tolerance on TLR4-mediated TAK1 phosphorylation that results in a shift in its electrophoretic mobility (39) and correlates with TAK1 kinase activation (39, 47). LPS led to the appearance of slower migrating species of TAK1 and induced TAK1 kinase activity in control THP1 cells within 15–30 min (Fig. 5A), whereas it failed to activate TAK1 in endotoxin-tolerant cells. Likewise, LPS induced slower migrating TAK1 bands in control monocytes, whereas no shift in TAK1 electrophoretic mobility was observed in endotoxin-tolerant monocytes challenged with LPS (Fig. 5, B and C). The inhibitory effect of endotoxin tolerance on LPS-mediated TAK1 phosphorylation was confirmed by immunoblot analyses of whole cell lysates with α-p-TAK1 Ab (p-Thr184/187) (Fig. 5B). Consistent with the role of TAK1 phosphorylation in induction of kinase activity (47), inhibited TLR4-inducible TAK1 phosphorylation observed in endotoxin-tolerant cells correlated with impaired TAK1 kinase activity and with ablated LPS-mediated phosphorylation of p38 (Fig. 5, A and C). Different activation of signaling intermediates was not due to variations in protein loading, as evidenced by equal levels of total p38 and tubulin seen in the samples analyzed (Fig. 5).
FIGURE 5.
Deficient TLR4-induced TAK1 activation in LPS-tolerant cells. THP1 cells (A and B) and human monocytes (C) were pretreated for 20 h with medium or tolerized with 10 ng/ml LPS. The cells were washed and exposed to medium or 100 ng/ml LPS for the indicated time points (A and B) or for 15 min (C). TAK1 proteins were immunoprecipitated (IP) and analyzed by in vitro kinase assay (KA), using inactive MKK6 as a substrate (A and C, top blots), or subjected to immunoblot (IB) analyses with anti-p-TAK1 Ab (B). Whole cell lysates were analyzed by immunoblotting with Abs against p-TAK1 and TAK1 to determine TAK1 phosphorylation and total levels. Immunoblot analyses with Abs against IκB-α and p-p38 were performed to assess LPS activation/tolerance induction, and Abs against total p38 or tubulin were used to control for protein loading. Shown are the data of a representative (n = 3) experiment.
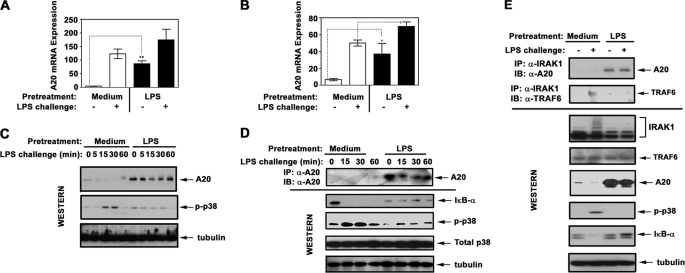
Induction of Endotoxin Tolerance in THP1 Cells and Human Monocytes Up-regulates A20 and Leads to Sustained A20-IRAK1 Association
Because A20 is a key deubiquitination enzyme that removes K63-linked polyUb chains from TRAF6 and IKKγ (30, 31, 48), we determined whether compromised K63-linked polyubiquitination of IRAK1, TRAF6, and IKKγ in endotoxin-tolerized cells correlates with increased levels of A20. LPS stimulation led to 75- and 9-fold induction of A20 mRNA in medium-pretreated THP1 cells and monocytes, respectively, which remained highly elevated in endotoxin-tolerized cells exhibiting reduced induction of TNF-α mRNA (Fig. 6, A and B and data not shown). To validate commercial α-A20 Ab for protein expression analyses, HEK293T cells were transfected with pcDNA3-HA-A20 or pcDNA3, and cell lysates were immunoprecipitated with α-HA or α-A20 Abs. Immunoblot analyses showed that the immune complexes from pcDNA3-HA-A20-transfected cells reacted with α-HA and α-A20 Abs and had identical electrophoretic mobilities, whereas no α-HA-immunoreactive bands were observed in pcDNA3-transfected samples (supplemental Fig. S1). Having validated α-A20 Ab, we examined the impact of endotoxin tolerance on A20 protein expression. Consistent with published data (48), LPS stimulation of control THP1 cells and human monocytes increased levels of A20 proteins (Fig. 6, C and D). Endotoxin tolerization suppressed LPS-mediated IκB-α degradation and p38 phosphorylation, which coincided with strong and sustained up-regulation of A20 protein levels (Fig. 6, C–E). Although LPS induced predominantly IRAK1-TRAF6 association in control THP1 cells, endotoxin-tolerant cells showed sustained and strong IRAK1-A20 interaction and negligible amounts of IRAK1-associated TRAF6 (Fig. 6E). Thus, endotoxin tolerance increases A20 expression and shifts the IRAK1 signalosome composition toward accumulation of IRAK1-A20 complexes.
FIGURE 6.
Induction of endotoxin tolerance increases A20 expression and shifts IRAK1 signalosome composition toward accumulation of IRAK1-A20 complexes. THP1 cells (A, C, and E) and human monocytes (B and D) were pretreated for 20 h with medium or 10 ng/ml LPS. Thereafter, the cells were washed and treated for the indicated time periods (A–D) or for 30 min (E) with medium or 100 ng/ml LPS. A and B, RNA was isolated, converted into cDNA, and analyzed by real time PCR with primers specific for hypoxanthine phosphoribosyltransferase and A20. C, A20 proteins were detected by immunoblot (IB) analyses of THP1 whole cell lysates with α-A20 Ab. D, A20 proteins were immunoprecipitated (IP) from whole cell lysates of monocytes with α-A20 Ab and subjected to Western blot analysis using α-A20 Ab. Whole cell lysates were fractionated by SDS-PAGE, electrotransferred onto PVDF membranes, and immunoblotted with Abs against p-p38 (to assess LPS-mediated cell activation/tolerization), total p38, and tubulin (to control for protein loading). The data of a representative experiment (n = 5) are depicted. E, IRAK1 proteins were immunoprecipitated from whole cell lysates and immunoblotted with Abs against A20 and TRAF6. Whole cell lysates were subjected to Western blot analysis with Abs against A20, TRAF6, and IRAK1 to determine their total levels. p-p38 IκB-α immunoblots were performed to examine cell activation/tolerance, and tubulin immunoblots were carried out to control protein loading. The results of a representative experiment are shown.
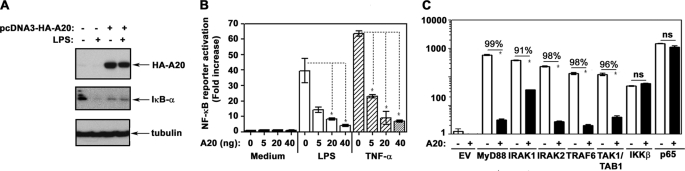
A20 Inhibits TLR4-inducible NF-κB Activation within the IRAK1/2-TRAF6-TAK1/TAB1 Signaling Axis
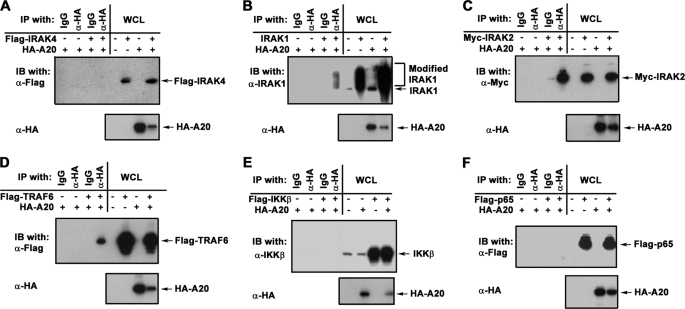
To discern functional significance of increased A20 expression in endotoxin-tolerant cells on TLR4 signaling, we studied the impact of A20 overexpression on LPS-mediated NF-κB activation. Overexpression of A20 in 293/TLR4/MD2 cells precluded LPS-mediated degradation of IκB-α (Fig. 7A) and dose-dependently inhibited NF-κB reporter activation in response to stimulation with LPS and TNF-α (Fig. 7B). Next, we overexpressed MyD88, IRAK1, IRAK2, TRAF6, TAK1, TAB1, IKKβ, and p65 in HEK293T cells to trigger NF-κB downstream of the respective transfected intermediates, with or without ectopic overexpression of HA-A20. A20 suppressed NF-κB reporter activation induced by MyD88, IRAK1, IRAK2, TAK1, and TAB1, whereas it fails to inhibit IKKβ- or p65-driven responses (Fig. 7C), analogous to a previous report (49). Co-immunoprecipitation showed A20 interactions with IRAK1, IRAK2, and TRAF6 but not with IRAK4, IKKβ, and p65 upon ectopic overexpression of the studied intermediates in HEK293T cells (Fig. 8). Our results indicate that increased levels of A20 inhibit TLR4-mediated activation of NF-κB within the IRAK1/2-TRAF6-TAK1 signaling cascade but do not affect IKKβ- and p65-driven NF-κB transactivation.
FIGURE 7.
A20 overexpression inhibits TLR4-inducible NF-κB activation within the MyD88-IRAK1-TRAF6-TAK1 axis but does not affect IKKβ- and p65-elicited responses. A, 293/TLR4/MD2 cells were transiently transfected with pcDNA3 or pcDNA3-HA-A20 (5 μg/dish each). After recovery, the cells were treated for 30 min with medium or 100 ng/ml LPS, and the cell extracts were prepared and immunoblotted with α-HA, α-IκB-α, and α-tubulin Abs. B, 293/TLR4/MD2 cells were transfected with pELAM-luciferase (500 ng/well) and pTK-RL (50 ng/well) in the presence of the indicated amounts of pcDNA3-HA-A20 or pcDNA3. After recovery, the cells were treated for 5 h with medium, LPS, or TNF-α (100 ng/ml each). C, HEK293T cells were transfected with pELAM-luciferase, pTK-RL alone, or in combination with the plasmids encoding AU1-MyD88, IRAK1, IRAK2, TRAF6, TAK1, TAB1, IKKβ (10 ng/well each), or p65 (1 ng/well) in the absence (empty vector) or in the presence of pcDNA3-HA-A20 (100 ng/well). After incubation, the cell lysates were prepared, and firefly versus Renilla luciferase activities were measured. The results (mean ± S.D.) of a representative experiment (n = 3) are shown. *, p < 0.05. The numbers above the lines indicate percentages of inhibition.
FIGURE 8.
A20 interacts with IRAK2, IRAK2, and TRAF6 but not with IRAK4, IKKβ and p65 upon ectopic overexpression. HEK293T cells were grown in six-well plates and transfected with pcDNA3-HA-A20 (1 μg/well) alone or in the presence of one of the following vectors: pRK5-IRAK1 (0.5 μg/well), pRK7-FLAG-IRAK4, pcDNA3-Myc-IRAK2, pcDNA3-FLAG-TRAF6, pCMV1-FLAG-TAK1, pcDNA3-HA-TAB1, and pcDNA3-FLAG-p65 (1 μg/well each) as shown (A–F). The total levels of the plasmids were adjusted to 2 μg/well, using pcDNA3 empty vector. After recovery for 24 h, whole cell lysates (WCL) were immunoprecipitated (IP) and immunoblotted (IB) as indicated. Shown are the results of a representative (n = 3) experiment.
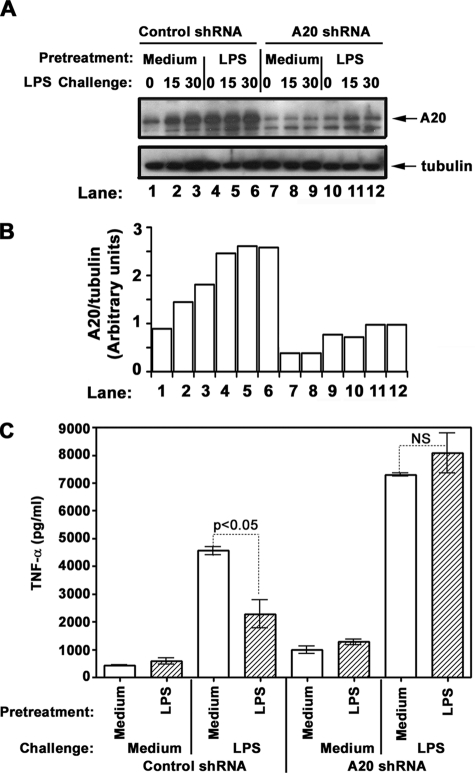
A20 Gene Knockdown Reverses Endotoxin Tolerization
To further delineate the role for A20 in endotoxin tolerance, we examined the impact of A20 deficiency on the capacity of cells to become endotoxin-tolerant. THP1 cells were transduced with lentiviral particles encoding A20 shRNA, control shRNA, or GFP; selected with puromycin; and analyzed for LPS tolerization/inducibility, using TNF-α secretion as a readout. Transduction with lentiviral particles encoding GFP yielded 83% GFP-expressing THP1 cells, indicating high transduction efficiency (supplemental Fig. S2). A20 shRNA knockdown significantly reduced A20 expression (Fig. 9, A and B), elevated basal TNF-α levels, and increased TNF-α secretion in response to LPS or PMA (Fig. 9C), consistent with the function of A20 as a negative regulator (48). Prior LPS exposure of THP1 cells transduced with control shRNA led to 51% reduction of TNF-α levels elicited by LPS challenge (Fig. 9C), coinciding with significant up-regulation of A20 (Fig. 9, A and B). Notably, LPS failed to induce endotoxin tolerance in A20 shRNA-transduced THP1 cells, as judged by comparable LPS-inducible TNF-α production observed in cells pretreated with medium versus tolerized with LPS (Fig. 9C). PMA stimulation caused comparable production of TNF-α in medium-treated and LPS-tolerized THP1 cells transduced with control or A20 shRNA (supplemental Fig. S3), indicating the TLR specificity of endotoxin tolerization procedure and the restriction of A20 negative effects to TLR signaling intermediates in our experimental settings. These data indicate that A20 gene silencing abolishes induction of endotoxin tolerance, providing a mechanistic link for A20 as an important regulator involved in endotoxin tolerance induction.
FIGURE 9.
A20 knockdown abolishes LPS tolerance induction in THP1 cells. THP1 cells were transduced with lentiviral particles encoding copGFP, control shRNA, or A20 shRNA and stable cell pools were selected with puromycin. Thereafter, the cells were pretreated for 20 h with medium or 10 ng/ml LPS, washed, and exposed to medium or 100 ng/ml LPS for the indicated time points (A) or treated for 24 with medium, 100 ng/ml LPS or 100 ng/ml PMA (C). A, protein levels of A20 and tubulin were determined by immunoblot analyses of whole cell lysates with the respective Abs. Similar results were obtained in other two independent experiments. B, densitometric quantification of A20 protein levels normalized to tubulin levels. C, supernatants were collected, and the levels of secreted TNF-α were analyzed by ELISA in triplicate samples. The data of a representative experiment (means ± S.D.) of a total of three experiments are shown.
DISCUSSION
Endotoxin tolerance is signified by decreased activation of molecules exerting positive regulation of TLR signaling, e.g. deficient tyrosine phosphorylation of TLR4 and Mal (26, 32), recruitment of MyD88 and TRIF to TLR4 (20, 25), and activation of IRAK1 and TBK1 (20, 25–27, 50, 51). Enhanced expression of negative regulators, such as Toll-interacting protein, suppressor of cytokine signaling-1, IRAK-M, SH2-containing inositol-5′-phosphatase-1, sterile α and HEAT-Armadillo motifs-containing adapter (SARM), and suppressor of IκB-ϵ also underlies tolerance (20, 28, 29, 52–54). In this paper, we present new findings related to these two important mechanisms of endotoxin tolerance. We report that endotoxin tolerization of THP1 cells and human monocytes inhibits LPS-induced recruitment of IRAK4 to TLR4 and IRAK4 activation. Because IRAK4 activates IRAK1 (6, 8, 10, 12), impaired LPS-inducible IRAK4 activation in endotoxin-tolerant cells is likely to account for tolerance-associated, impaired phosphorylation of IRAK1, and suppressed induction of IRAK1 kinase activity (25, 27). Deficiencies in IRAK4-TLR4 assembly and IRAK4 activation were not due to a decrease in total levels of TLR4 and IRAK4, indicating that decreased IRAK4 expression is not the primary mechanism. Our data support earlier observations of sustained IRAK4 expression found in mouse macrophages and HEK293 cells stimulated via IL-1R or TLRs (12, 40, 55, 56) but differ with another report on decreased IRAK4 levels in LPS-tolerant mouse macrophages (57). It is plausible that altered tyrosine phosphorylation of TLR4 and Mal caused by endotoxin tolerization (26, 32) impairs assembly of TLR4-Mal docking platforms and interferes with recruitment of MyD88. MyD88 recruitment to TLR4 enables IRAK4 and IRAK1 to interact with the TLR4-Mal-MyD88 complex to assemble the “Myddosome signaling tower” (7, 9). Indeed, induction of endotoxin tolerance in monocytes decreases LPS-induced MyD88 recruitment to TLR4 (25) and impairs MyD88-IRAK1 association (27). Thus, interference with MyD88 assembly would be expected to underlie impaired recruitment and activation of IRAK4.
We demonstrate that induction of endotoxin tolerance ablates LPS-inducible K63-linked polyubiquitination of IRAK1 without affecting K48-linked polyubiquitination. Although K48-linked polyubiquitination targets IRAK1 for proteasomal degradation and provides a negative feedback loop, K63-linked polyubiquitination of IRAK1 promotes its associations with TRAF6 and IKKγ, positively regulating TLR4 signaling (45). Preferential K63-linked polyubiquitination of IRAK1 and only weak K48-linked polyubiquitination revealed in this paper support similar findings in mouse macrophages and THP1 cells reported by other groups (15, 58). Even though IRAK1 may be degraded upon a longer LPS stimulation in control cells (59), we observed comparable expression of unmodified IRAK1 in LPS-tolerant and control monocytes, suggesting that IRAK1 undergoes resynthesis in LPS-tolerant cells. Likewise, induction of endotoxin tolerance in mice in vivo with a low tolerizing dose of LPS did not result in IRAK1 degradation (52), supporting our findings in human monocytes. We observed a similar extent of inhibition of LPS-inducible TNF-α mRNA expression in endotoxin-tolerant wild-type and IRAK1−/− immortalized bone marrow-derived macrophages,5 consistent with an earlier report (60). These data highlight the importance of deficient K63-linked polyubiquitination of IRAK1 but not IRAK1 degradation in the induction of endotoxin tolerance.
This paper reveals that deficiency in IRAK1 post-translational modifications underlies decreased interactions of TRAF6 and IKKγ with K63-polyubiquitinated IRAK1 in endotoxin-tolerized THP1 cells and monocytes. Our data support the prerequisite role for phosphorylation and K63-linked polyubiquitination of IRAK1 in promoting IRAK1 assembly with TRAF6 and IKKγ reported previously (15, 46, 61). IRAK1-TRAF6 interactions are important in the induction of E3 Ub ligase activity of TRAF6 and its K63-linked polyubiquitination (39, 62). Similar to the IRAK1 ubiquitination patterns, we found that endotoxin tolerization inhibits only LPS-inducible K63-linked polyubiquitination of TRAF6 but does not affect the weak K48-linked polyubiquitination response and does not reduce total TRAF6 levels. Because TRAF-interacting protein with a forkhead-associated domain (TIFA) facilitates oligomerization and K63-linked polyubiquitination of TRAF6 (63), we cannot exclude the impact of endotoxin tolerance on a TIFA-dependent mechanism involving altered expression or activities of TIFA.
We report inhibited LPS-inducible TAK1 phosphorylation and activation in endotoxin-tolerant THP1 cells and human monocytes, consistent with ablated TLR4-mediated K63-linked polyubiquitination of TRAF6 and the importance of recognition of K63-polyubiquitinated TRAF6 by TAB2/TAB3/TAK1 complex in mediating in TAK1 activation. TAK1 triggers the MAPK and IKK signaling cascades (64, 65), and inhibition of TAK1 may account for severely suppressed MAPK and NF-κB activation found in tolerant cells (20, 25, 26, 32, 66). Because two pathways of NF-κB activation have been described, TAK1- and MEKK3-dependent (55, 58), decreased TAK1-dependent NF-κB activation may shift the equilibrium toward preferential activation of the MEKK3-driven NF-κB pathway. The MEKK3-dependent pathway has nonredundant aspects in NF-κB activation compared with the TAK1-dependent pathway (67) and may underlie increased expression of RelB and p50 found in LPS-tolerant macrophages (18, 24, 68, 69). Studies are underway to delineate whether deficient TAK1 activation in endotoxin-tolerant monocytes signifies increased involvement of the MEKK3 pathway, leading to differential induction of the NF-κB subunits and inflammatory genes.
This paper reveals that increased expression of A20 is essential for the induction of endotoxin tolerance in THP1 cells and human monocytes. A20 negatively regulates TLR and TNFR signaling pathways by removing K63-linked Ub chains from its substrates TRAF6, receptor-interacting protein 1 (RIP-1), and IKKγ (30, 31, 48). Induction of endotoxin tolerance in THP1 cells and human monocytes coincides with significant and sustained increases in A20 mRNA and protein compared with those seen in control cells. To mimic increased A20 levels in endotoxin-tolerized cells, we overexpressed A20 in 293/TLR4/MD2 cells and found that it inhibits LPS-inducible IκB-α degradation and NF-κB reporter activation. Furthermore, we show that A20 gene silencing in THP1 cells prevents endotoxin tolerization, providing a mechanistic link between elevated A20 expression and induction of endotoxin tolerance in human macrophages. Previous studies on the role of A20 in endotoxin tolerance using mouse macrophages and human intestinal epithelial cells have yielded controversial results. Boone et al. (48) reported only slightly elevated levels of IL-6 upon LPS challenge of A20−/− mouse macrophages pretreated with LPS compared with the response of endotoxin-tolerant wild-type cells, arguing that A20 is not involved in endotoxin tolerance. Oshima et al. (70) found that A20 gene knockdown in human intestinal epithelial cell lines fails to affect tolerance induction to flagellin. In contrast, Wang et al. (71) found that endotoxin tolerization of primary human entherocytes elevates levels of A20 mRNA and protein and A20 gene knockdown completely abrogates endotoxin tolerization. The differences between our results and data by Boone et al. (48) could be due to species-specific differences between mouse and human cells. This is a known feature reported previously, e.g. with regard to different functions for SARM. SARM functions as a negative TRIF regulator in humans, whereas no apparent role for SARM in TLR signaling was observed in mice where SARM is expressed in neurons and acts by activating cell death upon stress (6, 72, 73). The reason for a striking difference in A20 functions in mediating TLR5 (flagellin) and TLR4 (LPS) tolerance in intestinal epithelial cells is unknown but could be related to different TLRs studied, as well as differences between intestinal epithelial cell lines and primary entherocytes. Our data uncover a principal role for A20 in endotoxin tolerizaton of THP1 cells and human monocytes, supporting previous data on A20 involvement in LPS tolerance in human entherocytes (71).
Since A20 removes K63-linked polyUb chains from TRAF6 (30, 31), increased A20 expression in LPS-tolerant THP1 cells and human monocytes could account for decreased TRAF6 activation. Because recognition of K63-linked Ub moieties on TRAF6 by the TAB2/TAB3/TAK1 complex promotes TAK1 activation (2, 3, 6), A20-mediated TRAF6 deubiquitination is likely to contribute to impaired TAK1 activation in endotoxin-tolerized THP1 cells and human monocytes. K63-polyubiquitinated RIP-1 and IRAK1 share a similar function to directly recruit IKKγ in the case of TNFR and TLR signaling, respectively. Therefore, because A20 deubiquitinates RIP-1, IRAK1 was suggested as a target for A20 (31). Interestingly, we demonstrate that induction of endotoxin tolerance not only increases total levels of A20 but also shifts the composition of IRAK1 signalosome toward accumulation of IRAK1-A20 complexes, with a significant decrease in amounts of IRAK1-TRAF6. This paper shows that A20 inhibits NF-κB reporter activation driven by overexpressed MyD88, IRAK1/2, TRAF6, and TAK1/TAB1 while not affecting IKKβ- and p65-mediated responses. Our data reveal a new feature of A20 to associate with IRAK1 signaling complexes and confirm its ability to interact with TRAF6, but not with IKKβ, as reported previously (31, 49). A20 associates with IKKγ and TRAF6 (31, 49, 74), and because these intermediates interact with modified IRAK1, one can suggest that A20-IRAK1 associations could be indirect because of the presence of TRAF6 and/or IKKγ in the complex. However, very low, if any, amounts of TRAF6 and IKKγ were associated with IRAK1 in endotoxin-tolerant THP1 cells and monocytes, suggesting direct A20 interactions with IRAK1. Studies are in progress to substantiate whether A20 directly interacts with K63-polyubiquitinated IRAK1, leading to removal of K63-linked polyUb, or interferes with IRAK1 associations with Pellinos, altering Pellino-mediated ubiquitination of IRAK1. A20 interacts with and removes K63-linked polyUb from IKKγ, inhibiting NF-κB activation (49). This phenomenon could account for the ability of A20 to inhibit TAK1-driven NF-κB activation shown in ectopic overexpression experiments herein and in a previous report (75), suggesting an additional negative feedback mechanism operating in endotoxin tolerance.
In summary, we provide several new findings to support the notion that deficient LPS-inducible recruitment, post-translational modifications, and activation of proximal kinases and adapters, as well as increased expression of negative TLR4 regulators are central in endotoxin tolerance. These include suppressed recruitment and activation of IRAK4, ablated K63-linked polyubiquitination of IRAK1 and TRAF6 translated into deficient IRAK1-TRAF6 and IRAK1-IKKγ interactions and suppressed TAK1 activation. This paper reveals increased A20 expression and strong, sustained A20-IRAK1 association in endotoxin-tolerant THP1 cells and monocytes. It provides a mechanistic insight into an essential role of A20 in endotoxin tolerization of THP1 cells by showing reversal of the LPS-tolerant phenotype by A20 shRNA gene knockdown. Studies are in progress to further determine mechanisms underlying reprogramming of the TLR4 pathway in endotoxin tolerance.
Supplementary Material
Acknowledgments
We are grateful to Drs. Katherine Fitzgerald, Douglas T. Golenbock (University of Massachusetts Medical School, Worcester, MA), Jun Ninomiya-Tsuji (North Carolina State University, Raleigh, NC), and Antonio Leonardi (University of Naples, Naples, Italy) for providing us with stable cell lines and expression vectors.
This work was supported, in whole or in part, by National Institutes of Health Grant AI-059524 (to A. E. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Y. Xiong and A. Medvedev, unpublished observation.
- TLR
- Toll-like receptor
- PAMPs
- pathogen-associated molecular patterns
- TIR
- Toll-IL-1 receptor
- MyD
- myeloid differentiation response gene
- TRIF
- TIR domain containing adapter-inducing interferon
- IRAK
- IL-1 receptor-associated kinase
- TAK
- TGF-β-activated kinase
- IKK
- IκB kinase
- TRAF
- TNF receptor-associated factor
- TBK
- TRAF-associated NF-κB activator-binding kinase
- Ab
- antibody
- Ub
- ubiquitin
- p
- phospho
- SARM
- sterile α and HEAT-Armadillo motifs-containing adapter
- TIFA
- TRAF-interacting protein with a forkhead-associated domain
- TAB
- TAK-binding protein
- PMA
- phorbol 12-myristate 13-acetate
- MKK6
- MAP kinase kinase 6
- MBP
- myelin basic protein
- RIP-1
- receptor-interacting protein 1.
REFERENCES
- 1. Medzhitov R., Janeway C. A., Jr. (1997) Cell 91, 295–298 [DOI] [PubMed] [Google Scholar]
- 2. O'Neill L. A. (2006) Curr. Opin. Immunol. 18, 3–9 [DOI] [PubMed] [Google Scholar]
- 3. Beutler B. (2009) Immunol. Rev. 227, 248–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kawai T., Akira S. (2010) Nat. Immunol. 11, 373–384 [DOI] [PubMed] [Google Scholar]
- 5. O'Neill L. A., Bowie A. G. (2007) Nat. Rev. Immunol. 7, 353–364 [DOI] [PubMed] [Google Scholar]
- 6. O'Neill L. A. (2008) Immunity 29, 12–20 [DOI] [PubMed] [Google Scholar]
- 7. Motshwene P. G., Moncrieffe M. C., Grossmann J. G., Kao C., Ayaluru M., Sandercock A. M., Robinson C. V., Latz E., Gay N. J. (2009) J. Biol. Chem. 284, 25404–25411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li S., Strelow A., Fontana E. J., Wesche H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5567–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin S. C., Lo Y. C., Wu H. (2010) Nature 465, 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheng H., Addona T., Keshishian H., Dahlstrand E., Lu C., Dorsch M., Li Z., Wang A., Ocain T. D., Li P., Parsons T. F., Jaffee B., Xu Y. (2007) Biochem. Biophys. Res. Commun. 352, 609–616 [DOI] [PubMed] [Google Scholar]
- 11. Qian Y., Commane M., Ninomiya-Tsuji J., Matsumoto K., Li X. (2001) J. Biol. Chem. 276, 41661–44167 [DOI] [PubMed] [Google Scholar]
- 12. Kawagoe T., Sato S., Matsushita K., Kato H., Matsui K., Kumagai Y., Saitoh T., Kawai T., Takeuchi O., Akira S. (2008) Nat. Immunol. 9, 684–691 [DOI] [PubMed] [Google Scholar]
- 13. Keating S. E., Maloney G. M., Moran E. M., Bowie A. G. (2007) J. Biol. Chem. 282, 33435–33443 [DOI] [PubMed] [Google Scholar]
- 14. Takaesu G., Ninomiya-Tsuji J., Kishida S., Li X., Stark G. R., Matsumoto K. (2001) Mol. Cell Biol. 21, 2475–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Conze D. B., Wu C. J., Thomas J. A., Landstrom A., Ashwell J. D. (2008) Mol. Cell Biol. 28, 3538–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kagan J. C., Su T., Horng T., Chow A., Akira S., Medzhitov R. (2008) Nat. Immunol. 9, 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., Maniatis T. (2003) Nat. Immunol. 4, 491–496 [DOI] [PubMed] [Google Scholar]
- 18. McCall C. E., Yoza B. K. (2007) Am. J. Respir. Crit. Care Med. 175, 763–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Foster S. L., Hargreaves D. C., Medzhitov R. (2007) Nature 447, 972–978 [DOI] [PubMed] [Google Scholar]
- 20. Piao W., Song C., Chen H., Diaz M. A., Wahl L. M., Fitzgerald K. A., Li L., Medvedev A. E. (2009) J. Leukocyte Biol. 86, 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Medvedev A. E., Sabroe I., Hasday J. D., Vogel S. N. (2006) J. Endotoxin Res. 12, 133–150 [DOI] [PubMed] [Google Scholar]
- 22. Shnyra A., Brewington R., Alipio A., Amura C., Morrison D. C. (1998) J. Immunol. 160, 3729–3736 [PubMed] [Google Scholar]
- 23. Dobrovolskaia M. A., Medvedev A. E., Thomas K. E., Cuesta N., Toshchakov V., Ren T., Cody M. J., Michalek S. M., Rice N. R., Vogel S. N. (2003) J. Immunol. 170, 508–519 [DOI] [PubMed] [Google Scholar]
- 24. Medvedev A. E., Kopydlowski K. M., Vogel S. N. (2000) J. Immunol. 164, 5564–5574 [DOI] [PubMed] [Google Scholar]
- 25. Medvedev A. E., Lentschat A., Wahl L. M., Golenbock D. T., Vogel S. N. (2002) J. Immunol. 169, 5209–5216 [DOI] [PubMed] [Google Scholar]
- 26. Medvedev A. E., Piao W., Shoenfelt J., Rhee S. H., Chen H., Basu S., Wahl L. M., Fenton M. J., Vogel S. N. (2007) J. Biol. Chem. 282, 16042–16053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li L., Cousart S., Hu J., McCall C. E. (2000) J. Biol. Chem. 275, 23340–23345 [DOI] [PubMed] [Google Scholar]
- 28. Kobayashi K., Hernandez L. D., Galán J. E., Janeway C. A., Jr., Medzhitov R., Flavell R. A. (2002) Cell 110, 191–202 [DOI] [PubMed] [Google Scholar]
- 29. Sly L. M., Rauh M. J., Kalesnikoff J., Song C. H., Krystal G. (2004) Immunity 21, 227–239 [DOI] [PubMed] [Google Scholar]
- 30. Shembade N., Ma A., Harhaj E. W. (2010) Science 327, 1135–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Coornaert B., Carpentier I., Beyaert R. (2009) J. Biol. Chem. 284, 8217–8221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Piao W., Song C., Chen H., Wahl L. M., Fitzgerald K. A., O'Neill L. A., Medvedev A. E. (2008) J. Biol. Chem. 283, 3109–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Medvedev A. E., Thomas K., Awomoyi A., Kuhns D. B., Gallin J. I., Li X., Vogel S. N. (2005) J. Immunol. 174, 6587–6591 [DOI] [PubMed] [Google Scholar]
- 34. Geleziunas R., Ferrell S., Lin X., Mu Y., Cunningham E. T., Jr., Grant M., Connelly M. A., Hambor J. E., Marcu K. B., Greene W. C. (1998) Mol. Cell Biol. 18, 5157–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martin M., Katz J., Vogel S. N., Michalek S. M. (2001) J. Immunol. 167, 5278–5285 [DOI] [PubMed] [Google Scholar]
- 36. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 37. Sun J., Fegan P. E., Desai A. S., Madara J. L., Hobert M. E. (2007) Am. J. Physiol. Gastrointest Liver Physiol. 292, G767–G778 [DOI] [PubMed] [Google Scholar]
- 38. Kajino T., Ren H., Iemura S., Natsume T., Stefansson B., Brautigan D. L., Matsumoto K., Ninomiya-Tsuji J. (2006) J. Biol. Chem. 281, 39891–39896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jiang Z., Ninomiya-Tsuji J., Qian Y., Matsumoto K., Li X. (2002) Mol. Cell Biol. 22, 7158–7167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim T. W., Staschke K., Bulek K., Yao J., Peters K., Oh K. H., Vandenburg Y., Xiao H., Qian W., Hamilton T., Min B., Sen G., Gilmour R., Li X. (2007) J. Exp. Med. 204, 1025–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kollewe C., Mackensen A. C., Neumann D., Knop J., Cao P., Li S., Wesche H., Martin M. U. (2004) J. Biol. Chem. 279, 5227–5236 [DOI] [PubMed] [Google Scholar]
- 42. Li X., Commane M., Jiang Z., Stark G. R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4461–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lamothe B., Besse A., Campos A. D., Webster W. K., Wu H., Darnay B. G. (2007) J. Biol. Chem. 282, 4102–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang K., Zhu J., Sun S., Tang Y., Zhang B., Diao L., Wang C. (2004) Biochem. Biophys. Res. Commun. 324, 432–439 [DOI] [PubMed] [Google Scholar]
- 45. Skaug B., Jiang X., Chen Z. J. (2009) Annu. Rev. Biochem. 78, 769–796 [DOI] [PubMed] [Google Scholar]
- 46. Windheim M., Stafford M., Peggie M., Cohen P. (2008) Mol. Cell Biol. 28, 1783–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kishimoto K., Matsumoto K., Ninomiya-Tsuji J. (2000) J. Biol. Chem. 275, 7359–7364 [DOI] [PubMed] [Google Scholar]
- 48. Boone D. L., Turer E. E., Lee E. G., Ahmad R. C., Wheeler M. T., Tsui C., Hurley P., Chien M., Chai S., Hitotsumatsu O., McNally E., Pickart C., Ma A. (2004) Nat. Immunol. 5, 1052–1060 [DOI] [PubMed] [Google Scholar]
- 49. Mauro C., Pacifico F., Lavorgna A., Mellone S., Iannetti A., Acquaviva R., Formisano S., Vito P., Leonardi A. (2006) J. Biol. Chem. 281, 18482–18488 [DOI] [PubMed] [Google Scholar]
- 50. Li C. H., Wang J. H., Redmond H. P. (2006) J. Leukocyte Biol. 79, 867–875 [DOI] [PubMed] [Google Scholar]
- 51. Sato S., Takeuchi O., Fujita T., Tomizawa H., Takeda K., Akira S. (2002) Int. Immunol. 14, 783–791 [DOI] [PubMed] [Google Scholar]
- 52. van 't Veer C., van den Pangaart P. S., van Zoelen M. A., de Kruif M., Birjmohun R. S., Stroes E. S., de Vos A. F., van der Poll T. (2007) J. Immunol. 179, 7110–7120 [DOI] [PubMed] [Google Scholar]
- 53. Kinjyo I., Hanada T., Inagaki-Ohara K., Mori H., Aki D., Ohishi M., Yoshida H., Kubo M., Yoshimura A. (2002) Immunity 17, 583–591 [DOI] [PubMed] [Google Scholar]
- 54. Nakagawa R., Naka T., Tsutsu H. I., Fujimoto M., Kimura A., Abe T., Seki E., Sato S., Takeuchi O., Takeda K., Akira S., Yamanishi K., Kawase I., Nakanishi K., Kishimoto T. (2002) Immunity 17, 677–687 [DOI] [PubMed] [Google Scholar]
- 55. Qin J., Yao J., Cui G., Xiao H., Kim T. W., Fraczek J., Wightman P., Sato S., Akira S., Puel A., Casanova J. L., Su B., Li X. (2006) J. Biol. Chem. 281, 21013–21021 [DOI] [PubMed] [Google Scholar]
- 56. Qin J., Jiang Z., Qian Y., Casanova J. L., Li X. (2004) J. Biol. Chem. 279, 26748–26753 [DOI] [PubMed] [Google Scholar]
- 57. De Nardo D., Nguyen T., Hamilton J. A., Scholz G. M. (2009) Cell Signal. 21, 246–252 [DOI] [PubMed] [Google Scholar]
- 58. Xiao H., Qian W., Staschke K., Qian Y., Cui G., Deng L., Ehsani M., Wang X., Qian Y. W., Chen Z. J., Gilmour R., Jiang Z., Li X. (2008) J. Biol. Chem. 283, 14654–14664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hu J., Jacinto R., McCall C., Li L. (2002) J. Immunol. 168, 3910–3914 [DOI] [PubMed] [Google Scholar]
- 60. Berglund M., Thomas J. A., Hörnquist E. H., Hultgren O. H. (2008) Scand J. Immunol. 67, 473–479 [DOI] [PubMed] [Google Scholar]
- 61. Abbott D. W., Yang Y., Hutti J. E., Madhavarapu S., Kelliher M. A., Cantley L. C. (2007) Mol. Cell Biol. 27, 6012–6025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dong W., Liu Y., Peng J., Chen L., Zou T., Xiao H., Liu Z., Li W., Bu Y., Qi Y. (2006) J. Biol. Chem. 281, 26029–26040 [DOI] [PubMed] [Google Scholar]
- 63. Ea C. K., Sun L., Inoue J., Chen Z. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15318–15323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sato S., Sanjo H., Takeda K., Ninomiya-Tsuji J., Yamamoto M., Kawai T., Matsumoto K., Takeuchi O., Akira S. (2005) Nat. Immunol. 6, 1087–1095 [DOI] [PubMed] [Google Scholar]
- 65. Shim J. H., Xiao C., Paschal A. E., Bailey S. T., Rao P., Hayden M. S., Lee K. Y., Bussey C., Steckel M., Tanaka N., Yamada G, Akira S., Matsumoto K., Ghosh S. (2005) Genes Dev. 19, 2668–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nomura F., Akashi S., Sakao Y., Sato S., Kawai T., Matsumoto M., Nakanishi K., Kimoto M., Miyake K., Takeda K., Akira S. (2000) J. Immunol. 164, 3476–3479 [DOI] [PubMed] [Google Scholar]
- 67. Yao J., Kim T. W., Qin J., Jiang Z., Qian Y., Xiao H., Lu Y., Qian W., Gulen M. F., Sizemore N., DiDonato J., Sato S., Akira S., Su B., Li X. (2007) J. Biol. Chem. 282, 6075–6089 [DOI] [PubMed] [Google Scholar]
- 68. El Gazzar M., Yoza B. K., Hu J. Y., Cousart S. L., McCall C. E. (2007) J. Biol. Chem. 282, 26857–26864 [DOI] [PubMed] [Google Scholar]
- 69. Ziegler-Heitbrock H. W., Wedel A., Schraut W., Ströbel M., Wendelgass P., Sternsdorf T., Bäuerle P. A., Haas J. G., Riethmüller G. (1994) J. Biol. Chem. 269, 17001–17004 [PubMed] [Google Scholar]
- 70. Oshima N., Ishihara S., Rumi M. A., Aziz M. M., Mishima Y., Kadota C., Moriyama I., Ishimura N., Amano Y., Kinoshita Y. (2010) Clin. Exp. Immunol. 159, 185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang J., Ouyang Y., Guner Y., Ford H. R., Grishin A. V. (2009) J. Immunol. 183, 1384–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Carty M., Goodbody R., Schröder M., Stack J., Moynagh P. N., Bowie A. G. (2006) Nat. Immunol. 7, 1074–1081 [DOI] [PubMed] [Google Scholar]
- 73. Kim Y., Zhou P., Qian L., Chuang J. Z., Lee J., Li C., Iadecola C., Nathan C., Ding A. (2007) J. Exp. Med. 204, 2063–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Heyninck K., Beyaert R. (1999) FEBS Lett. 442, 147–150 [DOI] [PubMed] [Google Scholar]
- 75. Zetoune F. S., Murthy A. R., Shao Z., Hlaing T., Zeidler M. G., Li Y., Vincenz C. (2001) Cytokine 15, 282–298 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.