Abstract
D-type cyclins regulate cellular outcomes in part through cyclin-dependent, kinase-independent mechanisms that modify transcription factor action, and recent in vivo studies showed that cyclin D1 associates with a large number of transcriptional regulators in cells of the retina and breast. Given the frequency of cyclin D1 alterations in cancer, it is imperative to delineate the molecular mechanisms by which cyclin D1 controls key transcription factor networks in human disease. Prostate cancer was used as a paradigm because this tumor type is reliant at all stages of the disease on androgen receptor (AR) signaling, and cyclin D1 has been shown to negatively modulate AR-dependent expression of prostate-specific antigen (KLK3/PSA). Strategies were employed to control cyclin D1 expression under conditions of hormone depletion, and the effect of cyclin D1 on subsequent androgen-dependent gene expression was determined using unbiased gene expression profiling. Modulating cyclin D1 conferred widespread effects on androgen signaling and revealed cyclin D1 to be a selective effector of hormone action. A subset of androgen-induced target genes, known to be directly regulated by AR, was strongly suppressed by cyclin D1. Analyses of AR occupancy at target gene regulatory loci of clinical relevance demonstrated that cyclin D1 limits AR residence after hormone stimulation. Together, these findings reveal a new function for cyclin D1 in controlling hormone-dependent transcriptional outcomes and demonstrate a pervasive role for cyclin D1 in regulating transcription factor dynamics.
Keywords: Cell Cycle, Chromatin, Cyclins, Microarray, Transcription, Androgen Receptor, Prostate Cancer
Introduction
The D-type cyclins (cyclins D1, D2, and D3) utilize pleiotropic functions to elicit cellular outcomes and are frequently altered in the course of human cancer (1–4). A well characterized function of D-cyclins in many model systems is their ability to associate with and activate cyclin-dependent kinase 4 or 6 (CDK4 or -6)2 to initiate proliferative phenotypes (5–7). Evidence has revealed that reconstituting individual D-cyclins in fibroblasts lacking cyclins D1, D2, and D3 may result in distinct functions (8). Interestingly, in this system, cyclin D1 failed to confer significant CDK4 kinase activity, suggesting that cyclin D1 may have functions in addition to cell cycle control (9). These findings are consistent with robust in vitro and in vivo findings that revealed the existence of “kinase-independent” cyclin D1 activities (1).
The kinase-independent functions of cyclin D1 have significant consequence for both tissue development and tumor biology (2, 4, 10, 11). First, it is notable that D-type cyclins and associated CDKs are dispensable for cellular proliferation (12, 13). Second, retinal and mammary hypoplasia observed in cyclin D1−/− mice can be rescued by knock-in of a mutant allele, defective in the ability to activate CDK4, indicating that selected developmental requirements for cyclin D1 may be kinase-independent (14). Third, recent unbiased, in vivo analysis of cyclin D1 complexes showed that endogenous cyclin D1 is found in complex with a large number of sequence specific transcription factors (15). In fact, transcriptional regulators represented the most prevalent class of protein found in association with cyclin D1. Subsequent ChIP-chip analyses showed that in the retina, cyclin D1 is found associated with chromatin and that disruption of cyclin D1 function results in critical, tissue-specific effects on gene transcription. These findings have drawn significant interest and support previous studies demonstrating that perturbation of cyclin D1-mediated transcriptional control impacts human cancers. For example, the ability of cyclin D1 to bind and regulate C/EBPβ impacts clinical outcomes in breast cancer (16). In the context of PCa, cyclin D1 has been shown to influence the response to anoikis through association with FOXO1 (17). Cell cycle progression can also be altered through kinase-independent mechanisms because cyclin D1 antagonizes the antiproliferative effects of DMP1 through direct association (18). Last, cyclin D1 has been shown to interact with and modulate several nuclear receptors of critical importance for hormone-dependent cancers, including estrogen receptor (19, 20), thyroid hormone receptor (21), peroxisome proliferator-activated receptor γ (22), and the androgen receptor (AR) (23, 24). Taken together, these observations indicate that cyclin D1 plays an important role in regulating transcriptional factor activity.
Previous investigation revealed that cross-talk between AR and cyclin D1 serves as a rheostat to modulate mitogen-mediated AR signaling (22) and that this process may be disrupted in PCa (25–27). Ligand-activated AR initiates signaling events that result in the mTOR-dependent induction of cyclin D1 translation (26, 28). Accumulated cyclin D1 protein acts both to initiate CDK4 activation (promoting G1-S transition) and to dampen further AR activation through direct and CDK-independent association with the receptor. Through these means, cyclin D1 appears to serve as a mechanism to control the strength and duration of mitogenic signaling in the presence of androgen. The ability of cyclin D1 to govern AR transcriptional activity has been extensively studied using the well known AR target gene KLK3/PSA (29). Molecular analyses demonstrated that cyclin D1 engages at least two mechanisms to suppress ligand-dependent AR activity. First, cyclin D1 binds to the FXXLF motif of AR to block ligand-induced conformational changes in the receptor (N-C-terminal interaction) that foster transactivation potential (30). Second, cyclin D1 is known to associate with a select group of histone deacetylases (21, 31–33), and this activity is essential for robust suppression of ligand-stimulated AR activity (34).
The importance of cyclin D1-mediated AR regulation is underscored by recent studies addressing both the cellular and clinical relevance. Investigation of human prostatic adenocarcinomas showed a large percentage of specimens with low or undetectable cyclin D1 expression, and tumors lacking cyclin D1 have been shown to be associated with elevated serum PSA (27), suggestive of increased AR activity. Strikingly, a significant subset of tumors examined had elevated cyclin D1b (a variant of cyclin D1) (26), which has compromised AR-regulatory capacity (25). Thus, the ability of cyclin D1 to suppress AR activity appears to be diminished in PCa, consistent with the role of AR in promoting tumor development and progression (35). Conversely, introduction of the isolated cyclin D1 domain (repressor domain) responsible for transcriptional regulation of AR revealed that this functional motif is sufficient to attenuate ligand-dependent AR activity, cooperate with AR-directed therapeutics, and reduce cell viability in AR-dependent PCa cells (36). Combined, these findings identify cyclin D1 as a major effector of AR function and cellular outcomes in PCa.
Given the importance of cyclin D1 as a transcriptional regulator of AR in PCa, an unbiased approach was utilized to assess the overall impact of cyclin D1 on androgen-responsive gene expression and AR function at endogenous target gene sites. These studies unexpectedly revealed that cyclin D1 serves as a selective modifier of androgen activity, capable of both suppressing and facilitating androgen-dependent gene expression. However, genes that are known to be directly regulated by AR were suppressed by cyclin D1, indicating that this is a primary means of AR modulation. Subsequent analyses identified that cyclin D1 limits ligand-induced AR residence on chromatin, thus illuminating additional mechanisms of cyclin D1 action. Together, these findings provide critical insight into the means by which cyclin D1 controls AR function and the response to androgen stimulation.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatments
The androgen-dependent prostate cancer cell lines (LNCaP and VCaP) were maintained as previously described (37). To examine transcriptional outcome, LNCaP or VCaP (2.9 × 104/cm2) cells were plated on poly-l-lysine in 5% charcoal-dextran-treated serum (HyClone) for 72 h. Cells were transduced (12 h) with either Ad-GFP control or Ad-cyclin D1 and subsequently treated (18 h) with ethanol (0.1%) or a physiological dose of dihydrotestosterone (DHT) (1 nm) (38). Cyclin D1-transduced cells treated with ethanol were included in the validation experiments to assess the impact of cyclin D1 on basal transcription. RNA was isolated using the standard TRIzol method and was either subjected directly to microarray analysis or converted to cDNA for gene expression analysis. RNA interference (RNAi) was performed using LNCaP (8.6 × 104/cm2) cells plated on poly-l-lysine and maintained (24 h) in standard growth conditions. Then cells were transfected overnight (16 h) in serum-free conditions with a control or CCND1 siRNA (D-001810-10-20 or L-003210-00-0020, respectively; Thermo Scientific) according to the manufacturer's specifications and then incubated with standard growth conditions and harvested for analysis at the indicated times.
Microarray Analysis and Bioinformatics
Microarray analysis was performed as follows. Total RNA samples (0.5 μg) for each treatment condition (n = 3), as described above, were labeled using the standard labeling protocol (small scale protocol version 2.0) and hybridized to HG-U133plus2 GeneChips (Affymetrix). GeneChips were quantified with an Affymetrix Gene Array Scanner (software version 1.4, default settings), and then “CEL” files were generated using Affymetrix Microarray Suite 5.0. Individual samples were normalized using the robust multichip analysis algorithm as implemented in Bioconductor/R. Normalized data were refined using a custom chip definition file based on target definitions (Hs133 REFSEQ version 8, represented by 26,183 transcripts) to provide a more accurate interpretation of the expression data (39). The data set (.CEL files) is available in the online Gene Expression Omnibus (GEO) repository (accession number GSE26483). All statistical comparisons and visualizations were performed using GeneSpring GX version 7.3.1 (Agilent). Androgen-regulated transcripts were identified using a t test (p ≤ 0.05) between control-transduced LNCaP cells treated with ethanol or DHT. Androgen-regulated transcripts were filtered using a 1.2-fold cut-off and then overlaid with the corresponding expression values in the presence of cyclin D1 and DHT. To identify expression patterns, the transcripts were empirically assigned to clusters using the k-means clustering algorithm. Statistically overrepresented functional annotations were identified using the GOterm biological processes setting in the Web-based Database for Annotation, Visualization, and Integrated Discovery (DAVID). Assessment of the presence or absence of androgen receptor-occupied regions (ARORs) within 50 kb of transcriptional start sites (TSSs) was performed by uploading a published (40) and publicly available ChIP-seq data set from LNCaP cells into the University of California Santa Cruz Genome Browser on the NCBI36/Hg18 (March 2006) assembly (41). The TSS for individual transcripts from the microarray expression data set was determined by submitting the transcript accession numbers in batch mode to MatchMiner (42).
Gene Expression Analysis
Independent validation of the microarray expression profile was performed with cDNA generated from RNA (5 μg) using the Superscript system (Invitrogen). Conventional PCR analysis and oligonucleotides for KLK3/PSA and GAPDH have been described previously (43). Briefly, conventional PCR for KLK3/PSA and GAPDH was performed at 26 cycles. Products were resolved on agarose (2%) and visualized with ethidium bromide. The quantitative PCR method and Taqman assays for KLK3/PSA have been described previously (26), whereas the relative expression of all other transcripts normalized to GAPDH (oligonucleotides are described in supplemental Table 2 except for the TMPRSS2 primers that have been previously described (44)) was performed using Power SYBR Green and a StepOne Machine (Applied Biosystems). Validation of transcripts is represented as the mean -fold change ± S.E. of 3–4 individual experiments where each condition within an experiment is the average of two technical replicates. Statistics were determined by analysis of variance, and significance (p ≤ 0.05) was calculated using Tukey's multiple comparison test using GraphPad Prism version 4.
Immunoblot Analysis
Representative LNCaP cell lysates (40 μg), treated as described above, were separated by polyacrylamide gel electrophoresis to evaluate cyclin D1 protein expression. Gels were transferred to PVDF and immunoblotted (1:1000) for cyclin D1 (NeoMarkers, catalog no. AB-3), GFP (Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), catalog no. SC-9996), and loading control β-Tubulin (Santa Cruz Biotechnology, Inc., catalog no. SC-5274).
ChIP Analysis
ChIP assays for AR occupancy were performed according to a method described previously (40). LNCaP cells were treated as described above, except cells were stimulated with 10 nm DHT for 1–3 h. Genomic DNA was used for conventional PCR, as described above, with oligonucleotides for the enhancer regions of KLK3/PSA (ARE III) and TMPRSS2 (ARE V) as described previously (45). Quantitative PCR was performed, as described above, except using ExpressSYBR® Green-ER/ROX mix (Invitrogen). Relative occupancy was calculated according to the following: ΔCt = Ct (of immunoprecipitation or input) − Ct (of IgG); ΔΔCt = ΔCt (of treated) − ΔCt (of control); occupancy = 2−ΔΔCt.
RESULTS
Cyclin D1 Expression and Function Can Be Reconstituted after Hormone Depletion
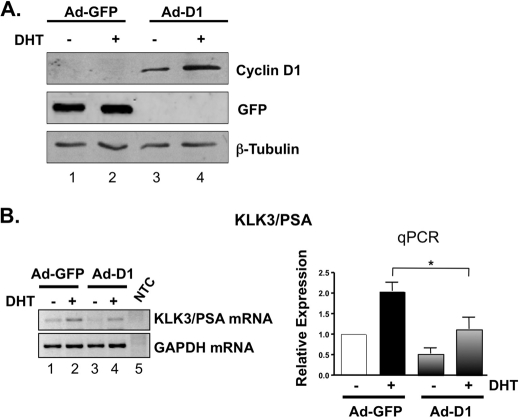
To discern the role of cyclin D1 in controlling the transcriptional response to androgen, model systems were developed to rigorously control cyclin D1 expression (26). Androgen-dependent, AR-positive prostate cancer cells (LNCaP) were utilized because these cells lack cyclin D2 (46–49) and arrest tightly in G0/G1 after hormone depletion with accompanying loss of cyclin D1 and cyclin D3 expression (28, 37, 50). The impact of hormone depletion on D-cyclin expression was recapitulated herein (supplemental Fig. 1, A and B, lanes 1 and 2). Suppression of endogenous D-cyclins is critical because loss of a single D-type cyclin can result in partial compensation by remaining family members (51). Following hormone deprivation, cells were transduced with adenovirus encoding either GFP control (Fig. 1A, lanes 1 and 2) or cyclin D1 (lanes 3 and 4) and then stimulated with vehicle (ethanol) control or physiologic levels of androgen (DHT) (38). Stimulation with androgen after reconstitution restored cyclin D1 levels in the absence of androgen and prior to DHT-mediated accumulation of endogenous cyclin D1 expression, thereby allowing assessment of cyclin D1 function in the absence of the other D-type cyclins. The impact of cyclin D1 reconstitution was determined by monitoring mRNA levels of the AR target gene KLK3/PSA because the role of cyclin D1 in suppressing androgen-induced KLK3/PSA expression has been well established (29). As expected, DHT stimulation resulted in marked induction of KLK3/PSA expression (Fig. 1B, left, compare lanes 1 and 2). Notably, DHT-mediated induction of KLK3/PSA expression was attenuated upon cyclin D1 reconstitution (45.2%, p < 0.05), as determined by quantitative PCR (Fig. 1B, right). In contrast, cyclin D1 had minimal impact on basal KLK3/PSA expression, reinforcing the postulate that cyclin D1 can alter the transcriptional response to androgen. Similar results were observed using transfected, rather than transduced, cyclin D1 (supplemental Fig. 1B), similar to previous reports (25, 52). The suppressive effect of cyclin D1 on KLK3/PSA expression was not limited to a single cell type because similar results were observed in a second androgen-dependent, AR-positive PCa model system, VCaP (supplemental Fig. 2). Thus, rapid cyclin D1 reconstitution (either by transduction or transfection) under hormone-depleted conditions can be used as a means to assess the impact of individual D-type cyclins on transcriptional outcomes.
FIGURE 1.
Cyclin D1 regulates prostate-specific tumor marker expression. To examine the cyclin D1-regulated transcriptional outcome in response to androgen, LNCaP prostate cancer cells were incubated in 5% charcoal-dextran-treated serum to naturally deplete D-type cyclins and then transduced with cyclin D1 (Ad-D1) or control (Ad-GFP) and subsequently treated for 18 h with a physiological dose of androgen (DHT; 1 nm) or ethanol (EtOH) control. A, to evaluate cyclin D1 protein expression, representative LNCaP cell lysates treated as described above were immunoblotted for cyclin D1, GFP, and loading control β-tubulin. Note that the post-transcriptional induction of cyclin D1 protein by androgen is maintained under cyclin D1-reconstituted conditions (lanes 3 and 4). B, KLK3/PSA expression was determined by conventional PCR (left) and Taqman-based qPCR (right) to assess the ability of cyclin D1 to regulate prostate-specific tumor marker expression. NTC, non-template control. The bar graph shows the mean-fold change ± S.E. (error bars) of three independent experiments, where each control sample (Ad-GFP + EtOH) is set to 1. Significant (p < 0.05) down-regulation by cyclin D1 of androgen-induced expression is indicated by an asterisk.
Cyclin D1 Differentially Regulates Androgen-sensitive Gene Expression
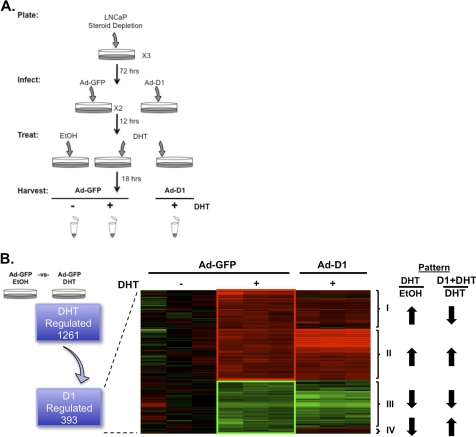
Because the data above demonstrated that cyclin D1 activity can be effectively reconstituted after hormone deprivation, this model system afforded the opportunity to discern the overall impact of cyclin D1 on androgen-responsive gene expression in an unbiased manner. Such analyses are crucial because the current understanding of cyclin D1-mediated control of AR function has been largely limited to assessment of KLK3/PSA regulation. To determine the overall impact of cyclin D1, cells were cultured for 72 h in the absence of androgen to deplete D-cyclins and then, following cyclin D1 reconstitution, were stimulated with 1 nm DHT for 18 h (as depicted in Fig. 2A). Validation of cyclin D1 mRNA levels pre- and post-transduction was conducted (supplemental Fig. 3A), and gene expression analysis was performed using biological replicates on the Affymetrix Human Genome U133plus2 platform. Following confirmation of cyclin D1 RNA levels on the microarray (supplemental Fig. 3B), a custom GeneChip library file based on REFSEQ target definitions was used to provide accurate interpretation of GeneChip data (39). Initially, 1,261 transcripts were identified that were significantly (p < 0.05) altered by androgen stimulation compared with control, GFP-transduced cells (Fig. 2B, left). However, upon cyclin D1 reconstitution, only a subset of androgen-responsive transcripts (n = 393 transcripts, 257 up-regulated and 136 down-regulated) proved sensitive to cyclin D1 status. The complete list of androgen-sensitive, cyclin D1-regulated transcripts is provided in supplemental Fig. 4. Combined, these findings provided the first indication that cyclin D1 regulates a distinct subset of androgen-responsive genes in the context of PCa.
FIGURE 2.
Cyclin D1 modulates androgen-dependent gene expression. A, experimental design for unbiased gene expression array analyses to identify androgen-regulated transcripts that are sensitive to cyclin D1. Treatment conditions are indicated and fully described under “Experimental Procedures”and “Results.” Microarray analysis was performed in triplicate for each treatment condition on the HG-U133plus2 platform (Affymetrix). Individual samples were normalized and evaluated using a custom GeneChip library file to provide a more accurate interpretation of the expression data. All statistical comparisons and visualizations were performed using GeneSpring GX version 7.3.1 (Agilent). B, schematic, to identify androgen-regulated transcripts responsive to cyclin D1, a statistical (p < 0.05) comparison between GFP-transduced LNCaP cells treated with EtOH or DHT was performed. Transcripts were then selected using a 1.2-fold cut-off, and the corresponding expression values in the presence of cyclin D1 and DHT are shown. Heat map, to identify expression patterns (as indicated, Patterns I–IV), transcripts were empirically assigned to clusters using a k-means algorithm. Red and green indicate up-regulated and down-regulated transcripts, respectively. The androgen response and influence of cyclin D1 on the androgen response are indicated by arrows.
Closer examination of the 393 androgen-regulated, cyclin D1-sensitive transcripts was facilitated by k-means clustering analyses. As depicted in the heat map (Fig. 2B, right), four distinct cyclin D1-responsive patterns were identified. Unexpectedly, these studies revealed that cyclin D1 could antagonize (Patterns I and IV, n = 127 and 54, respectively) or act in concert (Patterns II and III, n = 130 and 82, respectively) with androgen to regulate gene networks. Gene ontology analyses of all 393 androgen-regulated and cyclin D1-sensitive transcripts (using the DAVID Bioinformatics Resource) revealed significant associations with diverse biological processes (supplemental Table 1), many of which are expected, based on known functions of cyclin D1 (i.e. regulation of cell cycle). Gene ontology analysis of the individual patterns was limited by the small number of transcripts identified in Patterns III and IV (supplemental Fig. 4), thus confounding the ability to assess the overall impact of cyclin D1 on androgen signaling. Therefore, the top regulated transcripts for each pattern were assessed to identify potential commonality (Table 1). Interestingly, among the top androgen-regulated transcripts sensitive to cyclin D1, Pattern I contained a number of known AR target genes (i.e. KLK genes), consistent with the role of cyclin D1 in negatively regulating KLK3/PSA expression (29).
TABLE 1.
Top transcripts regulated by DHT and cyclin 1
a-Fold change: Ad-GFP + EtOH versus Ad-GFP + DHT.
b -Fold change: Ad-GFP + DHT versus Ad-D1 + DHT.
Cyclin D1 Antagonizes Androgen-dependent Up-regulation of AR Target Genes
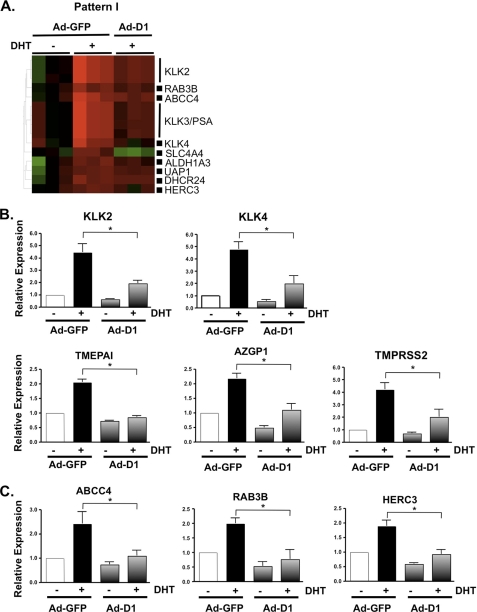
Because the data above suggest that cyclin D1 may selectively inhibit AR function, it became necessary to validate these findings through quantitative assessment of other known and putative AR target genes (Fig. 3A). As an additional control, expression of these transcripts was measured in non-DHT-stimulated cells to determine the relevance of cyclin D1-mediated repression on basal AR activity. Consistent with observations for KLK3/PSA expression (Fig. 1B), cyclin D1 repressed the androgen-dependent induction but not basal expression of the other kallikrein family members KLK2 and KLK4 by 56.8 and 58.7%, respectively (Fig. 3B). The kallikrein genes are located in the same genomic cluster on chromosome 19 (53); to determine whether the effects of cyclin D1 on known AR target genes were specific to this chromosomal location, the impact of cyclin D1 status on expression of established AR target genes residing on distinct loci (TMEPAI, AZGP1, and TMPRSS2; chromosomes 20 (54), 7 (55, 56), and 21 (57), respectively) was determined. Each was significantly induced by androgen, consistent with previous studies (56, 58–61). Furthermore, ligand-induced gene expression was reduced in the presence of cyclin D1 (58.5, 49.5, and 52%, respectively). Together, these data indicate that the repressive capacity of cyclin D1 on androgen-induced expression of known AR target genes is not specific to one gene cluster and/or chromosomal position.
FIGURE 3.
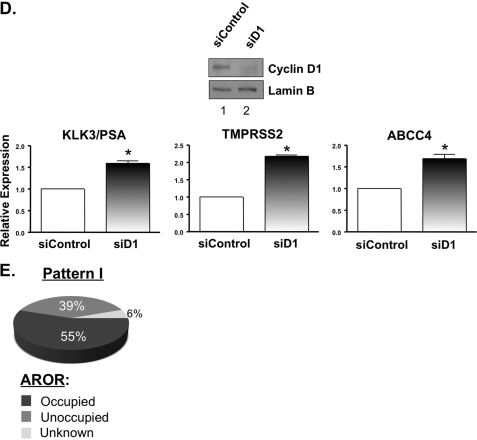
Cyclin D1 attenuates AR-dependent gene expression. A, to determine the role of cyclin D1 on AR-dependent gene expression, heat map analysis was performed on selected Pattern I transcripts that are frequently observed as androgen/AR-regulated transcripts. The relative expression of known AR target genes KLK2, KLK4, TMEPAI, AZGP1, and TMPRSS2 (B) and putative AR target genes ABCC4, RAB3B, and HERC3 (C) was performed by SYBR-based qPCR from three or four independent experiments and presented as described in the legend to Fig. 1. Cyclin D1-transduced cells treated with ethanol were included in the validation to assess the impact of cyclin D1 on basal transcription. All transcripts tested were validated as androgen-dependent; however, only the transcripts that demonstrated a significant (p < 0.05) difference in the presence of cyclin D1 are indicated by an asterisk. D, knockdown of cyclin D1, in LNCaP cells cultured under standard growth conditions (i.e. 5% FBS), was performed to further validate the influence of cyclin D1 on AR target gene expression. A representative immunoblot for cyclin D1 knockdown is provided. The relative expression of KLK3/PSA, TMPRSS2, and ABCC4 was determined by qPCR, from three individual experiments, and plotted as described above. E, the transcripts from Pattern I were assessed, bioinformatically, for ARORs within 50 kb of the TSS using a publicly available data set, as described under “Experimental Procedures,” to determine the overall potential of AR to regulate these gene loci.
Last, quantitative analyses were performed for genes induced by androgen and suspected to be directly regulated by AR (62). Importantly, each gene has been recently shown to have potential AR binding sites within regions capable of altering gene expression (40) and may have significance in PCa. ABCC4 (which encodes a ATP-binding cassette transporter) expression is increased in PCa (63, 64) and, as a multidrug resistance protein (MRP) family member, may contribute to drug resistance (65). RAB3B (which encodes a vesicular transport protein of the RAS oncogene family) and HERC3 (which encodes a HECT domain E3 ubiquitin-protein ligase) were also investigated. As shown in Fig. 3C, androgen-induced expression of ABCC4, RAB3B, and HERC3 was diminished by cyclin D1 reconstitution (54.6, 61.6, and 51.2%, respectively). Analysis of four other androgen/AR-dependent transcripts identified in Pattern I (DHCR24, ALDH1A3, UAP1, and SLC4A4) revealed a consistent trend for cyclin D1-mediated suppression of androgen-dependent expression (data not shown). Importantly, expression of HERC3 and ABCC4 (both of which are induced by androgen in the VCaP model) was significantly inhibited by cyclin D1 (supplemental Fig. 5A). Interestingly, only weak induction of RAB3B was observed upon androgen treatment in the VCaP cells (data not shown), indicating potential differences in AR function between model systems. Conversely, siRNA-mediated ablation of endogenous cyclin D1 in LNCaP cells was sufficient to deregulate AR target gene expression (Fig. 3D), further indicating the importance of cyclin D1 in modulating AR function.
A crucial step in AR-dependent transcription is recruitment of the receptor to chromatin, and a recent study revealed that the majority of ARORs are located within 50 kb of the TSS of a gene (40). These data prompted a bioinformatic evaluation of the Pattern I transcripts to further define which loci of the cyclin D1-sensitive transcripts have ARORs near the TSS. Interestingly, more than half (55%) of Pattern I contained ARORs within 50 kb of the TSS (Fig. 3E), suggesting that the majority of these cyclin D1-sensitive transcripts are probably regulated by AR in a direct fashion. Combined, these analyses not only identify a gene-selective cyclin D1-responsive signature but also demonstrate that cyclin D1 significantly attenuates ligand-induced gene expression of AR target genes with potential PCa importance.
AR Residence on Chromatin Is Regulated by Cyclin D1
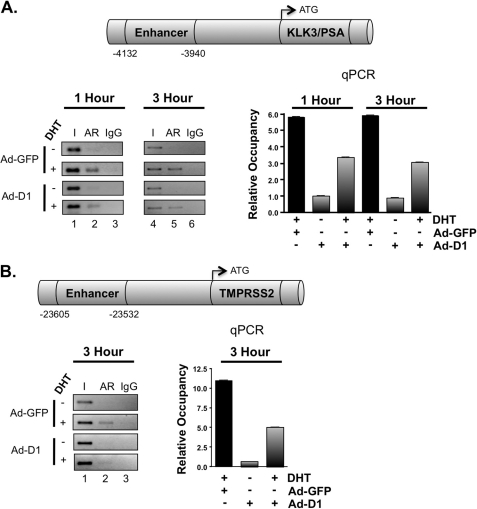
To further explore the mechanisms by which cyclin D1 specifically acts to suppress AR target gene expression, ChIP assays were performed. Despite the increasingly large number of potential androgen-regulated genes with ARORs (40, 66–69), only a few genes have been validated for functional output due to AR binding (70). The KLK3/PSA regulatory locus was initially analyzed (Fig. 4A), wherein AR recruitment to the KLK3/PSA enhancer region has been documented to occur with an initial periodicity of 60–90 min (71–74). Consistent with these findings, DHT induced AR recruitment after 1 h, and AR occupancy was maintained at the 3 h time point (5.8- and 5.9-fold over vehicle control, respectively). Importantly, restoration of cyclin D1 reduced androgen-induced AR occupancy at both time points (42.4 and 48.5%, respectively). These data are consistent with the magnitude of KLK3/PSA expression changes observed by qPCR analysis (Fig. 1B) and microarray expression profiling (Fig. 3A). Similar results were observed in the VCaP model (supplemental Fig. 5B). However, a more modest reduction in AR occupancy was observed after cyclin D1 transduction, as might be expected because VCaP cells harbor amplification of the AR locus and express higher levels of the receptor (75, 76). Together, these data yielded the first indication that cyclin D1 alters AR association with chromatin. To assess these findings further, the impact of cyclin D1 on AR occupancy was performed using the TMPRSS2 locus (Fig. 4B), whose AR-dependent regulatory region has been recently identified in a chromosomal translocation event of high significance in PCa (77). Of the five potential AREs associated with the TMPRSS2 locus, the enhancer region (ARE V) is the predominant site regulating androgen responsiveness and AR recruitment (45). Similar to observations at the KLK3/PSA locus, cyclin D1 suppressed DHT-stimulated AR occupancy at the TMPRSS2 enhancer by 54.3%. Together, these data demonstrate that a predominant transcriptional consequence of cyclin D1 with regard to AR target gene regulation is to suppress DHT-induced AR occupancy and target gene expression and provide new insight into the potential consequences of aberrant cyclin D1 expression in human disease.
FIGURE 4.
Cyclin D1 displaces AR occupancy at target gene loci. ChIP analysis was performed to determine the influence of cyclin D1 on AR occupancy. LNCaP cells were treated as described in the legend to Fig. 2, except cells were stimulated with 10 nm DHT for 1–3 h. Bar graphs represent the relative occupancy ± S.D. (error bars) from a representative AR ChIP, where each condition is a biological triplicate. A, schematic, KLK3/PSA locus showing the location of the well characterized, AR-responsive enhancer region upstream of the TSS. Bar graph, qPCR analysis for the enhancer region from AR ChIP assays at 1 and 3 h. Representative conventional PCR is provided (left). B, schematic, TMPRSS2 locus showing the location of the AR-responsive enhancer region. Bar graph, qPCR analysis for the enhancer region from an AR ChIP assay at 3 h. Representative conventional PCR is provided (left).
DISCUSSION
It is now apparent that a major function of cyclin D1 in vivo is to bind and regulate transcription factor action. Findings supporting this contention are robust, and multiple studies have validated the importance of cyclin D1-mediated transcriptional regulation with regard to cellular and in vivo outcomes (15, 16). In PCa, previous studies established a paradigm whereby cyclin D1 attenuates AR-mediated KLK3/PSA expression and established that this ability to suppress AR function is subverted in human disease through multiple mechanisms (29). Despite these advances, previous reports have been limited to a small subset of AR target genes, and the overall consequence of cyclin D1 on androgen-dependent gene expression has remained elusive. Here, an unbiased approach was used to illuminate three critical facets of cyclin D1 function. First, it was observed that cyclin D1 selectively regulates androgen-dependent programming. Unexpectedly, cyclin D1 was able to oppose and enhance androgen function in a gene-selective manner, thus providing a new understanding of how altered cyclin D1 expression and/or function can rewire the cellular response to androgen stimulation. Second, it was shown that androgen-induced transcripts that are direct AR target genes were suppressed by cyclin D1, thus providing unbiased evidence that a primary function of cyclin D1 is to limit AR activity induced by ligand. Third, it was demonstrated that cyclin D1 markedly reduced AR residence on clinically relevant gene loci, thus identifying a new mechanism of cyclin D1 action. Taken together, these studies identify the transcriptional regulatory functions of cyclin D1 as critical effectors of androgen-dependent signaling and AR-associated chromatin dynamics.
The capacity of cyclin D1 to coordinate mitogenic signals through the G1-S cell cycle machinery is exceedingly well understood and is frequently cited as an important driver of tumorigenesis (3). However, cyclin D1 also interacts with and modulates multiple transcription factors, including prominent members of the nuclear hormone receptor superfamily: estrogen receptor α (20), thyroid hormone receptor (21), peroxisome proliferator-activated receptor γ (33), and AR (29). Little is known concerning the overall impact of cyclin D1 on the transcriptional regulatory networks of these nuclear receptors. The prostate is a unique model to study the transcriptional consequence of cyclin D1 because androgens are important for the growth and survival of PCa cells. The mechanisms by which cyclin D1 elicits transcriptional repression have been preliminarily characterized (23, 30, 32, 34, 36), and the clinical importance has been suggested because human PCa specimens that lack cyclin D1 are associated with elevated serum PSA (27). The current study was conducted using physiological concentrations of androgen, and the number of overall androgen-regulated transcripts identified is consistent with previous reports (78–81). The present study is one of the first to determine the global impact of cyclin D1 on hormone-dependent gene expression, and the transcriptional patterns identified probably impact a broad range of cellular processes, especially cell cycle control and metabolism, both of which are consistent with the ability androgens to influence growth and differentiation. Further studies will be required to understand the complex role cyclin D1 plays with regard to these biological functions. Clearly, these observations indicate that cyclin D1 regulates a complex, androgen-dependent gene expression profile.
The finding that cyclin D1 can both antagonize and synergize with androgen-regulated gene programming was unexpected. As shown, k-means analyses clustered an overwhelming majority of putative and known AR target genes, including KLK3/PSA, into Pattern I (androgen-induced, cyclin D1-repressed). Many of the AR target genes that were validated in the current study are involved in processes such as catabolism (i.e. KLK genes, TMEPAI, AZGP1, and HERC3) or transport (i.e. ABCC4 and RAB3B), consistent with the ability of androgens to regulate growth and metabolic phenotypes, as was recently indicated through combined gene expression and proteomic profiling (82). Genome-wide association data in LNCaP cells suggested that AR binding is enriched at androgen-activated but not androgen-repressed genes (40). Consistent with this notion, overlay of genome-wide data with Pattern I genes demonstrated that AR occupancy was enriched in this subset of androgen-induced transcripts. Currently, it remains to be determined if other potential AR-regulated genes within Pattern I also contribute to the growth and differentiation phenotype. However, with regard to those androgen-induced and AR-mediated transcriptional events, these data are consistent with the model that cyclin D1 negatively regulates AR function.
In contrast, Pattern II (androgen-induced and cyclin D1-induced) contained a paucity of known or putative AR target genes, suggesting that the observed synergy between cyclin D1 and androgen is probably the result of secondary transcriptional effects that may include CDK-dependent functions. Consonantly, many Pattern II transcripts are also regulated by the E2F family of transcription factors, and similar results were observed in previous studies using the murine liver, wherein deregulation of cyclin D1 resulted in transcriptional changes in known E2F genes, including CDC6, CDT1, RRM2, MCM2, MCM4, and MCM5 (83). Alternatively, p21Cip1 can facilitate the assembly of cyclin D1 with CDKs and regulate their subsequent nuclear localization (84–86), and it has been shown that p21Cip1 interacts with estrogen receptor α and behaves as a transcriptional co-activator in a gene-specific manner (87). Thus, p21Cip1 may serve to facilitate the simultaneous androgen and cyclin D1-mediated activation of Pattern II genes, which could potentially explain the perplexing PCa clinical data indicating that increased p21Cip1 is associated with decreased survival (88–91). Overall, these findings demonstrate that cyclin D1 is a selective modifier of androgen-induced gene expression, wherein cyclin D1 utilizes disparate mechanisms to potentiate a subset of androgen-induced genes but preferentially suppresses genes directly regulated by AR.
The finding that cyclin D1 alters AR residence on chromatin of well characterized AR target genes within Pattern I suggests that a mechanism of cyclin D1-mediated transcriptional control occurs through altering transcription factor-chromatin interactions. Notably, cyclin D1 is known to suppress androgen-induced N-C-terminal interaction in the AR (30), which has been hypothesized to facilitate chromatin binding (92). Similarly, the ability of cyclin D1 to inhibit KLK3/PSA gene expression through histone deacetylase involvement (32, 34) suggests that alterations in the chromatin microenvironment probably contribute to the observed effects within Pattern I. Currently, only a few genes have been extensively characterized with regard to AR binding, and future directions will challenge the concept that cyclin D1 influences the association of AR with chromatin. Moreover, in keeping with the notion that cyclin D1 alters transcription factor-chromatin interactions, recent analyses demonstrated that cyclin D1 serves a significant transcriptional role during mouse retinal development (15). Thus, it will be of future interest to utilize genome-wide methods to characterize the extent of cyclin D1 chromatin association in the prostate.
In summary, cyclin D1 has been shown previously to be aberrantly regulated in PCa (27). In the current study, unbiased gene expression profiling illuminated new and unexpected functions for cyclin D1 in this tissue type. These data identify a “signature” of cyclin D1 activity that impinges on androgen-dependent signaling and demonstrate that AR target genes of clinical relevance are suppressed by cyclin D1. Further assessment of cyclin D1 function revealed a potential new mechanism of action, wherein cyclin D1 limits AR occupancy at endogenous loci. However, a subset of androgen-induced genes were potentiated by cyclin D1, thus demonstrating that cyclin D1 exerts complex, pleiotropic effects on hormone action. Together, these data identify cyclin D1 as a selective effector of androgen-dependent signaling and AR-associated chromatin dynamics.
Supplementary Material
Acknowledgments
We thank the members of the Knudsen lab and the group of E. Knudsen for insightful discussions and critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant CA099996 (to K. E. K.). This work was also supported by Department of Defense New Investigator Award PC094507 (to C. E. S. C.) and Department of Defense Predoctoral Fellowships PC094195 (to M. J. S.) and PC094596 (to M. A. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–3.
- CDK
- cyclin-dependent kinase
- PCa
- prostate cancer
- AR
- androgen receptor
- PSA
- prostate-specific antigen
- AROR
- AR-occupied region
- ARE
- androgen response element
- DHT
- dihydrotestosterone
- TSS
- transcription start site
- Ad
- adenovirus
- qPCR
- quantitative PCR.
REFERENCES
- 1. Ewen M. E., Lamb J. (2004) Trends Mol. Med. 10, 158–162 [DOI] [PubMed] [Google Scholar]
- 2. Fu M., Wang C., Li Z., Sakamaki T., Pestell R. G. (2004) Endocrinology 145, 5439–5447 [DOI] [PubMed] [Google Scholar]
- 3. Kim J. K., Diehl J. A. (2009) J. Cell. Physiol. 220, 292–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Knudsen K. E. (2006) Cell Div. 1, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bates S., Bonetta L., MacAllan D., Parry D., Holder A., Dickson C., Peters G. (1994) Oncogene 9, 71–79 [PubMed] [Google Scholar]
- 6. Matsushime H., Ewen M. E., Strom D. K., Kato J. Y., Hanks S. K., Roussel M. F., Sherr C. J. (1992) Cell 71, 323–334 [DOI] [PubMed] [Google Scholar]
- 7. Meyerson M., Harlow E. (1994) Mol. Cell. Biol. 14, 2077–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yu Q., Ciemerych M. A., Sicinski P. (2005) Oncogene 24, 7114–7119 [DOI] [PubMed] [Google Scholar]
- 9. Ciemerych M. A., Yu Q., Szczepanska K., Sicinski P. (2008) Int. J. Dev. Biol. 52, 299–305 [DOI] [PubMed] [Google Scholar]
- 10. Bernards R. (1999) Biochim. Biophys. Acta 1424, M17–22 [DOI] [PubMed] [Google Scholar]
- 11. Coqueret O. (2002) Gene 299, 35–55 [DOI] [PubMed] [Google Scholar]
- 12. Kozar K., Ciemerych M. A., Rebel V. I., Shigematsu H., Zagozdzon A., Sicinska E., Geng Y., Yu Q., Bhattacharya S., Bronson R. T., Akashi K., Sicinski P. (2004) Cell 118, 477–491 [DOI] [PubMed] [Google Scholar]
- 13. Malumbres M., Sotillo R., Santamaría D., Galán J., Cerezo A., Ortega S., Dubus P., Barbacid M. (2004) Cell 118, 493–504 [DOI] [PubMed] [Google Scholar]
- 14. Landis M. W., Pawlyk B. S., Li T., Sicinski P., Hinds P. W. (2006) Cancer Cell 9, 13–22 [DOI] [PubMed] [Google Scholar]
- 15. Bienvenu F., Jirawatnotai S., Elias J. E., Meyer C. A., Mizeracka K., Marson A., Frampton G. M., Cole M. F., Odom D. T., Odajima J., Geng Y., Zagozdzon A., Jecrois M., Young R. A., Liu X. S., Cepko C. L., Gygi S. P., Sicinski P. (2010) Nature 463, 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lamb J., Ramaswamy S., Ford H. L., Contreras B., Martinez R. V., Kittrell F. S., Zahnow C. A., Patterson N., Golub T. R., Ewen M. E. (2003) Cell 114, 323–334 [DOI] [PubMed] [Google Scholar]
- 17. Gan L., Liu P., Lu H., Chen S., Yang J., McCarthy J. B., Knudsen K. E., Huang H. (2009) Cell Death Differ. 16, 1408–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Inoue K., Sherr C. J. (1998) Mol. Cell. Biol. 18, 1590–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Neuman E., Ladha M. H., Lin N., Upton T. M., Miller S. J., DiRenzo J., Pestell R. G., Hinds P. W., Dowdy S. F., Brown M., Ewen M. E. (1997) Mol. Cell. Biol. 17, 5338–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zwijsen R. M., Wientjens E., Klompmaker R., van der Sman J., Bernards R., Michalides R. J. (1997) Cell 88, 405–415 [DOI] [PubMed] [Google Scholar]
- 21. Lin H. M., Zhao L., Cheng S. Y. (2002) J. Biol. Chem. 277, 28733–28741 [DOI] [PubMed] [Google Scholar]
- 22. Wang C., Pattabiraman N., Zhou J. N., Fu M., Sakamaki T., Albanese C., Li Z., Wu K., Hulit J., Neumeister P., Novikoff P. M., Brownlee M., Scherer P. E., Jones J. G., Whitney K. D., Donehower L. A., Harris E. L., Rohan T., Johns D. C., Pestell R. G. (2003) Mol. Cell. Biol. 23, 6159–6173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Knudsen K. E., Cavenee W. K., Arden K. C. (1999) Cancer Res. 59, 2297–2301 [PubMed] [Google Scholar]
- 24. Reutens A. T., Fu M., Wang C., Albanese C., McPhaul M. J., Sun Z., Balk S. P., Jänne O. A., Palvimo J. J., Pestell R. G. (2001) Mol. Endocrinol. 15, 797–811 [DOI] [PubMed] [Google Scholar]
- 25. Burd C. J., Petre C. E., Morey L. M., Wang Y., Revelo M. P., Haiman C. A., Lu S., Fenoglio-Preiser C. M., Li J., Knudsen E. S., Wong J., Knudsen K. E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2190–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Comstock C. E., Augello M. A., Benito R. P., Karch J., Tran T. H., Utama F. E., Tindall E. A., Wang Y., Burd C. J., Groh E. M., Hoang H. N., Giles G. G., Severi G., Hayes V. M., Henderson B. E., Le Marchand L., Kolonel L. N., Haiman C. A., Baffa R., Gomella L. G., Knudsen E. S., Rui H., Henshall S. M., Sutherland R. L., Knudsen K. E. (2009) Clin Cancer Res. 15, 5338–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Comstock C. E., Revelo M. P., Buncher C. R., Knudsen K. E. (2007) Br. J. Cancer 96, 970–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu Y., Chen S. Y., Ross K. N., Balk S. P. (2006) Cancer Res. 66, 7783–7792 [DOI] [PubMed] [Google Scholar]
- 29. Burd C. J., Morey L. M., Knudsen K. E. (2006) Endocr. Relat. Cancer 13, 979–994 [DOI] [PubMed] [Google Scholar]
- 30. Burd C. J., Petre C. E., Moghadam H., Wilson E. M., Knudsen K. E. (2005) Mol. Endocrinol. 19, 607–620 [DOI] [PubMed] [Google Scholar]
- 31. Li J., Lin Q., Wang W., Wade P., Wong J. (2002) Genes Dev. 16, 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Petre-Draviam C. E., Williams E. B., Burd C. J., Gladden A., Moghadam H., Meller J., Diehl J. A., Knudsen K. E. (2005) Oncogene 24, 431–444 [DOI] [PubMed] [Google Scholar]
- 33. Fu M., Rao M., Bouras T., Wang C., Wu K., Zhang X., Li Z., Yao T. P., Pestell R. G. (2005) J. Biol. Chem. 280, 16934–16941 [DOI] [PubMed] [Google Scholar]
- 34. Petre C. E., Wetherill Y. B., Danielsen M., Knudsen K. E. (2002) J. Biol. Chem. 277, 2207–2215 [DOI] [PubMed] [Google Scholar]
- 35. Knudsen K. E., Scher H. I. (2009) Clin. Cancer Res. 15, 4792–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schiewer M. J., Morey L. M., Burd C. J., Liu Y., Merry D. E., Ho S. M., Knudsen K. E. (2009) Oncogene 28, 1016–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Knudsen K. E., Arden K. C., Cavenee W. K. (1998) J. Biol. Chem. 273, 20213–20222 [DOI] [PubMed] [Google Scholar]
- 38. de Launoit Y., Veilleux R., Dufour M., Simard J., Labrie F. (1991) Cancer Res. 51, 5165–5170 [PubMed] [Google Scholar]
- 39. Dai M., Wang P., Boyd A. D., Kostov G., Athey B., Jones E. G., Bunney W. E., Myers R. M., Speed T. P., Akil H., Watson S. J., Meng F. (2005) Nucleic Acids Res. 33, e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang Q., Li W., Zhang Y., Yuan X., Xu K., Yu J., Chen Z., Beroukhim R., Wang H., Lupien M., Wu T., Regan M. M., Meyer C. A., Carroll J. S., Manrai A. K., Jänne O. A., Balk S. P., Mehra R., Han B., Chinnaiyan A. M., Rubin M. A., True L., Fiorentino M., Fiore C., Loda M., Kantoff P. W., Liu X. S., Brown M. (2009) Cell 138, 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., Haussler D. (2002) Genome Res. 12, 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bussey K. J., Kane D., Sunshine M., Narasimhan S., Nishizuka S., Reinhold W. C., Zeeberg B., Ajay W., Weinstein J. N. (2003) Genome Biol. 4, R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wetherill Y. B., Petre C. E., Monk K. R., Puga A., Knudsen K. E. (2002) Mol. Cancer Ther. 1, 515–524 [PubMed] [Google Scholar]
- 44. Link K. A., Balasubramaniam S., Sharma A., Comstock C. E., Godoy-Tundidor S., Powers N., Cao K. H., Haelens A., Claessens F., Revelo M. P., Knudsen K. E. (2008) Cancer Res. 68, 4551–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang Q., Li W., Liu X. S., Carroll J. S., Jänne O. A., Keeton E. K., Chinnaiyan A. M., Pienta K. J., Brown M. (2007) Mol. Cell 27, 380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Henrique R., Costa V. L., Cerveira N., Carvalho A. L., Hoque M. O., Ribeiro F. R., Oliveira J., Teixeira M. R., Sidransky D., Jerónimo C. (2006) J. Mol. Med. 84, 911–918 [DOI] [PubMed] [Google Scholar]
- 47. Müller I., Wischnewski F., Pantel K., Schwarzenbach H. (2010) BMC Cancer 10, 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peng Y., Chen F., Melamed J., Chiriboga L., Wei J., Kong X., McLeod M., Li Y., Li C. X., Feng A., Garabedian M. J., Wang Z., Roeder R. G., Lee P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5236–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wissmann C., Wild P. J., Kaiser S., Roepcke S., Stoehr R., Woenckhaus M., Kristiansen G., Hsieh J. C., Hofstaedter F., Hartmann A., Knuechel R., Rosenthal A., Pilarsky C. (2003) J. Pathol. 201, 204–212 [DOI] [PubMed] [Google Scholar]
- 50. Olshavsky N. A., Groh E. M., Comstock C. E., Morey L. M., Wang Y., Revelo M. P., Burd C., Meller J., Knudsen K. E. (2008) Oncogene 27, 3111–3121 [DOI] [PubMed] [Google Scholar]
- 51. Lam E. W., Glassford J., Banerji L., Thomas N. S., Sicinski P., Klaus G. G. (2000) J. Biol. Chem. 275, 3479–3484 [DOI] [PubMed] [Google Scholar]
- 52. Petre-Draviam C. E., Cook S. L., Burd C. J., Marshall T. W., Wetherill Y. B., Knudsen K. E. (2003) Cancer Res. 63, 4903–4913 [PubMed] [Google Scholar]
- 53. Obiezu C. V., Diamandis E. P. (2005) Cancer Lett. 224, 1–22 [DOI] [PubMed] [Google Scholar]
- 54. Xu L. L., Shanmugam N., Segawa T., Sesterhenn I. A., McLeod D. G., Moul J. W., Srivastava S. (2000) Genomics 66, 257–263 [DOI] [PubMed] [Google Scholar]
- 55. Pendás A. M., Matilla T., Uría J. A., Freije J. P., Fueyo A., Estivill X., López-Otín C. (1994) Cytogenet. Cell Genet. 66, 263–266 [DOI] [PubMed] [Google Scholar]
- 56. Nelson P. S., Clegg N., Arnold H., Ferguson C., Bonham M., White J., Hood L., Lin B. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11890–11895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lin B., Ferguson C., White J. T., Wang S., Vessella R., True L. D., Hood L., Nelson P. S. (1999) Cancer Res. 59, 4180–4184 [PubMed] [Google Scholar]
- 58. Chen C. D., Welsbie D. S., Tran C., Baek S. H., Chen R., Vessella R., Rosenfeld M. G., Sawyers C. L. (2004) Nat. Med. 10, 33–39 [DOI] [PubMed] [Google Scholar]
- 59. DePrimo S. E., Diehn M., Nelson J. B., Reiter R. E., Matese J., Fero M., Tibshirani R., Brown P. O., Brooks J. D. (2002) Genome Biol. 3, RESEARCH0032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Holzbeierlein J., Lal P., LaTulippe E., Smith A., Satagopan J., Zhang L., Ryan C., Smith S., Scher H., Scardino P., Reuter V., Gerald W. L. (2004) Am J. Pathol. 164, 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhao H., Kim Y., Wang P., Lapointe J., Tibshirani R., Pollack J. R., Brooks J. D. (2005) Prostate 63, 187–197 [DOI] [PubMed] [Google Scholar]
- 62. Comstock C. E., Burd C. J., Jessen W. J., Knudsen K. E. (2008) in Contemporary Endocrinology: Genomics in Endocrinology-DNA Microarray Analysis in Endocrine Health and Disease (Handwerger S., Aronow B. J. eds) pp. 83–113, Humana Press, New Jersey [Google Scholar]
- 63. Cai C., Omwancha J., Hsieh C. L., Shemshedini L. (2007) Prostate Cancer Prostatic Dis. 10, 39–45 [DOI] [PubMed] [Google Scholar]
- 64. Ho L. L., Kench J. G., Handelsman D. J., Scheffer G. L., Stricker P. D., Grygiel J. G., Sutherland R. L., Henshall S. M., Allen J. D., Horvath L. G. (2008) Prostate 68, 1421–1429 [DOI] [PubMed] [Google Scholar]
- 65. Kruh G. D., Belinsky M. G., Gallo J. M., Lee K. (2007) Cancer Metastasis Rev. 26, 5–14 [DOI] [PubMed] [Google Scholar]
- 66. Jia L., Berman B. P., Jariwala U., Yan X., Cogan J. P., Walters A., Chen T., Buchanan G., Frenkel B., Coetzee G. A. (2008) PLoS One 3, e3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lin B., Wang J., Hong X., Yan X., Hwang D., Cho J. H., Yi D., Utleg A. G., Fang X., Schones D. E., Zhao K., Omenn G. S., Hood L. (2009) PLoS One 4, e6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Massie C. E., Adryan B., Barbosa-Morais N. L., Lynch A. G., Tran M. G., Neal D. E., Mills I. G. (2007) EMBO Rep. 8, 871–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bolton E. C., So A. Y., Chaivorapol C., Haqq C. M., Li H., Yamamoto K. R. (2007) Genes Dev. 21, 2005–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Makkonen H., Kauhanen M., Paakinaho V., Jääskeläinen T., Palvimo J. J. (2009) Nucleic Acids Res. 37, 4135–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Clark E. L., Coulson A., Dalgliesh C., Rajan P., Nicol S. M., Fleming S., Heer R., Gaughan L., Leung H. Y., Elliott D. J., Fuller-Pace F. V., Robson C. N. (2008) Cancer Res. 68, 7938–7946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shi X. B., Xue L., Zou J. X., Gandour-Edwards R., Chen H., deVere White R. W. (2008) Prostate 68, 1816–1826 [DOI] [PubMed] [Google Scholar]
- 73. Kang Z., Jänne O. A., Palvimo J. J. (2004) Mol. Endocrinol. 18, 2633–2648 [DOI] [PubMed] [Google Scholar]
- 74. Kang Z., Pirskanen A., Jänne O. A., Palvimo J. J. (2002) J. Biol. Chem. 277, 48366–48371 [DOI] [PubMed] [Google Scholar]
- 75. Makkonen H., Kauhanen M., Jääskeläinen T., Palvimo J. J. (2011) Mol. Cell. Endocrinol. 331, 57–65 [DOI] [PubMed] [Google Scholar]
- 76. Waltering K. K., Helenius M. A., Sahu B., Manni V., Linja M. J., Jänne O. A., Visakorpi T. (2009) Cancer Res. 69, 8141–8149 [DOI] [PubMed] [Google Scholar]
- 77. Tomlins S. A., Rhodes D. R., Perner S., Dhanasekaran S. M., Mehra R., Sun X. W., Varambally S., Cao X., Tchinda J., Kuefer R., Lee C., Montie J. E., Shah R. B., Pienta K. J., Rubin M. A., Chinnaiyan A. M. (2005) Science 310, 644–648 [DOI] [PubMed] [Google Scholar]
- 78. Li H., Lovci M. T., Kwon Y. S., Rosenfeld M. G., Fu X. D., Yeo G. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20179–20184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mendiratta P., Mostaghel E., Guinney J., Tewari A. K., Porrello A., Barry W. T., Nelson P. S., Febbo P. G. (2009) J. Clin Oncol. 27, 2022–2029 [DOI] [PubMed] [Google Scholar]
- 80. Ngan S., Stronach E. A., Photiou A., Waxman J., Ali S., Buluwela L. (2009) Oncogene 28, 2051–2063 [DOI] [PubMed] [Google Scholar]
- 81. Wang G., Jones S. J., Marra M. A., Sadar M. D. (2006) Oncogene 25, 7311–7323 [DOI] [PubMed] [Google Scholar]
- 82. Vellaichamy A., Dezso Z., JeBailey L., Chinnaiyan A. M., Sreekumar A., Nesvizhskii A. I., Omenn G. S., Bugrim A. (2010) PLoS One 5, e10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mullany L. K., White P., Hanse E. A., Nelsen C. J., Goggin M. M., Mullany J. E., Anttila C. K., Greenbaum L. E., Kaestner K. H., Albrecht J. H. (2008) Cell Cycle 7, 2215–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Alt J. R., Gladden A. B., Diehl J. A. (2002) J. Biol. Chem. 277, 8517–8523 [DOI] [PubMed] [Google Scholar]
- 85. Cheng M., Olivier P., Diehl J. A., Fero M., Roussel M. F., Roberts J. M., Sherr C. J. (1999) EMBO J. 18, 1571–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. LaBaer J., Garrett M. D., Stevenson L. F., Slingerland J. M., Sandhu C., Chou H. S., Fattaey A., Harlow E. (1997) Genes Dev. 11, 847–862 [DOI] [PubMed] [Google Scholar]
- 87. Fritah A., Saucier C., Mester J., Redeuilh G., Sabbah M. (2005) Mol. Cell. Biol. 25, 2419–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lacombe L., Maillette A., Meyer F., Veilleux C., Moore L., Fradet Y. (2001) Int. J. Cancer 95, 135–139 [DOI] [PubMed] [Google Scholar]
- 89. Omar E. A., Behlouli H., Chevalier S., Aprikian A. G. (2001) Prostate 49, 191–199 [DOI] [PubMed] [Google Scholar]
- 90. Rigaud J., Tiguert R., Decobert M., Hovington H., Latulippe E., Laverdiere J., Larue H., Lacombe L., Fradet Y. (2004) Prostate 58, 269–276 [DOI] [PubMed] [Google Scholar]
- 91. Sarkar F. H., Li Y., Sakr W. A., Grignon D. J., Madan S. S., Wood D. P., Jr., Adsay V. (1999) Prostate 40, 256–260 [DOI] [PubMed] [Google Scholar]
- 92. Wong C. I., Zhou Z. X., Sar M., Wilson E. M. (1993) J. Biol. Chem. 268, 19004–19012 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.