Abstract
Proliferation of cerebellar granular neuronal precursors (CGNPs) is mediated by Sonic Hedgehog (Shh), which activates the Patched and Smoothened (Smo) receptor complex. Although its protein sequence suggests that Smo is a G protein coupled receptor (GPCR), the evidence that this receptor utilizes heterotrimeric G proteins as downstream effectors is controversial. In Drosophila, Gαi is required for Hedgehog (Hh) activity, but the involvement of heterotrimeric G proteins in vertebrate Shh signaling has not yet been established. Here, we show that Shh-induced proliferation of rat CGNPs is enhanced strongly by the expression of the active forms of Gαi/o proteins (Gαi1, Gαi2, Gαi3, and Gαo) but not by members of another class (Gα12) of heterotrimeric G proteins. Additionally, the mRNAs of these different Gαi members display specific expression patterns in the developing cerebellum; only Gαi2 and Gαi3 are substantially expressed in the outer external granular layer, where CGNPs proliferate. Consistent with this, Shh-induced proliferation of CGNPs is reduced significantly by knockdowns of Gαi2 and Gαi3 but not by silencing of other members of the Gαi/o class. Finally, our results demonstrate that Gαi2 and Gαi3 locate to the primary cilium when expressed in CGNP cultures. In summary, we conclude that the proliferative effects of Shh on CGNPs are mediated by the combined activity of Gαi2 and Gαi3 proteins.
Keywords: G proteins, Neurobiology, Neurodevelopment, Signal transduction, Tumor promoter, Brain tumors, Cerebellar Granular Neuron Precursors, G alpha inhibitory proteins, Medulloblastoma, Sonic Hedgehog
Introduction
Cerebellar granular neuronal precursors (CGNPs)2 are generated within the external germinal layer (EGL) during development of the cerebellar cortex. During cerebellum development, CGNPs exit the cell-cycle and migrate through the Purkinje cells to establish the three layers of the cerebellar cortex (1–3). Clonal expansion of CGNPs is achieved by the mitogenic activity of Sonic hedgehog (Shh) signaling emanating from the Purkinje cells to the EGL (4, 5). The binding of Shh to its membrane receptor Patched, located at the peri-ciliar area, promotes the accumulation of Smoothened (Smo) at the primary cilium and concomitant activation of this pathway (6, 7).
The Hedgehog (Hh) pathway can be regulated negatively by sequential phosphorylation of Gli2/3 by PKA, GSK3β and CK (8). These phosphorylation events target Gli2/3 to the proteasome, where removal of its transactivation domain generates a potent transcriptional repressor of the pathway (9, 10). Several lines of evidence suggest that inhibition of PKA is a universal requirement for Hh pathway activation. However, the precise details of this mechanism are not understood. Although PKA enzymatic activity is primarily controlled by cAMP levels, its sub-cellular distribution is determined by the association with specific members of protein kinase A anchoring protein (AKAP) protein family which permits local regulation of its activity (11, 12). In the basal state, the PKA catalytic subunit (C-PKA) is inactive due to the presence of a bound regulatory subunit of PKA. Increased levels of cAMP bind to and displace the regulatory subunit of PKA, thereby permitting active PKA to phosphorylate different substrates (13). The intracellular cAMP levels are regulated by stimulatory and inhibitory GTP-binding proteins (Gα proteins) which control the activity of adenylate cyclase (AC). Heterotrimeric G proteins are a family of intracellular proteins that are activated in response to G protein coupled receptors (GPCRs) and are involved in second messenger cascades. They are bound to the internal surface of the plasma membrane and consist of Gα and the tightly associated Gβγ subunits. There are four classes of Gα subunits: Gαs, Gαi/o, Gαq/11 and Gα12/13. They share similar mechanisms of activation but differ in the recognition of their effectors. Ligand binding induces a conformational change in the GPCR, allowing it to act as a guanine nucleotide exchange factor (GEF). The exchange of GDP for GTP at the Gα subunit causes dissociation from the Gβγ dimmer, thereby activating other signaling molecules. The Gα subunit has an inherent GTPase activity that hydrolyzes the GTP to GDP and thus, re-associates with the Gβγ dimer (14). The main role of proteins of the Gαi/o class is to decrease cAMP production through inhibition of AC.
Smo belongs to the GPCR family and has been reported to activate various members of Gαi/o class in reconstituted cell systems like frog melanophores (15) or SF9 insect cells (16). However, G protein independent transduction mechanisms have also been proposed for Smo (17). Recently, it has been shown that, at least in Drosophila, Smo functions as a canonical GPCR which signals through Gαi to regulate activation of the Hh pathway (18). Given that important differences in Hh signaling have recently been described between Drosophila and mammals, (19, 20) we studied whether Gαi also mediates mammalian Smo signal transduction. We demonstrate that Shh-induced proliferation of CGNPs was strongly enhanced by recombinant expression of the active forms of Gαi (Gαi1, Gαi2, Gαi3, and Gαo) but not by members of the G12 class of heterotrimeric G proteins. In addition, we show that the different Gαi/o class members display distinct expression patterns in the developing cerebellum. Interestingly, only Gαi2 and Gαi3 are substantially expressed in the outer external granular layer (oEGL) where CGNPs proliferate. In addition, the capacity of Shh to promote proliferation of CGNPs was reduced significantly by knockdowns of Gαi2 and Gαi3 but not other members of the Gαi class. Finally, we show that Gαi2 and Gαi3 locate to the primary cilium when expressed in CGNP cultures. Collectively, our results suggest that Shh-induced proliferation of CGNPs is mediated by the combined activity of Gαi2 and Gαi3 proteins.
EXPERIMENTAL PROCEDURES
Cerebellar Granular Neuronal Precursor Culture and Transfection
P7 rat cerebellar cultures and plate coating were performed as described previously (21). Cell cultures were either transfected with FuGENE 6 (Roche Diagnostics) or electroporated depending on the transfection efficiency requirements. For FuGENE transfection experiments, cells were plated in Neurobasal medium supplemented with B-27 (Invitrogen), 20 mm KCl, 2 mm glutamine, and Shh at 3 μg/ml for 24 h, and at the end of this period, the cells were transfected for 4 h following the manufacturer's guidelines. After removing the transfection mixture, the cultures were washed once, and fresh medium with or without Shh was added. The typical transfection efficiency of this procedure was 5–10%, making it the ideal method for experiments where an accurate cell counting is required. Electroporation was performed in suspension just after the tissue dispersion procedure with the Microporator MP-100 (Digital Bio, Seoul, Korea) according to the manufacturer's instructions, with a single pulse of 1700 mV for 20 ms. As above, electroporated cells were plated in Shh-containing media for 24 h prior to the start of different treatments. This procedure transfects up to 80% of the culture with minimal toxicity and is ideal for biochemistry or reporter assays. N-terminal Shh produced in our laboratory was used as indicated in each experiment.
Plasmid Preparation
Rat Gαi1, Gαi3, and the corresponding mutants were produced by PCR site-directed mutagenesis. Human Gαο, Gα2, and their mutants were obtained from Hae Young Suh. Human Gα12 and its mutant form were obtained from Silvio Gutkind. All Gα protein versions used in this study were subcloned into the bicistronic vector pCIG (22) that contains nuclear EGFP as a reporter. To produce the RNA probes used in the in situ hybridization experiment, the coding sequence of rat, Gαi1, Gαi2, Gαi3, and Gαio, kindly provided by Randall Reed, were subcloned into pBluescript IIKS. For mRNA knockdown experiments, the DNA primers designed to produce the different shRNA molecules, were cloned into pGHIN a GFP expressing version of pSUPER. The different shRNA target sequences were selected in accordance to the following rules: In brief, a size of 19 bp, not including the first or last nucleotides of the mRNA, flanked at 5′ by AA and, if possible, by TT nucleotides at 3′, the percentage of GC must be >50%, and finally, the sequence should not contain four or more A or T nucleotides together. The selected sequences were blasted against a rat cDNA database to ensure their specificity. We detected four different sequences with the above exposed characteristics in Gαi1 and Gαi2 3′-UTRs and one against the coding region. Although Gαi3 has a relatively large 3′-UTR, only three targeting sequences could be detected due to its high AT content. In addition, another sequence was found in the coding sequence. On the contrary, Gαo has a very short 3′-UTR and only two sequences were found, but none in the coding region. All targeting primers are listed in supplemental Table 1.
Semiquantitative RT-PCR Quantification of mRNA in Knockdown Experiments
Total cell RNA was purified following the method of Chomcynski and Sacchi (38). RNA integrity and concentration was assessed through spectral analysis with a NanoDrop Fotometer. Equal amounts of RNA were retrotranscribed with Omniscript reverse transcriptase (Qiagen). To avoid genomic DNA interferences, because all Gαi/o coding regions contained at least one large intronic sequence, we used primers spanning the entire coding region to measure Gαi/o mRNA concentrations. The optimal number of amplification cycles was established for each primer pair and cDNA preparation to ensure that no saturation occurred. GAPDH was used in all reactions as a control gene.
Immunofluorescence and Antibodies
Cells were grown in 24-well plates (Corning, Inc.), fixed with 4% paraformaldehyde, permeabilized with methanol, washed three times with PBS/BSA buffer containing 0.2% of Triton X-100, and incubated overnight at 4 °C with the corresponding primary antibodies in this same buffer. For BrdU incorporation experiments, cells were treated with BrdU for 6 h before fixation, and an additional treatment with DNase II (SIGMA) for 15 min was included just after the methanol step. After three washes, cells were incubated for 1 h at room temperature with anti-mouse or anti-rabbit secondary antibodies labeled with Alexa Fluor 488, 594, or 555 (Invitrogen) or Cy5 (Jackson ImmunoResearch Laboratories), and nuclei were stained with DAPI (Molecular Probes). The monoclonal anti-BrdU antibody was obtained from Developmental Studies Hybridoma Bank, rabbit anti-C-PKA was from Cell Signaling, monoclonal antibodies anti-FLAG (M2) and anti-acetylated tubulin were from Sigma, rabbit anti-adenylate cyclase III (C-20) was from Santa Cruz Biotechnology, mouse monoclonal anti-trans-Golgi (TGN38) was from BD Biosciences, mouse polyclonal anti-HA, rabbit polyclonal anti-FLAG, anti-HA, and anti-GFP antibodies were produced in our laboratory. For confocal images, cells were grown on coated coverslips, which were mounted with Mowiol (CalBiochem). Inmunofluorescent images were obtained either with a Leica inverted fluorescence microscope or a Leica Sp5 confocal spectral scanning microscope.
In Situ Hybridization
P7 rat cerebellum were fixed overnight with 4% of paraformaldehyde and washed three times for 10 min with PBT (DEPC-PBS1x, 0.1% Triton). 50-μm Slices from 10% sacarose/5% agarose blocks were sectioned with a vibratome (Leica) and dehydrated with increasing concentrations of methanol/PBT (25, 50, 75, and 100%). Hybridization was performed at 70 °C using standard procedures and revealed by alkaline phosphatase-coupled anti-digoxigenin Fab fragments (Roche Applied Science). Rat Gαi/o RNA probes were produced with T7 enzyme from the corresponding cDNA sequences cloned in Bluescript-II-KS.
Proliferation Assays and [3H]Thymidine Incorporation
Electroporated cells were plated for 48 h and then pulsed for 2 h with [3H]thymidine (1μCi in each well, Amersham Biosciences, Buckinghamshire, UK). Cells were then lysed in 0.04% SDS, and proliferation was measured with a Wallac scintillation counter (Wallac-Perkin Elmer, Quebec, Canada).
Proliferation Assays and BrdU Incorporation, and Statistical Analysis
Cell proliferation was calculated as the percentage of BrdU-positive cells among the transfected population (GFP-expressing cells). Duplicate wells from at least three different cultures were counted (minimum of six wells), lack of coherence between duplicate wells always dismissed both. The number of cells counted per well ranged from 350 to 1500 with an average of 750, mainly depending on the transfection efficiency and the internal data dispersion of each experiment. Quantitative data were expressed as the mean ± S.D. Significant differences among groups were tested by two-way analysis of variance followed by the Tukey's test.
RESULTS
Capacity of Shh to Induce Proliferation of CGNPs Is Strongly Enhanced by Active Forms of Gαi/o Class Members but Not by Gα12
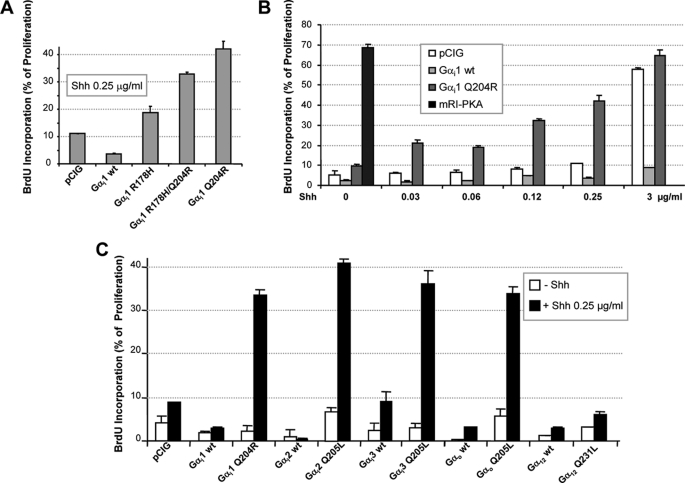
Given the apparent discrepancy in CGNPs between the strong proliferative effect of a dominant negative form of PKA (mRI-PKA) and the lack of an effect of Shh on cAMP levels (11), we studied the relevance of Gαi activity in mammalian Hh signal transduction. In Drosophila, the inhibitory Gαi/o class is comprised of only one Gαi and one Gαo isoform. In mammals, this family is much more complex and is formed by three Gαi, two Gαo, three Gαt, and one Gαz (23) (UniProtKB/SwissProt). However, considering that Gαt function is mostly restricted to taste transduction and that Gαz activation by Smo was only partially reversed by cyclopamine (16), we restricted our study to Gαi and Gαo isoforms. Thus, we created three putative Gαi1 constitutive active forms based on known Gαs mutations (24). These mutations were designed to impair the endogenous Gα GTPase, thereby extending their active state (Gα-GTP). When transfected into CGNPs, all three mutants, Gαi1H178R, Gαi1Q204R (Gαi1QR), and Gαi1H178R/Q204R, increased the proliferation rate of CGNPs as measured by BrdU or [3H]thymidine incorporation (Fig. 1A and supplemental Fig. 1A, respectively), although the most potent effect was obtained with Gαi1QR. However, the proliferative effects were dependent on the presence of Shh, demonstrating that, although this mutation prolongs Gαi1QR activity, priming of the system by Smo was still required. Conversely, the mRI-PKA mutant potently increased (68%) cell proliferation even in the absence of Shh, due to its ligand-independent nature (Fig. 1B). Interestingly, Gαi1QR enhanced proliferation progressively as the subproliferative amounts of Shh in the culture were increased, reaching up to 42% with 0.25 μg/ml Shh. When CGNPs were cultured with a saturating concentration of Shh (3 μg/ml), the proliferation rates between cells transfected with either empty vector or Gαi1QR were very similar (59 and 64%, respectively), suggesting that when the Shh pathway is fully activated, endogenous Gαi activity is sufficient to maintain a maximum proliferation rate (Fig. 1B). It is worth noting that the wild-type form of Gαi1 displayed a marked anti-proliferative effect, even in the presence of saturating concentrations of Shh, but this was not observed with the wild-type forms of Gαi2, Gαi3, Gαο, or Gα12 (supplemental Fig. 1B).
FIGURE 1.
Shh-induced proliferation of CGNPs is strongly enhanced by the active forms of members of the Gαi/o class but not by G12. CGNP cells were plated in a saturating concentration of Shh (3 μg/ml) and transfected with different wild-type or active mutant forms of Gα cloned into pCIG, a nuclear GFP-expressing bicistronic vector. After 24 h, cells were washed and treated for an additional 48 h with the indicated amounts of Shh, and proliferation was analyzed by BrdU incorporation. A, comparison of different Gαi1-activating mutations at 0.25 μg/ml Shh. B, proliferation was studied in cells transfected with wild-type Gαi1, Gαi1Q204R, or mRI-PKA and treated with increasing amounts of Shh (0, 0.03, 0.06, 0.12, 0.25, and 3 μg/ml). C, comparison of the proliferation induced by expression of wild-type or active mutant forms of Gαi1, Gαi2, Gαi3, Gαo, and Gα12 in CGNPs maintained in 0.25 μg/ml Shh.
With the aim of identifying which Gαi/o class members mediate Shh-induced proliferation in CGNPs, we next studied the proliferation induced by transfecting GTPase-deficient mutants of Gαi2 (Q204L), Gαi3 (Q204L), Gαο (Q205L), and Gα12 (Q231L) in cultures growing in a sub-proliferative concentration of Shh (0.25 μg/ml). Although Gα12 does not belong to the Gαi/o class, it was included as it has been reported to participate in Shh signaling through Rho-kinase activation (25). Unexpectedly, all Gαi/o class members increased proliferation rates at similar levels when transfectd in CGNPs cultured with a low Shh concentration (0.25 μg/ml). In contrast, under these same conditions, the proliferation rate of cells transfected with the active form of G12 was lower than that of cells transfected with empty vector (Fig. 1C). Collectively, theses results suggest that Gαi/o members do indeed mediate Shh signaling in CGNPs.
Gαi2 and Gαi3 Are Specifically Enriched in Outer EGL of Developing Cerebellum
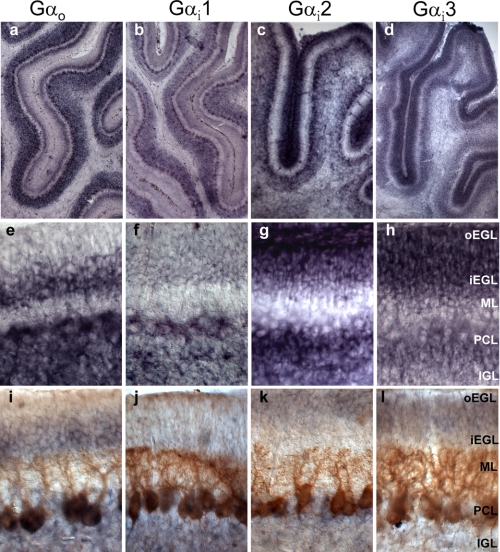
Given that the results with the activating mutants suggested that Shh signaling in CGNPs could be mediated indiscriminately by all Gαi/o members, we next explored the expression patterns of Gαo, Gαi1, Gαi2, and Gαi3 mRNAs in P7 rat cerebellum (Fig. 2). Using in situ hybridization, very distinct layer distributions could be appreciated at low magnification (Fig. 2, a–d). The expression pattern of each Gαi/o member in the different cerebellum layers was confirmed by comparing in situ hybridization alone (Fig. 2, e–h) with in situ hybridization plus immunohistochemistry performed with the Purkinje cell marker, Calbindin (Fig. 2, i–l). Thus, commencing from the innermost layer, we observed that the mRNAs of all four members of the Gαi/o class were significantly expressed at the internal granular layer, which contains primarily differentiated granular cells and Golgi cells in much lower abundance. Gαo and Gαi1 were predominant forms expressed in the Purkinje cell layer, which is basically occupied by the huge cell bodies of Purkinje cells. In the adult cerebellum, the molecular layer is formed exclusively by the Purkinje cell dendrites, the granular cell axons, and low abundance interneurons, and although at P7, it also contains migrating granular cells, it is typically an area of low reactivity for most mRNAs probes due to its acellular nature. The internal part of the external granular layer contains the granular cells that have stopped dividing and initiated migration. In this layer, only Gαo expression was differentially augmented. Finally, the oEGL is where CGNPs intensively proliferate in response to Shh during early postnatal development (2 weeks in rat cerebellum). Interestingly, this layer expressed comparatively low levels of Gαio and Gαi1 mRNAs and high levels of Gαi2 and Gαi3. Thus, we concluded that each of the four Gαi/o class members displays a distinct expression pattern, suggestive of participation in different signaling pathways.
FIGURE 2.
In situ hybridization analysis of Gαi/o mRNA at P7 in rat cerebellum. mRNA levels of Gαo, Gαi1, Gαi2, and Gαi3 were studied in P7 rat cerebellum by in situ hybridization using rat specific riboprobes. a–d, low magnification images reveal the unique expression patterns of the mRNAs of the various Gαi/o class members. e–h, high magnification photos of the mRNA levels in the different cerebellar layers: oEGL, inner external granular layer (iEGL), molecular layer (ML), Purkinje cell layer (PCL), and internal granular layer (IGL). i–l, high magnification photos of the in situ hybridization of the Gαi/o class members combined with immuno-histochemistry against the Purkinje cell marker, calbindin, to further delineate the extension of each cerebellar layer. Only Gαi2 and Gαi3 were significantly expressed at the oEGL were CGNPs proliferate.
Knockdowns of Gαi2 and Gαi3 Significantly Reduce Shh-induced Proliferation in CGNP Cultures
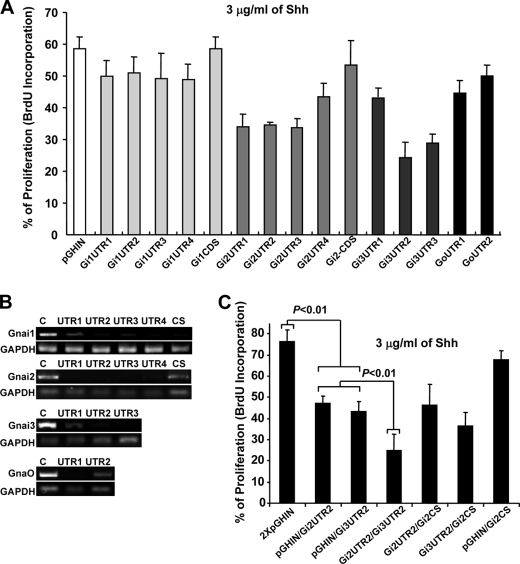
Encouraged by the unique expression patterns of the different Gαi/o class members in the developing cerebellum, we next asked whether knockdowns of each would differentially alter Shh-induced proliferation of CGNPs. Thus, we designed different DNA sequences encoding short hairpin inhibitory RNA molecules (shRNA), which would target either the 3′-UTR (UTR) or the coding sequence (CS) of the different members of the Gαi/o class (see “Experimental Procedures” for details). These were then cloned into pGHIN (a pSUPER-based GFP-expressing bicistronic vector). The shRNA-expressing constructs were transfected in CGNPs cultured with a saturating concentration of Shh (3 μg/ml), and 48 h later, proliferation was measured by BrdU incorporation (Fig. 3). In addition, half of each transfected culture was used to evaluate the targeting efficiency of each shRNA by RT-PCR. As previously observed, shRNAs directed to CS were less efficient at reducing expression (Fig. 3, A and B, and supplemental Fig. 2 for representative cell pictures). Although most shRNAs designed against the 3′-UTRs efficiently reduced the mRNA levels of their targets, only the shRNAs directed against Gαi2 and Gαi3 attenuated significantly Shh-induced proliferation in CGNPs cultures (Fig. 3, A and B). Similar results were obtained when the effect of expressing different shRNA constructs on [3H]thymidine incorporation, was tested in cultures treated with growing concentrations of Shh (supplemental Fig. 1C). These results agrees well with the expression patterns of the different Gαi/o class members in P7 rat cerebellum, where only Gαi2 and Gαi3 were appreciably expressed in the oEGL (Fig. 2). In an attempt to further define the individual roles played by Gαi2 and Gαi3 in mediating Shh-induced proliferation of CGNPs, we next tested the effects of reducing the expression of both molecules simultaneously. Using the experimental conditions described above, equal amounts of plasmid DNA encoding the most efficient shRNA for Gαi2 and Gαi3 (Gi2UTR2 and Gi3UTR2) were co-transfected into CGNPs. Additionally, different combinations of Gi2UTR2 or Gi3UTR2 with empty vector or the low performance Gi2CS were tested as controls. The combined effect of simultaneous knockdowns of Gαi2 and Gαi3 on Shh-induced proliferation was much greater than either of the individual knockdowns, whereas other combinations had no significant additional inhibitory effects (Fig. 3C). It is worth noting that no additional reduction of proliferation was ever observed when two shRNA against the same molecule were combined (data not shown). These results strongly suggest that Shh-induced proliferation in CGNPs is mediated by the additive effects of Gαi2 and Gαi3.
FIGURE 3.
Gαi2 and Gαi3 knockdowns significantly reduce Shh induced CGNP proliferation. Different DNA sequences encoding shRNA, targeting either the 3′UTR (UTR) or CS of the different Gαi/o class members, were cloned into pGHIN (a pSUPER-based GFP-expressing bicistronic vector). The number of shRNAs designed for each Gαi/o depended on the availability of adequate targeting motifs in the respective sequences (see “Experimental Procedures” for details). A, the shRNA expressing constructs were electroporated into freshly isolated CGNPs, which were then halved and grown in a saturating concentration of Shh (3 μg/ml) for 48 h. One culture was used to isolate mRNA for RT-PCR analysis and the other to measure proliferation by BrdU. B, the targeting efficiency of each shRNA was evaluated by RT-PCR. A direct correlation between targeting efficiency and proliferation reduction was observed only for shRNAs targeting Gαi2 and Gαi3. C, the most efficient shRNAs against Gαi2 and Gαi3 (Gi2UTR2 and Gi3UTR2) were assayed together or in combination with Gi2CS, a low efficiency shRNA which was used as a control. The total amount of DNA was equivalent for all transfections. Proliferation was significantly reduced compared with empty vector when either Gi2UTR2 or Gi3UTR2 were transfected (p < 0.01). Double transfection of Gi2UTR2 and Gi3UTR2 significantly reduced proliferation as compared with single transfections or to doubles with Gi2CS (p < 0.01).
Ectopic Expression of Different Gαi/o Class Proteins, but Not Gα12, Rescues Impaired Proliferation Caused by Gαi2/Gαi3 Knockdown
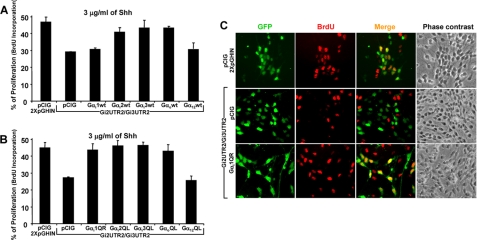
Although sequence analysis predicted that all shRNAs used in the previous experiment were 100% specific for their indicated targets, cross-reactivity with other genes cannot be totally excluded. Thus, to demonstrate specificity, we tested whether the anti-proliferative effects induced by Gαi2/Gαi3 knockdowns could be restored by co-transfecting shRNA-resistant expression constructs (coding sequences with no UTRs) of these molecules. Hence, CGNP cultures were transfected with mixtures containing Gi2UTR2/Gi3UTR2 and constructs to express wild-type (Fig. 4A) or active forms (Fig. 4B) of the different members of the Gαi/o class. Interestingly, Shh-induced proliferation was rescued by both wild-type and active forms of Gαi1, Gαi2, Gαi3, and Gαo but not by Gα12. This result was consistent with the experiments shown in Fig. 1C, where similar levels of cell proliferation were induced by overexpressing the individual members of the Gαi/o class but not with Gα12. Moreover, wild-type Gαi1 did not rescue proliferation, in agreement with its anti-proliferative effect observed in Fig. 1B and supplemental Fig. 1B. Selected images of these experiments are shown in Fig. 4C, and a complete set of images is presented in supplemental Fig. 3. These observations reinforce the notion that although Gαi/o class molecules display rather specific expression patterns in the developing cerebellum, their function is interchangeable, at least, in transmitting Shh-induced proliferation of CGNPs under our experimental conditions.
FIGURE 4.
Expression of different Gαi/o class members but not Gα12 rescues the impaired proliferation caused by Gαi2/Gαi3 knockdowns. Freshly isolated CGNPs were electroporated with mixtures of cDNA containing Gi2UTR2/Gi3UTR2 and constructs including wild-type (A) or active forms (B) of the different Gαi/o class members. Cells were cultured in a saturating concentration of Shh (3 μg/ml) for 48 h, and proliferation was measured by BrdU incorporation. Selected images of transfected cells are shown in C. A more complete panel of images is presented in supplemental Fig. 3.
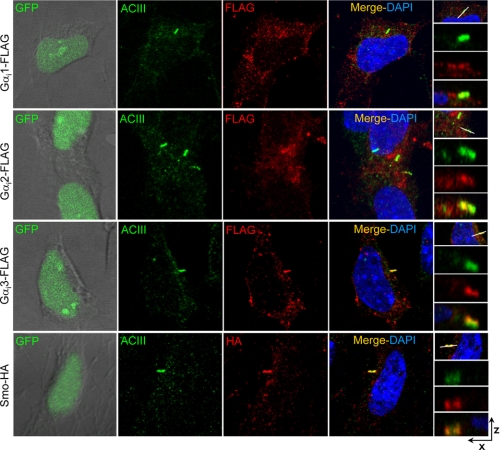
Three Gαi Members Are Distributed Differently between Base and Shaft of Primary Cilium
Various lines of evidence have demonstrated that the primary cilium acts as the main integration center for Shh signaling, because the majority of molecules involved in Shh signaling have been observed in the vicinity of the cilium. On the other hand, members of the Gαi family transduce signals from a wide range of membrane receptors in addition to Smo. Thus, we studied the cellular distribution of Gαi isoforms in the cilium and although the knockdown experiments suggested the potential importance of Gαi2 and Gαi3 subunits, we also evaluated the expression pattern of Gαi1. To avoid complications of cross-reactivity or nonspecific staining observed for most of the commercial antibodies tested, we cloned FLAG-tagged constructs for the different Gαi subunits in pCIG, a nuclear-GFP expressing bicistronic vector that permitted us to assess Gαi distribution in transfected cells. We generated Z-stacks from confocal images of cells double-stained with anti-FLAG and anti-adenylate cyclase III (ACIII), a cilium shaft marker (11, 26). Although positive staining was detected throughout cells transfected with the individual constructs, a periciliar (below and surrounding the cilium shaft) accumulation was observed for each of the three subunits in CGNPs. However, we observed marked differences with respect to their expression in the cilium shaft. Gαi1 was detected at the periciliar area but never in the shaft (Fig. 5A). Although Gαi2 was also detected in most cells below the cilium, low levels of this protein were detected in the cilium shaft of some cells (see Fig. 5B, which depicts two neighbor cells with similar Gαi2 periciliar expression, one of which also expresses Gαi2 in the cilium shaft). Finally, Gαi3 was detected mainly in the cilium shaft rather than underneath it, where it accumulates similarly to Smo (Fig. 5C and supplemental Fig. 4C). Thus, although these results suggested that Gαi3 localization could be mediated through its binding to Smo, we did not observe the required stable association between these two molecules (supplemental Fig. 4A). Moreover, Gαi3 location to the cilium was not affected by Shh deprivation, which prevents Smo accumulation at the cilium (supplemental Fig. 4B), neither by the trasnsfection of SmoM2, an active form of Smo that constitutively accumulates at the cilium (supplemental Fig. 4C). To further define the periciliar location of Gαi, we double-stained FLAG-Gαi1 transfected cells with anti-FLAG and anti-C-PKA or anti-TGN38, a trans-Golgi marker. We have previously shown that C-PKA in CGNPs locates at the cilium base surrounding the centrioles (11). The present results revealed that Gαi1 distributes around the area positive for anti-C-PKA, coincident with the vesicles of the trans-Golgi network (supplemental Fig. 5).
FIGURE 5.
The three Gαi members are distributed differently between the base and the shaft of the primary cilium. CGNP cultures were transfected with FLAG tagged constructs of Gαi1, Gαi2, and Gαi3 subunits cloned in pCIG, a nuclear-GFP expressing bicistronic vector that permitted us to compare the Gαi distribution in similarly transfected cells. Transfection of Smo-HA was included as a positive control. Cells were cultured for 48 h with 3 μg/ml of Shh, fixed, and double-stained with rabbit anti-ACIII (used as a cilium marker) and mouse anti-FLAG or anti-HA. The first column depicts images of direct GFP fluorescence combined with phase contrast pictures, demonstrating the transfection levels of each cell. ACIII staining was detected with anti-rabbit Cy5 (far red 675) but has been colored green for clarity. FLAG or HA tags were detected with anti-mouse Alexa Fluor 555 (red). The last column contains Z-stacks generated from the confocal images.
DISCUSSION
Smo Signals through Gα Inhibitory Proteins in CGNPs
The downstream components of Smo signaling have long been a question of intense debate. Although its protein sequence clearly predicts that Smo belongs to the GPCR family, the evidence that this receptor uses heterotrimeric G proteins as downstream effectors is inconclusive. Early experiments in zebrafish embryos demonstrated that inhibition of Gαi-mediated events with pertussis toxin (PTX) produced phenotypes, which resembled those obtained by inhibition of the Hh pathway, suggesting a possible role for Gαi in Smo signaling (27). Similarly, expression of human Smo in frog melonophores caused a phenotype of persistent pigment aggregation that could be blocked by PTX (15). However, other reports have shown that neither PTX nor Gαi Q205L has effects on Hh-mediated events in chicken embryos such as spinal cord patterning and Gli3 processing, suggesting that G proteins may not be required for all Hh-dependent signaling. Additionally, silencing of a wide spectrum of G protein subunits in cultured Drosophila cells failed to disrupt Hh signaling (28). Nevertheless, a more recent report has demonstrated a direct activation of different Gαi/o class members by Smo in a reconstituted SF9 cell system (16). These authors additionally showed that, in NIH-3T3 cells, PTX inhibits Gli-dependent luciferase activity stimulated either by addition of Shh or the transfection of an oncogenic form of Smo (SmoM2). Another line of evidence to support the involvement of heterotrimeric G proteins in Smo signaling has come from the embryonic fibroblasts of Patched-1 knock-out mice where transcription of Hh target genes appears to depend on Gαi-mediated signal transduction (29). Finally, it has recently been shown that at least in Drosophila, Smo functions as a canonical GPCR, which signals through Gαi to regulate Hh pathway activation (18). In the present work, we demonstrate that Shh-induced proliferation of rat CGNPs is enhanced by recombinant expression of active forms of Gαi/o class. Conversely, when endogenous Gαi/o proteins were knocked down by shRNA transfection, Shh-mediated proliferation was reduced significantly. Our results agree completely with the results obtained in Drosophila (18) and reflect the first study to implicate Gαi proteins in a physiologic process (proliferation of CGNPs) mediated by Hh activity in mammals. It is worth noting that the signal transduction associated with the different Hh-mediated processes, such as proliferation and patterning, may have different molecular components. When attempting to reconcile all the data mentioned above, one must bear in mind that studies performed with PTX must be interpreted with caution because not all cell types can process this toxin (23). Likewise, is important to note that, although the Gαi Q205L mutation used in particular studies (29) extends the period in which Gαi is active (Gαi-GTP), it is not a constitutively active mutant and thus, still requires an upstream signal for activation. Consistent with this, the data we present in Fig. 1B demonstrates that this same mutant could enhance proliferation of CGNPs but only if a minimum of Shh was present in the culture medium.
Smo Specificity among Gαi/o Class Members
Relatively few types of G proteins transduce signals from a vast number of GPCRs. Regulating the specificity of these interactions is important for proper signal transduction. Therefore, the receptor-G protein interface must contain important information that determines which G proteins can interact with a particular receptor. However, although many of the contact sites that comprise this interface have been identified, the connections that define the coupling between receptors and specific G protein family members remain poorly defined (14). Although it is now clear that the three components of the G protein trimer (α,β,γ) may participate in the receptor-G protein binding, it is still not possible to predict which class of G proteins will interact with a particular GPCR (14). In the present work, we found that all Gαi/o class members, but not Gα12, were able to mediate Shh signaling. Similarly, Riobo and colleagues (16) demonstrated that Smo-induced GTP loading was restricted to Gαi/o class members. Kasai et al. (25), however, have reported that Shh-induced activation of Gli in HEK-293 cells was repressed by the inhibitors of Gα12 or Rho, a downstream effector of Gα12. It is possible that Gli-dependent transcription requires Rho activity, but our current results strongly suggest that Gα12 is not essential for Smo activity in CGNPs. Similar to other studies (16), we observed that the capacity to promote Shh-induced proliferation in CGNPs was similar among the members of the Gαi/o class. These results suggest that, at least in terms of cell proliferation, Smo signals can be mediated by any member of this class of G proteins when overexpressed. Alternatively, a Gβγ-dependent effect could be considered in this way; it has been reported that the Gβγ complex can modulate local cAMP production through regulating the activity of specific AC isoforms (30). Nevertheless, the fact that proliferation was increased by the active forms of the Gαi/o class and not by the wild-type molecules and that GTPase-deficient Gαi/o mutants decrease Gβ/γ turnover instead of augmenting it, strongly suggests that the lack of specificity can be a direct consequence of the overexpression itself. On the other hand, only the silencing of Gαi2 and/or Gαi3 significantly impaired proliferation, indicating that, although all class members can mediate Smo-dependent proliferation, only Gαi2 and Gαi3 are expressed appropriately for the transduction of Shh proliferation signals in CGNPs. These results are consistent with the mRNA expression patterns of the Gαi/o members in the developing cerebellum (Fig. 2); only Gαi2 and Gαi3 were expressed at the oEGL where the CGNPs proliferate.
Gαi Subcellular Distribution and Function
Local regulation of cAMP production at the primary cilium may function as a key event in Shh signal transduction (11, 31). Recently, we have demonstrated that the activity of PKA, which accumulated at the cilium base, was directly regulated by Shh signaling (11). Therefore, we hypothesized that cAMP production at the cilium may be locally controlled by Smo. Consistent with this, high levels of ACIII were detected in the cilium shaft, although participation of this specific isoform of AC in Shh signaling has not yet been established. According to this working hypothesis, signal transduction from Smo to AC would require Gαi in the cilium. With the present study, we now show that Gαi3 clearly accumulates in the cilium shaft of rat CGNPs. In addition, knockdown of Gαi3 reduced more potently the Shh-induced proliferation of CGNPs as compared with the knockdowns of the other Gαi proteins. Moreover, a direct correlation between distribution to the cilium and impaired proliferation could be observed, suggesting that the localization of Gαi proteins to the cilium is a requirement for their participation in Shh signal transduction. On the other hand, we have also observed that the constitutive mutants of Gαi1 and Gαi2 increased cell proliferation to the same extent as Gαi3, although Gαi1 and Gαi2 were detected predominantly at the cilium base rather than in the shaft. Additionally, the knockdown experiments of Fig. 4 clearly demonstrate that reduction of both Gαi2 and Gαi3 expression has an additive effect on Smo signal transduction, although Gαi2 was localized mainly out of the cilium shaft. These results, along with the surprising antiproliferative activity demonstrated by the wild-type Gαi1, suggest a more complex mechanism than simple regulation by distribution to the cilium. It was recently demonstrated that the control Smo entrance into the cilium is a rather a complicated mechanism (32) that depends on a Septin diffusion barrier at the cilium base (33); however, a participation of Gαi on ciliar Smo location seems not to occur because we have observed it to be independent of Gαi signaling (supplemental Fig. 4D). Proper anterograde and retrograde intraflagellar transport are both required for cilium formation, maintenance, and signaling of cilium-dependent pathways (34). Likewise, vesicular traffic from the trans-Golgi network to the cilium has been considered as the main source of proteins entering the cilium (35). Conversely, Milenkovic et al. (32), using innovative SNAP-tag technology, have recently demonstrated that at least for Smo, lateral diffusion and local recycling are the main sources of Smo entering the cilium upon Shh stimulation. In agreement with this study, the expression of a dominant negative form of the Rab23, a small GTPase previously implicated in vesicular traffic, caused a significant decrease of Smo recycling pathway when measured by fluorescence recovery after photobleaching of cilium yellow fluorescent protein-SmoA1 (36). Consequently, the cilium shaft and base should not be considered as separate compartments but rather as a continuous conveyor belt where the different signaling molecules cycle and interact with each other in a dynamic manner. Thus, Gαi at the cilium base could interact with the Smo exiting the cilium, thereby inhibiting the ACIII pool in the lower part of the shaft. This proposed model could explain the fact that, although the active form of Gαi1 was detected mainly out of the cilium shaft, it still mediated Shh-induced proliferation activity in CGNPs. Interestingly, Gαi has also been reported to control microtubule dynamics through an AC-independent mechanism (37). Thus, although the experiments mentioned in Introduction clearly indicate that control of AC is the main task performed by Gαi in Shh signaling, AC-independent activities cannot be excluded for the different Gαi proteins. Due to the complexities of the system, which cannot be captured by static methods, a complete understanding of the details of Smo signal transduction mechanisms will require simultaneous real-time recordings of Smo, ACIII, and the different Gαi.
Supplementary Material
Acknowledgments
We wish to thank the invaluable research assistance of Anghara Menendez. We are grateful to all of the researchers who sent us the cDNAs as detailed under “Experimental Procedures.” We kindly thank Dr. Deborah Burks for editing the manuscript.
This work was supported by Spanish Ministry of Education Grant BFU2008-02424/BFI.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–5.
- CGNP
- cerebellar granular neuronal precursor
- Smo
- Smoothened
- Shh
- Sonic Hedgehog
- GPCR
- G protein-coupled receptor
- Hh
- Hedgehog
- AC
- adenylate cyclase
- CS
- coding sequence
- ACIII
- adenylate cyclase III
- PTX
- pertussis toxin
- PKA
- protein kinase A
- C-PKA
- PKA catalytic subunit
- Gαi1QR
- Gαi1Q204R.
REFERENCES
- 1. Altman J. A., Bayer S. A. (1997) Development of the Cerebellar System in Relation to Its Evolution, Structure, and Functions, CRC Press, Boca Raton, FL [Google Scholar]
- 2. Hatten M. E., Heintz N. (1995) Annu. Rev. Neurosci. 18, 385–408 [DOI] [PubMed] [Google Scholar]
- 3. Ramon y Cajal S. (1911) Histologie du Systeme Nerveux de l′Homme et des Vertebrates, Malonie, Paris, France [Google Scholar]
- 4. Dahmane N., Ruiz i., Altaba A. (1999) Development 126, 3089–3100 [DOI] [PubMed] [Google Scholar]
- 5. Wechsler-Reya R. J., Scott M. P. (1999) Neuron 22, 103–114 [DOI] [PubMed] [Google Scholar]
- 6. Corbit K. C., Aanstad P., Singla V., Norman A. R., Stainier D. Y., Reiter J. F. (2005) Nature 437, 1018–1021 [DOI] [PubMed] [Google Scholar]
- 7. Rohatgi R., Milenkovic L., Scott M. P. (2007) Science 317, 372–376 [DOI] [PubMed] [Google Scholar]
- 8. Tempé D., Casas M., Karaz S., Blanchet-Tournier M. F., Concordet J. P. (2006) Mol. Cell. Biol. 26, 4316–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang B., Fallon J. F., Beachy P. A. (2000) Cell 100, 423–434 [DOI] [PubMed] [Google Scholar]
- 10. Wang B., Li Y. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 33–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barzi M., Berenguer J., Menendez A., Alvarez-Rodriguez R., Pons S. (2010) J. Cell Sci. 123, 62–69 [DOI] [PubMed] [Google Scholar]
- 12. McConnachie G., Langeberg L. K., Scott J. D. (2006) Trends Mol. Med. 12, 317–323 [DOI] [PubMed] [Google Scholar]
- 13. Iyer G. H., Moore M. J., Taylor S. S. (2005) J. Biol. Chem. 280, 8800–8807 [DOI] [PubMed] [Google Scholar]
- 14. Oldham W. M., Hamm H. E. (2007) Adv. Protein Chem. 74, 67–93 [DOI] [PubMed] [Google Scholar]
- 15. DeCamp D. L., Thompson T. M., de Sauvage F. J., Lerner M. R. (2000) J. Biol. Chem. 275, 26322–26327 [DOI] [PubMed] [Google Scholar]
- 16. Riobo N. A., Saucy B., Dilizio C., Manning D. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12607–12612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ruiz-Gómez A., Molnar C., Holguín H., Mayor F., Jr., de Celis J. F. (2007) Biochim. Biophys. Acta 1768, 901–912 [DOI] [PubMed] [Google Scholar]
- 18. Ogden S. K., Fei D. L., Schilling N. S., Ahmed Y. F., Hwa J., Robbins D. J. (2008) Nature 456, 967–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gerdes J. M., Davis E. E., Katsanis N. (2009) Cell 137, 32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Varjosalo M., Li S. P., Taipale J. (2006) Dev. Cell 10, 177–186 [DOI] [PubMed] [Google Scholar]
- 21. Rios I., Alvarez-Rodríguez R., Martí E., Pons S. (2004) Development 131, 3159–3168 [DOI] [PubMed] [Google Scholar]
- 22. Megason S. G., McMahon A. P. (2002) Development 129, 2087–2098 [DOI] [PubMed] [Google Scholar]
- 23. Riobo N. A., Manning D. R. (2007) Biochem. J. 403, 369–379 [DOI] [PubMed] [Google Scholar]
- 24. Landis C. A., Masters S. B., Spada A., Pace A. M., Bourne H. R., Vallar L. (1989) Nature 340, 692–696 [DOI] [PubMed] [Google Scholar]
- 25. Kasai K., Takahashi M., Osumi N., Sinnarajah S., Takeo T., Ikeda H., Kehrl J. H., Itoh G., Arnheiter H. (2004) Genes Cells 9, 49–58 [DOI] [PubMed] [Google Scholar]
- 26. Bishop G. A., Berbari N. F., Lewis J., Mykytyn K. (2007) J. Comp Neurol. 505, 562–571 [DOI] [PubMed] [Google Scholar]
- 27. Hammerschmidt M., McMahon A. P. (1998) Dev. Biol. 194, 166–171 [DOI] [PubMed] [Google Scholar]
- 28. Lum L., Yao S., Mozer B., Rovescalli A., Von Kessler D., Nirenberg M., Beachy P. A. (2003) Science 299, 2039–2045 [DOI] [PubMed] [Google Scholar]
- 29. Low W. C., Wang C., Pan Y., Huang X. Y., Chen J. K., Wang B. (2008) Dev. Biol. 321, 188–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Diel S., Klass K., Wittig B., Kleuss C. (2006) J. Biol. Chem. 281, 288–294 [DOI] [PubMed] [Google Scholar]
- 31. Milenkovic L., Scott M. P. (2010) Sci. Signal. 3, e14. [DOI] [PubMed] [Google Scholar]
- 32. Milenkovic L., Scott M. P., Rohatgi R. (2009) J. Cell Biol. 187, 365–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hu Q., Milenkovic L., Jin H., Scott M. P., Nachury M. V., Spiliotis E. T., Nelson W. J. (2010) Science 329, 436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pedersen L. B., Rosenbaum J. L. (2008) Curr. Top. Dev. Biol. 85, 23–61 [DOI] [PubMed] [Google Scholar]
- 35. Wang Y., Zhou Z., Walsh C. T., McMahon A. P. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2623–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Boehlke C., Bashkurov M., Buescher A., Krick T., John A. K., Nitschke R., Walz G., Kuehn E. W. (2010) J. Cell Sci. 123, 1460–1467 [DOI] [PubMed] [Google Scholar]
- 37. Yu J. Z., Dave R. H., Allen J. A., Sarma T., Rasenick M. M. (2009) J. Biol. Chem. 284, 10462–10472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chomczynski P., Sacchi N. (1987) Anal. Biochem. 162, 156–159 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.