Abstract
Angiopoietin-1 (Ang1) regulates both vascular quiescence and angiogenesis through the receptor tyrosine kinase Tie2. We and another group previously showed that Ang1 and Tie2 form distinct signaling complexes at cell-cell and cell-matrix contacts. We further demonstrated that the former up-regulates Notch ligand delta-like 4 (Dll4) only in the presence of cell-cell contacts. Because Dll4/Notch signal restricts sprouting angiogenesis and promotes vascular stabilization, we investigated the mechanism of how the Ang1/Tie2 signal induces Dll4 expression to clarify the role of the Dll4/Notch signal in Ang1/Tie2 signal-mediated vascular quiescence. Under confluent endothelial cells, the basal Notch signal was observed. Ang1, moreover, induced Dll4 expression and production of the Notch intracellular domain (NICD). Ang1 stimulated transcriptional activity of β-catenin through phosphoinositide 3-kinase (PI3K)/AKT-mediated phosphorylation of glycogen synthase kinase 3β (GSK3β). Correspondingly, the GSK3β inhibitor up-regulated Dll4, whereas depletion of β-catenin by siRNA blocked Ang1-induced Dll4 expression, indicating the indispensability of β-catenin in Ang1-mediated up-regulation of Dll4. In addition, Dll4 expression by the GSK3β inhibitor was only observed in confluent cells, and was impeded by DAPT, a γ-secretase inhibitor, implying requirement of the Notch signal in β-catenin-dependent Dll4 expression. Consistently, we found that either Ang1 or NICD up-regulates Dll4 through the RBP-J binding site within intron 3 of the DLL4 gene and that β-catenin forms a complex with NICD/RBP-J to enhance Dll4 expression. Ang1 induced the deposition of extracellular matrix that is preferable for basement membrane formation through Dll4/Notch signaling. Collectively, the Ang1/Tie2 signal potentiates basal Notch signal controlling vascular quiescence by up-regulating Dll4 through AKT-mediated activation of β-catenin.
Keywords: AKT PKB, β-Catenin, Endothelium, Extracellular Matrix Proteins, Glycogen Synthase Kinase 3, Notch Pathway, Tie2, Angiopoietin-1, Δ-Like 4, Vascular Quiescence
Introduction
Angiopoietin-1 (Ang1)3 is a ligand for endothelium-specific receptor tyrosine kinase Tie2. Ang1/Tie2 signaling is essential for developmental vascular formation, as evidenced by the gene-targeting analyses of either Ang1 or Tie2 in mice (1–3). In quiescent adult vasculature, Ang1 secreted from mural cells induces Tie2 activation in endothelial cells to maintain mature blood vessels by enhancing vascular integrity and endothelial survival (4–6). Ang1/Tie2 signaling also plays an important role in physiological and pathological angiogenesis, as opposed to its function in quiescent vasculature (7–9). As to the dual functions of Ang1/Tie2 signaling, we and Saharinen et al. (11) have previously reported that Ang1 assembles distinct Tie2 signaling complexes in the presence or absence of endothelial cell-cell junctions, thereby regulating both vascular quiescence and angiogenesis (10). Ang1 induces trans-association of Tie2 in the presence of cell-cell contacts, whereas Tie2 is anchored to the cell-substratum contacts through extracellular matrix-bound Ang1 in the isolated endothelial cells. Trans-associated Tie2 and extracellular matrix-anchored Tie2 stimulate AKT and extracellular signal-regulated kinase 1/2 pathways preferable for vascular quiescence and angiogenesis, respectively.
By performing DNA microarray analyses, we have revealed that trans-associated Tie2, but not extracellular matrix-anchored Tie2, regulates the expression of genes involved in vascular quiescence, which include Krüppel-like factor 2, delta-like 4 (Dll4), TIS11d, and connexin-40 (10). We have extended the studies on Ang1-mediated vascular quiescence and have shown that Ang1-induced Krüppel-like factor 2 expression occurs through the phosphoinositide 3-kinase (PI3K)/AKT pathway-mediated activation of myocyte enhancer factor 2, and counteracts vascular endothelial growth factor (VEGF)-mediated inflammatory responses (12).
Dll4 is a type 1 membrane protein belonging to the Delta/Serrate/Lag2 family of Notch ligands. Notch signaling is an evolutionally conserved pathway involved in cell fate specification during embryonic and postnatal development, and plays crucial roles in multiple aspects of vascular development such as arterial venous cell fate determination and tip/stalk cell specification during sprouting angiogenesis (13–15). Delta/Serrate/Lag2 ligands bind to Notch family receptors in a cell-cell contact-dependent manner, leading to cleavage of the Notch intracellular domain (NICD). NICD cleaved from Notch enters into the nucleus, associates with transcription factor RBP-J, and regulates the expression of Hes (Hairy/Enhancer of Slit) and Hey (Hes related with YRPW, also known as HesR, HRT and HERP) family of transcriptional repressors (16).
During the tip-stalk cell communication during sprouting angiogenesis, the Dll4/Notch signal is well characterized (17–21). VEGF up-regulates Dll4 expression in endothelial tip cells, which in turn leads to Notch activation in adjacent stalk cells. The stalk cells subsequently lose their responsiveness to VEGF through down-regulation of VEGF receptors such as VEGFR2 and Neuropilin-1 (22), thereby maintaining a quiescent and stabilized phenotype. Similarly, Dll4/Notch signaling is reported to be involved in tumor angiogenesis. In tumor vasculature, tumor-derived VEGF induces Dll4 expression in endothelial cells, which acts as a negative regulator of tumor angiogenesis, but is required for formation of functional vascular network (23–25). Indeed, blockade of Dll4/Notch signaling in tumor vasculature inhibits tumor growth by promoting non-productive angiogenesis associated with excessive sprouting from tumor vessels. The effect of Dll4/Notch signaling on tumor vasculature is reminiscent of that of Ang1/Tie2 signaling. Ang1/Tie2 signaling is also capable of inducing normalization of tumor vasculature by reducing excessive endothelial sprouting and promoting pericyte coverage (26–29).
The Notch signaling not only restricts angiogenesis but also maintains vascular quiescence (15, 30). It has been reported that conditional deletion of RBP-J, the key transcription factor downstream of the Notch receptor, induces spontaneous angiogenesis in quiescent adult vasculature (31). Similarly, Tie2 is activated in the endothelium of quiescent adult vasculature, and is believed to be involved in the maintenance of vascular quiescence (6, 32). Furthermore, both Ang1/Tie2 and Dll4/Notch signaling promote recruitment of mural cells to the vessel wall and induce deposition of basement membrane proteins around the vessels, both of which are important for vascular stabilization (26, 33–37).
Besides the role for the Dll4/Notch signal in the tip-stalk communication, the Dll4/Notch signal appears to function in mature blood vessels with tight interendothelial cell-cell contacts. Functional similarity between the Ang1/Tie2 signal and Dll4/Notch signal and our previous data that Ang1 induced Dll4 expression prompted us to test our hypothesis that the Ang1/Tie2 signal may promote vascular stabilization through the Dll4/Notch signal and to investigate how Dll4 is induced by Ang1/Tie2 signaling. In this study, we found that Ang1 induces activation of β-catenin through PI3K/AKT pathway-mediated inhibition of glycogen synthase kinase 3β (GSK3β) and that the stabilized β-catenin subsequently enhances Notch signal-induced Dll4 expression by forming a complex with NICD/RBP-J on the Dll4 intron3 enhancer, thereby potentiating the Dll4/Notch signal leading to vascular stabilization.
EXPERIMENTAL PROCEDURES
Reagents, Antibodies, and siRNAs
Ang1, angiopoietin-2, (Ang2), and cartilage oligomeric matrix protein (COMP)-Ang1 were prepared as described (38). Other reagents were purchased as follows: wortmannin, AKT inhibitor IV, and N-(N-(3,5-difluorophenacetyl)-l-alanyl)-S-phenylglycine t-butyl ester (DAPT) from Calbiochem; SB216763 from Sigma; lithium chloride (LiCl) from Wako Pure Chemical Industries; and basic fibroblast growth factor (bFGF) from PeproTech. Antibodies were purchased as follows: anti-Dll4, anti-cleaved Notch1 (Val1744) (anti-NICD), anti-AKT, anti-phospho-AKT, anti-GSK3β, and anti-phospho-GSK3β (Ser9) from Cell Signaling Technology; anti-tubulin from Sigma; anti-β-catenin from BD Biosciences; anti-RBP-J (K0043) from Tokusyu-meneki Laboratory; anti-collagen type IV from Millipore; rhodamine-phalloidin from Invitrogen Corp.; and horseradish peroxidase-coupled sheep anti-mouse and anti-rabbit IgG from GE Healthcare. Stealth small interfering RNAs (siRNAs) targeting the genes indicated were purchased from Invitrogen Corp.: human Dll4 (HSS123068, HSS182569) and human β-catenin (VHS50819, VHS50822).
Cell Culture, Transfection, siRNA-mediated Protein Knockdown, and Adenovirus Infection
Human umbilical vein endothelial cells (HUVECs) and human aortic endothelial cells were purchased from Kurabo, maintained as described previously (10, 39), and used for the experiments before passages 8 and 10, respectively. Human dermal microvascular endothelial cells were purchased from Kurabo, and maintained in HuMedia-MvG with a growth additive set. The cells were placed on collagen-coated plates at densities of 2,000 and 40,000 cells/cm2, and cultured overnight to obtain sparse and confluent cell cultures, respectively. HUVECs were transfected using Lipofectamine 2000 (Invitrogen) and Lipofectamine Plus reagents (Invitrogen) according to the manufacturer's instructions. For siRNA-mediated gene silencing, HUVECs were transfected with 20 nm siRNA duplexes using Lipofectamine RNAi MAX reagent (Invitrogen), and cultured for 36–48 h. As a control, siRNA duplexes with irrelevant sequences were used. Then, the cells were harvested, re-plated on collagen-coated plates, and cultured for an additional 24 h before experiments. HUVECs were infected with adenovirus vectors at the appropriate multiplicity of infection. Forty-eight h after infection, the cells were used for experiments.
Plasmids and Adenoviruses
The DNA, including intron 3 of the human DLL4 gene, was amplified by PCR using the genomic DNA extracted from HUVECs as a template and the following primer set: 5′-gtgagtagctcgctccgc-3′ and 5′-ctgagggggcagagggtc-3′. The amplified DNA was cloned into pGL3 Promoter vector (Promega Corporation) to construct the Dll4-Int3-Luc reporter plasmid. To generate the Dll4-Int3mut-Luc reporter plasmid, the RBP-J binding site was mutated using the QuikChange Site-directed Mutagenesis kit (Stratagene) with the Dll4-Int3-Luc plasmid as a template. To construct the p3xFLAG-NICD plasmid encoding FLAG-tagged NICD, a DNA fragment encoding the Notch1 intracellular domain was excised from the pcDNA-FLAG-Notch1-ICD vector, a gift from M. Kurabayashi (Gunma University), and subcloned into p3xFLAG-CMV10 vector (Sigma). A cDNA encoding human Foxc2 was amplified by PCR using human heart cDNAs as a template, and cloned into pERed-NLS vector, a gift from M. Matsuda (40), namely pERed-NLS-Foxc2 plasmid. A 3.7-kb fragment of the mouse Dll4 promoter (−3631/+76) cloned in the pGL3 Basic vector (Promega Corporation) has already been reported (41). An expression plasmid encoding the constitutively active form of β-catenin (CA-βCat) in which Ser37 is replaced with Ala, was kindly provided by J. S. Gutkind (National Institute of Health). Other vectors are purchased as follows: pRL-SV40 and pRL-TK from Promega Corporation and TOPflash reporter plasmid from Millipore Corporation. Recombinant adenovirus vectors encoding LacZ and the constitutively active form of AKT (CA-AKT) were kindly provided by M. Matsuda (Kyoto University) and Y. Fujio (Osaka University), respectively.
Real Time Reverse Transcription-PCR
Endothelial cells placed on collagen-coated plates under either sparse or confluent culture conditions were starved in medium 199 containing 1% BSA for 12 h, and stimulated with either 400 ng/ml of COMP-Ang1 or 10 μm SB216763 as described in the figure legends. After stimulation, total RNA was purified using TRIzol (Invitrogen). Quantitative real time reverse transcription (RT)-PCR was carried out using the QuantiFast SYBR Green RT-PCR kit (Qiagen) as described (12). For each reaction, 100 ng of total RNA was transcribed for 10 min at 50 °C, followed by a denaturing step at 95 °C for 5 min and 40 cycles of 10 s at 95 °C and 30 s at 60 °C. Fluorescence data were collected and analyzed using Mastercycler ep realplex (Eppendorf). The primers used for amplification were as follows: human Dll4, 5′-tccaactgcccttcaatttcac-3′ and 5′-ctggatggcgatcttgctga-3′; for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′-atggggaaggtgaaggtcg-3′ and 5′-ggggtcattgatggcaacaata-3′. For normalization, expression of human GAPDH was determined in parallel as an endogenous control.
Immunoprecipitation and Western Blot Analysis
Confluent and sparse HUVECs plated on a collagen-coated dish were starved in medium 199 containing 1% BSA for 12 h, and stimulated as described in the figure legends. After stimulation, the cells were lysed in ice-cold lysis buffer containing 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 20 mm sodium fluoride, 1 mm sodium vanadate, and 1× protease inhibitor mixture (Roche Applied Science), and centrifuged at 15,000 × g for 20 min at 4 °C. The supernatant was used as precleared cell lysate. To detect the Dll4 protein expression and the NICD production, the cell lysates were subjected to SDS-PAGE and Western blot analysis with anti-Dll4 and anti-NICD antibodies. To evaluate phosphorylation of AKT and GSK3β, aliquots of cell lysate were subjected to Western blot analysis with anti-phospho-AKT and anti-phospho-GSK3β antibodies, respectively. The total contents of AKT and GSK3β in each cell lysate were also assayed in a parallel run using corresponding antibodies. To detect interaction between NICD and β-catenin, NICD was immunoprecipitated with anti-NICD antibody from the precleared lysates. Immunoprecipitated NICD and aliquots of cell lysate were subjected to SDS-PAGE and Western blot analysis with anti-β-catenin and anti-NICD antibodies, respectively.
Luciferase Reporter Assay
Luciferase reporter assay was performed as described previously (42). Briefly, HUVECs plated on a collagen-coated dish were transfected with different expression vectors, together with reporter plasmids as described in the figure legends. The total amount of plasmid DNA was adjusted with empty vector. The cells were harvested, and re-plated on the collagen-coated 24-well plate in confluent culture conditions. Luciferase activity was assayed 48 h after the transfection. To examine the effect of COMP-Ang1, the cells were starved and stimulated as described in the figure legends. The cells were lysed using passive lysis buffer (Promega Corporation), and luciferase activities in cell extract were determined using a dual luciferase assay system (Promega Corporation).
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) assay was performed using the EZ ChIPTM kit (Millipore Corporation) according to the manufacturer's instructions. Confluent HUVECs plated on collagen-coated dishes were starved in medium 199 containing 1% BSA for 12 h, and stimulated with 400 ng/ml of COMP-Ang1 for 30 min. After stimulation, genomic DNA and protein were cross-linked by addition of formaldehyde (1% final concentration) directly to culture medium, and incubated for 10 min at room temperature (RT). The cells were then harvested, lysed, and sonicated to generate 0.3–1.0-kb DNA fragments. After centrifugation, the cleared supernatant was incubated with anti-RBP-J, anti-NICD, anti-β-catenin, and control antibodies for immunoprecipitation. Co-immunoprecipitated and input DNA were used as a template for PCR amplification. PCR amplifications were carried out using the primers specific for intron 3 of the human Dll4 gene (5′-gacgcttagcttggcctggagctg-3′ and 5′-tgtaaaatacaggaaggggcccgtcag-3′). PCR sensitivity was evaluated with serial dilutions of input DNA collected after sonication. Amplified DNA was separated on 2% agarose gels and visualized with ethidium bromide.
Endothelial Cell Tube Formation Assay
Endothelial cell tube formation assay was performed according to the method of Davis and co-workers (43). HUVECs were suspended in 2.5 mg/ml of collagen type I matrices (Nitta Gelatin) at a density of 2 × 106 cells/ml, and incubated at 37 °C for 48 h in medium 199 containing reduced serum supplement, bFGF at 40 ng/ml, and ascorbic acid at 50 μg/ml. During the incubation, the cells were stimulated with or without COMP-Ang1 in the presence or absence of 20 μm DAPT. The cultures were fixed in PBS containing 2% paraformaldehyde for 2 h at RT, and blocked with PBS containing 1% BSA for 12 h at 4 °C. To detect extracellular deposition of collagen type IV, the cultures were stained with anti-collagen type IV antibody for 12 h at 4 °C, and visualized with Alexa 488-labeled donkey anti-goat IgG. To visualize filamentous actin, the cultures were subsequently permeabilized with 0.1% Triton X-100 for 1 h at RT, and stained with rhodamine-phalloidin for 12 h at 4 °C. Fluorescence images of Alexa 488 and rhodamine were recorded with a FV1000 confocal microscope (Olympus Corporation) with a ×20 water immersion objective lens. To quantify the extracellular deposition of collagen type IV, fluorescence intensity of Alexa 488 within the areas of the rhodamine-marked tube structures was determined using FluoView software (Olympus Corporation). Data were expressed as average pixel intensity in the areas of tube structures.
Statistical Analysis
The values are expressed as mean ± S.D. Statistical significance was determined using one-way analysis of variance or unpaired t test. p values < 0.05 were considered statistically significant.
RESULTS
Ang1 Induces Notch Signaling by Up-regulating Dll4 under Confluent, But Not Sparse Cultures of HUVECs
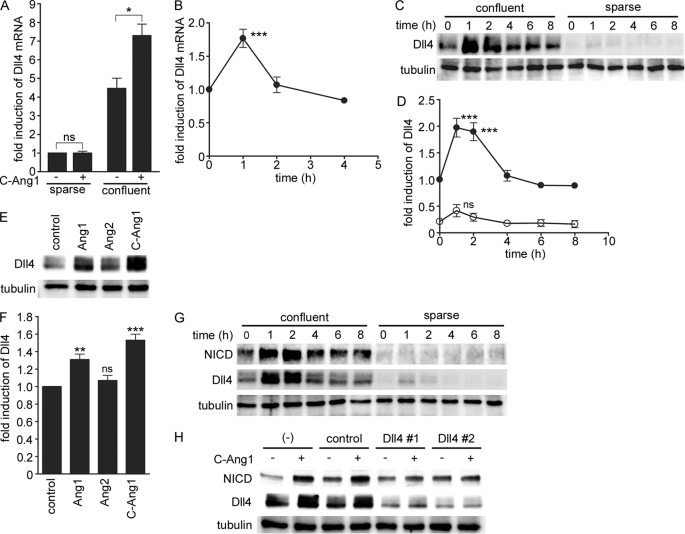
We previously found that Dll4 expression was up-regulated in confluent HUVECs stimulated with Ang1 by the microarray analyses (10). To first confirm whether Dll4 expression is increased by Ang1 in the HUVECs with cell-cell contacts, HUVECs were stimulated with COMP-Ang1, a potent Ang1 variant, under either confluent or sparse culture conditions. Before the stimulation, Dll4 mRNA was ∼4 times higher in confluent HUVECs than in the sparse cells, indicating that the basal Notch signal is present in the HUVECs with cell-cell contacts (Fig. 1A). COMP-Ang1 significantly increased Dll4 mRNA in the confluent HUVECs, which peaked at 1 h after the stimulation and immediately declined to the basal level by 2 h (Fig. 1, A and B). Similarly, DLL4 mRNA levels were increased by stimulation with COMP-Ang1 in human aortic endothelial cells and human dermal microvascular endothelial cells under confluent culture conditions (supplemental Fig. S1). However, in the sparse HUVECs, DLL4 mRNA was not affected by the stimulation with COMP-Ang1 (Fig. 1A). Consistently, Dll4 in the confluent HUVECs was higher than that in the sparse cells, and was increased in response to COMP-Ang1 (Fig. 1, C and D). In contrast, COMP-Ang1 did not induce Dll4 in the sparse cells (Fig. 1, C and D). Dll4 was induced by native Ang1 as well as COMP-Ang1, but not by Ang2, an antagonist for Tie2 (Fig. 1, E and F). These results suggest that Dll4 up-regulation by Ang1 depends on the cell-cell contacts that allow activation of Notch signaling, and is dependent on the specific signal downstream of the trans-associated Tie2 by Ang1.
FIGURE 1.
Ang1 induces Dll4 expression leading to activation of Notch signaling in confluent endothelial cells. A, sparse and confluent HUVECs were starved in medium 199 containing 1% BSA for 12 h, and stimulated with vehicle (−) or COMP-Ang1 at 400 ng/ml (C-Ang1) for 1 h. (COMP-Ang1 was used at the concentration of 400 ng/ml throughout the following experiments.) After stimulation, total RNA was extracted and subjected to real time RT-PCR analysis to determine the expression of Dll4 mRNA as described under “Experimental Procedures.” Bar graphs show relative mRNA levels of Dll4 mRNA normalized to that of GAPDH. Data are expressed as fold-induction relative to that in the vehicle-treated sparse cells, and shown as mean ± S.D. of three independent experiments. B, confluent HUVECs starved for 12 h were stimulated with COMP-Ang1 for the periods indicated at the bottom (h). Dll4 mRNA levels were analyzed by real time RT-PCR as described in A. Values are expressed as fold-induction relative to that in the unstimulated cells, and shown as mean ± S.D. of five independent experiments. C, confluent and sparse HUVECs were starved in medium 199 containing 1% BSA for 12 h, and stimulated with COMP-Ang1 for the periods indicated at the top (h). Cell lysates were subjected to Western blot analysis with anti-Dll4 (Dll4) and anti-tubulin (tubulin) antibodies. D, the relative expression of Dll4 observed in C are quantified by normalizing the expression of Dll4 by that of tubulin. Values are expressed as fold-induction relative to that observed in the confluent unstimulated cells, and shown as mean ± S.D. of three independent experiments. E, confluent HUVECs starved for 12 h were stimulated with vehicle (control), 600 ng/ml of Ang1 (Ang1), 600 ng/ml of Ang2 (Ang2) and COMP-Ang1 (C-Ang1) for 1 h. Cell lysates were subjected to Western blot analysis with anti-Dll4 (Dll4) and anti-tubulin (tubulin) antibodies. F, expression of Dll4 protein observed in E are quantified as described in D. Values are expressed as fold-induction relative to that observed in the control cells, and shown as mean ± S.D. of four independent experiments. G, confluent and sparse HUVECs were starved in medium 199 containing 1% BSA for 12 h, and stimulated with COMP-Ang1 for the periods indicated at the top (h). Cell lysates were subjected to Western blot analysis with anti-NICD (NICD), anti-Dll4 (Dll4), and anti-tubulin (tubulin) antibodies. H, confluent HUVECs transfected without (−) or with either control siRNA (control) or two independent siRNAs targeting Dll4 (Dll4#1 and Dll4#2) were stimulated with COMP-Ang1 as described in A. Cell lysates were subjected to Western blot analysis with anti-NICD (NICD), anti-Dll4 (Dll4), and anti-tubulin (tubulin) antibodies. Significant differences between two groups (A) or from the control (B, D, and F) are indicated as: *, p < 0.05; **, p < 0.01; or ***, p < 0.001. n.s. indicates no significance between two groups or from the control.
To further investigate whether Dll4 expression by trans-associated Tie2 leads to activation of Notch signaling, we examined the amount of NICD. COMP-Ang1 increased NICD in parallel with Dll4 up-regulation under confluent culture conditions, which peaked at 1–2 h after stimulation and declined to basal levels by 4 h, although NICD was not induced by COMP-Ang1 in the sparse cells (Fig. 1G). In addition, depletion of Dll4 by siRNA blocked increase in NICD by COMP-Ang1 (Fig. 1H). Collectively, these results indicate that Tie2 activation in the presence of cell-cell contacts results in activation of Notch signaling by up-regulating Dll4 expression.
A PI3K/AKT Pathway Is Involved in Ang1-induced Dll4 Expression
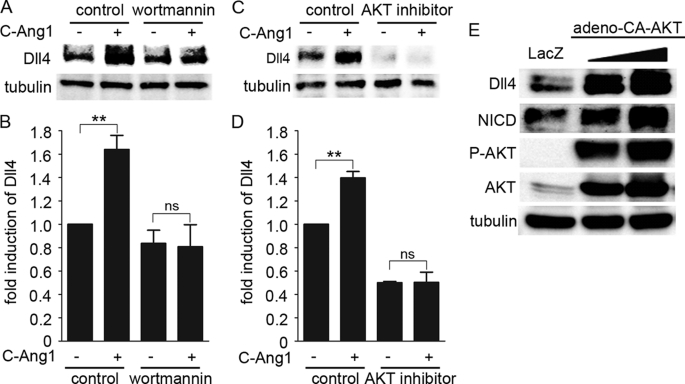
To understand the molecular mechanism underlying Dll4 expression by Ang1/Tie2 in confluent cells, we focused on the downstream signaling of Tie2 in the presence of cell-cell contacts. We previously demonstrated that the PI3K/AKT signal is preferentially activated by trans-associated Tie2 (10). Thus, we next investigated involvement of the PI3K/AKT pathway in Ang1-induced Dll4 expression by using specific inhibitors for PI3K (wortmannin) and AKT (AKT inhibitor). Either inhibitor prevented not only COMP-Ang1-induced AKT activation but also COMP-Ang1-induced Dll4 expression (Fig. 2, A–D, and supplemental Fig. S2, A and B), indicating the requirement of the PI3K/AKT pathway for Ang1-induced Dll4 expression. We further tested whether activation of the PI3K/AKT pathway is sufficient to induce Dll4 expression by infecting HUVECs with adenovirus-encoding CA-AKT, an active mutant of AKT. Overexpression of CA-AKT led to the increase in both Dll4 and NICD (Fig. 2E). These findings indicate that Ang1 activates Notch signaling through the PI3K/AKT pathway-mediated Dll4 expression.
FIGURE 2.
Ang1 induces Dll4 expression through a PI3K/AKT pathway. A, confluent HUVECs starved for 12 h were pretreated with vehicle (control) or 60 nm wortmannin for 30 min, and subsequently stimulated with vehicle (−) or COMP-Ang1 (+) for 1 h. Dll4 protein expression was examined by Western blot analysis as described in the legend of Fig. 1C. B, expression of Dll4 protein observed in A are quantified as described in legend of Fig. 1D. Values are expressed as fold-induction relative to that in the wortmannin-untreated cells stimulated with vehicle, and shown as mean ± S.D. of five independent experiments. C, confluent HUVECs starved for 12 h were pretreated with vehicle (control) or 8 μm AKT inhibitor for 10 min and subsequently stimulated with vehicle (−) or COMP-Ang1 (+) for 1 h. Dll4 protein expression was analyzed as described in A. D, expression of Dll4 protein observed in C are quantified as described in legend of Fig. 1D. Values are expressed as fold-induction relative to that in the AKT inhibitor-untreated cells stimulated with vehicle, and shown as mean ± S.D. of four independent experiments. E, confluent HUVECs were infected with adenoviruses encoding LacZ or with two different titers of adenoviruses encoding AKT-CA for 48 h. Cell lysates were subjected Western blot analysis with anti-Dll4 (Dll4), anti-NICD (NICD), anti-phospho-AKT (P-AKT), anti-AKT (AKT), and anti-tubulin (tubulin) antibodies. Significant differences between two groups (B and D) are indicated as: **, p < 0.01. n.s. indicates no significance between two groups.
GSK3β Is a Downstream Target of AKT Responsible for Ang1-induced Dll4 Expression
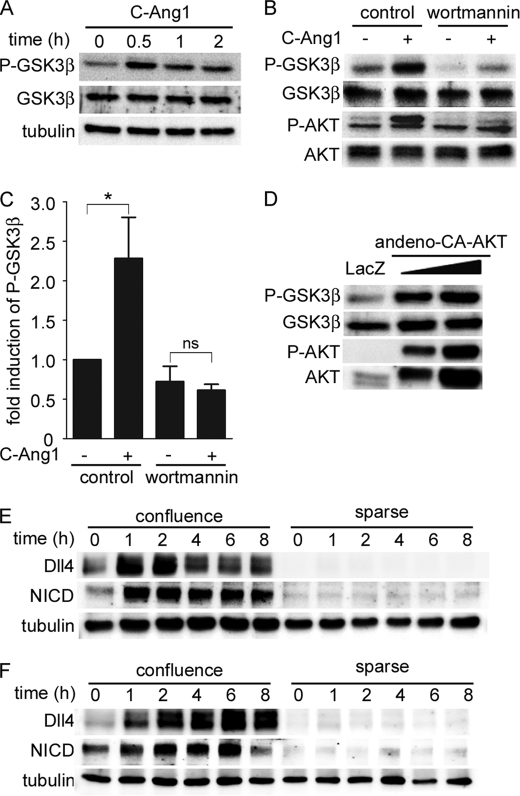
Among the substrates of AKT, GSK3β is a well documented downstream target of the PI3K/AKT pathway that regulates various cellular functions (44). Thus, we investigated whether GSK3β acts downstream of the PI3K/AKT pathway to mediate Ang1-induced Dll4 expression. AKT phosphorylates GSK3β on Ser9 to render it inactive (44). Therefore, we examined the effect of Ang1 on GSK3β phosphorylated at Ser9 by using the anti-phospho-GSK3β antibody. COMP-Ang1 induced an increase in the phosphorylation of GSK3β, which peaked at 30 min after the stimulation (Fig. 3A). COMP-Ang1-induced GSK3β phosphorylation was inhibited by wortmannin (Fig. 3, B and C). Consistently, adenovirus-mediated overexpression of CA-AKT resulted in increased GSK3β phosphorylation in HUVECs (Fig. 3D). These results indicate that Ang1 inhibits GSK3β through phosphorylation by AKT. We further tested whether GSK3β inactivation is sufficient to induce Dll4 expression. Confluent but not sparse HUVECs treated with GSK3β inhibitors, LiCl and SB216763, exhibited up-regulation of Dll4 and an increase in NICD (Fig. 3, E and F, and supplemental Fig. S3). Collectively, these findings reveal that Ang1 induces Dll4 expression through AKT-mediated inactivation of GSK3β.
FIGURE 3.
Ang1 induces Dll4 expression through AKT-mediated inhibition of GSK3β. A, confluent HUVECs starved for 12 h were stimulated with COMP-Ang1 for the periods indicated at the top (h). Cell lysates were subjected to Western blot analysis with anti-phospho-GSK3β (P-GSK3β), anti-GSK3β (GSK3β), and anti-tubulin (tubulin) antibodies. B, confluent HUVECs starved for 12 h were pretreated with vehicle (control) or 60 nm wortmannin for 30 min, and subsequently stimulated with vehicle (−) or COMP-Ang1 (+) for 30 min. Cell lysates were subjected to Western blot analysis with anti-phospho-GSK3β (P-GSK3β), anti-GSK3β (GSK3β), anti-phospho-AKT (P-AKT), and anti-AKT (AKT) antibodies. C, phosphorylated GSK3β levels observed in B are quantified by normalizing the expression of phosphorylated GSK3β by that of total GSK3β. Values are expressed as fold-induction relative to that in the wortmannin-untreated cells stimulated with vehicle, and shown as mean ± S.D. of three independent experiments. D, confluent HUVECs were infected with adenoviruses encoding either LacZ or CA-AKT. Cell lysates were subjected to Western blot analysis as described in B. E, confluent and sparse HUVECs were starved in medium 199 containing 1% BSA for 12 h, and treated with 10 μm SB216763 for the periods indicated at the top (h). Cell lysates were subjected to Western blot analysis with anti-Dll4 (Dll4), anti-NICD (NICD), and anti-tubulin (tubulin) antibodies. F, the effect of 20 mm LiCl on the expression of Dll4 and NICD was analyzed as described in E. In C, a significant difference between two groups is indicated as: *, p < 0.05. n.s. indicates no significance between two groups.
β-Catenin Is Required for Ang1-induced Dll4 Expression
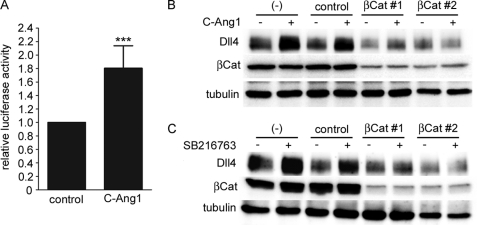
β-Catenin is one of the major substrates of GSK3β, and undergoes proteasomal degradation through GSK3β-mediated phosphorylation (45). Recently, Corada et al. (46) have reported that the Wnt/β-catenin pathway up-regulates Dll4 transcription through the TCF-binding site located 706 bp upstream from the transcription start site of mouse Dll4 gene. Considering these evidences, we hypothesized that stabilization of β-catenin through AKT-mediated inactivation of GSK3β is involved in Ang1-induced Dll4 expression. To address this possibility, HUVECs were transfected with a β-catenin-responsive luciferase reporter construct containing four native TCF binding sites (TOPflash). COMP-Ang1 significantly induced luciferase activity driven by TOPflash reporter (Fig. 4A), indicating the ability of Ang1 to induce β-catenin-dependent transcription. We further clarified the requirement of β-catenin in Ang1-induced Dll4 expression by transfecting HUVECs with two independent siRNAs targeting β-catenin. Depletion of β-catenin by siRNAs completely abolished COMP-Ang1-induced Dll4 expression (Fig. 4B). Similarly, Dll4 expression induced by SB216763 did not occur in the absence of β-catenin (Fig. 4C). These results suggest that Ang1 stimulates β-catenin-dependent transcriptional activity through AKT-mediated inhibition of GSK3β, thereby inducing Dll4 expression.
FIGURE 4.
Ang1 induces Dll4 expression through activation of β-catenin. A, confluent HUVECs were transfected with TOPflash reporter plasmid together with pRL-TK vector. After transfection, the cells were starved in medium 199 containing 1% BSA for 4 h, and stimulated with vehicle (control) or COMP-Ang1 (C-Ang1) for 4 h. After stimulation, the cells were collected, and the lysates were assayed for firefly and Renilla luciferase activities as described under “Experimental Procedures.” The data represent firefly luciferase activity normalized by the Renilla luciferase activity present in each cellular lysate. Values are expressed relative to that observed in the cells treated with vehicle, and shown as mean ± S.D. of four independent experiments. B, confluent HUVECs were transfected without (−) or with either control siRNA (control) or two independent siRNAs targeting β-catenin (βCat#1 and βCat#2). Then, the cells were starved and stimulated with vehicle (−) or COMP-Ang1 (+) for 1 h. Cell lysates were subjected to Western blot analysis with anti-Dll4 (Dll4), anti-β-catenin (βCat), and anti-tubulin (tubulin) antibodies. C, confluent HUVECs transfected with siRNAs as described in B were starved, and treated with vehicle (−) or 10 μm SB216763 (+) for 2 h. Cell lysates were subjected to Western blot analysis as described in B. In A, a significant difference between two groups is indicated as: ***, p < 0.001.
We next investigated whether Ang1 stimulates Dll4 transcription through the TCF-binding site located 706 bp upstream from the transcription initiation site of the mouse Dll4 gene. For that, HUVECs were transfected with the luciferase reporter plasmid in which the reporter is driven by the 3.7-kb mouse Dll4 promoter (Dll4–3.7k-Luc). COMP-Ang1 did not activate the 3.7-kb mouse Dll4 promoter, although Foxc2 significantly stimulated the Dll4–3.7k-Luc reporter activity as previously reported (supplemental Fig. S4, A and B) (41). However, CA-βCat, an active mutant of β-catenin, did not induce luciferase expression driven by the Dll4–3.7k-Luc reporter gene, although the TOPflash reporter activity was significantly enhanced by CA-βCat (supplemental Fig. S4, B and C). These results indicate that Ang1 stimulates Dll4 transcription independently of the TCF-binding element located in the proximal Dll4 promoter.
Cell-Cell Contact-dependent Notch Signaling Is Required for Ang1-induced Dll4 Expression
Expression of Dll4 is higher in confluent endothelial cells than in sparse cells (Fig. 1, C, D, and G). In addition, either COMP-Ang1 or GSK3β inhibitor induced Dll4 up-regulation only in the confluent but not sparse endothelial cells (Figs. 1, A and C, and 3, E and F). These results imply that the cell-cell contact-dependent signal induces Dll4 expression and is required for β-catenin-mediated Dll4 up-regulation. Recently, Yamamizu et al. (47) have reported that β-catenin forms a complex with NICD on the RBP-J binding sites of genes that determine the arterial fate of endothelial cells. Importantly, they also identified the RBP-J binding site within intron 3 of both mouse Dll4 gene and human DLL4 gene by performing in silico analysis of the cis-acting elements (Fig. 5A). These findings prompted us to hypothesize that the Notch signal is a cell-cell contact-dependent signal responsible for Ang1-induced Dll4 expression. To address this possibility, we examined the effect of depletion of NICD by DAPT, a γ-secretase inhibitor, on Ang1-induced Dll4 expression. Treatment of confluent HUVECs with DAPT not only depleted NICD but also reduced basal Dll4 expression. In addition, DAPT prevented COMP-Ang1-induced Dll4 expression and subsequent NICD production (Fig. 5, B and C). Dll4 up-regulation induced by SB216763 was also inhibited by treatment with DAPT (Fig. 5, D and E). These results indicate that cell-cell contact-dependent Notch signaling contributes to basal Dll4 expression and is indispensable for Ang1-induced Dll4 up-regulation through β-catenin.
FIGURE 5.
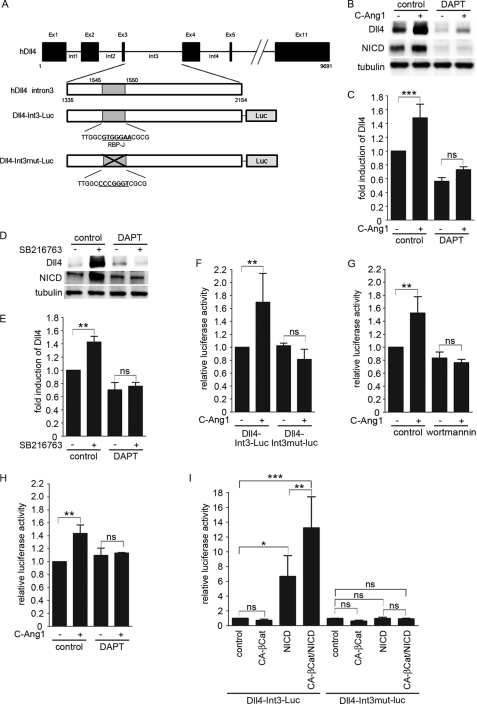
Ang1 stimulates the enhancer activity of the DLL4 intron 3 in a Notch signal-dependent manner. A, the exon-intron organization of the human DLL4 gene and the structures of luciferase reporter constructs. Note that human DLL4 intron 3 contains the RBP-J binding site. The Dll4-Int3-Luc reporter plasmid expresses the firefly luciferase reporter gene under control of the human DLL4 intron 3. In the Dll4-Int3mut-Luc reporter construct, the RBP-J binding site is disrupted. B, confluent HUVECs starved for 12 h were pretreated with vehicle (control) or 10 μm DAPT for 8 h, and subsequently stimulated with vehicle (−) or COMP-Ang1 (+) for 1 h. Western blot analysis was performed as described in the legend of Fig. 1G. C, the expression of Dll4 observed in B are quantified as described in legend of Fig. 1D. Values are expressed as fold-induction relative to that in the DAPT-untreated cells stimulated with vehicle, and shown as mean ± S.D. of five independent experiments. D, confluent HUVECs pretreated with DAPT as described in B were stimulated with vehicle (−) or SB216763 (+) for 2 h. Western blot analysis was performed as described in B. E, the expression of Dll4 observed in D are quantified as described in legend of Fig. 1D. Values are expressed as described in C, and shown as mean ± S.D. of 4 independent experiments. F, confluent HUVECs transfected with either Dll4-Int3-Luc or Dll4-Int3mut-Luc reporter constructs together with pRL-SV40 vector were starved in medium 199 containing 1% BSA for 12 h, and stimulated with vehicle (control) or COMP-Ang1 (+) for 3 h. After the stimulation, the cells were collected, and the lysates were assayed for firefly and Renilla luciferase activities as described in the legend of Fig. 4A. Values are expressed relative to that observed in control cells expressing the Dll4-Int3-Luc plasmid, and shown as mean ± S.D. of three independent experiments. G, confluent HUVECs co-transfected with the Dll4-Int3-Luc reporter plasmid and pRL-SV40 vector were starved in medium 199 containing 1% BSA for 12 h, pretreated with vehicle (control) or 60 nm wortmannin for 30 min, and stimulated with vehicle (−) or COMP-Ang1 (+) for 3 h. After stimulation, the cells were collected, and the lysates were assayed for firefly and Renilla luciferase activities as described in the legend of Fig. 4A. Values are expressed relative to that observed in the wortmannin-untreated cells stimulated with vehicle, and shown as mean ± S.D. of four independent experiments. H, confluent HUVECs co-expressing both Dll4-Int3-Luc plasmid and pRL-SV40 vector were starved for 12 h, pretreated with vehicle (control) or 10 μm DAPT for 1 h, and stimulated with vehicle (−) or COMP-Ang1 (+) for 3 h. Firefly and Renilla luciferase activities were assayed as described in the legend of Fig. 4A. Values are expressed relative to that in the DAPT-untreated cells stimulated with vehicle, and shown as mean ± S.D. of three independent experiments. I, confluent HUVECs were transfected with either Dll4-Int3-Luc or Dll4-Int3mut-Luc reporter construct together with pRL-SV40 vector and the empty vector (control) or the plasmid encoding either CA-βCat or NICD. Cell lysates were assayed for firefly and Renilla luciferase activities as described in the legend of Fig. 4A. Values are expressed relative to that observed in the control cells expressing the Dll4-Int3-Luc reporter plasmid, and shown as mean ± S.D. of five independent experiments. Significant differences between two groups (C and E–I) are indicated as: *, p < 0.05; **, p < 0.01; ***, p < 0.001. n.s. indicates no significance between two groups.
To further investigate whether intron 3 of the DLL4 gene containing the RBP-J binding site acts as an Ang1-responsive enhancer element, HUVECs were transfected with either a plasmid expressing the luciferase reporter gene under control of the human DLL4 intron 3 (Dll4-Int3-Luc) or its mutant plasmid in which the RBP-J binding site is mutated (Dll4-Int3mut-Luc) (Fig. 5A). COMP-Ang1 significantly stimulated Dll4-Int3-Luc reporter activity, which was inhibited by wortmannin (Fig. 5, F and G). In contrast, the Dll4-Int3mut-Luc reporter was not activated by COMP-Ang1 (Fig. 5F). In addition, inhibition of Notch signaling by DAPT abolished COMP-Ang1-induced activation of the Dll4-Int3-Luc reporter (Fig. 5H). These results indicate that Ang1 stimulates the enhancer activity of DLL4 intron 3 in a Notch signal-dependent manner.
Because β-catenin was essential for Ang1-induced Dll4 expression (Fig. 4B), we assumed that β-catenin and NICD might cooperatively stimulate the enhancer activity of the DLL4 intron 3. To address this possibility, HUVECs were transfected with either Dll4-Int3-Luc or the Dll4-Int3mut-Luc reporter together with the plasmid encoding CA-βCat and/or that expressing NICD. NICD stimulated Dll4-Int3-Luc but not Dll4-Int3mut-Luc reporter activity (Fig. 5I). Although CA-βCat did not stimulate both reporter genes, it potently augmented NICD-stimulated Dll4-Int3-Luc reporter activity (Fig. 5I). However, Dll4-Int3mut-Luc reporter activity did not increase even if CA-βCat and NICD were co-expressed (Fig. 5I). Collectively, these results indicate that NICD stimulates the enhancer activity of DLL4 intron 3 via the RBP-J binding site and that β-catenin potentiates the NICD-induced stimulation of the enhancer activity.
Ang1 Recruits β-Catenin to the NICD·RBP-J Complexes on the Dll4 Intron 3
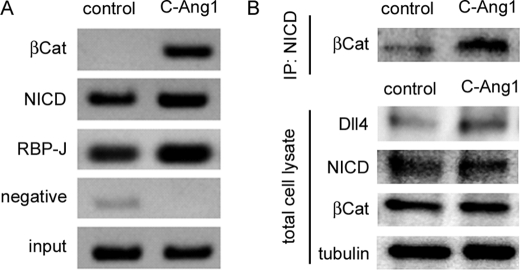
To understand how β-catenin potentiates Notch signal-mediated Dll4 expression, we examined the complex formation of β-catenin, NICD, and RBP-J on the Dll4 intron 3 enhancer region by performing a ChIP assay. Binding of NICD and RBP-J to the DLL4 intron 3 was detected in confluent HUVECs irrespective of the presence and absence of COMP-Ang1 (Fig. 6A). Although β-catenin did not exist in DLL4 intron 3 in the unstimulated confluent cells, COMP-Ang1 potently induced binding of β-catenin to the DLL4 intron 3 (Fig. 6A). Together with the results of Dll4-Int3-Luc reporter assays, these findings suggest that the Ang1/Tie2 signal recruits β-catenin to the NICD·RBP-J complexes on Dll4 intron 3. To confirm it, we carried out a co-immunoprecipitation assay using the anti-NICD antibody. Only a small fraction of β-catenin interacted with NICD in confluent HUVECs (Fig. 6B). However, stimulation with COMP-Ang1 enhanced the association between β-catenin and NICD without affecting the expression of Dll4 and NICD (Fig. 6B). Collectively, these findings indicate that the Ang1/Tie2 signal recruits β-catenin to the NICD·RBP-J complexes on the enhancer region of Dll4 intron 3, thereby inducing Dll4 up-regulation.
FIGURE 6.
β-Catenin is recruited to the NICD·RBP-J complexes on Dll4 intron 3 in response to Ang1. A, confluent HUVECs starved for 12 h were stimulated with vehicle (control) or COMP-Ang1 (C-Ang1) for 30 min. After stimulation, the cells were fixed with formaldehyde, and the cross-linked chromatin was immunoprecipitated with anti-β-catenin (βCat), anti-NICD (NICD), anti-RBP-J (RBP-J), and control (negative) antibodies. Input (input) and co-immunoprecipitated DNA were used as a template for PCR amplification. PCR amplification was performed using the primers specifically targeting Dll4 intron 3. B, confluent HUVECs starved for 12 h were stimulated with COMP-Ang1 as described in A. Cell lysates were immunoprecipitated with anti-NICD antibody. Immunoprecipitates (IP: NICD) and aliquots of cell lysates (total cell lysate) were subjected to Western blot analysis with anti-β-catenin (βCat), anti-Dll4 (Dll4), anti-NICD (NICD), and anti-tubulin (tubulin) antibodies as indicated at the left.
Ang1 Induces Extracellular Deposition of Collagen Type IV through Dll4/Notch Signaling
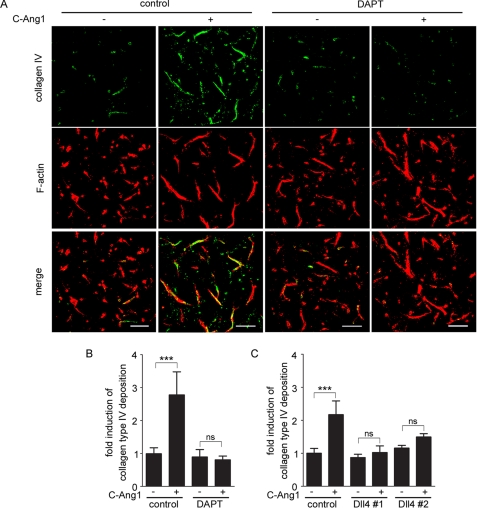
Both Ang1/Tie2 and Dll4/Notch signaling are known to induce formation of the vascular basement membrane (33, 36, 37), which is a hallmark of vascular stabilization. Therefore, we investigated whether Ang1 induces deposition of collagen type IV, a major basement membrane component, during endothelial cell tube formation in three-dimensional collagen matrices. Extracellular deposition of collagen type IV was markedly increased by stimulation with COMP-Ang1 (Fig. 7, A and B). However, inhibition of Notch signaling by treatment with DAPT inhibited COMP-Ang1-induced deposition of collagen type IV (Fig. 7, A and B). Consistently, collagen type IV deposition was not induced by COMP-Ang1 in Dll4-depleted cells (Fig. 7C and supplemental Fig. S5). These findings suggest that Ang1 induces basement membrane formation through Dll4/Notch signaling.
FIGURE 7.
Ang1 induces extracellular deposition of collagen type IV via DII4/Notch signaling. A, HUVECs were cultured to form tube structures in three-dimensional collagen matrices for 48 h. During this period, the cells were stimulated with vehicle (−) or COMP-Ang1 (+) in the presence (DAPT) or absence (control) of 20 μm DAPT as indicated at the top. To detect the extracellular deposition of collagen type IV, the cultures were fixed, immunostained with anti-collagen type IV antibody, and visualized with Alexa 488-conjugated secondary antibody. After permeabilization, the cells were further stained with rhodamine-phalloidin to visualize filamentous actin. Alexa 488 and rhodamine images were obtained through a confocal microscope. Alexa 488 (collagen IV) and rhodamine (F-actin) images and the merged images (merge) are shown as indicated at the left. Scale bar, 100 μm. B, extracellular deposition of collagen type IV was quantified as described under “Experimental Procedures.” Values are expressed as fold-induction relative to that observed in DAPT-untreated cells stimulated with vehicle, and shown as mean ± S.D. of five different fields. Similar results were obtained in four independent experiments. C, HUVECs transfected with either control siRNA (control) or two independent siRNAs targeting Dll4 (Dll4#1 and Dll4#2) were cultured to form tube structures in three-dimensional collagen matrices for 48 h. During this period, the cells were stimulated with vehicle (−) or COMP-Ang1 (+) as indicated at the bottom. The extracellular deposition of collagen type IV was detected and quantified as described in A and B. Values are expressed as fold-induction relative to that observed in control siRNA-transfected cells stimulated with vehicle, and shown as mean ± S.D. of five different fields. In B and C, significant differences between two groups are indicated as: ***, p < 0.001. n.s. indicates no significance between two groups.
DISCUSSION
We here explored how Ang1 induces Dll4 expression and suggested its contribution to Ang1-regulated vascular quiescence. Ang1 assembles distinct Tie2 signaling complexes in the presence or absence of cell-cell junctions, thereby regulating both vascular quiescence and angiogenesis. In the presence of cell-cell junctions, Ang1 induces formation of trans-associated Tie2, which induces expression of the genes involved in vascular stabilization, which include Notch ligand Dll4. Because the Dll4/Notch signal is known to restrict sprouting angiogenesis and promote vascular stabilization (15, 17–21, 30, 31), we hypothesized that the Dll4/Notch signal is involved in Ang1/Tie2 signal-mediated vascular quiescence. To address this possibility, we decided to delineate the signaling pathways underlying Ang1-induced Dll4 expression. We found that the Ang1/Tie2 signal induces activation of β-catenin through AKT-mediated inhibition of GSK3β and that β-catenin resistant to degradation enhances Notch signal-mediated Dll4 expression by forming a complex with NICD/RBP-J on the RBP-J binding site in Dll4 intron 3, thereby potentiating the Dll4/Notch signal leading to vascular quiescence (Fig. 7).
Basal Dll4 expression and NICD is higher in confluent cells than in the sparse cells (Fig. 1G), consistent with the previous report that cell-cell contact-dependent Notch signaling induces Dll4 expression (48). Importantly, either Ang1- or GSK3β inhibitor-induced Dll4 expression requires endothelial cell-cell contacts, and is sensitive to DAPT, suggesting that Notch signaling is a prerequisite for β-catenin-mediated Dll4 expression.
Augmentation of Dll4 expression by Ang1 is dependent on β-catenin. We have previously shown that the Ang1/Tie2 signal preferentially activates PI3K/AKT signaling (10). Although phosphorylation of β-catenin by GSK3β leads to its degradation, β-catenin is stabilized by inhibition of GSK3β by AKT. Dll4 expression by Ang1 was inhibited by depletion of β-catenin and inhibition of either PI3K or AKT (Figs. 2, A–D, and 4B), indicating the essential role of β-catenin for Ang1-induced Dll4 expression. Thus, we further extend the study on the transcriptional regulation of Dll4 by β-catenin.
We first analyzed the −3.7-kb promoter region of the mouse Dll4 gene, because this region contains transcription factor binding sites for forkhead transcription factors and TCF. The forkhead transcription factors, Foxc1 and Foxc2, are the first transcription factors identified to regulate Dll4 expression during vascular development (41). Mouse embryos deficient in both Foxc1 and Foxc2 exhibit arteriovenous malformation and lack of expression of arterial genes, such as Dll4 and ephrinB2. Consistently, Foxc1 and Foxc2 directly activate the Dll4 promoter via the forkhead binding element located ∼3.7 kb upstream from the transcription initiation site (41). Thus, Dll4 induction responsible for arterial-venous cell fate determination appears to be mediated by Foxc genes. However, Ang1 did not stimulate the −3.7-kb Dll4 promoter containing the forkhead binding element, suggesting that Foxc1 and Foxc2 are not involved in Ang1-induced Dll4 expression. In addition, Corada et al. (46) have recently reported that β-catenin up-regulates Dll4 transcription through the TCF-binding site located 706 bp upstream from the transcription initiation site of the mouse Dll4 gene. However, in our experiments, neither Ang1 nor CA-βCat activated the −3.7-kb mouse Dll4 promoter containing the corresponding TCF binding site. Instead, our luciferase reporter assays and ChIP experiments performed in this study revealed that Dll4 intron 3 is an enhancer element responsible for Ang1-induced Dll4 transcription through β-catenin. Currently, the reason for this discrepancy remains unclear, but it may be due to the different cell types used for the experiments. We performed the experiments with HUVECs, whereas they used endothelial cells isolated from mouse embryos (46). Consistent with this idea, VEGF-induced Dll4 expression occurs only in arterial endothelial cell, but not in venous cells (49). Thus, the signaling pathways leading to Dll4 expression may vary in different endothelial cell types and in different upstream mediators.
Ang1 induces recruitment of β-catenin to NICD·RBP-J complexes on the RBP-J binding site in Dll4 intron 3, which enhances NICD-mediated Dll4 expression. Thus, Ang1/Tie2 and Notch signaling converges into β-catenin·NICD·RBP-J complexes on the Dll4 intron 3 enhancer to cooperatively induce Dll4 expression. Consistently, functional interaction between NICD and β-catenin has recently been reported (47, 50). In arterial, but not venous, endothelial cells, β-catenin·NICD·RBP-J complexes are formed on the RBP-J binding sites of arterial genes, thereby regulating their expression leading to arterial fate specification (47). In addition, it has also been shown that β-catenin·NICD·RBP-J complexes on the Hes1 promoter induce Hes1 expression to suppress the differentiation of neural precursor cells (50). Thus, the functional interaction between Notch and β-catenin signaling may be involved in a variety of biological processes.
Both Ang1/Tie2 and Notch signal are known to regulate vascular quiescence. Functional similarity between them and our present evidence that Ang1 induces Dll4 expression leading to Notch activation imply the role of the Dll4/Notch signal in Ang1/Tie2-mediated vascular quiescence. We further revealed that Ang1 induces extracellular deposition of collagen type IV, a major component of basement membrane, during endothelial cell tube formation. This Ang1-mediated deposition of collagen type IV is dependent of the Dll4/Notch signal, as demonstrated by evidence that inhibition of the Notch signal by DAPT and deletion of Dll4 by siRNA prevented this effect (Fig. 7). Because basement membrane matrix assembly is a crucial step for vascular maturation and stabilization (51), these findings suggest that the Ang1/Tie2 signal might promote vascular stabilization through activation of Dll4/Notch signal.
The Dll4/Notch signal is also involved in tip/stalk cell specification (15). Activation of the Notch signal in the stalk cells restricts their angiogenic behavior, thereby maintaining a quiescent and stabilized phenotype of stalk cells. Interestingly, Yana et al. (37) have found by using an ex vivo angiogenesis system that Tie2 is specifically expressed in stalk cells and is involved in vessel maturation. Thus, the Ang1/Tie2 signal may also regulate Dll4 expression in the stalk cells, leading to the maturation of neovessels. However, the in vivo study must be required to clarify the role of cross-talk between the Ang1/Tie2 and Dll4/Notch signal in vascular stabilization.
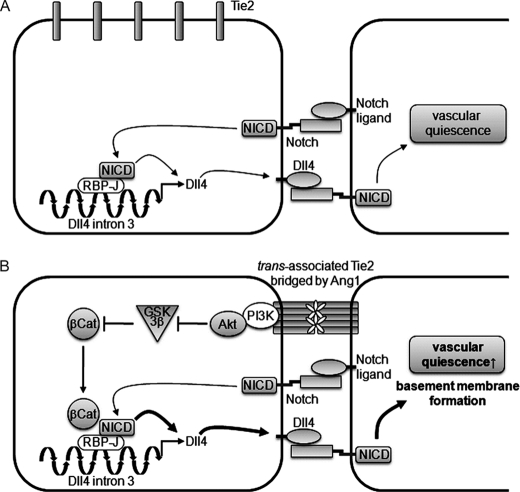
In conclusion, we found that the Ang1/Tie2 signal induces activation of β-catenin through the PI3K/AKT pathway-mediated inhibition of GSK3β in the presence of cell-cell contacts, and that the undegraded β-catenin subsequently potentiates the Notch signal-mediated Dll4 expression by forming a complex with NICD/RBP-J on the RBP-J binding site in Dll4 intron 3, which in turn up-regulates the Dll4/Notch signal. In addition, we also revealed that the Dll4/Notch signal augmented by the Ang1/Tie2 signal promotes formation of vascular basement membrane leading to vascular stabilization (Fig. 8).
FIGURE 8.
Schematic representation of a proposed model for how Ang1/Tie2 signal induces Dll4 expression to potentiate Notch signal. A, in the confluent endothelial cells, cell-cell contact-dependent Notch signaling induces production of NICD, which subsequently binds to the RBP-J binding site in Dll4 intron 3, leading to Dll4 expression. B, in confluent cells, the Ang1/Tie2 signal stimulates the transcriptional activity of β-catenin through the PI3K/AKT pathway-mediated inhibition of GSK3β. The stabilized β-catenin enhances NICD-mediated Dll4 expression by forming a complex with NICD and RBP-J on Dll4 intron 3, which augments the Notch signal. Dll4/Notch signal augmented by the Ang1/Tie2 signal promotes formation of vascular basement membrane leading to vascular quiescence. In the absence of cell-cell contacts, Dll4 expression is very low due to the lack of Notch signaling. Even if the cells are stimulated with Ang1 under this condition, Dll4 up-regulation does not occur, because the Ang1/Tie2 signal is unable to induce Dll4 expression in the absence of Notch signaling (not described in this figure).
Supplementary Material
Acknowledgments
We are grateful to J. S. Gutkind (National Institute of Health) for the CA-βCat plasmid, M. Kurabayashi (Gunma University) for the Notch1 ICD plasmid, M. Matsuda (Kyoto University) and Y. Fujio (Osaka University) for the adenovirus encoding LacZ and CA-AKT, respectively. We also thank K. Hiratomi, M. Sone, M. Minamimoto, and Y. Matsuura for technical assistance, and N. Takakura (Osaka University), J. K. Yamashita (Kyoto University), K. Yamamizu (Kyoto University), and M. Masuda for helpful advice.
This work was supported in part by grants from the Ministry of Education, Science, Sports and Culture of Japan (to S. F. and N. M.), the Ministry of Health, Labor, and Welfare of Japan (to N. M.), and the Program for the Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (to S. F. and N. M.), grants from the Naito Foundation (to S. F.), Takeda Science Foundation (to S. F. and N. M.), the Sagawa Foundation for Promotion of Cancer Research (to S. F.), Mochida Memorial Foundation for Medical and Pharmaceutical Research (to S. F.), Kowa Life Science Foundation (to S. F.), Kanae Foundation for the Promotion of Medical Science (to S. F.), The Novartis Foundation (Japan) for the Promotion of Science (to S. F.), Senri Life Science Foundation (to S. F.), the Mitsubishi Foundation (to N. M.), and a AstraZeneca Research Grant (to N. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- Ang1
- angiopoietin-1
- Dll4
- delta-like 4
- VEGF
- vascular endothelial growth factor
- NICD
- Notch intracellular domain
- GSK3β
- glycogen synthase kinase 3β
- Ang2
- angiopoietin-2
- COMP
- cartilage oligomeric matrix protein
- DAPT
- N-(N-(3,5-difluorophenacetyl)-l-alanyl)-S-phenylglycine t-butyl ester
- HUVECs
- human umbilical vein endothelial cells
- CA-βCat
- constitutively active mutant of β-catenin
- CA-AKT
- constitutively active mutant of AKT.
REFERENCES
- 1. Dumont D. J., Gradwohl G., Fong G. H., Puri M. C., Gertsenstein M., Auerbach A., Breitman M. L. (1994) Genes Dev. 8, 1897–1909 [DOI] [PubMed] [Google Scholar]
- 2. Sato T. N., Tozawa Y., Deutsch U., Wolburg-Buchholz K., Fujiwara Y., Gendron-Maguire M., Gridley T., Wolburg H., Risau W., Qin Y. (1995) Nature 376, 70–74 [DOI] [PubMed] [Google Scholar]
- 3. Suri C., Jones P. F., Patan S., Bartunkova S., Maisonpierre P. C., Davis S., Sato T. N., Yancopoulos G. D. (1996) Cell 87, 1171–1180 [DOI] [PubMed] [Google Scholar]
- 4. Brindle N. P., Saharinen P., Alitalo K. (2006) Circ. Res. 98, 1014–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peters K. G., Kontos C. D., Lin P. C., Wong A. L., Rao P., Huang L., Dewhirst M. W., Sankar S. (2004) Recent Prog. Horm. Res. 59, 51–71 [DOI] [PubMed] [Google Scholar]
- 6. Wong A. L., Haroon Z. A., Werner S., Dewhirst M. W., Greenberg C. S., Peters K. G. (1997) Circ. Res. 81, 567–574 [DOI] [PubMed] [Google Scholar]
- 7. Asahara T., Chen D., Takahashi T., Fujikawa K., Kearney M., Magner M., Yancopoulos G. D., Isner J. M. (1998) Circ. Res. 83, 233–240 [DOI] [PubMed] [Google Scholar]
- 8. Eklund L., Olsen B. R. (2006) Exp. Cell Res. 312, 630–641 [DOI] [PubMed] [Google Scholar]
- 9. Lin P., Polverini P., Dewhirst M., Shan S., Rao P. S., Peters K. (1997) J. Clin. Invest. 100, 2072–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fukuhara S., Sako K., Minami T., Noda K., Kim H. Z., Kodama T., Shibuya M., Takakura N., Koh G. Y., Mochizuki N. (2008) Nat. Cell. Biol. 10, 513–526 [DOI] [PubMed] [Google Scholar]
- 11. Saharinen P., Eklund L., Miettinen J., Wirkkala R., Anisimov A., Winderlich M., Nottebaum A., Vestweber D., Deutsch U., Koh G. Y., Olsen B. R., Alitalo K. (2008) Nat. Cell. Biol. 10, 527–537 [DOI] [PubMed] [Google Scholar]
- 12. Sako K., Fukuhara S., Minami T., Hamakubo T., Song H., Kodama T., Fukamizu A., Gutkind J. S., Koh G. Y., Mochizuki N. (2009) J. Biol. Chem. 284, 5592–5601 [DOI] [PubMed] [Google Scholar]
- 13. Artavanis-Tsakonas S., Rand M. D., Lake R. J. (1999) Science 284, 770–776 [DOI] [PubMed] [Google Scholar]
- 14. Gridley T. (1997) Mol. Cell. Neurosci. 9, 103–108 [DOI] [PubMed] [Google Scholar]
- 15. Phng L. K., Gerhardt H. (2009) Dev. Cell 16, 196–208 [DOI] [PubMed] [Google Scholar]
- 16. Iso T., Kedes L., Hamamori Y. (2003) J. Cell. Physiol. 194, 237–255 [DOI] [PubMed] [Google Scholar]
- 17. Hellström M., Phng L. K., Hofmann J. J., Wallgard E., Coultas L., Lindblom P., Alva J., Nilsson A. K., Karlsson L., Gaiano N., Yoon K., Rossant J., Iruela-Arispe M. L., Kalén M., Gerhardt H., Betsholtz C. (2007) Nature 445, 776–780 [DOI] [PubMed] [Google Scholar]
- 18. Leslie J. D., Ariza-McNaughton L., Bermange A. L., McAdow R., Johnson S. L., Lewis J. (2007) Development 134, 839–844 [DOI] [PubMed] [Google Scholar]
- 19. Lobov I. B., Renard R. A., Papadopoulos N., Gale N. W., Thurston G., Yancopoulos G. D., Wiegand S. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3219–3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Siekmann A. F., Lawson N. D. (2007) Nature 445, 781–784 [DOI] [PubMed] [Google Scholar]
- 21. Suchting S., Freitas C., le Noble F., Benedito R., Bréant C., Duarte A., Eichmann A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3225–3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Williams C. K., Li J. L., Murga M., Harris A. L., Tosato G. (2006) Blood 107, 931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li J. L., Sainson R. C., Shi W., Leek R., Harrington L. S., Preusser M., Biswas S., Turley H., Heikamp E., Hainfellner J. A., Harris A. L. (2007) Cancer Res. 67, 11244–11253 [DOI] [PubMed] [Google Scholar]
- 24. Noguera-Troise I., Daly C., Papadopoulos N. J., Coetzee S., Boland P., Gale N. W., Lin H. C., Yancopoulos G. D., Thurston G. (2006) Nature 444, 1032–1037 [DOI] [PubMed] [Google Scholar]
- 25. Ridgway J., Zhang G., Wu Y., Stawicki S., Liang W. C., Chanthery Y., Kowalski J., Watts R. J., Callahan C., Kasman I., Singh M., Chien M., Tan C., Hongo J. A., de Sauvage F., Plowman G., Yan M. (2006) Nature 444, 1083–1087 [DOI] [PubMed] [Google Scholar]
- 26. Augustin H. G., Koh G. Y., Thurston G., Alitalo K. (2009) Nat. Rev. Mol. Cell Biol. 10, 165–177 [DOI] [PubMed] [Google Scholar]
- 27. Falcón B. L., Hashizume H., Koumoutsakos P., Chou J., Bready J. V., Coxon A., Oliner J. D., McDonald D. M. (2009) Am. J. Pathol. 175, 2159–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hawighorst T., Skobe M., Streit M., Hong Y. K., Velasco P., Brown L. F., Riccardi L., Lange-Asschenfeldt B., Detmar M. (2002) Am. J. Pathol. 160, 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Machein M. R., Knedla A., Knoth R., Wagner S., Neuschl E., Plate K. H. (2004) Am. J. Pathol. 165, 1557–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Phng L. K., Potente M., Leslie J. D., Babbage J., Nyqvist D., Lobov I., Ondr J. K., Rao S., Lang R. A., Thurston G., Gerhardt H. (2009) Dev. Cell 16, 70–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dou G. R., Wang Y. C., Hu X. B., Hou L. H., Wang C. M., Xu J. F., Wang Y. S., Liang Y. M., Yao L. B., Yang A. G., Han H. (2008) FASEB J. 22, 1606–1617 [DOI] [PubMed] [Google Scholar]
- 32. Fukuhara S., Sako K., Noda K., Zhang J., Minami M., Mochizuki N. (2010) Histol. Histopathol. 25, 387–396 [DOI] [PubMed] [Google Scholar]
- 33. Benedito R., Trindade A., Hirashima M., Henrique D., da Costa L. L., Rossant J., Gill P. S., Duarte A. (2008) BMC Dev. Biol. 8, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Iivanainen E., Nelimarkka L., Elenius V., Heikkinen S. M., Junttila T. T., Sihombing L., Sundvall M., Maatta J. A., Laine V. J., Yla-Herttuala S., Higashiyama S., Alitalo K., Elenius K. (2003) FASEB J. 17, 1609–1621 [DOI] [PubMed] [Google Scholar]
- 35. Kobayashi H., DeBusk L. M., Babichev Y. O., Dumont D. J., Lin P. C. (2006) Blood 108, 1260–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trindade A., Kumar S. R., Scehnet J. S., Lopes-da-Costa L., Becker J., Jiang W., Liu R., Gill P. S., Duarte A. (2008) Blood 112, 1720–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yana I., Sagara H., Takaki S., Takatsu K., Nakamura K., Nakao K., Katsuki M., Taniguchi S., Aoki T., Sato H., Weiss S. J., Seiki M. (2007) J. Cell Sci. 120, 1607–1614 [DOI] [PubMed] [Google Scholar]
- 38. Cho C. H., Kammerer R. A., Lee H. J., Steinmetz M. O., Ryu Y. S., Lee S. H., Yasunaga K., Kim K. T., Kim I., Choi H. H., Kim W., Kim S. H., Park S. K., Lee G. M., Koh G. Y. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 5547–5552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fukuhara S., Sakurai A., Sano H., Yamagishi A., Somekawa S., Takakura N., Saito Y., Kangawa K., Mochizuki N. (2005) Mol. Cell. Biol. 25, 136–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aoki K., Nakamura T., Fujikawa K., Matsuda M. (2005) Mol. Biol. Cell 16, 2207–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Seo S., Fujita H., Nakano A., Kang M., Duarte A., Kume T. (2006) Dev. Biol. 294, 458–470 [DOI] [PubMed] [Google Scholar]
- 42. Fukuhara S., Marinissen M. J., Chiariello M., Gutkind J. S. (2000) J. Biol. Chem. 275, 21730–21736 [DOI] [PubMed] [Google Scholar]
- 43. Koh W., Stratman A. N., Sacharidou A., Davis G. E. (2008) Methods Enzymol. 443, 83–101 [DOI] [PubMed] [Google Scholar]
- 44. Frame S., Cohen P. (2001) Biochem. J. 359, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu D., Pan W. (2010) Trends Biochem. Sci. 35, 161–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Corada M., Nyqvist D., Orsenigo F., Caprini A., Giampietro C., Taketo M. M., Iruela-Arispe M. L., Adams R. H., Dejana E. (2010) Dev. Cell 18, 938–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yamamizu K., Matsunaga T., Uosaki H., Fukushima H., Katayama S., Hiraoka-Kanie M., Mitani K., Yamashita J. K. (2010) J. Cell Biol. 189, 325–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Benedito R., Roca C., Sörensen I., Adams S., Gossler A., Fruttiger M., Adams R. H. (2009) Cell 137, 1124–1135 [DOI] [PubMed] [Google Scholar]
- 49. Liu Z. J., Shirakawa T., Li Y., Soma A., Oka M., Dotto G. P., Fairman R. M., Velazquez O. C., Herlyn M. (2003) Mol. Cell. Biol. 23, 14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shimizu T., Kagawa T., Inoue T., Nonaka A., Takada S., Aburatani H., Taga T. (2008) Mol. Cell. Biol. 28, 7427–7441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Davis G. E., Senger D. R. (2005) Circ. Res. 97, 1093–1107 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.