Abstract
Candida albicans binds and utilizes human complement inhibitors, such as C4b-binding protein (C4BP), Factor H, and FHL-1 for immune evasion. Here, we identify Candida pH-regulated antigen 1 (Pra1) as the first fungal C4BP-binding protein. Recombinant Pra1 binds C4BP, as shown by ELISA and isothermal titration calorimetry, and the Pra1-C4BP interaction is ionic in nature. The Pra1 binding domains within C4BP were localized to the complement control protein domain 4 (CCP4), CCP7, and CCP8. C4BP bound to Pra1 maintains complement-inhibitory activity. C4BP and Factor H bind simultaneously to Candida Pra1 and do not compete for binding at physiological levels. A Pra1-overexpressing C. albicans strain, which had about 2-fold Pra1 levels at the surface acquired also about 2-fold C4BP to the surface, compared with the wild type strain CAI4. A Pra1 knock-out strain showed ∼22% reduced C4BP binding. C4BP captured by C. albicans from human serum inhibits C4b and C3b surface deposition and also maintains cofactor activity. In summary, Candida Pra1 represents the first fungal C4BP-binding surface protein. Pra1, via binding to C4BP, mediates human complement control, thereby favoring the immune and complement evasion of C. albicans.
Keywords: Complement; Fungi; Immunology; Innate Immunity; Plasma; Complement Inhibition; Complement Evasion; Fungal Biology; Candida Pra1, C4BP
Introduction
Candida albicans is a dimorphic human pathogenic fungus, which causes superficial as well as systemic infections most frequently in patients undergoing immunosuppressive therapy or long term catheterization (1). Despite currently applied antifungal therapies, both mortality and morbidity mediated by C. albicans are still unacceptably high (2–4). Therefore, new prophylactic and therapeutic strategies are needed to prevent fungal spreading and tissue damage. The identification of novel yeast virulence factors that contribute to pathogenicity is necessary for approaching new strategies to fight and interfere with Candida infections. In this study, we aimed to identify surface proteins that are central for C. albicans innate immune escape.
The human complement system forms the first defense line of innate immunity. Upon infection, microbes are immediately attacked by this highly efficient human immune system (5, 6). Complement can be activated via three major pathways. The classical pathway (CP)2 is mainly induced by antibodies bound to target structures or by C-reactive protein, and the lectin pathway (LP) is activated by binding of mannose-binding lectin or ficolins to mannan-containing structures on surfaces (7–9). The alternative pathway is initiated spontaneously and continuously by randomly generated C3b, and activated C3b can bind directly to any surface and initiate an amplification loop of the alternative pathway (10, 11).
Progression of the complement cascade is controlled by fluid phase inhibitors, which are distributed in plasma and body fluid, or surface-bound inhibitors. These inhibitors include C4BP, a 570-kDa plasma glycoprotein, which is the major fluid phase CP and LP inhibitor (12), as well as Factor H and FHL-1 (Factor H-like protein 1), which are the major fluid phase alternative pathway inhibitors (13–17). The CP/LP inhibitor C4BP is formed by one β-chain and seven identical α-chains, all of which are composed of complement control protein (CCP) domains. The α-chain consists of eight CCPs, and the β-chain consists of three CCPs (18, 19). C4BP regulates complement by binding to C4b via the N terminus of each α-chain (20), thereby making C4b susceptible to degradation by the plasma serine protease Factor I and by accelerating the decay of the CP/LP C3-convertase C4bC2b (21, 22). C4BP also inhibits the activity of the alternative pathway C3-convertase in a fluid phase and acts as a cofactor in Factor I-mediated cleavage of C3b (23).
Microbial pathogens mimic human surfaces; acquire complement inhibitors, including C4BP, Factor H, and FHL-1, to their surface; and utilize the attached human regulators for complement evasion (6). These kinds of microbial pathogens include fungi, like C. albicans and Aspergillus fumigatus (24–27); Gram-positive bacteria, such as Streptococcus pyogenes (28–31) and Streptococcus pneumoniae (32–34); and Gram-negative bacteria like Borrelia burgdorferii (35, 36), Haemophilus influenzae (37–39) and Neisseria gonorrhoeae (40, 41).
Pra1 (pH-regulated antigen 1) of C. albicans is a glycosylated fungal protein composed of 299 amino acids. Pra1 is located on the fungal surface and is also released by both yeast and hyphae of C. albicans into the culture supernatant (25, 42–44). As a surface protein, Pra1 binds human plasma proteins Factor H, FHL-1, and plasminogen. Released to a supernatant, soluble Pra1 binds back to the fungal surface (25) and also binds to human phagocytes via the integrin CR3 receptor (43).
Here we identify Candida Pra1 as the first fungal C4BP-binding protein. Via binding to C4BP, Pra1 controls the classical and lectin pathway complement attack, such as C3b and C4b surface deposition, as well as C4b cleavage, thereby favoring C. albicans infection.
EXPERIMENTAL PROCEDURES
C. albicans Strains and Growth Conditions
The C. albicans wild type strains SC5314 (45), CAI4 (46), and RM1000 as well as a Pra1-overexpressing strain3 and a Pra1 knock-out C. albicans strain (47) were cultivated in YPD medium (2% (w/v) glucose, 2% (w/v) peptone, 1% (w/v) yeast extract) at 30 °C. Yeast cells were collected by centrifugation and counted with a hemocytometer (Fein-Optik, Bad Blankenburg, Germany).
Antibodies, Proteins, and Serum
Polyclonal Pra1 antiserum was raised in rabbits by immunization with purified recombinant Pra1 (25). A polyclonal goat C4 (Calbiochem) antiserum was used for detection of C4b degradation products and C4b deposited on the fungal surface. A polyclonal goat C3 (CompTech) antiserum was used to determine C3b surface deposition. Alexa Fluor®-647-labeled goat anti-rabbit, Alexa Fluor®-488-labeled goat anti-rabbit, and Alexa Fluor®-647-labeled rabbit anti-goat sera (Molecular Probes) were used as the secondary antisera for flow cytometry and confocal microscopy. Horseradish peroxidase (HRP)-conjugated rabbit anti-goat and HRP-conjugated rabbit anti-mouse as well as HRP-conjugated swine anti-rabbit sera were obtained from Dako. Monoclonal antibody (mAb) 104 and mAb 67, which recognize CCP1 or CCP4 of C4BP, respectively, recombinant C4BP, and its deletion constructs lacking one CCP each time were generated as described (48). Pra1 was expressed in Pichia pastoris strains (25). Native, plasma-purified C4BP, Factor H, Factor I, and C4b were obtained from CompTech. Gelatin was purchased from Merck, and BSA was from Sigma. Normal human serum (NHS) was collected from five healthy individuals, pooled, and stored at −80 °C until use. Factor B-depleted serum was bought from Calbiochem.
Binding Assays
Pra1 or native, plasma-purified C4BP, collagen type I (Calbiochem), and collagen type III and type IV (BD Biosciences) (0.5 μg/well in carbonate-bicarbonate buffer) were immobilized onto microtiter plates (MaxiSorb, Nunc) at 4 °C overnight. After washing, nonspecific binding sites were blocked with gelatin (0.2% in Dulbecco's phosphate-buffered saline (DPBS)) for 2 h at room temperature. After washing, full-length recombinant C4BP, various C4BP deletion constructs lacking one CCP domain at a time (ΔCCP1, ΔCCP2, ΔCCP3, ΔCCP4, ΔCCP5, ΔCCP6, ΔCCP7, and ΔCCP8), or Pra1 was added (1 μg/well in 100 μl of DPBS) and incubated for 1.5 h at room temperature. Wells were washed with DPBS-T buffer (DPBS containing 0.05% Tween 20), and specific antibody (anti-C4BP or anti-Pra1) was added for 1 h at room temperature. After washing with DPBS-T, HRP-conjugated secondary antisera were added to the wells and incubated for 1 h at room temperature. After the addition of substrate o-phenylenediamine dihydrochloride (Dako), the interaction was stopped by 2 m H2SO4. Absorbance signals were measured at 492 nm in a microtiter plate reader (SpektraMax 190, Molecular Devices). To further prove Pra1-C4BP interaction, isothermal titration calorimetry (ITC200, GE Healthcare) was performed. C4BP purified from human plasma and recombinant Pra1 were both dialyzed in the same buffer, DPBS. C4BP (300 μl with a concentration of 15 μm (i.e. 2.6 mg)) was loaded into the sample cells, and Pra1 (60 μl at a concentration of 352 μm (1.3 mg)) was loaded into the syringe. The interaction was assayed at 25 °C, using a reference power of 5 μCal/s under low feedback mode. A total of 17 initial injections were performed. The volume of the first injection was 0.3 μl, and that of the following 16 injections was 2.4 μl. After the whole interaction, the data were analyzed with a MicroCal LLC ITC200 isothermal titration calorimeter.
Competition Assays
Pra1 (0.5 μg/well) was immobilized onto microtiter plates (MaxiSorb, Nunc) overnight at 4 °C. After washing, nonspecific binding sites were blocked with 0.2% gelatin for 2 h at room temperature. After washing, Factor H (2.5 μg/ml) and C4BP (Factor H/C4BP ratios of 1:0, 1:1, 1:2, 1:5, 1:10, and 1:20) or C4BP (10 μg/ml) and heparin (C4BP/heparin ratios of 1:0, 1:1, 1:2, 1:4, and 1:8) or C4BP (10 μg/ml) and C4b (C4BP/C4b ratios of 1:0, 1:1, 1:2, 1:4, and 1:8) diluted in DPBS with the indicated mass ratios were added and incubated for 1.5 h. After washing, bound Factor H or C4BP was detected by polyclonal goat Factor H antiserum or monoclonal mouse C4BP (mAb 67) antibody, followed by HRP-conjugated secondary antisera.
Cofactor Assay
Cofactor activity of C4BP bound to Pra1 in the solid phase was assayed as described (24). Briefly, different amounts of Pra1 were immobilized onto the surface of a microtiter plate (MaxiSorb, Nunc) overnight at 4 °C. After blocking, C4BP dissolved in DPBS (0.4 μg/well) was added. Following extensive washing with DPBS, C4b (0.4 μg/well) together with Factor I (0.8 μg/well) was applied in DPBS and incubated for 30 min at 37 °C. To show the cofactor activity of C. albicans recruited C4BP, C. albicans yeast cells (wild type SC5314, RM1000, and the corresponding Pra1 knock-out mutant) (1 × 106/well) were coated onto the ELISA plate in PRMI1640 medium overnight at 37 °C. After blocking with blocking buffer I (purified casein with NaCl and Tween 20, pH, 7.2) (Applichem) for 2 h at room temperature, complement-active, Factor H-depleted human serum (ΔFactor H-HS) diluted in DPBS, supplemented with EDTA (10 mm), was added to C. albicans for 1 h at 37 °C (49). Following washing, C4b (5 μg/ml) together with Factor I (0.5 μg/ml) were added, and the reaction was incubated for 30 min at 37 °C. The supernatant was treated under reducing buffer, separated by SDS-PAGE, and transferred to a PDVF membrane (Roth), and C4b degradation products were visualized by polyclonal goat C4 antiserum, followed by a secondary HRP-conjugated rabbit anti-goat serum.
Flow Cytometry and Laser-scanning Microscopy
To compare the Pra1 surface expression level on different C. albicans strains, wild type strains CAI4 and SC5314 and a Pra1 overexpression strain were cultivated in YPD medium overnight at 30 °C. Cells were washed and incubated with polyclonal Pra1 antiserum (1:200) for 30 min on ice, followed by an Alexa Fluor®-647-labeled goat anti-rabbit serum as a secondary antiserum for another 30 min on ice. After washing, fluorescence signal was measured by flow cytometry (LSR II, BD Biosciences). For C4BP binding to C. albicans, wild type (CAI4) and a Pra1-overexpressing C. albicans strain were incubated with EDTA-NHS (1:3 dilution of NHS in DPBS, supplemented with 10 mm EDTA) for 1 h at room temperature, and then C. albicans cell pellets were washed with DPBS containing 1% BSA and incubated with polyclonal rabbit C4BP antiserum for 30 min on ice, followed by Alexa Fluor®-488-labeled goat anti-rabbit serum as a secondary antiserum for 30 min on ice. After washing, the nuclei of C. albicans were stained by 4′,6-diamidino-2-phenylindole (DAPI; 10 μg/ml) for 15 min at room temperature. Then the samples were washed and examined either by flow cytometry or by laser-scanning microscopy (LSM 510, Carl Zeiss). To compare binding of human complement inhibitors C4BP and Factor H as well as plasminogen to a Pra1 knock-out mutant and a corresponding wild type C. albicans strain RM1000, both strains were cultivated in YPD medium. After washing, the cell pellets were incubated with NHS-EDTA (30% NHS in DPBS, supplemented with 10 mm EDTA) for 1 h at room temperature. After washing, binding of C4BP, Factor H, and plasminogen to both strains was analyzed by flow cytometry using polyclonal rabbit C4BP, polyclonal goat Factor H, or polyclonal goat plasminogen antisera, followed by Alexa Fluor®-647-labeled goat anti-rabbit or rabbit anti-goat sera.
C4b and C3b Surface Deposition
C. albicans yeast cells (SC5314), RM1000, and the Pra1 knock-out C. albicans strain, as well as CAI4 and a Pra1-overexpressing strain were cultivated overnight in YPD medium. Then cells were washed in DPBS and coated onto an ELISA plate in RPMI1640 medium overnight at 37 °C (1 × 106/well). Wells were blocked using milk powder (4%) supplemented with BSA (1%) and Tween 20 (0.05%) in DPBS for 2 h at 37 °C. After washing with DPBS, different amounts of heat-inactivated normal human serum (Hi-NHS) diluted in DPBS were added to C. albicans. The mixtures were incubated for 40 min at 37 °C (24). After washing, complement-active, Factor B-depleted human serum (ΔFactor B-HS) (7.5%) diluted in GVB++ buffer (CompTech) or buffer alone was added to C. albicans. Following further incubation for 1 h at 37 °C and washing with DPBS, supplemented with Tween 20 (0.05%), C4b and C3b surface deposition was followed by staining with polyclonal goat C4 (1:5000) or goat C3 (1:5000) antiserum and by horseradish peroxidase-conjugated rabbit anti-goat sera as secondary antisera.
RESULTS
Candida Pra1 Binds C4BP
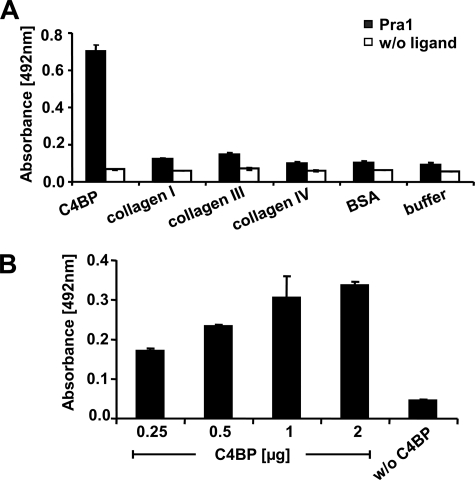
In order to identify additional ligands for Pra1, we asked whether Pra1, which has collagen-like sequences (50), binds to human collagen type I, type III, or type IV or to the human complement inhibitor C4BP. Pra1 did not bind to any of the three collagens tested but bound to immobilized C4BP, as demonstrated by ELISA (Fig. 1A). In addition, the Pra1-C4BP interaction was confirmed in a reverse setting by ELISA. Recombinant C4BP bound to immobilized Pra1 in a dose-dependent manner (Fig. 1B). Thus, Candida Pra1 is a C4BP-binding protein.
FIGURE 1.
Candida Pra1 binds C4BP. A, binding of Candida Pra1 to immobilized C4BP and collagen type I, type III, and type IV was assayed by ELISA. Pra1 was detected with a specific rabbit Pra1 antiserum, followed by secondary HRP-conjugated anti-rabbit serum. Pra1 binds to C4BP but not to collagens. B, C4BP binds to immobilized Pra1 in a dose-dependent manner. Pra1 was immobilized onto a microtiter plate, increasing amounts of recombinant C4BP were added, and bound C4BP was detected by mAb 67, followed by secondary HRP-conjugated rabbit anti-mouse serum. The data shown represent the mean values ± S.D. (error bars) of three separate experiments.
Candida Pra1 Binds C4BP as Revealed by ITC
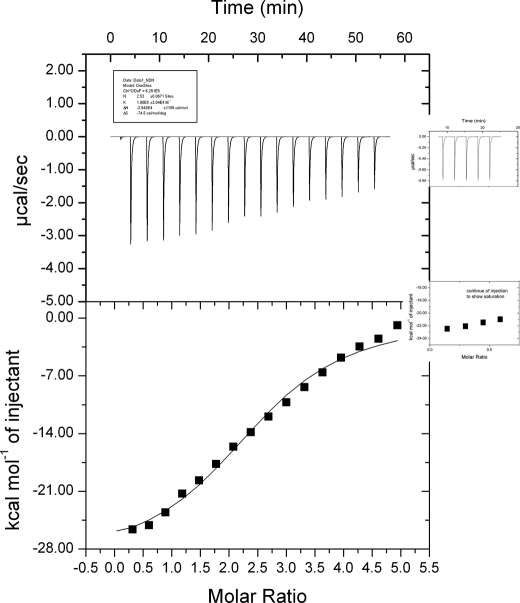
Plasma-purified C4BP and recombinant Pra1 were prepared and dialyzed in the same buffer, DPBS. C4BP with a concentration of 15 μm was loaded into the sample cells, and Pra1 at a concentration of 352 μm was applied in the syringe. The interaction was followed at 25 °C, with a reference power of 5 μcal/s under low feedback mode. A total of 17 sequential cycles of injection were applied over a period of 60 min. After 17 sequential injections, the profile was close to saturation (Fig. 2). In order to confirm that binding was almost saturated, additional Pra1 was added to the syringe, and injections were continued at the same setting. The next five injections did not change the profile significantly. The release of the power is stable (Fig. 2, right inset panels), thus confirming that saturation was almost obtained after the first 17 injections. After interaction, the data were analyzed by a MicroCal ITC200. Pra1-C4BP interaction is an exothermic reaction with an affinity (Ka) of 1.90E5 ± 3.04E4 m−1. The ITC experiment also confirmed multiple interaction sites for Pra1 sites within C4BP (n = 2.5 ± 0.07 sites) (Fig. 2).
FIGURE 2.
Candida Pra1 binds C4BP as shown by ITC. Plasma purified C4BP and recombinant Pra1 were prepared and dialyzed in the same buffer DPBS. C4BP (300 μl with a concentration of 15 μm, 2.6 mg) was loaded into the sample cell, and Pra1 (60 μl at 353 μm, 1.3 mg) was applied into the syringe. The interaction was assayed at 25 °C, with a reference power of 5 μcal/s under low feedback mode. A total of 17 sequential injections were performed; the first injection was 0.3 μl, and the following 16 injection were 2.4 μl. After the interaction, the data were analyzed by a MicroCal ITC200. After 17 injections, the reaction appeared close to saturation. However to confirm that the reaction had reached saturation, additional Pra1 was applied in the syringe, and five subsequent infections were performed (inset panels on the right). During each injection, the heat of release is stable, representing background signals based on the ligand-buffer interaction. These inset panels further show the saturation of Pra1-C4BP interaction.
Effect of NaCl on C4BP Binding to Candida Pra1
To characterize the nature of the Pra1-C4BP interaction, the effect of NaCl on binding of plasma-purified or recombinant C4BP to immobilized Pra1 was assayed. Native, plasma-derived C4BP bound to Pra1 at NaCl concentrations of 0–100 mm. NaCl at a physiological concentration of 150 mm reduced C4BP binding to Pra1 by 69.7%, compared with 0 mm NaCl. The inhibitory effect by NaCl was even stronger at higher concentrations (Fig. 3, black bars). Moreover, recombinant C4BP bound to Pra1 with lower intensity compared with plasma-purified C4BP. Binding was increased at 50 mm NaCl and again decreased at higher salt concentrations (Fig. 3, hatched bars). Thus, Pra1-C4BP interaction is ionic in nature, and the binding intensity is influenced by the salt concentration.
FIGURE 3.
NaCl affects Pra1 binding to C4BP. Pra1 was immobilized. The effect of different concentrations of NaCl on binding of either plasma purified- (black bars) or recombinant C4BP (hatched bars) to Pra1 was assayed. Bound C4BP was detected with mAb 67. Pra1-C4BP interaction is ionic in nature. Data shown represent the mean values ± S.D. (error bars) of three separate experiments.
Competition of C4BP and Factor H for Binding to Candida Pra1
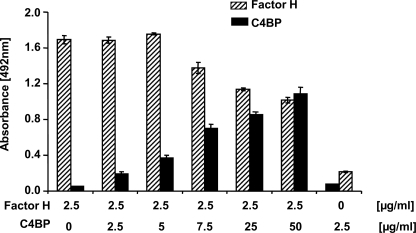
Pra1 binds C4BP and also Factor H (25). Therefore, we asked if C4BP and Factor H bind simultaneously to Candida Pra1 and if these two human inhibitors bind to overlapping or to separate sites within the Pra1 molecule. To this end, Pra1 was coated onto microtiter plates, a constant amount of Factor H together with increasing concentrations of C4BP were added, and bound C4BP and Factor H were detected with specific antibodies. Using a constant amount of Factor H (2.5 μg/ml), C4BP binding was increased upon increasing concentrations of C4BP (Fig. 4, black bars). Factor H binding remained constant when C4BP levels were less than 5 μg/ml (Fig. 4, hatched bars 1–3). Factor H binding slightly decreased using a concentration of 7.5 μg/ml C4BP (5 times the Factor H amount) and was further reduced by less than 50% at a C4BP concentration of 50 μg/ml (20 times the Factor H concentration). The latter ratios exceed the physiological level of the two inhibitors in human plasma, where the concentration of C4BP is about 50% of that of Factor H. These results show that C4BP and Factor H bind simultaneously to Candida Pra1 protein and suggest that the two human inhibitors bind to distinct sites.
FIGURE 4.
Independent binding of the human complement inhibitors C4BP and Factor H to C. albicans Pra1. Pra1 was immobilized, and a constant amount of Factor H (2.5 μg/ml) together with increasing concentrations of C4BP used at different mass ratios (1:0, 1:1, 1:2, 1:5, 1:10, and 1:20) were added. Bound Factor H (hatched bars) was detected with polyclonal goat Factor H antiserum, and bound C4BP (black bars) was detected with mouse C4BP antibody (mAb 67). Data shown represent the mean values ± S.D. (error bars) of three separate experiments.
Localization of Pra1 Binding Domains within C4BP
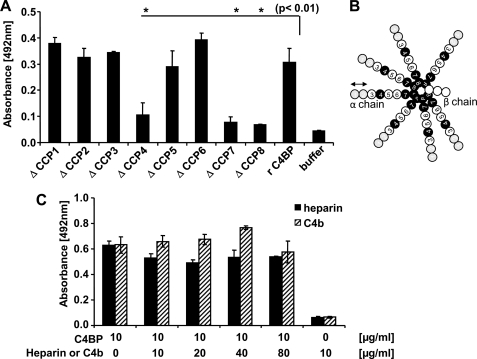
To localize Pra1 binding domains within C4BP, binding of recombinant C4BP and its deletion mutants to immobilized Pra1 was determined. Full-length C4BP, as well as C4BP deletion mutants ΔCCP1, ΔCCP2, ΔCCP3, ΔCCP5, and ΔCCP6 bound to Pra1 with similar intensities, whereas binding of constructs ΔCCP4, ΔCCP7, and ΔCCP8 to Pra1 was reduced (Fig. 5A). These results show that C4BP contacts Pra1 via three domains (i.e. CCP4, CCP7, and CCP8) (Fig. 5B, domains labeled in black).
FIGURE 5.
Localization of the binding domains within C4BP. A, localization of Pra1 binding domains within C4BP. Pra1 was immobilized, and binding of recombinant C4BP and C4BP deletion mutants lacking one single CCP domain each time was detected with the monoclonal C4BP antibody (mAb 67 for ΔCCP1, ΔCCP2, ΔCCP3, ΔCCP5, ΔCCP6, ΔCCP7, and ΔCCP8 or mAb 104 for ΔCCP4). B, spider-like conformation of the native C4BP protein. The native C4BP protein is composed of seven α-chains and one β-chain. Each α-chain is composed of eight CCP elements, and the β-chain is composed of three. The CCPs of the α-chain that mediate contact with Candida Pra1 are highlighted in black. C, effect of heparin and C4b on C4BP binding to Pra1. Pra1 was immobilized, and C4BP (10 μg/ml) together with increasing concentrations of heparin (C4BP/heparin ratio of 1:0, 1:1, 1:2, 1:4, and 1:8) (black bars) or C4b (C4BP/C4b ratio of 1:0, 1:1, 1:2, 1:4, and 1:8) (hatched bars) were added, and bound C4BP was detected with mAb 67. Data shown represent the mean values ± S.D. (error bars) of three separate experiments.
Initially, when binding of C4BP deletion mutants to intact yeast cells was analyzed, CCP1 and CCP2 were identified as the major contact domains (24) (Fig. 5B, domains marked in gray). CCPs1–3 also interact with heparin and C4b. To confirm that domains CCP1 to CCP3 are not relevant for Pra1-C4BP interaction, Pra1 was immobilized, and C4BP was added together with increasing amounts of heparin or C4b. C4BP binding to Pra1 was affected neither by heparin (Fig. 5C, black bars) nor by C4b (Fig. 5C, hatched bars). Thus, C4BP-Pra1 interaction is independent of domains CCP1 to CCP3.
C4BP Bound to Pra1 Displays Complement-inhibitory Activity
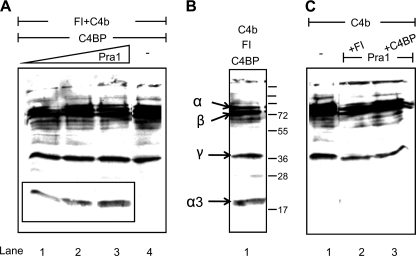
To analyze whether C4BP bound to Pra1 is functionally active, Pra1 was immobilized at different concentrations, and C4BP was added. After intensive washing, purified C4b and Factor I were added. Following further incubation, supernatants were collected, separated by SDS-PAGE, and transferred to a membrane, and C4b as well as C4b cleavage fragments were identified by Western blotting. C4BP bound to Pra1 showed cofactor activity, as indicated by the appearance of a 20-kDa α3-fragment of C4b (Fig. 6A, lanes 1–3). No cleavage products were observed when Pra1 was absent (Fig. 6A, lane 4) or when C4BP or Factor I was absent (Fig. 6C, lanes 1 and 2). C4BP, together with Factor I and C4b, was used as a positive control (Fig. 6B). Thus, C4BP bound to Pra1 maintains the cofactor activity for Factor I-mediated cleavage of C4b.
FIGURE 6.
C4BP bound to Pra1 displays cofactor activity. A, Pra1 was immobilized at a concentration of 0.5 (lane 1), 1.0 (lane 2), or 2.0 μg (lane 3), and C4BP was added. After blocking and intensive washing, purified Factor I and C4b were added. Following incubation, the reaction mixture was separated by SDS-PAGE and transferred to a membrane, and C4b as well as C4b cleavage fragments were identified by Western blotting using polyclonal goat C4 antiserum. Cofactor activity of C4BP bound to Pra1 is indicated by the appearance of the 20-kDa α3-fragment (A, lanes 1–3). No cleavage products were identified when Pra1 was absent (A, lane 4). C4BP incubated with Factor I and C4b was used as a positive control to identify C4b cleavage fragments (B). No cleavage occurred in the absence of C4BP or Factor I (C, lanes 2 and 3) or C4b alone (C, lane 1). A representative result is shown of three independent experiments.
Pra1 Overexpression Increases C4BP Binding to C. albicans Surface
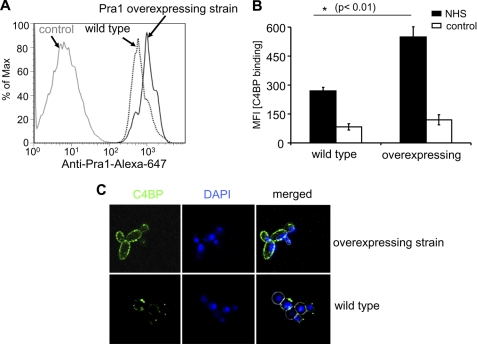
To verify whether surface-expressed Pra1 is relevant for C4BP acquisition to intact yeast cells, binding of C4BP to a Pra1-overexpressing C. albicans strain was studied. This strain has about 2-fold levels of Pra1 at the surface, compared with the wild type strain CAI4 (Fig. 7A). Upon incubation in NHS-EDTA, C4BP binding to the Pra1-overexpressing strain was also about 2-fold as compared with the wild type strain (MFI; 549 versus 269) (Fig. 7B). This enhanced binding was confirmed by confocal microscopy. Following incubation in NHS-EDTA, C4BP-specific fluorescence was stronger for the Pra1-overexpressing strain as compared with the wild type strain (Fig. 7C). Thus, surface-expressed Pra1 mediates C. albicans to acquire the human classical pathway inhibitor C4BP from NHS, thereby evading the classical pathway complement activation.
FIGURE 7.
Binding of C4BP to a Pra1-overexpressing C. albicans strain. A, Pra1 expression levels on different C. albicans strains. The C. albicans wild type CAI4 (dotted black curve) and a Pra1-overexpressing strain (black curve) were incubated with polyclonal Pra1 antiserum, followed by an Alexa Fluor®-647-labeled goat anti-rabbit serum. Preimmune serum (gray curve) was used as a negative control. The fluorescence signal was recorded by flow cytometry. Shown is one representative experiment of three separate assays. B, C4BP derived from NHS binds strongly to the Pra1-overexpressing strain, as compared with the wild type strain CAI4 shown. Both C. albicans strains were incubated in NHS-EDTA. After washing, bound C4BP was detected by flow cytometry using polyclonal rabbit C4BP antiserum, followed by an Alexa Fluor®-488-labeled goat anti-rabbit serum. The data represent the mean values ± S.D. (error bars) of three separate experiments. C, binding of C4BP to both Pra1-overexpressing and wild type strains was confirmed by laser-scanning microscopy. Again, the Pra1-overexpressing (top) and wild type (bottom) strains were incubated with NHS-EDTA. After washing, the cells were incubated with polyclonal rabbit C4BP antiserum, followed by an Alexa Fluor®-488-labeled goat anti-rabbit serum. Nuclei were stained with DAPI (10 μg/ml) and examined by confocal microscopy. The figures show one representative experiment of three performed.
Binding of C4BP, Factor H, and Plasminogen from NHS to a Pra1 Knock-out Strain
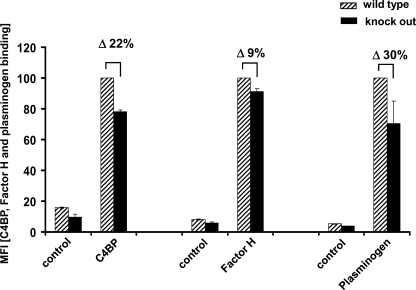
To further prove that surface-expressed Pra1 mediates C. albicans in acquiring the complement inhibitors as well as plasminogen from NHS for complement evasion and tissue invasion, binding of C4BP, Factor H, and plasminogen to a Pra1 knock-out strain and to the corresponding wild type C. albicans strain RM1000 was analyzed by flow cytometry. Both strains were incubated in NHS-EDTA. After washing, bound C4BP, Factor H, and plasminogen were analyzed by specific antisera. C4BP, Factor H, and plasminogen binding to the Pra1 knock-out mutant were reduced by ∼22, 9, and 30%, respectively (Fig. 8, black bars), compared with the wild type C. albicans strain (Fig. 8, hatched bars). Thus, native surface expressed Pra1 binds C4BP, Factor H, and plasminogen, thereby mediating complement evasion and degradation of extracellular matrices. However, additional C4BP, Factor H, and plasminogen-binding fungal proteins exist at the surface of C. albicans, explaining the partial inhibition.
FIGURE 8.
Binding of C4BP, Factor H, and plasminogen to a Pra1 knock-out C. albicans strain. Both Pra1 knock-out and the corresponding wild type C. albicans strain RM1000 were incubated with NHS-EDTA. After washing, bound C4BP, Factor H, or plasminogen was identified with polyclonal rabbit C4BP, polyclonal goat Factor H, or polyclonal goat plasminogen antiserum, respectively, followed by Alexa Fluor®-647-labeled goat anti-rabbit or rabbit anti-goat serum. Binding of C4BP, Factor H, and plasminogen to the Pra1 knock-out strain (black bars) was reduced by about 22, 9, and 30%, respectively, compared with the wild type strain (hatched bars). The data shown represent the mean values ± S.D. (error bars) of three separate experiments.
Function of C. albicans-recruited C4BP from Human Serum
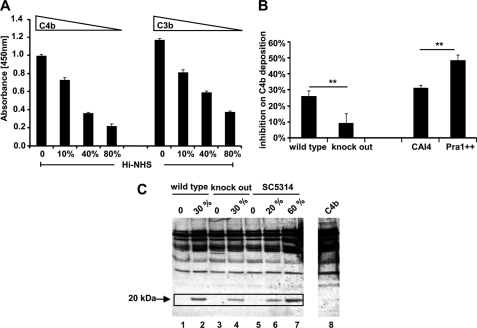
To show that C4BP recruited to the surface of C. albicans by Pra1 has complement-inhibitory activity, the effect of recruited C4BP on C3b/C4b surface deposition was analyzed. C. albicans strain SC5314 was treated with different amounts of Hi-NHS to allow acquisition of C4BP to the surface. Then the cells were incubated in complement-active ΔFactor B-HS. After washing, surface-deposited C4b as well as C3b were detected with polyclonal goat C4 or C3 antiserum. Surface-attached C4BP inhibited C4b and also C3b deposition at the fungal surface in a dose-dependent manner. This effect was observed upon complement activation of the classical and the lectin pathway by using ΔFactor B-HS. This effect is different from that described previously for the alternative pathway via Factor H acquisition. The effect was dose-dependent; the more C4BP that was recruited, the stronger the inhibitory effects on C4b and C3b deposition. When 80% heat-inactive human serum was used, recruited C4BP inhibited C4b and C3b surface deposition by 78 and 68%, respectively (Fig. 9A).
FIGURE 9.
Function of C. albicans-recruited C4BP from normal human serum. A and B, C. albicans-recruited C4BP blocks C4b and C3b surface deposition. C. albicans yeast cells (SC5314) were cultivated overnight in YPD medium at 30 °C, washed by DPBS, and immobilized onto an ELISA plate in RPMI1640 medium overnight at 37 °C (1 × 106/well). Wells were blocked using milk powder (4%), supplemented with BSA (1%) and Tween 20 (0.05%) in DPBS for 2 h at 37 °C. After washing with DPBS, different amounts of Hi-NHS diluted in DPBS were added to the wells. The mixture was incubated for 40 min at 37 °C. After washing, ΔFactor B-HS (7.5%) (Calbiochem) diluted in GVB++ buffer or buffer alone (background control) was added to C. albicans. The mixture was further incubated for 1 h at 37 °C. Following washing with DPBS plus Tween 20 (0.05%), C4b and C3b surface deposition were determined using polyclonal goat anti-C4 (1:5000) or goat anti-C3 (1:5000) serum, followed by horseradish peroxidase conjugated-rabbit anti-goat serum as a secondary antiserum (1:2000). Surface-attached C4BP inhibited C4b and C3b deposition onto the fungal surface in a dose-dependent manner (A). When 30% Hi-NHS was used to incubate with different C. albicans strains, C4b surface deposition was inhibited on wild type RM1000 strain by about 26%. The inhibitory effect on the knock-out strain was about 9% and thus lower. Moreover, C4b surface deposition was more efficiently inhibited on the Pra1-overexpressing strain as compared with the wild type strain (CAI4) (B). Data represent the mean values ± S.D. (error bars) of three separate experiments. C, cofactor activity of C. albicans recruited C4BP from human serum. C. albicans yeast cells (wild type RM1000 and the corresponding Pra1 knock-out mutant and the laboratory reference strain SC5314) (1 × 106/well) were coated onto the ELISA plate in PRMI1640 medium overnight at 37 °C. After blocking with blocking buffer I for 2 h at room temperature, ΔFactor H-HS diluted in DPBS, supplemented with EDTA (10 mm), was added to C. albicans for 1 h at 37 °C. After washing, C4b (5 μg/ml) together with Factor I (0.5 μg/ml) were added. After incubation for 30 min at 37 °C, the supernatant was separated by SDS-PAGE under reducing conditions and transferred onto the membrane. The C4b cleavage fragments were detected by Western blotting using polyclonal goat C4 antiserum (1:1000). C4BP bound to C. albicans displays cofactor activity, indicated as the appearance of the 20-kDa cleavage product (boxed, lanes 1 and 2, for the C. albicans wild type strain RM1000, lanes 3 and 4 for the corresponding Pra1 knock-out mutant, and lanes 5–7 for the reference strain SC5314). Lane 8 shows the C4b protein alone. The data show a representative result of three separate experiments.
In order to compare the effect of recruited C4BP by different C. albicans strains on C4b surface depositions, RM1000 and the Pra1 knock-out C. albicans strain as well as CAI4 and Pra1 overexpression strains were treated with 30% Hi-NHS to acquire C4BP onto the surface. Then the cells were incubated in ΔFactor B-HS. After washing, C4b surface deposition was detected by polyclonal C4 antiserum. C4BP inhibited C4b surface deposition by about 26% on wild type RM1000 strain and by about 9% on the Pra1 knock-out strain. Moreover, C4b surface deposition was more efficiently inhibited on the Pra1-overexpressing strain, as compared with that on the wild type strain CAI4 (Fig. 9B).
In addition, cofactor activity of C. albicans recruited C4BP was assayed. C. albicans yeast cells (wild type RM1000 and the corresponding Pra1 knock-out mutant and also the laboratory reference strain SC5314) (1 × 106/well) were coated onto the ELISA plate in PRMI1640 medium overnight at 37 °C. After blocking, complement-active ΔFactor H-HS diluted in DPBS and supplemented with EDTA (10 mm) was added to the yeast cells for 1 h at 37 °C. Following washing, C4b (5 μg/ml) together with Factor I (0.5 μg/ml) was added and incubated for 30 min at 37 °C. Then the supernatant was separated by SDS-PAGE under reducing conditions and transferred onto the membrane. The C4b cleavage fragments were detected by Western blotting using polyclonal goat C4 antiserum (1:1000) (Calbiochem). C4BP bound to C. albicans displayed cofactor activity, as indicated by the appearance of the 20-kDa cleavage product (Fig. 9C). For the C. albicans wild type strain RM1000, which bound more C4BP than the corresponding Pra1 knock-out strain, the intensity of the 20 kDa band was more pronounced (Fig. 9C, boxed, lanes 2 and 4). When using the wild type strain SC5314, the intensity of the 20 kDa band correlates with the amount of ΔFactor H-HS that was added (Fig. 9C, boxed, lanes 5–7).
DISCUSSION
In the current study, we identify Candida Pra1 as the first fungal C4BP-binding protein, localize the Pra1 binding domains within C4BP, and show that bound C4BP maintains complement-inhibitory activity, such as C4b cleavage, as well as C3b and C4b surface deposition. Thus, Pra1, by acquiring human complement inhibitor C4BP to C. albicans surface, controls the classical pathway of complement and favors fungal infection. The identification of this additional novel function makes the fungal virulence factor Candida Pra1 an attractive candidate for immune intervention.
C4BP acquisition is a general immune evasion strategy and is used by many pathogens. Pra1 is the first fungal C4BP-binding protein identified from C. albicans. Candida Pra1 binds both plasma-purified and recombinant C4BP (Fig. 1, A and B). Pra1-C4BP interaction is an exothermic reaction and has an affinity (Ka) of 1.90E5 ± 3.04E4 m−1 as determined by ITC (Fig. 2). Pra1-C4BP interaction is ionic in nature and is affected by NaCl (Fig. 3). This type of interaction is similar to the Pra1-Factor H interaction (25) but different from that of the streptococcal M protein-C4BP interaction, which is hydrophobic in nature (30). C4BP binds Candida Pra1 via domains CCP4, CCP7, and CCP8. Thus, C4BP attached to Candida Pra1 still has the complement interaction domains (i.e. CCP1 to CCP3) are accessible and expose the functional regions to the outside (20, 23). ITC data identify multiple binding sites (n = 2.5 ± 0.07) within C4BP for Pra1. ELISAs with various purified recombinant deletion constructs of the α chain of C4BP revealed three separated interaction sites, which were located to CCP4, CCP7, and CCP8. Thus, the difference between the ELISA and the ITC data indicates that some binding sites are hidden and not accessible for Pra1 in the intact C4BP protein. C4BP-Pra1 interaction shows a rather similar attachment to other microbial proteins. The ubiquitous surface proteins A1 (Usp A1) and Usp A2 of Moraxella catarrhalis bind C4BP via domains CCP2 and CCP7 (51), and outer membrane protein A (OmpA) of Escherichia coli K1 and Haemophilus influenzae (38) binds C4BP via domains CCP3 and CCP8 (52). In contrast, streptococcal M protein (30) and N. gonorrhoeae porins (53) bind to C4BP via the N-terminal CCP1. Filamentous hemagglutinin of Bordetella pertussis (54) binds to C4BP via CCP1 and CCP2. Similarly, both yeast and hyphae forms of intact C. albicans bind to C4BP mainly via CCP1 and CCP2 (24). This difference between C4BP binding to Pra1 and to intact C. albicans cells indicates the existence of an additional C4BP ligand(s) at the surface of C. albicans.
Candida Pra1 binds both C4BP and Factor H. The acquired human inhibitors have a functional relevance. At physiological levels, both C4BP and Factor H bind at the same time to Pra1, and Factor H binding is not affected upon increasing C4BP levels. However, at non-physiological C4BP levels, Factor H binding was only mildly affected by C4BP binding. This weak inhibitory effect of C4BP is explained by steric effects due to the large size of the C4BP protein of 550 kDa. Thus, C4BP and Factor H bind simultaneously to Candida Pra1, and the two human inhibitors bind to distinct sites within the Candida Pra1 protein at the physiological conditions.
The Pra1-overexpressing C. albicans strain binds C4BP more efficiently (about 2-fold) as compared with wild type strain CAI4 (Fig. 7). Furthermore, a Pra1 knock-out mutant shows reduced (∼22%) but not a complete lack of C4BP binding (Fig. 8). These results show that Pra1 at the yeast surface binds C4BP and also suggest the existence of additional C4BP-binding proteins at the C. albicans surface. C. albicans utilizes surface Pra1 to acquire human complement inhibitor C4BP. Bound C4BP maintains complement-inhibitory activity and acts as a cofactor for Factor I-mediated cleavage of C4b (Figs. 6 and 9). This C4b inactivation will inhibit formation of the classical pathway C3 convertase (C4bC2b) at the fungal surface and consequently blocks further progression of the complement cascade, generation of proinflammatory products, and C4b and C3b surface opsonization (Fig. 9). The lower number of C4b/C3b on the fungal surface reduces adhesion to and uptake of the C. albicans by human macrophages (supplemental Fig. S2). There is a clear correlation between the number of deposited C4b/C3b molecules on the fungal surface and better adhesion and phagocytosis.
At present, the domain structure of Candida Pra1 is not known. So far, a collagen-like motive was identified in the Pra1 sequence, which may be involved in the anchoring, attaching, and colonization of C. albicans onto the extracellular matrices (50). However, the binding sites for the various human complement inhibitors have not been mapped in this fungal protein.
Candida Pra1 is an effective fungal virulence factor that is present on the surface of the pathogen and that is also secreted. Pra1 displays multiple biological roles at these different locations. As a yeast surface protein, Pra1 binds several human complement inhibitors, including the classical pathway inhibitor C4BP and the alternative pathway regulators Factor H and FHL-1, as well as plasminogen. The Pra1-bound human regulators are functionally active, inhibit complement activation at the fungal surface, and degrade extracellular matrix components (25). As a secreted protein, Pra1 complexes C3 in solution, blocks cleavage of C3 by a C3 convertase, and thus inhibits further complement activation and immune effector functions (55). In addition, secreted Pra1 enhances Factor H-mediated complement control in the fluid phase (25). This effect on cofactor activity is specific for the alternative pathway but is not observed for the classical pathway (supplemental Fig. S1). In addition, secreted Pra1 can bind back to the surface of C. albicans, which can replenish surface function of Pra1. Secreted Pra1 binds to human CR3-expressing cells like neutrophils and monocytes. The addition of Pra1 to co-cultures of C. albicans and human neutrophils blocks CR3-mediated uptake of the pathogen and enhances fungal survival (43, 56). Thus, by binding to the integrin receptor CR3, Pra1 may act as a decoy to block CR3 signaling or CR3-mediated fungal uptake.
In summary, Candida Pra1 harbors multiple functions to control human immune responses and provides an example of the multiple and complex immune escape reactions that are all mediated by one single fungal virulence factor. A detailed understanding of these multiple roles of Pra1 could also allow the creation of new strategies to interfere with and fight against C. albicans infection.
Supplementary Material
Acknowledgment
We thank Anne Marcil for generating the Pra1 knock-out mutant and proving the parental C. albicans strain RM1000.
This work was supported by the International Leibniz Research School for Microbial and Biomolecular Interactions Jena and the Jena School for Microbial Communication, Priority Program 1160 of the Deutsche Forschungsgemeinschaft (Zi 432, Ru 608), and the Swedish Research Council, Swedish Foundation for Strategic Research, and Foundations of Osterlund, King Gustav V's 80th Anniversary, Greta and Johan Kock, Knut and Alice Wallenberg, and Inga-Britt and Arne Lundberg. The data for this work were presented in part at the XXIII International Complement Workshop in New York in 2010.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
S. Rupp, manuscript in preparation.
- CP
- classical pathway
- LP
- lectin pathway
- CCP
- complement control protein
- C4BP
- C4b-binding protein
- DPBS
- Dulbecco's phosphate-buffered saline
- NHS
- normal human serum
- Hi-NHS
- heat-inactivated normal human serum
- ITC
- isothermal titration calorimetry.
REFERENCES
- 1. Jong A. Y., Stins M. F., Huang S. H., Chen S. H., Kim K. S. (2001) Infect. Immun. 69, 4536–4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alonso-Valle H., Acha O., García-Palomo J. D., Fariñas-Alvarez C., Fernández-Mazarrasa C., Fariñas M. C. (2003) Eur. J. Clin. Microbiol. Infect. Dis. 22, 254–257 [DOI] [PubMed] [Google Scholar]
- 3. Gudlaugsson O., Gillespie S., Lee K., Vande Berg J., Hu J., Messer S., Herwaldt L., Pfaller M., Diekema D. (2003) Clin. Infect. Dis. 37, 1172–1177 [DOI] [PubMed] [Google Scholar]
- 4. Pappas P. G., Rex J. H., Lee J., Hamill R. J., Larsen R. A., Powderly W., Kauffman C. A., Hyslop N., Mangino J. E., Chapman S., Horowitz H. W., Edwards J. E., Dismukes W. E. (2003) Clin. Infect. Dis. 37, 634–643 [DOI] [PubMed] [Google Scholar]
- 5. Zipfel P. F., Skerka C. (2009) Nat. Rev. Immunol. 9, 729–740 [DOI] [PubMed] [Google Scholar]
- 6. Zipfel P. F., Würzner R., Skerka C. (2007) Mol. Immunol. 44, 3850–3857 [DOI] [PubMed] [Google Scholar]
- 7. Mihlan M., Stippa S., Józsi M., Zipfel P. F. (2009) Cell Death Differ. 16, 1630–1640 [DOI] [PubMed] [Google Scholar]
- 8. Garred P., Honoré C., Ma Y. J., Munthe-Fog L., Hummelshøj T. (2009) Mol. Immunol. 46, 2737–2744 [DOI] [PubMed] [Google Scholar]
- 9. Zhang M. X., Kozel T. R. (1998) Infect. Immun. 66, 4845–4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zipfel P. F., Mihlan M., Skerka C. (2007) Adv. Exp. Med. Biol. 598, 80–92 [DOI] [PubMed] [Google Scholar]
- 11. Zipfel P. F., Heinen S., Józsi M., Skerka C. (2006) Mol. Immunol. 43, 97–106 [DOI] [PubMed] [Google Scholar]
- 12. Blom A. M., Nandakumar K. S., Holmdahl R. (2009) Ann. Rheum. Dis. 68, 136–142 [DOI] [PubMed] [Google Scholar]
- 13. Friese M. A., Hellwage J., Jokiranta T. S., Meri S., Peter H. H., Eibel H., Zipfel P. F. (1999) Mol. Immunol. 36, 809–818 [DOI] [PubMed] [Google Scholar]
- 14. Zipfel P. F., Jokiranta T. S., Hellwage J., Koistinen V., Meri S. (1999) Immunopharmacology 42, 53–60 [DOI] [PubMed] [Google Scholar]
- 15. Zipfel P. F., Skerka C., Hellwage J., Jokiranta S. T., Meri S., Brade V., Kraiczy P., Noris M., Remuzzi G. (2002) Biochem. Soc. Trans. 30, 971–978 [DOI] [PubMed] [Google Scholar]
- 16. Zipfel P. F. (2009) Immunol. Lett. 126, 1–7 [DOI] [PubMed] [Google Scholar]
- 17. Zipfel P. F., Hellwage J., Friese M. A., Hegasy G., Jokiranta S. T., Meri S. (1999) Mol. Immunol. 36, 241–248 [DOI] [PubMed] [Google Scholar]
- 18. Scharfstein J., Ferreira A., Gigli I., Nussenzweig V. (1978) J. Exp. Med. 148, 207–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blom A. M., Villoutreix B. O., Dahlbäck B. (2004) Mol. Immunol. 40, 1333–1346 [DOI] [PubMed] [Google Scholar]
- 20. Blom A. M., Kask L., Dahlbäck B. (2001) J. Biol. Chem. 276, 27136–27144 [DOI] [PubMed] [Google Scholar]
- 21. Blom A. M., Zadura A. F., Villoutreix B. O., Dahlbäck B. (2000) Mol. Immunol. 37, 445–453 [DOI] [PubMed] [Google Scholar]
- 22. Gigli I., Sorvillo J., Halbwachs-Mecarelli L. (1985) J. Immunol. 135, 440–444 [PubMed] [Google Scholar]
- 23. Blom A. M., Kask L., Dahlbäck B. (2003) Mol. Immunol. 39, 547–556 [DOI] [PubMed] [Google Scholar]
- 24. Meri T., Blom A. M., Hartmann A., Lenk D., Meri S., Zipfel P. F. (2004) Infect. Immun. 72, 6633–6641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luo S., Poltermann S., Kunert A., Rupp S., Zipfel P. F. (2009) Mol. Immunol. 47, 541–550 [DOI] [PubMed] [Google Scholar]
- 26. Behnsen J., Hartmann A., Schmaler J., Gehrke A., Brakhage A. A., Zipfel P. F. (2008) Infect. Immun. 76, 820–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Poltermann S., Kunert A., von der Heide M., Eck R., Hartmann A., Zipfel P. F. (2007) J. Biol. Chem. 282, 37537–37544 [DOI] [PubMed] [Google Scholar]
- 28. Perez-Casal J., Okada N., Caparon M. G., Scott J. R. (1995) Mol. Microbiol. 15, 907–916 [DOI] [PubMed] [Google Scholar]
- 29. Blackmore T. K., Fischetti V. A., Sadlon T. A., Ward H. M., Gordon D. L. (1998) Infect. Immun. 66, 1427–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thern A., Stenberg L., Dahlbäck B., Lindahl G. (1995) J. Immunol. 154, 375–386 [PubMed] [Google Scholar]
- 31. Jenkins H. T., Mark L., Ball G., Persson J., Lindahl G., Uhrin D., Blom A. M., Barlow P. N. (2006) J. Biol. Chem. 281, 3690–3697 [DOI] [PubMed] [Google Scholar]
- 32. Dieudonné-Vatran A., Krentz S., Blom A. M., Meri S., Henriques-Normark B., Riesbeck K., Albiger B. (2009) J. Immunol. 182, 7865–7877 [DOI] [PubMed] [Google Scholar]
- 33. Hammerschmidt S., Agarwal V., Kunert A., Haelbich S., Skerka C., Zipfel P. F. (2007) J. Immunol. 178, 5848–5858 [DOI] [PubMed] [Google Scholar]
- 34. Janulczyk R., Iannelli F., Sjoholm A. G., Pozzi G., Bjorck L. (2000) J. Biol. Chem. 275, 37257–37263 [DOI] [PubMed] [Google Scholar]
- 35. Kraiczy P., Skerka C., Kirschfink M., Brade V., Zipfel P. F. (2001) Eur. J. Immunol. 31, 1674–1684 [DOI] [PubMed] [Google Scholar]
- 36. Pietikäinen J., Meri T., Blom A. M., Meri S. (2010) Mol. Immunol. 47, 1299–1305 [DOI] [PubMed] [Google Scholar]
- 37. Hallström T., Zipfel P. F., Blom A. M., Lauer N., Forsgren A., Riesbeck K. (2008) J. Immunol. 181, 537–545 [DOI] [PubMed] [Google Scholar]
- 38. Hallström T., Jarva H., Riesbeck K., Blom A. M. (2007) J. Immunol. 178, 6359–6366 [DOI] [PubMed] [Google Scholar]
- 39. Hallström T., Trajkovska E., Forsgren A., Riesbeck K. (2006) J. Immunol. 177, 430–436 [DOI] [PubMed] [Google Scholar]
- 40. Ram S., Cullinane M., Blom A. M., Gulati S., McQuillen D. P., Monks B. G., O'Connell C., Boden R., Elkins C., Pangburn M. K., Dahlbäck B., Rice P. A. (2001) J. Exp. Med. 193, 281–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ram S., McQuillen D. P., Gulati S., Elkins C., Pangburn M. K., Rice P. A. (1998) J. Exp. Med. 188, 671–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Casanova M., Lopez-Ribot J. L., Monteagudo C., Llombart-Bosch A., Sentandreu R., Martinez J. P. (1992) Infect. Immun. 60, 4221–4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Soloviev D. A., Fonzi W. A., Sentandreu R., Pluskota E., Forsyth C. B., Yadav S., Plow E. F. (2007) J. Immunol. 178, 2038–2046 [DOI] [PubMed] [Google Scholar]
- 44. Sentandreu M., Elorza M. V., Sentandreu R., Fonzi W. A. (1998) J. Bacteriol. 180, 282–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gillum A. M., Tsay E. Y., Kirsch D. R. (1984) Mol. Gen. Genet. 198, 179–182 [DOI] [PubMed] [Google Scholar]
- 46. Fonzi W. A., Irwin M. Y. (1993) Genetics 134, 717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Marcil A., Gadoury C., Ash J., Zhang J., Nantel A., Whiteway M. (2008) Infect. Immun. 76, 4345–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Blom A. M., Berggârd K., Webb J. H., Lindahl G., Villoutreix B. O., Dahlbäck B. (2000) J. Immunol. 164, 5328–5336 [DOI] [PubMed] [Google Scholar]
- 49. Józsi M., Licht C., Strobel S., Zipfel S. L., Richter H., Heinen S., Zipfel P. F., Skerka C. (2008) Blood 111, 1512–1514 [DOI] [PubMed] [Google Scholar]
- 50. Sepúlveda P., Murgui A., López-Ribot J. L., Casanova M., Timoneda J., Martínez J. P. (1995) Infect. Immun. 63, 2173–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nordström T., Blom A. M., Forsgren A., Riesbeck K. (2004) J. Immunol. 173, 4598–4606 [DOI] [PubMed] [Google Scholar]
- 52. Prasadarao N. V., Blom A. M., Villoutreix B. O., Linsangan L. C. (2002) J. Immunol. 169, 6352–6360 [DOI] [PubMed] [Google Scholar]
- 53. Jarva H., Ngampasutadol J., Ram S., Rice P. A., Villoutreix B. O., Blom A. M. (2007) J. Immunol. 179, 540–547 [DOI] [PubMed] [Google Scholar]
- 54. Berggård K., Johnsson E., Mooi F. R., Lindahl G. (1997) Infect. Immun. 65, 3638–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Luo S., Hartmann A., Dahse H. M., Skerka C., Zipfel P. F. (2010) J. Immunol. 185, 2164–2173 [DOI] [PubMed] [Google Scholar]
- 56. Agarwal V., Asmat T. M., Luo S., Jensch I., Zipfel P. F., Hammerschmidt S. (2010) J. Biol. Chem. 285, 23486–23495 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.