Abstract
The Alzheimer BACE1 enzyme cleaves numerous substrates, with largely unknown physiological consequences. We have previously identified the contribution of elevated BACE1 activity to voltage-gated sodium channel Nav1.1 density and neuronal function. Here, we analyzed physiological changes in sodium channel metabolism in BACE1-null mice. Mechanistically, we first confirmed that endogenous BACE1 requires its substrate, the β-subunit Navβ2, to regulate levels of the pore-forming α-subunit Nav1.1 in cultured primary neurons. Next, we analyzed sodium channel α-subunit levels in brains of BACE1-null mice at 1 and 3 months of age. At both ages, we found that Nav1.1 protein levels were significantly decreased in BACE1-null versus wild-type mouse brains, remaining unchanged in BACE1-heterozygous mouse brains. Interestingly, levels of Nav1.2 and Nav1.6 α-subunits also decreased in 1-month-old BACE1-null mice. In the hippocampus of BACE1-null mice, we found a robust 57% decrease of Nav1.1 levels. Next, we performed surface biotinylation studies in acutely dissociated hippocampal slices from BACE1-null mice. Hippocampal surface Nav1.1 levels were significantly decreased, but Nav1.2 surface levels were increased in BACE1-null mice perhaps as a compensatory mechanism for reduced surface Nav1.1. We also found that Navβ2 processing and Nav1.1 mRNA levels were significantly decreased in brains of BACE1-null mice. This suggests a mechanism consistent with BACE1 activity regulating mRNA levels of the α-subunit Nav1.1 via cleavage of cell-surface Navβ2. Together, our data show that endogenous BACE1 activity regulates total and surface levels of voltage-gated sodium channels in mouse brains. Both decreased Nav1.1 and elevated surface Nav1.2 may result in a seizure phenotype. Our data caution that therapeutic BACE1 activity inhibition in Alzheimer disease patients may affect Nav1 metabolism and alter neuronal membrane excitability in Alzheimer disease patients.
Keywords: Alzheimer Disease, Neurodegeneration, Presenilin, Secretases, Sodium Channels, BACE1, Nav1.1, Nav1.2, Voltage-gated Sodium Channel
Introduction
Alzheimer disease (AD)3 is a devastating neurodegenerative disorder characterized by progressive memory loss. In the late stages of the disease, neuropsychiatric symptoms include depression, aggressiveness, agitation, and generalized anxiety (1). Pathological hallmarks of AD patient brains consist of extracellular amyloid deposits (senile plaques), neurofibrillary tangles, and loss of neurons and synapses in the hippocampus and the cerebral cortex (2, 3). The main component of amyloid deposits is a short amyloid β (Aβ) peptide most commonly of 40–42 amino acids, which derives from the amyloid precursor protein (APP). To generate Aβ, APP is first cleaved by β-site APP-cleaving enzyme 1 (BACE1) and subsequently by γ-secretase. Because of their central role in Aβ generation, it is not surprising that the two secretases are among the prime drug targets for AD treatment (4, 5).
BACE1 is a transmembrane aspartic protease that is ubiquitously expressed with the highest levels in the brain (6, 7). BACE1 levels and activity are increased in AD patient brains (8–10). The reason why BACE1 is elevated in AD brains is unknown, although it has been shown that depletion of the adaptor protein GGA3 and phosphorylation of the translation initiation factor eIF2α increase BACE1 levels (11, 12). Cellular and oxidative stresses as well as energy starvation have an effect on the enzyme function (11, 13, 14). Currently, only a few proteins are known as physiological substrates of BACE1 (15–19). To understand the physiological function of BACE1, it is essential to characterize BACE1-mediated cleavage of its neuronal substrates.
Voltage-gated sodium channels (Nav1s) consist of one pore-forming α-subunit, associated with one or two β-accessory subunits (20). Currently, 10 α- and 4 β-subunits have been reported (21). The β-subunits are type I single-transmembrane proteins that regulate the localization, trafficking, and inactivation of the α-subunits by direct interaction (20, 22–24). Of the four β-subunits, the β2-subunit (Navβ2) appears to have a major role in the regulation of the total and cell surface density of sodium channels in neurons (25–27). Navβ2-null mice show altered response to pain and a reduced threshold for seizures because of defects in electrical excitability in neurons (26, 27).
BACE1-null mice have revealed abnormalities in behavior, cognitive, and emotional functions (28–31). An imbalance in sodium channel function may contribute to some of these phenotypes. Indeed, the auxiliary β2-subunit of voltage-gated sodium channels undergoes sequential processing by BACE1 and γ-secretase similar to APP (18, 19). We found that BACE1-mediated cleavage of Navβ2 alters neuronal activity (32). Moreover, elevated levels of the pore-forming α-subunit Nav1.1 are observed in AD brains with elevated BACE1 expression and also in mice overexpressing human BACE1 (32). The β2-intracellular domain (β2-ICD), derived from BACE1 and γ-secretase-mediated cleavage of β2, regulates Nav1.1 expression. However, Nav1.1 trafficking to the cell membrane is impaired resulting in reduced Nav1.1 surface levels, which leads to reduced sodium current densities (32).
Here, we asked whether endogenous BACE1 activity regulates Nav1 metabolism in neuronal cells under physiological conditions. First, we found that Navβ2 is essential for BACE1 activity-dependent regulation of Nav1.1 mRNA and protein levels in cultured embryonic neurons. Next, we also found that endogenous BACE1 activity regulates total and surface levels of Nav1.1 especially in adult hippocampus. Interestingly, Nav1.2 appears to compensate for decreased surface levels of Nav1.1.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
N-[N-3,5-Difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester and BACE inhibitor IV were purchased from Calbiochem. BACE1 monoclonal antibody (3D5) was a kind gift from Dr. Robert Vassar (Northwestern University), and anti-Navβ2 directed against the C terminus of endogenous Navβ2 was from Dr. Nobuyuki Nukina (RIKEN Brain Science Institute). The following antibodies were purchased for this study: anti-GAPDH (Chemicon); anti-Nav1.1 antibody (University of California, Davis/National Institutes of Health NeuroMab Facility); anti-Nav1.2, anti-Nav1.3, and anti-Nav1.6 (Alomone Labs); anti-Nav1s (Sigma); anti-N-cadherin (BD Transduction Laboratories); and anti-synaptophysin (StressGen).
Primary Neuronal Cultures
Mouse primary hippocampal/cortical neuronal cultures were prepared and maintained in Neurobasal media (Invitrogen) supplemented with B27 and 0.5 mm l-glutamine as described previously (18). The neuronal cultures were maintained at 37 °C in a humidified 5% CO2 atmosphere for 14 days. For rat primary neuronal culture, cryopreserved rat hippocampal/cortical regions were purchased from BrainBits (Springfield, IL), dissociated, and plated on poly-d-lysine/laminin-coated plates. The neuronal cultures were maintained for 14–17 days in Neurobasal media (Invitrogen) supplemented with B27 and 0.5 mm l-glutamine.
Breeding
All animal procedures were approved by the Subcommittee on Research Animal Care on the Use and Care of Animals, Massachusetts General Hospital. BACE1-null and wild-type control mice (C57BL/6J background) were purchased from The Jackson Laboratory (Bar Harbor, ME). BACE1-heterozygous knock-out mice were generated by crossing BACE1-null and wild-type control mice. Genotypes were determined by genomic PCR using the Allele-in-One mouse tail direct PCR system (Allele Biotechnology, San Diego) using the following primers: 5′-AGGCAGCTTTGTGGAGATGGTG-3′, 5′-CGGGAAATGGAAAGGCTACTCC-3′, and 5′-TGGATGTGGAATGTGTGCGAG-3′. Navβ2-null and control mice were kindly provided by Dr. Lori L. Isom (University of Michigan). Generation and genotyping of Navβ2-null mice were described previously (26).
Preparation of Mouse Brain Lysates
One- to 3-month-old mice were sacrificed, and brains were immediately removed and stored at −80 °C until use. To prepare total brain lysates, the frozen brains were powdered under liquid nitrogen with the use of mortar and pestle. The aliquots were stored at −80 °C for individual experiments. For total brain extraction, 50–100 μg of brain powder was extracted in 5 volumes of SDS extraction buffer (10 mm Tris-HCl (pH 6.8), 2% SDS, 1 mm EDTA, 150 mm NaCl, a protease inhibitor mixture (Roche Applied Science), 2 mm 1,10-phenanthroline, 2 μm MG132, and 1 mm PMSF). After incubation on ice for 10 min, samples were sonicated at 4 °C for 5 min, passed through 0.22-μm pore-sized filter in spin-x centrifuge tubes (Costar, Corning, NY), and centrifuged for 45 min at 14,000 rpm. To prepare crude brain membrane fractions, the brain powder was resuspended in TBS extraction buffer (10 mm Tris-HCl (pH 6.8), 1 mm EDTA, 150 mm NaCl, a protease inhibitor mixture (Roche Applied Science), 2 mm PNT, 2 μm MG132, and 1 mm PMSF) and passed through 22-gauge needle three times. After brief centrifugation (500 × g, 5 min), the supernatant was then centrifuged again at 10,000 × g for 1 h. The membrane pellet was then resuspended in TBS extraction buffer, and the protein level was determined by BCA assay. For SDS-PAGE analysis, the crude membrane fractions were directly mixed with 4× lithium dodecyl sulfate sample buffer (Invitrogen) supplemented with 8% (v/v) β-mercaptoethanol and heated at 95 °C for 5 min. To prepare hippocampal brain lysates, brains were microdissected under microscope immediately after the brains were removed. Hippocampi were then directly lysed in SDS extraction buffer (10 mm Tris-HCl (pH 6.8), 1.5% SDS, 1 mm EDTA, 150 mm NaCl, a protease inhibitor mixture (Roche Applied Science), 2 mm PNT, 2 μm MG132, and 1 mm PMSF). RNeasy mini kit (Qiagen) was used to extract total RNA from the brains. Approximately 30 mg of brain powder was dissolved in 600 μl of buffer RLT containing 1% β-mercaptoethanol by vortexing. Lysates were applied to QIAshredder (Qiagen). The instructions for RNeasy mini kit (Qiagen) were followed.
Confocal Immunofluorescence Staining of Hippocampal Sections
BACE1-null, BACE1 heterozygote knock-out, and wild-type control mice were perfused with 12 ml of PBS and 1% paraformaldehyde, 1% sucrose solution, respectively. Brains were rapidly removed, further fixed in 1% paraformaldehyde, 1% sucrose solution for 3 h at 4 °C, transferred to 30% sucrose solution, and incubated for 48 h at 4 °C. 40-μm sections were cut, washed three times with PBS, and incubated with blocking solution containing 0.1% Triton X-100 and 5% purified goat IgG (Jackson ImmunoResearch) for 12 h. The sections were then incubated with primary antibody solution containing anti-Nav1.1 antibody (monoclonal, 1:20 dilution, University of California, Davis/National Institutes of Health NeuroMab Facility), 0.1% Triton X-100, and 5% goat IgG for 24 h 4 °C. After washing three times, sections were incubated with secondary antibody solution with anti-mouse Alexa Fluor488 (Invitrogen) for 3 h at room temperature. Immunostained sections were mounted and analyzed by Olympus IX70 fluorescence microscope equipped with a confocal Disk Spinning Unit (Olympus). Images were captured and analyzed by IPlab software.
Western Blot Analysis
20–100 μg of protein were resolved on 4–12% gradient BisTris gels, 12% BisTris gels, or 16% Tricine gels (Invitrogen), depending on the individual experiment as described. The blots were visualized by enhanced chemiluminescence (ECL). The images were captured by using BioMax film (Eastman Kodak Co.) or VersaDoc imaging system (Bio-Rad) and quantitated by using QuantityOne software (Bio-Rad).
Quantitative RT-PCR
Levels of sodium channel genes were analyzed by quantitative RT-PCR performed in a Light Cycler PCR system (Bio-Rad). 1 μg of total RNA was used for the synthesis of cDNA using oligo(dT) primers following the protocol of the SuperScript first-strand synthesis system for RT-PCR (Invitrogen). Relative quantification of mRNA expression of Scn1a (Nav1.1) compared with endogenous expression of GAPDH was analyzed by TaqMan real time PCR. Primers were purchased from Applied Biosystems (catalogue numbers Mn01329052-m1, Mn00488110-m1, and Mn01270368-m1). Expression analysis was performed with 40 two-step cycles for 30 s at 95 °C and 60 s 60 °C as described in the supplier's protocol (Applied Biosystems) using a Bio-Rad iCycler. Sodium channel mRNA was normalized to GAPDH mRNA by the comparative CT method.
Acute Hippocampal Slice Preparation
Hippocampal neurons from BACE1-transgenic and control mice (1–2 month) were acutely isolated using standard procedures (26, 33). Mice were decapitated under deep isoflurane anesthesia, and brains were rapidly removed and iced. Slices (400 μm) were cut and transferred to a low calcium buffer containing 15 mm HEPES (pH 7.4), 140 mm sodium isethionate, 2 mm KCl, 2 mm MgCl2, 0.1 mm CaCl2, and 23 mm glucose. Slices were incubated for more than 1 h in NaHCO3-buffered Earle's balanced salt solution (Sigma) bubbled with 95% O2 and 5% CO2 at room temperature.
Surface Biotinylation
Surface biotinylation of acutely prepared hippocampal slices was performed according to a method described by Thomas-Crusells et al. (34) with slight modifications. The slices were prepared as described earlier and incubated with 100 μm Sulfo-NHS-Biotin (Pierce) in Earle's balanced salt solution bubbled with 95% O2 and 5% on ice for 80 min under dark conditions. The reaction was stopped by washing three times with cold Earle's balanced salt solution containing 100 μm l-lysine. Hippocampal regions were rapidly separated from the individual slices under a dissecting microscope. The isolated hippocampi were extracted with a lysis buffer containing 10 mm Tris-HCl (pH 6.8), 1 mm EDTA, 150 mm NaCl, 0.2% SDS, 0.5% sodium deoxycholate, 1% Triton X-100, and protease inhibitors, followed by a spin at 16,000 × g. Biotinylated proteins were captured by Neutravidin beads (Pierce) at 4 °C overnight.
Statistical Analyses
All statistical analyses were performed using a two-tailed Student's t test or one-way ANOVA followed by a post hoc Tukey's test. Error bars represented in graphs denote the means ± S.E.
RESULTS
Navβ2 Mediates the Effect of BACE1 on Nav1.1 Levels in Primary Neurons
Previously, we found that BACE1 and γ-secretase sequentially cleave Navβ2 to release the intracellular domain of Navβ2 (β2-ICD). In neuroblastoma cells, β2-ICD is localized to the nucleus and regulates mRNA and protein levels of Nav1.1 (32). To test whether endogenous BACE1 activity regulates Nav1.1 through the processing of Navβ2 in vivo, we treated cultured rat cortical/hippocampal neurons with BACE1 inhibitors and analyzed Nav1.1 levels. We found that DR9 treatment decreased Nav1.1 levels up to 50% in a dose-dependent manner (Fig. 1A). Interestingly, we also found that Nav1.2 levels were dose-dependently increased upon DR9 treatment, suggesting potential compensatory changes (supplemental Fig. 1, A and B). Previously, we have shown that recombinant β2-ICD expression significantly elevates Scn1a (Nav1.1) mRNA levels in rat neuroblastoma cells (32). Similarly to Nav1.1, Nav1.2 may also be regulated at the level of β2-ICD. Therefore, we tested the effect of recombinant β2-ICD on Scn2a (Nav1.2) mRNA levels. Unlike Scn1a (Nav1.1), Scn2a (Nav1.2) mRNA levels did not change in a statistically meaningful way under the same conditions (supplemental Fig. 1C). These results suggest that a potential compensatory change of Nav1.2 may not be directly regulated by β2-ICD.
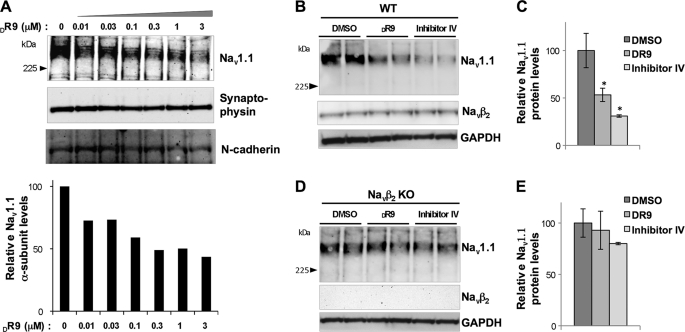
FIGURE 1.
Navβ2 is required for BACE1 activity-dependent regulation of Nav1.1 levels. Primary neuronal culture (14 days in vitro) from Navβ2-null mice (Navβ2 KO), wild-type control mice (WT), and wild-type rat were treated with control vehicle (DMSO) or BACE inhibitors (DR9 and/or inhibitor IV) for 48–72 h. Nav1.1 protein levels were analyzed by Western blot. A, upper panel, Nav1.1 Western blot analysis of rat primary neurons (14 days in vitro) treated with various concentrations of DR9 and control vehicle DMSO for 72 h; lower panel, quantitative analysis of Nav1.1 protein levels. B, representative Western blot showing changes in Nav1.1 levels upon BACE1 inhibitor treatment in cultured WT neurons. C, quantitative analysis of Nav1.1 protein levels upon BACE1 inhibitor treatment in cultured WT neurons. D, representative Western blot showing Nav1.1 levels of cultured Navβ2 KO neurons treated with DMSO, DR9, or inhibitor IV. E, quantitative analysis of Nav1.1 protein levels of cultured Navβ2 KO neurons treated with DMSO, DR9, or inhibitor IV. One-way ANOVA followed by a post hoc Tukey's test was used for statistical analysis (*, p < 0.05; n = 3 per each condition).
To explore whether Navβ2 is required for BACE1 activity-dependent Nav1.1 decrease, we took advantage of primary neuronal cultures from Navβ2-null mice (26). Cultured hippocampal/cortical neurons (14 days in vitro) from wild-type and Navβ2-null mice were treated with BACE1 inhibitors, followed by analysis of Nav1.1 mRNA and protein levels. As expected, in wild-type neurons we found that two different BACE inhibitors, DR9 or inhibitor IV, decrease Nav1.1 protein levels by 42–70% (Fig. 1, B and C). Scn1a (Nav1.1) mRNA levels were also decreased by DR9 or inhibitor IV (supplemental Fig. 1D). These data show that BACE1 activity is required for maintaining Nav1.1 levels in cultured rat and mouse neurons. In contrast, in Navβ2-null neurons Nav1.1 levels were unaffected by the same BACE1 inhibitor treatments (Fig. 1, D and E). These data show that Navβ2 is required for BACE1-dependent regulation of Nav1.1 levels in cultured embryonic neurons.
Decreased Nav1 Levels in BACE-null Mice
Next, we investigated whether endogenous BACE1 activity is required for maintaining normal Nav1 α-subunit levels in adult brains. Brain samples were prepared from 1- and 3-month-old BACE1-null (BACE1-KO), BACE1-heterozygous knock-out (BACE1-HE), and aged-matched wild-type control mice (WT). One month of age was chosen because it is the minimum age for analyzing adult Nav1s and is also age-compatible with acute preparation of hippocampal slices without inducing significant neuronal death. All animals were healthy; however, BACE1-null mice were generally smaller in size than wild-type and BACE1-HE animals as reported previously (28). Furthermore, increased mortality was observed among BACE1-null mice.
We compared total brain Nav1 α-subunits levels in 1-month-old BACE1-null and BACE1-heterozygous knock-out with age-matched wild-type mice. To avoid degradation of protease-prone Nav1 α-subunits, brains were rapidly removed, powdered under liquid nitrogen, aliquoted, and stored at −80 °C until extraction. Because Nav1 α-subunits are hydrophobic integral membrane proteins, extraction conditions with strong ionic detergents were required to solubilize α-subunits from insoluble membrane fractions. We compared several extraction conditions, and we found that extraction with 2% SDS showed the most consistent results, tightly comparable with a method based on direct boiling with SDS-PAGE sample loading buffer (Fig. 2; for detailed buffer formulation and conditions, please see “Experimental Procedures”).
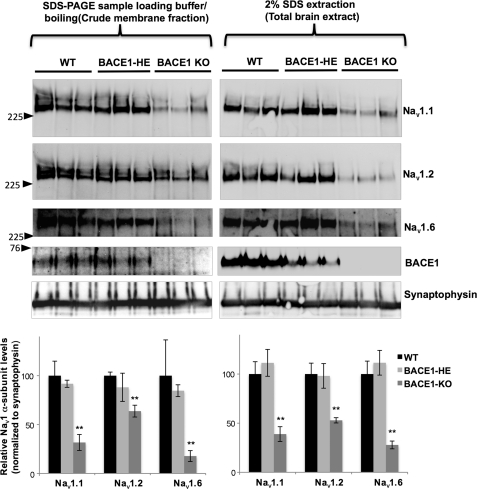
FIGURE 2.
Nav1 α-subunit levels are decreased in brains from BACE1-null mice. Upper left panel, Western blot showing Nav1.1, Nav1.2, and Nav1.6 α-subunit levels in brain membrane fractions prepared from 1-month-old wild-type (WT), BACE1-heterozygous knock-out (BACE1-HE), and BACE1-null mice (BACE1-KO). Lower left panel, graph showing normalized Nav1.1 protein levels in WT, BACE1-HE, and BACE1-KO normalized against synaptophysin (ANOVA followed by a post hoc Tukey's test; *, p < 0.05; **, p < 0.01; n = 3 for WT, BACE1-HE, and BACE1-KO, respectively). Upper right panel, Western blot showing Nav1.1, Nav1.2 and Nav1.6 α-subunit levels in total brain extract prepared from 1-month-old WT, BACE1-HE, and BACE1-KO; 2% SDS was used to extract total brain homogenates. Lower right panel, a graph showing normalized Nav1.1 protein levels in WT, BACE1-HE, and BACE1-KO normalized against synaptophysin (ANOVA followed by a post hoc Tukey's test; **, p < 0.01; n = 3 for WT, BACE1-HE, and BACE1-KO, respectively).
Western blot analysis showed more than 50% decrease of Nav1.1 levels in brain extracts from BACE1-null as compared with age-matched wild-type control mice, although no significant difference was detected between BACE1-heterozygous knock-out and wild-type controls (Fig. 2). These data suggest that more than 50% decrease in BACE1 expression is required for decreasing Nav1.1 levels in adult mouse brains. Interestingly, we also observed significant decreases in Nav1.2 and Nav1.6 levels, two additional major CNS Nav1 α-subunits (Fig. 2). Levels of Nav1.3, an embryonic Nav1 α-subunit, did not change (data not shown). The supplemental Fig. 2 shows a littermate analysis of BACE1-null and BACE1-heterozygous knock-out mice. As expected, we found more than 50% decrease of Nav1.1 levels in BACE1-null mice as compared with BACE1-heterozygous knock-out littermate controls at the same age.
Finally, we investigated whether the changes in Nav1.1 levels were also seen in older mice. For this purpose, Nav1 levels were analyzed in whole brain extracts from 3-month-old BACE1-null and wild-type control mice (Fig. 3A). Similar to 1-month-old mice, we observed a significant ∼20% decrease in Nav1.1 and Nav1.2 levels in BACE1-null mice but no significant changes in Nav1.6 levels (Fig. 3B). Interestingly, decreases in Nav1 α-subunit levels were less pronounced at 3 months versus 1 month of age, suggesting the possibility that the lack of BACE1 activity affects Nav1 α-subunit levels more prominently at early ages (Fig. 3, A and B). High expression of BACE1 in early postnatal stages may also explain the strong effects of BACE1 knock-out on Nav1 α-subunit levels in young mouse brains (15). Nav1.3 levels were hardly detectable, because this subunit is predominantly expressed in early developmental stages (data not shown). Together, these data indicate that endogenous BACE1 activity regulates Nav1 α-subunit levels in adult neurons.
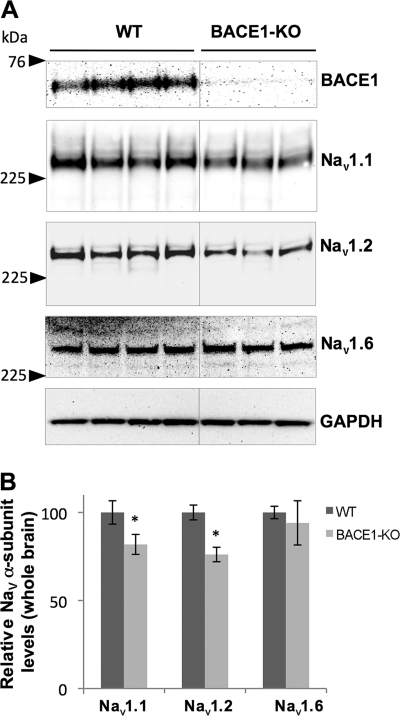
FIGURE 3.
Nav1.1 and Nav1.2 are decreased in 3-month-old BACE1-null mice. A, Western blot showing Nav1.1, Nav1.2, and Nav1.6 α-subunit levels in total brain extracts from 3-month-old wild-type (WT) and BACE1-null mice (BACE1-KO). B, quantitative analysis showing normalized Nav1.1, Nav1.2, and Nav1.6 protein levels in WT and BACE1-KO (Student t test; *, p < 0.05; n = 4 for both WT and BACE1-KO).
Nav1.1 Protein Levels in the Hippocampus of BACE-null Mice
We then investigated the effect of BACE1 ablation in the hippocampus, which is a highly affected brain region in patients with Alzheimer disease. Hippocampal regions were dissected from the brains of 3-month-old BACE1-null and wild-type control mice, extracted, and analyzed for Nav1 subunits by Western blot analysis. We observed a robust 57% decrease of Nav1.1 levels in BACE1-null as compared with wild-type control mice (Fig. 4, A and B). Interestingly, we could not detect any significant changes in Nav1.2 or Nav1.6 levels in these samples, suggesting that Nav1.1 is the major Nav1 α-subunit affected by BACE1 activity in the hippocampal region (Fig. 4, A and B).
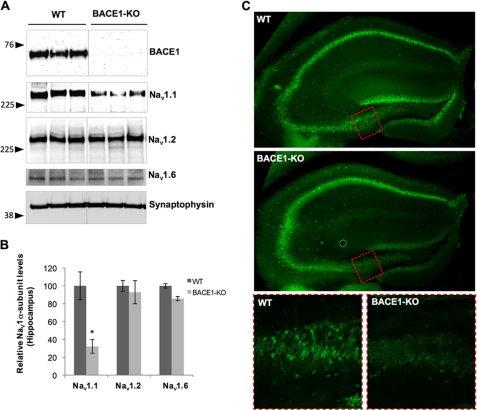
FIGURE 4.
Hippocampal Nav1.1 levels are decreased in BACE1-null mice. A, Western blot analysis of Nav1.1, Nav1.2, and Nav1.6 from WT and BACE1-KO mice. B, quantitative analysis of Nav1.1, Nav1.2, and Nav1.6 levels in hippocampal brain extracts from BACE1-KO and WT mice (Student's t test; *, p < 0.01; n = 3 for WT and n = 4 for BACE1-KO). C, confocal immunofluorescence analysis of Nav1.1 (green) in hippocampal regions of 3-month-old WT or BACE1-KO mice. Top and middle panels, low magnification (×50) images of hippocampal region stained with Nav1.1 staining (green). The red dotted box indicates the area of high magnification shown in the bottom panels. Bottom panels, high magnification (×200) images with Nav1.1 staining (bottom left panel (WT); bottom right panel, BACE1-KO).
We also investigated Nav1.1 distribution in the hippocampal region of 3-month-old BACE1-null mice by immunofluorescence (Fig. 4C). Using a monoclonal Nav1.1 antibody (Neuromab), we mainly detected Nav1.1 in neuronal cell bodies of hippocampal neurons (35, 36). As shown in Fig. 4C, we found that Nav1.1 levels were significantly decreased in the dentate gyrus, hilus, and CA3 regions (Fig. 4C, upper and middle panels). At higher magnification, we confirmed that somatic Nav1.1 staining is specifically decreased in BACE1-null mice in the same regions (Fig. 4C, bottom panels). We also found similar but more dramatic decreases of Nav1.1 in 1-month-old BACE1-null mice as compared with age-matched controls (supplemental Fig. 3). These data show that ablation of BACE1 significantly decreases Nav1.1 levels in adult hippocampal neurons.
Altered Surface Nav1 Levels in BACE1-null Brain Slices
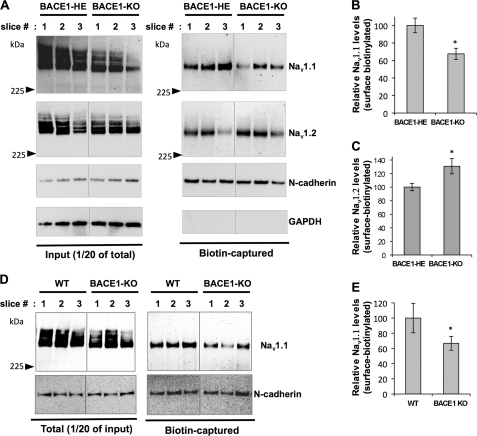
In neurons, newly synthesized Nav1 α-subunits accumulate inside the cell body, and only a small fraction of active channels reaches the cell surface (22). To explore whether endogenous BACE1 activity modulates surface expression of Nav1 α-subunits, we performed surface biotinylation analysis using acute hippocampal slices prepared from BACE1-HE and BACE1-null mice as described previously (32). The biotinylated surface proteins were captured, and total and surface Nav1 α-subunit levels were analyzed by Western blot analysis (Fig. 5A). As expected, total levels of Nav1.1 were largely decreased in BACE1-null as compared with BACE1-HE (Fig. 5A, left panel). Similar to the total Nav1.1 levels, surface levels of Nav1.1 also largely and significantly decreased in BACE1-null as compared with BACE1-HE (Fig. 5A, right panel). When quantitated, we found 32% decrease in BACE1-null as compared with BACE1-HE control mice (Fig. 5B). Surface expression of Nav1.2 showed a significant increase, in contrast with Nav1.1 (Fig. 5, A and C). This suggests a compensatory increase in Nav1.2 sodium channel density in adult mice. We also compared Nav1.1 surface levels between BACE1-null with wild-type control mice and found a similar 35% decrease (Fig. 5, D and E). Our data show that endogenous BACE1 activity regulates surface expression of Nav1 α-subunit levels in adult neurons and suggest a compensatory regulation between Nav1.1 and Nav1.2 surface levels.
FIGURE 5.
Surface Nav1 α-subunit levels are altered in BACE1-null hippocampal slices. Surface biotinylation experiments were performed on acutely prepared hippocampal slices from WT, BACE1-HE, and BACE1-KO mice. 2–3 matching slice pairs from similar locations of the brains were selected for the experiments. The slices with the same numbers are matching pairs. A, representative Western blot showing total and biotin-captured surface Nav1.1 and Nav1.2 levels in hippocampal slices (Slice #1–3) from BACE1-HE and BACE1-KO. B, representative Western blot showing total and biotin-captured surface Nav1.1 levels in hippocampal slices (Slice #1–3) from wild-type control (WT) and BACE1-KO. C, graph showing quantitated surface Nav1.1 levels in BACE1-HE and BACE1-KO (Student's t test; *, p < 0.01; n = 7 slices from three BACE1-HE and n = 8 from three BACE1-KO mice). Surface N-cadherin levels were used to normalize the surface Nav1.1 and Nav1.2 levels. D, graph showing quantitated surface Nav1.2 levels in BACE1-HE and BACE1-KO (Student's t test; *, p < 0.01; n = 7 slices from three BACE1-HE and n =8 from three BACE1-KO mice). E, graph showing quantitated surface Nav1.1 levels in WT and BACE1-KO (Student t test; *, p < 0.01; n = 8 slices from three WT and BACE1-KO mice, respectively).
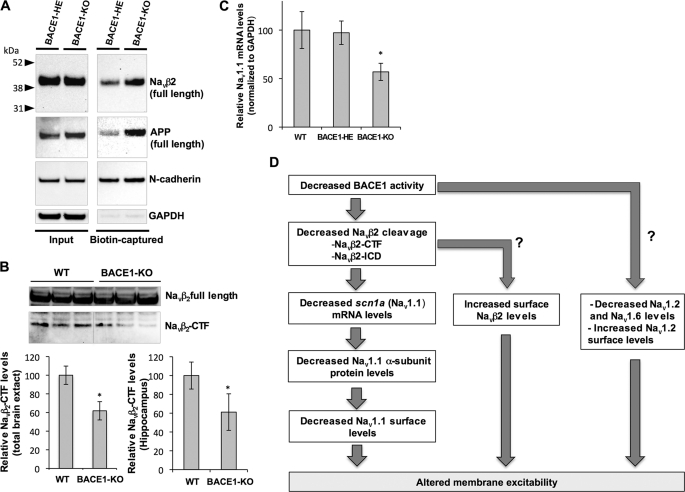
In addition to Nav1 α-subunit surface levels, we also investigated surface expression of full-length Navβ2 in acute brain slices from BACE1-HE and BACE1-null mice. Interestingly, we found that surface levels of full-length Navβ2 increased by 1.92-fold in BACE1-null as compared with BACE1-HE (±0.06, Fig. 6A). Similarly, the APP, another well known physiological substrate of BACE1, also showed increased surface expression of its full-length mature form in BACE1-null mice (Fig. 6A). These surface level changes suggest that ablation of BACE1 decreased the cleavage of these substrates at the cell surface because BACE1 is known to cleave its substrates at the cell surface or in the endosomal recycling compartment (37).
FIGURE 6.
Navβ2 processing and scn1a (Nav1. 1) mRNA levels are decreased in brains of BACE1-null mice. A, surface Navβ2 and APP levels are largely decreased in brain slices from BACE1-null (BACE1-KO) as compared with BACE1-heterozygous knock-out (BACE1-HE). Slice biotinylation experiments were performed as described in Fig. 5. B, upper panel, representative Western blot showing full-length Navβ2 and C-terminal fragment (Navβ2-CTF) levels in hippocampal brain extracts prepared from WT and BACE1-KO mice. Lower panel, quantitative analysis of Navβ2-CTF levels in total brain extract from 1-month-old WT and BACE1-KO (left) and hippocampal extracts from 3-month-old WT and BACE1-KO WT and BACE1-KO mice (right, Student's t test; *, p < 0.01; n = 3 per each condition). C, TaqMan real time RT-PCR analysis of scn1a (Nav1.1) mRNA levels in 1-month-old WT, BACE1-HE, and BACE1-KO (ANOVA followed by a post hoc Tukey's test; *, p < 0.05; n = 5 for WT, n =8 for BACE1-HE, and n =6 for BACE1 KO). GAPDH levels were used to normalize Scn1a (Nav1.1) mRNA levels. D, diagram summarizing altered sodium channel metabolism in BACE1-null mice.
Previously, we found that BACE1 and γ-secretase sequentially cleave Navβ2 to release β2-ICD (32). As shown in our previous study and supplemental Fig. 1C, β2-ICD is localized to the nucleus and regulates mRNA and protein levels of Nav1.1 in neuroblastoma cells. To test whether decreased Nav1.1 proteins levels corresponded to reduced Navβ2 processing, we assessed levels of the BACE1 cleavage product Navβ2 C-terminal fragment (Navβ2-CTF). Consistent with previous reports (18, 19), we found that ablation of BACE1 reduced generation of Navβ2-CTF by 40% both in total brain extract from 1-month-old BACE1-KO and in hippocampal regions from 3-month-old BACE1-KO mice (Fig. 6B). We were not able to directly detect β2-ICD because the Navβ2 antibody was not sensitive enough to detect β2-ICD, which was present in very low levels. However, together these data strongly suggest that endogenous BACE1 activity modulates Navβ2 processing, and ablation of BACE1 decreases Navβ2-CTF and possibly β2-ICD.
Next, we investigated whether decreased Navβ2 processing also results in reduced Scn1a (Nav1.1) mRNA in brains of BACE1-null mice (32). In brain samples from 1-month-old WT, BACE1-HE, and BACE1-null, we measured Scn1a (Nav1.1) mRNA levels by the TaqMan real time RT-PCR method. Similar to its protein levels, we found a significant decrease of Scn1a (Nav1.1) mRNA in BACE1-null as compared with wild-type mice (Fig. 6C). Interestingly, we were not able to detect significant changes of Scn2a (Nav1.2) or Scn8a (Nav1.6) mRNA levels, suggesting that changes at the protein level may be responsible for decreases in Nav1.2 or Nav1.6 in BACE1-null mice (supplemental Fig. 4). Together, our mechanistic data strongly suggest that decreased Navβ2 processing is responsible for the decrease of Nav1.1 protein levels by decreasing scn1a mRNA in BACE1-null mice.
DISCUSSION
Here, we show that expression levels of three major brain Nav1 channels are strongly affected by BACE1 deletion during early postnatal brain development. Levels of the Nav1.1 α-subunit showed the most dramatic decrease. These changes are predicted to have significant effects on the development of excitability. Ablation of BACE1 results in ∼40–50% decrease of Nav1.1 protein and mRNA levels in whole brains of 1-month-old mice and up to 56% in the hippocampus of 3-month-old mice (Figs. 2–4 and supplemental Figs. 2–4). We also found that the decrease in Nav1.1 in BACE1-null mice was less pronounced at 3 months versus 1 month of age, suggesting the possibility that the lack of BACE1 activity affects Nav1 α-subunit levels more prominently at early ages. Nav1.2 α-subunit levels were also decreased in whole brain samples from 1- and 3-month-old mice but not in the hippocampus. In addition, we found that Nav1.6 α-subunit levels were also decreased in brains of 1-month-old but not in 3-month-old mice. These data demonstrate that BACE1 regulates adult brain Nav1 α-subunit levels in physiological conditions, possibly in an age-dependent and/or region-specific manner.
Previously, we reported that BACE1/γ-secretase activities promote the release of the β2-ICD, which localizes to the nucleus and increases mRNA and protein levels of the pore-forming Nav1.1 α-subunit in neuroblastoma cells (18, 32). Elevated BACE1 activity also increases Navβ2 cleavages and Nav1.1 levels in brains of BACE1-transgenic mice and AD patients (32). Although BACE1 cleaves all four β-subunits in vitro, only Navβ2 and Navβ4 were reported as physiological substrates in mouse brains (19). Consistent with our previous findings, we found a significant decrease of Navβ2 C-terminal fragment levels in the hippocampus of BACE1-null mice (Fig. 6B). Therefore, endogenous BACE1 activity is essential for Navβ2 cleavage in the hippocampus in vivo. Fig. 1 also suggests a crucial role of Navβ2 in BACE1-dependent regulation of Nav1.1 levels at least in the embryonic neuronal cells. Although BACE1 inhibitors largely decreased Scn1a (Nav1.1) mRNA and protein levels in cultured cortical/hippocampal neurons, the same inhibitors did not change Nav1.1 levels in Navβ2-null neurons (Fig. 1 and supplemental Fig. 1, A, B, and D). Together, these data support the notion that Navβ2 and its processing are essential for regulating Nav1.1 levels under physiological conditions. Interestingly, we were not able to detect significant changes of Scn2a (Nav1.2) or Scn8a (Nav1.6) mRNA levels, although Nav1.2 and Nav1.6 protein levels are significantly decreased in the brains from 1-month-old BACE1-null mice. This suggests an alternative molecular pathway mediating the Nav1.2 or Nav1.6 decrease in BACE1-null mice (Fig. 6C and supplemental Fig. 4). Altered processing of other neuronal BACE1 substrates may contribute to these Nav1 α-subunit changes. Further studies will be required to fully characterize the underlying molecular mechanism of Nav1 α-subunit changes.
As shown in Fig. 5, ablation of BACE1 results in ∼40% decrease of surface Nav1.1 levels in hippocampal neurons, which is comparable with the decreased levels of total Nav1.1 in the same region. Therefore, surface expression of Nav1.1 is regulated by the decrease of total Nav1.1 levels in BACE1-null mice in physiological conditions. On the contrary, we found that Nav1.2 surface levels are significantly increased (Fig. 5, A and D). This suggests a compensatory mechanism between Nav1.1 and Nav1.2 surface levels. However, disconnected total versus cell surface distributions of Nav1.2 in BACE1-null mice are reminiscent of Nav1.1 distribution in BACE1-overexpressing systems. In neuroblastoma cells and adult hippocampal neurons from BACE1-transgenic mice, overexpressed BACE1 paradoxically reduced cell surface expression of Nav1 α-subunits, although total Nav1.1 α-subunits levels increased (32). The increased Nav1.1 was retained inside the cells and accumulated in HSP70-positive punctate structures, suggesting that Nav1.1 trafficking to cell surface was inhibited by BACE1 overexpression (32). Together, these data indicate that expression of Nav1 α-subunits on the cell surface is under tight regulation, and changes in intracellular α-subunit levels may not always reflect surface expression of each channel. Decreased BACE-mediated cleavage of surface Navβ2 may differentially affect Nav1.1 and Nav1.2 surface trafficking. Further studies will be required to clarify how BACE1-mediated cleavage of Navβ2 can differentially regulate the trafficking of each α-subunit.
Neuronal Nav1 channels are known to be up-regulated or redistributed upon nerve injury or demyelination (38–41). In experimental allergic encephalomyelitis, an animal model of multiple sclerosis, levels and distribution of Nav1.2 and Nav1.6 are altered in demyelinated axons within acute multiple sclerosis plaques (39–41). BACE1-null mice also show mild but significant hypomyelination in the CNS and peripheral nervous system, and these might contribute to the elevated Nav1.2 surface levels shown in BACE1-null mice (15, 31, 42). Interestingly, a recent study showed that Navβ2 also mediates demyelination-induced Nav1 α-subunit regulation in the same multiple sclerosis model (43). Together with our data, this study suggests the interesting possibility that in BACE1-null mice Navβ2 not only is an essential Nav1 β-subunit in regulating changes in Nav1 α-subunit levels via generation of its ICD but also indirectly via reduced myelination, which is a well characterized phenotype of BACE-null mice. Further studies are required to clarify whether Navβ2 also plays a role in regulating Nav1 α-subunit levels under mild hypomyelination in addition to demyelinating conditions.
Deletion of the Nav1.1 gene does not significantly affect sodium current density in hippocampal pyramidal neurons because of high expression of Nav1.2 and other α-subunits (35, 36, 44). However, Nav1.1 plays a major role in regulating sodium currents in hippocampal GABAergic interneurons. Indeed, 50% decrease of Nav1.1 dramatically reduced sodium current in GABAergic interneurons (36). Similarly, 40–50% decrease of total and surface Nav1.1 levels in BACE1-null mice may decrease sodium current in hippocampal GABAergic interneurons. This would result in electrical imbalances and the vulnerability to epileptic seizures as previously shown in Nav1.1-null mice (36). Increase of Nav1.2 surface expression may also contribute to the electrical imbalances by selectively increasing sodium currents in hippocampal pyramidal neurons.
While we were revising this manuscript, two independent groups reported that BACE1-null mice showed increased seizure susceptibility and spontaneous behavioral seizure phenotypes (45, 46). These results clearly support our prediction, based on altered Nav1.1 and Nav1.2 surface expression in BACE1-null mice (Fig. 5). Hu et al. (45) showed that surface levels of Nav1.2 are increased in cultured hippocampal neurons from BACE1-null mice, which is also consistent with our results on increased Nav1.2 surface levels in adult hippocampus from BACE1-null mice (Fig. 5). In addition to Nav1.2 changes, we also found reduced surface levels of Nav1.1, which allow us to propose a novel Nav1.1-based mechanism for the increased seizure susceptibility of BACE1-null mice (Fig. 6D). Indeed, mutations of the Nav1.1 α-subunit are frequently associated with inherited familial epileptic seizures in humans (44).
Early electrical deficits may also explain the enhanced lethality of BACE1/2-null mice at very early ages and their body weight loss (28, 46). Nav1.1-null and -heterozygous mice show severe and fatal seizure phenotypes (36, 44). A selective loss of Nav1.1-mediated sodium currents in inhibitory neurons is proposed as a molecular mechanism underlying these severe phenotypes. Interestingly, all the major Nav1 α-subunit levels are decreased in BACE1-null mice (Fig. 2) and may not be selective to one neuronal population. Therefore, neuronal excitability in BACE1-null mice may be generally reduced all over the brain rather than in specific circuits, which may not lead to a severe and fatal electrical imbalance in neural circuits. This may explain why BACE1-null mice do not show a fatal seizure phenotype as predicted by the drastic Nav1 α-subunit decreases at the early age (Fig. 2).
Nav1 α-subunits are large hydrophobic membrane proteins with multiple transmembrane domains (24). It is technically challenging to analyze total Nav1 α-subunits because they are highly insoluble in mild detergent extraction conditions. To overcome this technical problem, we used several extraction conditions and compared the results (Fig. 2). We found that 1.5–2% of SDS extraction conditions showed the most consistent results, tightly comparable with a method based on direct boiling of membrane fractions in the presence of SDS-PAGE sample loading buffer. The direct boiling method has been commonly used for sodium channel analysis by other groups (27, 36). In these conditions, we found that Nav1.1 and Nav1.2 levels are consistently decreased especially in young adult BACE1-null mice as compared with BACE-HE and wild-type controls (Fig. 2). These data are consistent with the results from Hu et al. and Hitt et al. (45, 46). However, Hitt et al. (46) reported that Nav1.2 and Nav1.6 levels were not significantly changed in BACE1-null mice (45). The discrepancy regarding Nav1.2 may derive from differential extraction conditions (2% SDS versus 1% Triton X-100) or different ages of the mice. Indeed, we found that the decrease in Nav1.2 is much less pronounced (∼20%) in 3-month-old mice as compared with 1-month-old mice (∼40–50%) (Figs. 2 and 3). Consistent with the data from Hitt et al. (46), we found that Nav1.2 levels were not significantly changed in the hippocampal region of 3-month-old mice, although Nav1.1 shows a dramatic decrease in the same samples (Fig. 4). However, Hitt et al. (46) did not analyze levels of Nav1.2 at the cell surface. Because the vast majority of sodium channels is retained inside the cells, total Nav1.2 levels may not reflect changes at the cell surface. We found that Nav1.1 levels decrease at the cell surface, although Nav1.2 levels increase in agreement with Hu et al. (45). Taken together, these data argue that the lack of BACE1 activity dynamically modulates changes in Nav1 α-subunit levels depending on age/brain region and that changes in cell surface Nav1 α-subunit levels are likely to cause epileptic seizures in BACE1-null mice. Further studies will be required to fully address BACE1 effects on Nav1 α-subunit levels in vivo.
BACE1 is one of the prime drug targets for therapeutic treatment of AD patents. Genetic ablation of BACE1 showed decreased Aβ levels and ameliorated cognitive function in animal models (47). Our findings that the ablation of BACE1 leads to decreased sodium channel levels suggest that complete block of BACE1 is likely to cause side effects through altered sodium current densities. However, our studies on Nav1 levels in BACE1-heterozygous mice show that 50% decrease of BACE1 activity does not significantly decrease Nav1.1 levels (Fig. 1), although partial reduction of BACE1 results in dramatic reduction in Aβ plaques, neuritic burden, and synaptic deficits in older mice (48). Therefore, it will be important to find a therapeutic window for inhibiting BACE1 activity to block Aβ burden, without significantly affecting Nav1 metabolism.
Supplementary Material
Acknowledgments
We thank Dr. Lori L. Isom (University of Michigan) for providing us Navβ2-null and control mice, Dr. Nobuyuki Nukina (RIKEN Brain Science Institute) for the gift of the C-terminal Navβ2 antibody, Dr. Robert Vassar (Northwestern University) for the BACE1 monoclonal antibody 3D5, and Dr. Jordan Tang (Oklahoma Medical Research Foundation) for DR9.
This work was supported, in whole or in part, by National Institutes of Health grants from NIA (to D. M. K. and D. Y. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- AD
- Alzheimer disease
- APP
- amyloid β-precursor protein
- Aβ
- amyloid β
- Nav1
- voltage-gated sodium channel
- Navβ2
- voltage-gated sodium channel β2 subunit
- Navβ2-CTF
- Navβ2 C-terminal fragment
- β2-ICD
- Navβ2 intracellular domain
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- ANOVA
- analysis of variance.
REFERENCES
- 1. Zayas E., Grossberg G. (1996) J. Clin. Psychiatry 57, Suppl. 7, 46–54 [PubMed] [Google Scholar]
- 2. de Leon M. J., Convit A., DeSanti S., Golomb J., Tarshish C., Rusinek H., Bobinski M., Ince C., Miller D. C., Wisniewski H. M. (1995) Neuroimaging Clin. N. Am. 5, 1–17 [PubMed] [Google Scholar]
- 3. Hardy J., Selkoe D. J. (2002) Science 297, 353–356 [DOI] [PubMed] [Google Scholar]
- 4. Vassar R. (2001) J. Mol. Neurosci. 17, 157–170 [DOI] [PubMed] [Google Scholar]
- 5. Wolfe M. S. (2008) Curr. Alzheimer Res. 5, 158–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sinha S., Anderson J. P., Barbour R., Basi G. S., Caccavello R., Davis D., Doan M., Dovey H. F., Frigon N., Hong J., Jacobson-Croak K., Jewett N., Keim P., Knops J., Lieberburg I., Power M., Tan H., Tatsuno G., Tung J., Schenk D., Seubert P., Suomensaari S. M., Wang S., Walker D., Zhao J., McConlogue L., John V. (1999) Nature 402, 537–540 [DOI] [PubMed] [Google Scholar]
- 7. Vassar R., Bennett B. D., Babu-Khan S., Kahn S., Mendiaz E. A., Denis P., Teplow D. B., Ross S., Amarante P., Loeloff R., Luo Y., Fisher S., Fuller J., Edenson S., Lile J., Jarosinski M. A., Biere A. L., Curran E., Burgess T., Louis J. C., Collins F., Treanor J., Rogers G., Citron M. (1999) Science 286, 735–741 [DOI] [PubMed] [Google Scholar]
- 8. Fukumoto H., Cheung B. S., Hyman B. T., Irizarry M. C. (2002) Arch. Neurol. 59, 1381–1389 [DOI] [PubMed] [Google Scholar]
- 9. Tyler S. J., Dawbarn D., Wilcock G. K., Allen S. J. (2002) Biochem. Biophys. Res. Commun. 299, 373–376 [DOI] [PubMed] [Google Scholar]
- 10. Yang L. B., Lindholm K., Yan R., Citron M., Xia W., Yang X. L., Beach T., Sue L., Wong P., Price D., Li R., Shen Y. (2003) Nat. Med. 9, 3–4 [DOI] [PubMed] [Google Scholar]
- 11. Tesco G., Koh Y. H., Kang E. L., Cameron A. N., Das S., Sena-Esteves M., Hiltunen M., Yang S. H., Zhong Z., Shen Y., Simpkins J. W., Tanzi R. E. (2007) Neuron 54, 721–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O'Connor T., Sadleir K. R., Maus E., Velliquette R. A., Zhao J., Cole S. L., Eimer W. A., Hitt B., Bembinster L. A., Lammich S., Lichtenthaler S. F., Hébert S. S., De Strooper B., Haass C., Bennett D. A., Vassar R. (2008) Neuron 60, 988–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zacchetti D., Chieregatti E., Bettegazzi B., Mihailovich M., Sousa V. L., Grohovaz F., Meldolesi J. (2007) Neurodegener Dis. 4, 117–126 [DOI] [PubMed] [Google Scholar]
- 14. Velliquette R. A., O'Connor T., Vassar R. (2005) J. Neurosci. 25, 10874–10883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Willem M., Garratt A., Novak B., Citron M., Kaufmann S., Rittger A., Destrooper B., Saftig P., Birchmeier C., Haass C. (2006) Science 314, 664–666 [DOI] [PubMed] [Google Scholar]
- 16. Kitazume S., Tachida Y., Oka R., Kotani N., Ogawa K., Suzuki M., Dohmae N., Takio K., Saido T. C., Hashimoto Y. (2003) J. Biol. Chem. 278, 14865–14871 [DOI] [PubMed] [Google Scholar]
- 17. Lichtenthaler S. F., Dominguez D. I., Westmeyer G. G., Reiss K., Haass C., Saftig P., De Strooper B., Seed B. (2003) J. Biol. Chem. 278, 48713–48719 [DOI] [PubMed] [Google Scholar]
- 18. Kim D. Y., Ingano L. A., Carey B. W., Pettingell W. H., Kovacs D. M. (2005) J. Biol. Chem. 280, 23251–23261 [DOI] [PubMed] [Google Scholar]
- 19. Wong H. K., Sakurai T., Oyama F., Kaneko K., Wada K., Miyazaki H., Kurosawa M., De Strooper B., Saftig P., Nukina N. (2005) J. Biol. Chem. 280, 23009–23017 [DOI] [PubMed] [Google Scholar]
- 20. Isom L. L. (2001) Neuroscientist 7, 42–54 [DOI] [PubMed] [Google Scholar]
- 21. Yu F. H., Catterall W. A. (2003) Genome Biol. 4, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schmidt J. W., Catterall W. A. (1986) Cell 46, 437–444 [DOI] [PubMed] [Google Scholar]
- 23. Catterall W. (2002) Novartis Found. Symp. 241, 206–218; discussion 218–232 [PubMed] [Google Scholar]
- 24. Catterall W. A. (2000) Neuron 26, 13–25 [DOI] [PubMed] [Google Scholar]
- 25. Isom L. L., Ragsdale D. S., De Jongh K. S., Westenbroek R. E., Reber B. F., Scheuer T., Catterall W. A. (1995) Cell 83, 433–442 [DOI] [PubMed] [Google Scholar]
- 26. Chen C., Bharucha V., Chen Y., Westenbroek R. E., Brown A., Malhotra J. D., Jones D., Avery C., Gillespie P. J., 3rd., Kazen-Gillespie K. A., Kazarinova-Noyes K., Shrager P., Saunders T. L., Macdonald R. L., Ransom B. R., Scheuer T., Catterall W. A., Isom L. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 17072–17077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lopez-Santiago L. F., Pertin M., Morisod X., Chen C., Hong S., Wiley J., Decosterd I., Isom L. L. (2006) J. Neurosci. 26, 7984–7994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dominguez D., Tournoy J., Hartmann D., Huth T., Cryns K., Deforce S., Serneels L., Camacho I. E., Marjaux E., Craessaerts K., Roebroek A. J., Schwake M., D'Hooge R., Bach P., Kalinke U., Moechars D., Alzheimer C., Reiss K., Saftig P., De Strooper B. (2005) J. Biol. Chem. 280, 30797–30806 [DOI] [PubMed] [Google Scholar]
- 29. Laird F. M., Cai H., Savonenko A. V., Farah M. H., He K., Melnikova T., Wen H., Chiang H. C., Xu G., Koliatsos V. E., Borchelt D. R., Price D. L., Lee H. K., Wong P. C. (2005) J. Neurosci. 25, 11693–11709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Savonenko A. V., Melnikova T., Laird F. M., Stewart K. A., Price D. L., Wong P. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5585–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hu X., Hicks C. W., He W., Wong P., Macklin W. B., Trapp B. D., Yan R. (2006) Nat. Neurosci. 9, 1520–1525 [DOI] [PubMed] [Google Scholar]
- 32. Kim D. Y., Carey B. W., Wang H., Ingano L. A., Binshtok A. M., Wertz M. H., Pettingell W. H., He P., Lee V. M., Woolf C. J., Kovacs D. M. (2007) Nat. Cell Biol. 9, 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Davis T. H., Chen C., Isom L. L. (2004) J. Biol. Chem. 279, 51424–51432 [DOI] [PubMed] [Google Scholar]
- 34. Thomas-Crusells J., Vieira A., Saarma M., Rivera C. (2003) J. Neurosci. Methods 125, 159–166 [DOI] [PubMed] [Google Scholar]
- 35. Westenbroek R. E., Merrick D. K., Catterall W. A. (1989) Neuron 3, 695–704 [DOI] [PubMed] [Google Scholar]
- 36. Yu F. H., Mantegazza M., Westenbroek R. E., Robbins C. A., Kalume F., Burton K. A., Spain W. J., McKnight G. S., Scheuer T., Catterall W. A. (2006) Nat. Neurosci. 9, 1142–1149 [DOI] [PubMed] [Google Scholar]
- 37. Vassar R., Kovacs D. M., Yan R., Wong P. C. (2009) J. Neurosci. 29, 12787–12794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Westenbroek R. E., Noebels J. L., Catterall W. A. (1992) J. Neurosci. 12, 2259–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Craner M. J., Newcombe J., Black J. A., Hartle C., Cuzner M. L., Waxman S. G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8168–8173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Waxman S. G., Craner M. J., Black J. A. (2004) Trends Pharmacol. Sci. 25, 584–591 [DOI] [PubMed] [Google Scholar]
- 41. Craner M. J., Damarjian T. G., Liu S., Hains B. C., Lo A. C., Black J. A., Newcombe J., Cuzner M. L., Waxman S. G. (2005) Glia 49, 220–229 [DOI] [PubMed] [Google Scholar]
- 42. Hu X., He W., Diaconu C., Tang X., Kidd G. J., Macklin W. B., Trapp B. D., Yan R. (2008) FASEB J. 22, 2970–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. O'Malley H. A., Shreiner A. B., Chen G. H., Huffnagle G. B., Isom L. L. (2009) Mol. Cell. Neurosci. 40, 143–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Catterall W. A., Kalume F., Oakley J. C. (2010) J. Physiol. 588, 1849–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hu X., Zhou X., He W., Yang J., Xiong W., Wong P., Wilson C. G., Yan R. (2010) J. Neurosci. 30, 8819–8829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hitt B. D., Jaramillo T. C., Chetkovich D. M., Vassar R. (2010) Mol. Neurodegener. 5, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ohno M., Sametsky E. A., Younkin L. H., Oakley H., Younkin S. G., Citron M., Vassar R., Disterhoft J. F. (2004) Neuron 41, 27–33 [DOI] [PubMed] [Google Scholar]
- 48. McConlogue L., Buttini M., Anderson J. P., Brigham E. F., Chen K. S., Freedman S. B., Games D., Johnson-Wood K., Lee M., Zeller M., Liu W., Motter R., Sinha S. (2007) J. Biol. Chem. 282, 26326–26334 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.