Abstract
We identified seven alternatively spliced forms of human 8-oxoguanine DNA glycosylase (OGG1) mRNAs, classified into two types based on their last exons (type 1 with exon 7: 1a and 1b; type 2 with exon 8: 2a to 2e). Types 1a and 2a mRNAs are major in human tissues. Seven mRNAs are expected to encode different polypeptides (OGG1–1a to 2e) that share their N terminus with the common mitochondrial targeting signal, and each possesses a unique C terminus. A 36-kDa polypeptide, corresponding to OGG1–1a recognized only by antibodies against the region containing helix-hairpin-helix-PVD motif, was copurified from the nuclear extract with an activity introducing a nick into DNA containing 8-oxoguanine. A 40-kDa polypeptide corresponding to a processed form of OGG1–2a was detected in their mitochondria using antibodies against its C terminus. Electron microscopic immunocytochemistry and subfractionation of the mitochondria revealed that OGG1–2a locates on the inner membrane of mitochondria. Deletion mutant analyses revealed that the unique C terminus of OGG1–2a and its mitochondrial targeting signal are essential for mitochondrial localization and that nuclear localization of OGG1–1a depends on the NLS at its C terminus.
INTRODUCTION
Oxidative phosphorylation in mitochondria is essential for eukaryotic organisms to produce energy to maintain life; however, ∼1–3% of consumed oxygen molecules are partially reduced by electrons leaked from the respiratory chain generating reactive oxygen species (ROS)1 such as superoxide, hydroxy peroxide, and hydroxyl radical (for a review see Kang et al., 1998). ROS are so highly reactive that they can readily oxidize macromolecules in living cells such as lipids, proteins, and nucleic acids, leading to various oxidative cellular damages such as cell death and mutagenesis (for reviews see Ames et al., 1993; Shigenaga et al., 1994).
Organisms are equipped with defense mechanisms to minimize accumulation of such ROS. For example, superoxide dismutase converts superoxide to oxygen and hydroxy peroxide, and the latter is further decomposed by catalase into water and oxygen. Mice lacking the SOD2 gene that encodes mitochondrial superoxide dismutase show severe abnormality in development and growth, including cardiomyopathy and neurodegeneration (Li et al., 1995; Lebovitz et al., 1996). Thus, once excessive ROS accumulates in cells, they can no longer avoid severe oxidative damages. Accumulation of oxidized macromolecules in human tissues gradually occurs during normal aging, even in the presence of functional superoxide dismutases; hence, oxidative damages have been implicated as one of the major causes of aging and degenerative diseases.
Among various oxidative damages in cellular macromolecules, damage to nucleic acid is particularly hazardous for cells because genetic information present in genomic DNAs, such as nuclear and mitochondrial DNAs, can be altered by such damage. Damage to genomic DNAs often causes cell death, the result being degenerative diseases, or mutations resulting in neoplasia and hereditary diseases (for a review see Friedberg et al., 1995). ROS cause various base or sugar modifications in DNA and free nucleotides and further introduce strand breaks in DNA (for a review see Demple and Harrison, 1994). Specifically, 8-oxoguanine (8-oxoG), one of the oxidized forms of guanine, is produced by ROS in a fairly large amount in both DNA and nucleotide pools and is somewhat stable and likely to accumulate in genomic DNAs in nuclei and mitochondria during normal aging (Kasai and Nishimura, 1984; Ames et al., 1993; Hayakawa et al., 1993). Accumulation of 8-oxoG in DNA attributable to either utilization of 8-oxo-dGTP in nucleotide pools as a precursor or direct oxidation of DNA increases the occurrence of A:T to C:G or G:C to T:A transversion mutation, respectively, because 8-oxoG forms a stable basepair with adenine as well as with cytosine (Shibutani et al., 1991; Cheng et al., 1992; Maki and Sekiguchi, 1992).
Studies on mutator mutants of Escherichia coli revealed that E. coli has several error-avoiding mechanisms to minimize the deleterious effects of 8-oxoG, in which MutT, MutM, and MutY proteins play important roles (Michaels and Miller, 1992; Sekiguchi, 1996). MutT protein hydrolyzes 8-oxo-dGTP to 8-oxo-dGMP and pyrophosphate (Maki and Sekiguchi, 1992) and thus avoids spontaneous occurrence of A:T to C:G transversion mutation during DNA synthesis, the rate of which in the mutT-deficient strain increases hundreds- to thousands-fold compared with the wild type (Tajiri et al., 1995). MutM protein, originally identified as a formamidopyrimidine DNA glycosylase, removes the 8-oxoG paired with cytosine and introduces a single-strand gap as a result of the accompanying apurinic/apyrimidinic (AP) lyase activity (Bailly et al., 1989; Michaels et al., 1992). MutY protein excises adenine paired with guanine or 8-oxoG by its DNA glycosylase activity (Au et al., 1989). The rate of spontaneous occurrence of G:C to T:A base change in the mutM- or mutY-deficient strain increases 10–50 times higher than that in the wild-type strain (Cabrera et al., 1988; Nghiem et al., 1988), and in double mutants of mutM and mutY it is equivalent to that of the mutT mutant.
Mammalian cells also come equipped with similar error-avoiding mechanisms. cDNAs encoding MutT homolog (MTH1) proteins with the 8-oxo-dGTPase activity have been cloned from human, mouse, and rat (Sakumi et al., 1993; Kakuma et al., 1995; Cai et al., 1997), and their genomic organizations and regulation of expression have been well characterized (Furuichi et al., 1994; Igarashi et al., 1997; Oda et al., 1997). Although an activity that excises adenine paired with guanine or 8-oxoG in DNA has been identified in human cells (Yeh et al., 1991), and a human gene encoding MutY homolog (MYH) protein also has been cloned (Slupska et al., 1996), biochemical identification of the MYH protein as the adenine DNA glycosylase remains to be elucidated. In contrast to the mutT and mutY gene families, there are at least two diverse genes in eukaryotes that encode 8-oxoG DNA glycosylase. One is the mutM homologue (AtMMH) identified in Arabidopsis thaliana (Ohtsubo et al., 1998); the other is a novel gene found in yeast, OGG1 (Nash et al., 1996; van der Kemp et al., 1996). Thereafter, on the basis of the retrieved information from the database of expressed sequence tags, human and mouse genes encoding homologous proteins to the yeast Ogg1 protein have been identified (Aburatani et al., 1997; Arai et al., 1997; Bjoras et al., 1997; Lu et al., 1997; Radicella et al., 1997; Roldán-Arjona et al., 1997; Rosenquist et al., 1997). Thus, mammalian cells have developed unique error-avoiding mechanisms against 8-oxoG by combining those evolved from ones in E. coli and yeast.
In eukaryotes, more than one genome in a single cell has to be maintained throughout their entire life, one in the nucleus and one in the mitochondrion, or the other in chloroplasts in the case of plants. Genomes in mitochondria are likely to be more susceptible to oxidative damages by ROS because of the high oxygen metabolism. We reported that MTH1 protein locates both in cytoplasm and mitochondria of human cells, indicating that errors caused by oxidation of DNA or nucleotides in mitochondria are avoided to maintain their genomes (Kang et al., 1995). On the basis of this observation, we extended our study to further examine error-avoiding mechanisms against oxidative DNA damages in human cells, and we characterized expression and intracellular localization of OGG1 protein.
We now report the first detection of the authentic OGG1 proteins in both mitochondria and nuclei in human cells, and mechanisms regulating their intracellular localization are also described
MATERIALS AND METHODS
Chemicals
[α-32P]dCTP was purchased from Amersham International (Buckinghamshire, United Kingdom). Recombinant Taq DNA polymerase, restriction enzymes, T4 DNA polymerase, and T4 DNA ligase were obtained from Takara Shuzo (Kyoto, Japan) and Toyobo Co. (Osaka, Japan). DNA-labeling kits were obtained from Nippon Gene (Toyama, Japan). DNA Ladder (1 kb) and RNA Ladder (0.24–9.5 kb) as size standards, 10× RPMI medium, DMEM, and formamide were purchased from Life Technologies (Gaithersburg, MD). Other chemicals were obtained from Wako Pure Chemical Industries Ltd. (Osaka, Japan). Sources of other materials are indicated at appropriate places in the text.
Antibodies
Anti-hemagglutinin (HA) mAb (clone 12CA5), which recognizes an HA epitope, was the product of Boehringer Mannheim (Mannheim, Germany), and anti-Bcl2 mAb (clone 124) was obtained from Dako A/S (Copenhagen, Denmark). Rabbit anti-mouse IgG was purchased from Wako Pure Chemical Industries Ltd., and alkaline phosphatase-conjugated mouse anti-rabbit IgG mAb was obtained from Promega (Madison, WI). Cy5-labeled goat anti-mouse IgG was obtained from Amersham International.
Oligonucleotides
The synthetic oligonucleotides listed below were purchased from Takara Shuzo, International Reagents Co. (Kobe, Japan) and Greiner Japan (Tokyo, Japan): HA-tag (5′-TACCCTTATGATGTGCCGGATTATGCAATTAATTGATTGATTATAATCTAGGATCC-3′), N1 (5′-GCGTGCGCAAGTACTTCCAGC-3′), N4 (5′-CCAGTGATGCGGG-CGATGTTG-3′), OG2 (5′-AGAATTGGGGTACGAAGC-3′), OG4 (5′-GATTTAGTAAATCTGGGGCAG-3′), OG7 (5′- CAGACCAACA-AGGAACTGG-3′), OG18 (5′-TAAAGGGAAGATAAAACCATC-3′), OG21 (5′-GCATCACATGACCAATTACTG-3′), GAD2 (5′-GTTTGGAATCACTACAGGGATGT-3′), GAD3 (5′-CAGTATCTACGA-TTCATAGATCTG-3′), SphI-primer (5′-TGCCTGCTGTGGGCAT-GCCTG-3′), ΔMTS2 (5′-CAAGCTTATGCCTTCTGGACAAT-3′), OG/HA (5′-AAGGGTAAAGTGCTAGTAAGCTG-3′), HA-primer (5′-AGCACTTTACCCTTATGATGTGC-3′), ΔNLS1F (5′-CAGCATACCCTTATGATGTGC-3′), ΔNLS1R (5′-AAGGGTATGCTGGTGGCTCCTG-3′), and 3-tag-2 (5′-CCGCTGGATCCTAGATTATAATCAAT-3′).
N-(3-fluoranthyl)maleimide (FAM)-16GO (5′-GGAATTCCTCGA-GGT<GO>GACGGTATCCGATGGCCGCT-3′), in which GO represents 8-oxoG, and a fluorescence dye, FAM, which was attached to its 5′-end, and c-16C (5′-AGCGGCCATCGGATACCGTCCACCT-CGAGGAATTCC-3′) were purchased from Greiner Japan. To prepare double-stranded substrates containing 8-oxoG for the nicking assay, FAM-16GO and c-16C (5 pmol each) were mixed in an annealing buffer containing 10 mM Tris-HCl, pH 8.0, and 20 mM NaCl in a total volume of 100 μl. The mixture was incubated at 65°C for 5 min, then gradually cooled to room temperature.
Isolation of Human OGG1 cDNAs and RT-PCR Analysis
Human OGG1 cDNAs were amplified by PCR using DNA prepared from the human leukemia MATCHMAKER cDNA library (HL4015AB, Clontech Laboratories, Palo Alto, CA) as a template, with several combinations of primers N1 and N4 for a human clone from the database of expressed sequence tags, N55394 (see Figure 1A), and primers GAD2 and GAD3 for multicloning sites of the vector, pGAD10, and were cloned into pT7Blue(R)T vector (Novagen, Madison, WI).
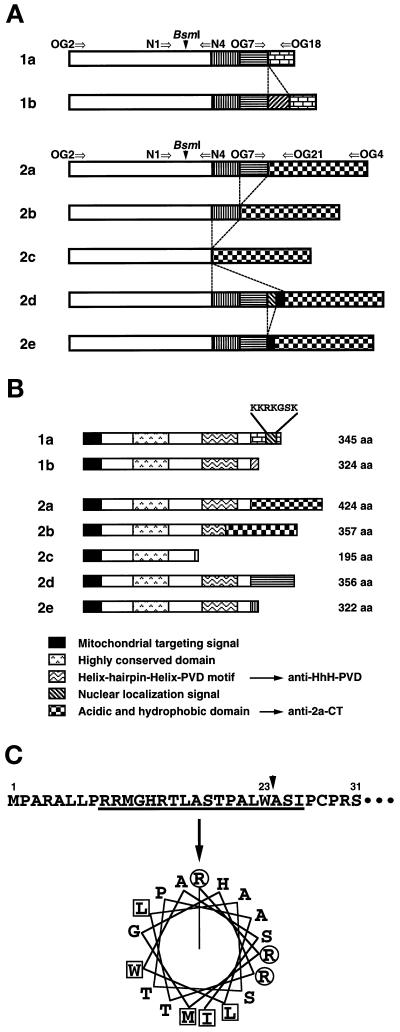
Figure 1.
Structures of multiple human OGG1 mRNAs and their products. (A) Alternatively spliced forms of OGG1 mRNAs. Each box represents a region alternatively joined in each OGG1 mRNA. The open box represents the sequence corresponding to exons 1 to 3 of the human OGG1 gene (Aburatani et al., 1997), in which alternative splicing was not observed. Arrows indicate primers used for RT-PCR analysis. These sequence data of human OGG1 cDNA are available from DNA Data Bank of Japan under the following accession numbers: 1b, AB019532; 2b, AB019528; 2c, AB019529; 2d, AB019530; and 2e, AB019531. (B) Structural features of different human OGG1 proteins. All forms of the OGG1 proteins contain a putative MTS at the common N-terminal region. There is a NLS only in the C-terminal end of OGG1–1a. The region exhibiting the highest homology to the corresponding region of yeast Ogg1 protein (aa 128–156) is shown as the highly conserved region. Anti–HhH-PVDrecognizes the region containing HhH-PVD motif (aa 221–290), and anti–2a-CT recognizes the C-terminal regions of both OGG1–2a (aa 322–424) and -2b (aa 255–357). (C) Putative mitochondrial targeting signal for OGG1 proteins. Using the MitoPlot II program, we predicted that the residues from 9 to 26 at the common N-terminal region of OGG1 proteins form an amphipathic helix (angle for the helical wheel is 95°), with three arginine residues but no acidic residue and values of 6.57 for hydrophobic moment (μH), 4.33 for maximum hydrophobicity (Hmax). The MTS may be processed at residue 23 (W) after being translocated into mitochondria (arrow). Basic residues (R) are shown in circles, and hydrophobic residues are in boxes.
RT-PCR was performed as follows. Total cellular RNA was prepared as described (Chirgwin et al., 1979), and poly(A)+ mRNA was purified using mRNA Purification Kits (Pharmacia Biotech, Uppsala, Sweden). First-strand cDNA, prepared using First-Strand cDNA Synthesis Kits (Pharmacia Biotech) according to the manufacturer’s instruction was subjected to PCR, which was performed in 50 μl of a reaction mixture containing 1 μl of the first-strand cDNA, 1 U of recombinant Taq DNA polymerase, 0.4 μM each primer, and 200 μM each deoxynucleotide triphosphate. The initial denaturation was performed at 95°C for 1 min, and then amplification was performed by 30 or 35 cycles of denaturation at 95°C for 20 s, annealing at 55°C for 20 s, extension at 72°C for 40 s, followed by additional extension at 72°C for 5 min.
Construction of Plasmids
Mammalian expression plasmids, pcDEBΔ::OGG1–1a:HA, pcDEBΔ:: OGG1–2a:HA, pcDEBΔ::ΔN31-OGG1–2a:HA, and pcDEBΔ::ΔC11-OGG1–1a:HA were constructed by inserting each cDNA fragment, to which a sequence for the HA epitope was introduced with the aid of PCR, into the HindIII–BamHI site of pcDEBΔ that carries the SRα promoter (Nakabeppu et al., 1993).
Plasmids for expression of recombinant OGG1 proteins in E. coli, pET3d::OGG1–1a, pET3d::OGG1–1b, and pET3d::OGG1–2a, were constructed by inserting each cDNA fragment prepared by the aid of PCR into the NcoI–BamHI site of pET3d, and in which each cDNA can be transcribed from the T7 phage promoter (Studier et al., 1990).
To construct a plasmid, pYN3103::TrpE-HhH-PVD coding a fusion protein, TrpE-HhH-PVD, a cDNA fragment encoding amino acid (aa) residues 221–290 of OGG1–1a protein was inserted into the SalI–BamHI site of pYN3103::TrpE (Nakabeppu and Nathans, 1991). A plasmid, pYN3103::TrpE-2a-CT, was generated by insertion of a cDNA fragment encoding aa 322–424 of OGG1–2a protein into the PstI–BamHI site of pYN3103::TrpE. Then, a NcoI–BamHI fragment prepared from each plasmid was transferred into the NcoI–BamHI site of pET3d to construct pET3d::TrpE-HhH-PVD and pET3d::TrpE-2a-CT.
DNA Sequencing
Nucleotide sequence was determined using Dye Terminator or Dye Primer Cycle Sequencing FS Ready Reaction Kits and model 373A automated DNA sequencer (Perkin Elmer-Cetus, Foster City, CA), according to the manufacturer’s instructions.
Northern Blot Analysis
Northern blot analysis was performed as described (Oda et al., 1997). RNAs prepared from various human tissues were purchased from Clontech Laboratories. A fragment of OGG1 cDNA for type 2a was prepared by PCR using a set of oligonucleotide primers (OG2 and OG4) from a plasmid carrying the cDNA insert and was labeled by [α-32P]dCTP using a DNA-labeling kit and then was used as a hybridization probe. Radioactivity on Northern blots was measured using BAS 2000 Bio-image analyzer (Fuji Photo Film Co. Ltd., Tokyo, Japan). To confirm the amounts of RNA loaded, the blot was reprobed with a 1.0-kb EcoRI–BamHI fragment of the 18S rRNA gene prepared from pRγcHE obtained from the Japanese Cancer Research Resources Bank.
Partial Purification of the Nuclear Form of OGG1 Protein
Nuclear extract was prepared from Jurkat cells (4 × 1010 cells) according to a method described by Dignam et al. (1983), but with a slight modification. All procedures were performed at 4°C. Nuclear extract (112 ml, 506 mg protein) was applied onto an activated Phosphocellulose (P11) column (bed volume 35 ml) (Whatman, Maidstone, England) equilibrated with buffer C (20 mM HEPES-KOH, pH 7.8, 15% glycerol, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF, 0.5 μg/ml pepstatin, 0.5 μg/ml leupeptin, 0.5 μg/ml chymostatin) containing 50 mM KCl, then the column was eluted with a 480-ml linear gradient of KCl (50–750 mM). Fractions containing an 8-oxoG–specific nicking enzyme, which eluted at 400–500 mM KCl, were pooled (150 ml). The pooled fraction was concentrated with Ficoll to 5 ml, then applied onto a HiLoad 26/60 Superdex 75-pg gel filtration column (bed volume 320 ml) (Pharmacia Biotech) equilibrated with buffer C containing 200 mM KCl. Active fractions were pooled (27 ml, 0.745 mg protein) and diluted by adding an equal volume of buffer C containing 50 mM KCl. The fraction was applied onto a RESOURCE Q column (bed volume 1 ml) (Pharmacia Biotech) equilibrated with buffer C containing 125 mM KCl. The flow-through fraction was collected (50 ml) and applied onto two RESOURCE S columns (bed volume 1 ml) (Pharmacia Biotech) equilibrated with the same buffer. The column was eluted with a 5-ml linear gradient of KCl (125–500 mM), and 200 μl of each fraction were collected.
Nicking Assay
Double-stranded oligonucleotides (FAM-16GO and c-16C; 300 fmol) were incubated in a reaction mixture containing 25 mM HEPES-KOH, pH 7.4, 0.5 mM EDTA, 0.5 mM DTT, 10 μM ZnCl2, 50 μg/ml BSA, and an enzyme fraction in a total volume of 15 μl, at 37°C for 1 h. Then, 15 μl of stop solution containing 80% formamide, 6 mg/ml BlueDextran (Sigma Chemical Co., St. Louis, MO), and 10 mM EDTA was added to each reaction mixture; then 6 μl of the mixture was fractionated on 6% polyacrylamide gel containing 7 M urea. Specifically nicked 15-base length oligomer labeled with FAM was detected using the model 373 automated DNA sequencer and quantified with 672 GENESCAN software (Perkin Elmer-Cetus), according to the manufacturer’s instruction. One unit of nicking activity was defined as the amount of enzyme that cleaves 1 fmol/h of substrate DNA.
Preparation of Antibodies against Human OGG1 Proteins
Rabbit polyclonal antibodies were raised against the fusion proteins TrpE-HhH-PVD and TrpE-2a-CT, as described (Nakabeppu and Nathans, 1991). Antibodies were purified with the aid of TrpE-HhH-PVD–Sepharose, TrpE-2a-CT–Sepharose, and TrpE–Sepharose columns (Nakabeppu and Nathans, 1991; Nakabeppu et al., 1993). The TrpE-HhH-PVD–Sepharose column, to which the antiserum against TrpE-HhH-PVD was loaded, was washed using buffer E (0.2 M glycine-HCl, pH 2.5, 0.1 M NaCl, 0.1% Triton X-100). TrpE-2a-CT–Sepharose column, to which the antiserum against TrpE-2a-CT was loaded, was washed using 10 mM sodium phosphate buffer, pH 7.6. The bound antibodies were then eluted using buffer E, pH 2.3. The eluted fraction was dialyzed against TBS and applied onto the TrpE–Sepharose column. The flow-through fractions were collected and designated as the anti–helix-hairpin-helix (HhH)–Pro/Val-rich stretch with nearby Asp (PVD) motif and the anti–2a-CT, respectively.
Preabsorption of polyclonal antibodies was performed as follows. Antibodies (20 μg) were incubated overnight at 4°C with an immobilized antigen on Sepharose (1 mg antigen equivalent) in 10 ml of TBS containing 0.05% Tween 20 and 5% BSA, with gentle shaking. Preabsorbed antibody solution was recovered and used for Western blotting and for immunocytochemistry studies.
Cell Cultures, DNA Transfection, Subcellular Fractionation, and Western Blotting
HeLa cells (MR and S3 strains) (Ishibashi et al., 1994) and Jurkat cells were maintained in DMEM and RPMI 1640 medium, respectively, at 37°C in a humidified atmosphere with 5% CO2. Each medium was supplemented with 100 μg/ml streptomycin, 100 U/ml penicillin, and 10% of FBS.
Transfection of plasmid DNA into HeLa MR cells was performed according to Chen and Okayama (1987). To establish stable cell lines, transformants were selected in medium containing 300 μg/ml hygromycin B, were replated twice for purification, and then were maintained in medium containing 100 μg/ml hygromycin B.
Subcellular fractionation and Western blot analysis were performed as described by Kang et al. (1995) and Nakabeppu et al. (1993), respectively. Antibodies bound to each blot were detected using 125I-protein A, and the radioactivity was measured using a BAS 2000 Bio-image analyzer.
Light Microscopic Immunocytochemistry
HeLa MR cells were plated on an eight-well collagen I-coated chamber slide (Becton Dickinson, Bedford, MA) one day before DNA transfection. Plasmid DNA was introduced into the cells according to Chen and Okayama (1987). Transfected cells, incubated for 45 min in the presence of 2 μM MitoTracker CMXRos-H2 (Molecular Probes, Eugene, OR), were fixed with 50% methanol and 50% acetone at room temperature for 10 min. Slides were subsequently washed with PBS and incubated with PBS containing 0.1% Tween 20, 3% BSA, and 10% normal goat serum at 37°C for 1 h in a humidified atmosphere. Next the slides were incubated overnight at 4°C with anti-HA and Cy5-labeled goat anti-mouse IgG at room temperature for 1 h, then the washed slides were mounted. Results were viewed under an Axiovert 100 photomicroscope (Carl Zeiss Co., Oberkochen, Germany), and digitized images were obtained using CELLscan system software (Scanalytics, Billerica, MA), then were processed for publication using Adobe Photoshop (Adobe Systems, San Jose, CA).
Electron Microscopic Immunocytochemistry
Thin sections of isolated mitochondria were prepared and processed for electron microscopic immunocytochemistry, as described (Kang et al., 1995).
Other Methods
The activity of lactate dehydrogenase was measured by the method of Bergmeyer et al. (1965). One unit of the activity was defined as the absorbance change of 0.01/min. The activity of succinate-cytochrome c reductase was measured as described (Schnaitman and Greenawalt, 1968), and one unit of its activity was defined as the absorbance change of 0.1/min. Protein concentration was determined using a Bio-Rad protein assay kit (Bio-Rad Laboratories, Hercules, CA) with BSA as a standard, according to the manufacturer’s instruction.
RESULTS
Seven Different Types of Human OGG1 mRNA Formed by Alternative Splicing and Their Predicted Products
By RT-PCR amplification, seven different types of human OGG1 cDNA were identified from poly(A)+ mRNA preparations of Jurkat cells, a human T cell leukemia cell line. Comparison with reported human OGG1 cDNAs and the genomic structure of the mouse OGG1 gene (Aburatani et al., 1997; Tani et al., 1998) suggested that all the sequences are derived from the human OGG1 gene and are formed by alternative splicing. We classified these OGG1 mRNAs into two groups based on their last exon (type 1: 1a, 1608-base, and 1b, 1852-base; type 2: 2a, 2085-base; 2b, 1884-base; 2c, 1702-base; 2d, 2185-base, and 2e, 2138-base) (Figure 1A). Although three forms of type 1 mRNAs (1a, 1b, and 1c) have been identified previously (Aburatani et al., 1997), we could not clone type 1c cDNA shown on the gel (Figure 2, B and C). Instead, we identified four of new type 2 mRNAs (2b, 2c, 2d, and 2e), in addition to the type 2a mRNA identified previously (Figure 2, B and C). Type 2b or 2c mRNAs are generated by skipping exons 5 and 6, or exons 4 to 6, respectively, whereas type 2d carries a 100-base insertion and type 2e a 53-base insertion between exons 6 and 8. The former is composed of the 53-base of the latter and an additional 47-base sequence, which have not been identified in human or mouse genomic sequences.
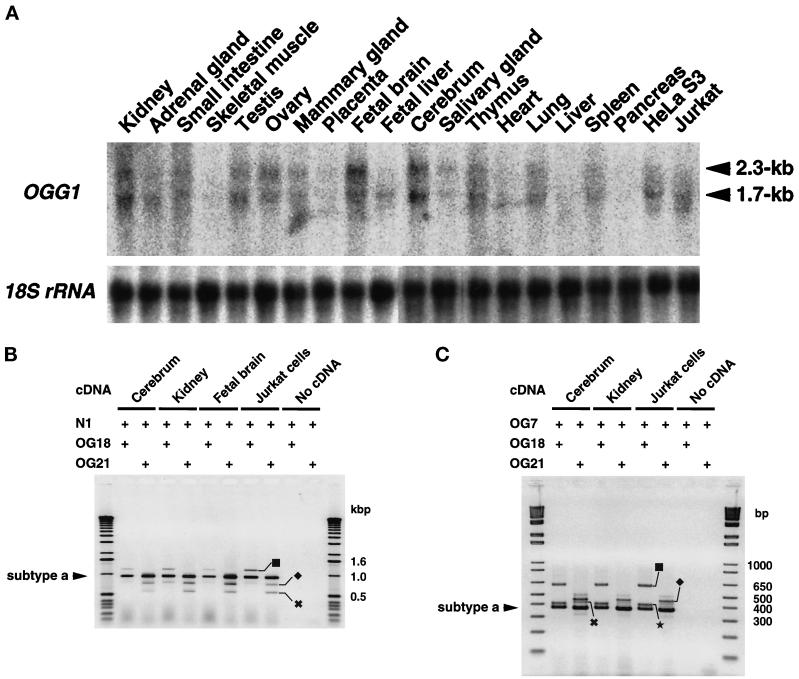
Figure 2.
Expression of OGG1 mRNAs in various human tissues and cell lines. (A) Northern blot analysis. Total RNAs (16 μg each) extracted from various human tissues and Jurkat and HeLa S3 cells were electrophoresed, transferred onto nitrocellulose membrane, and probed with a 32P-labeled 1891-bp fragment of type 2a cDNA. (B) RT-PCR analysis. cDNAs were synthesized from total RNA prepared from cerebrum, kidney, fetal brain, and Jurkat cells, using the oligo(dT)18 primer. The cDNAs were amplified using a common 5′ primer (N1) and 3′ specific primer for each type of OGG1 mRNA (type 1: OG18; type 2: OG21). PCR products were analyzed by 1% agarose gel electrophoresis. The presence of the primers in the reaction mixture is shown by +. ▪, Type 1b; ♦, type 2b; ✖, type 2c mRNA. (C) RT-PCR analysis. The cDNAs were amplified using a common 5′ primer (OG7) and 3′ specific primer for each type of OGG1 mRNA (type 1: OG18; type 2: OG21). PCR products were analyzed by 2% agarose gel electrophoresis. ▪, Type 1b; ♦, type 2d; ✖, type 2e mRNA; ★, a 0.4-kbp band that may correspond to the PCR product form type 1c mRNA.
OGG1 polypeptides encoded by these mRNAs share the first 190 aa in common, which are encoded by exons 1 through 3, and each has a unique C-terminal region, except that polypeptides encoded by type 2a and 2b mRNAs share the common C-terminal 108 aa (Figure 1B). Five polypeptides (OGG1–1a, 1b, 2a, 2d, and 2e) carry the HhH-PVD motif that seems to be essential for 8-oxoG DNA glycosylase activity (Girard et al., 1997; Nash et al., 1997). It has been shown that transiently expressed OGG1–1a, 1b, 1c, and 2a proteins were localized in mitochondria and only OGG1–1a among them was also translocated into nuclei of Cos7 cells (Takao et al., 1998), and a putative mitochondrial targeting signal (MTS) has been assigned at the common N-terminal region (Rosenquist et al., 1997; Takao et al., 1998). We predicted using a computational program, MitoProt II (Claros and Vincens, 1996), that the MTS, which is relatively poor, consists of the residues 9 to 26 at the common N-terminal region of OGG1 proteins and may be processed at residue 23 (W) after being translocated into mitochondria, as shown in Figure 1C. Among all the polypeptides, only OGG1–1a possesses NLS at the C-terminal end.
Expression of OGG1 mRNAs in Various Human Tissues
We examined levels of OGG1 mRNA in various human tissues and cell lines, using Northern blot analysis (Figure 2A). Two major bands corresponding to 2.3-kb and 1.7-kb mRNAs were detected in almost all the samples examined; however, radioactivities of the bands quantified using an image analyzer varied considerably. The largest amounts of OGG1 mRNAs were found in both adult and fetal brain, and levels of both 2.3-kb and 1.7-kb mRNAs were almost equal in the brain tissues. Although a relatively large amount of each mRNA was found in kidney, thymus, testis, ovary, and lung, levels of the 1.7-kb mRNA were higher than those of the 2.3-kb mRNA in these tissues, which was more evident in Jurkat and HeLa S3 cells.
RNAs from adult brain, kidney, fetal brain, and Jurkat cells were also analyzed by RT-PCR, using two sets of type-specific primers (Figure 2B). From all the samples, three bands (1.2, 1.0, and 0.8 kbp) were amplified with the set of primers for type 1 mRNAs, and sequence analysis of cloned fragments obtained from Jurkat cells revealed that two of them correspond to PCR products from type 1a (972 bp) and 1b (1216 bp) mRNA, respectively. The 0.8-kbp band may not correspond to the type 1c mRNA reported because size is expected to be 989 bp. On the other hand, RT-PCR with the set of primers for type 2 mRNAs also yielded three major bands (1.0, 0.8, and 0.6 kbp) from all samples, and their sequences indicate that each corresponds to the PCR product from type 2a (971 bp), 2b (770 bp), and 2c (588 bp) mRNA, respectively. For type 2d and 2e mRNAs, 1071-bp and 1024-bp PCR fragments were expected to be amplified, and a few clones for each were identified among 20 clones for type 2 cDNA obtained from Jurkat cells. To further confirm the results obtained by cDNA cloning, we performed RT-PCR with two more sets of specific primers, as shown in Figure 2C. As well as the PCR products from type 1a (374 bp) and 1b (618 bp), a 0.4-kbp band that may correspond to the product (391 bp) from type 1c mRNA was amplified in any sample examined, using the primers OG7 and OG18. With a set of primers (OG7 and OG21) for type 2 mRNA, bands corresponding to type 2a (373 bp), 2d (473 bp), and 2e (420 bp) with two more unidentified products were apparently amplified.
We concluded that in all the human tissues or cell lines analyzed, at least seven types of alternatively spliced OGG1 mRNAs are formed, that types 1a and 2a are major transcripts from the OGG1 gene, and that those correspond to the 1.7-kb and 2.3-kb mRNAs, respectively (Figure 2A).
Quantitative Detection of OGG1 Protein in Isolated Mitochondria from Human Cells
For immunological detection of human OGG1 protein, rabbit polyclonal antibodies were raised against the C-terminal region of OGG1–2a (anti–2a-CT) or a region containing the HhH-PVD motif (anti–HhH-PVD), and were purified using antigen-affinity columns (Figure 1B). Western blot analysis, in combination with 125I-labeled protein A, demonstrated that anti–2a-CT could specifically detect subnanograms of the recombinant OGG1–2a expressed in E. coli (Figure 3A, left panel), whereas anti–HhH-PVD required more than 10 ng of the recombinant OGG1–1a and -2a to give detectable signals (our unpublished results).
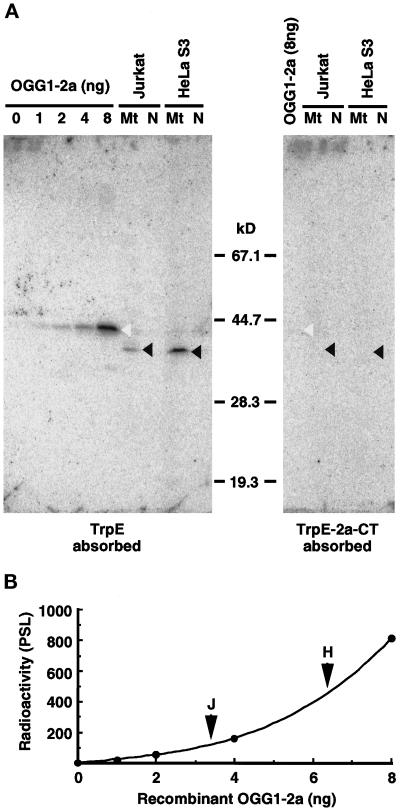
Figure 3.
Detection of the authentic OGG1 protein in human cell lines by Western blotting. (A) Western blotting. Various amounts of the recombinant OGG1–2a expressed in E. coli and isolated mitochondria (Mt) and nuclei (N) (equivalent to 100 μg of protein) from Jurkat and HeLa S3 cells were subjected to Western blot analysis using anti–2a-CT, which was preabsorbed by incubation with an immobilized nonspecific antigen (TrpE) or specific antigen (TrpE-2a-CT), respectively. Blots were probed with 125I-labeled protein A to detect bound antibody and scanned with a Bio-Image analyzer, BAS2000. Open arrowheads indicate bands corresponding to p43, and closed arrowheads indicate bands corresponding to p40, detected by the antibodies preabsorbed with TrpE. (B) Quantitative Western blot analysis of OGG1–2a. Correlation between the amounts of the recombinant OGG1–2a and the radioactivity measured was shown in the plot. Arrowheads indicate radioactivity for the band of p40 detected in each mitochondrial fraction (100 μg of protein) from Jurkat and HeLa S3 cells, respectively.
Isolated nuclei and mitochondria prepared from Jurkat and HeLa S3 cells were subjected to Western blot analysis. Although no band was detected in either fraction using the anti–HhH-PVD (our unpublished results), a single major band corresponding to a 40-kDa polypeptide (p40) was detected only in mitochondrial fractions from both cell lines, using the anti–2a-CT (Figure 3A, left panel, lane Mt).
The recombinant OGG1–2a (424 aa, calculated molecular mass 47,206) was detected as a band corresponding to a 43-kDa polypeptide (p43) in the Western blots (Figure 3A, left panel). To confirm that p40 in the mitochondrial fractions is specifically detected by the anti–2a-CT, we used anti–2a-CT preabsorbed with TrpE-2a-CT–Sepharose in Western blot analysis. With this preparation, neither p40 nor p43 was detected on the blot, which means that p40 shares the same epitope with the C-terminal region of the recombinant p43 (Figure 3A, right panel).
The amount of p40 in mitochondrial fractions of Jurkat and HeLa S3 cells was estimated to be ∼3.4 and 6.2 ng per 100 μg of total protein of each mitochondrial fraction; values were less than the limit of detection by the anti–HhH-PVD (Figure 3B).
Identification of the Nuclear Form of OGG1 Protein in Human Cells
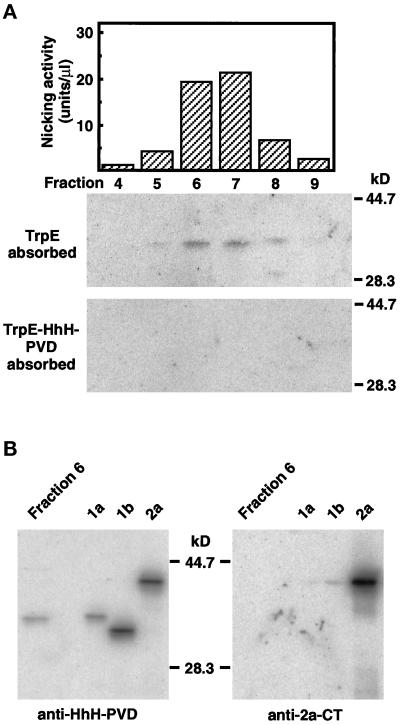
To obtain evidence for the existence of the nuclear form of OGG1 protein(s) in human cells, we partially purified an enzyme(s) that introduces a nick adjacent to 8-oxoG in double-stranded oligonucleotides in isolated nuclei prepared from Jurkat cells (Table 1). Such an enzyme was eluted from a RESOURCE S column (Figure 4A, top), and Western blot analysis revealed that the purified fractions contained a 36-kDa polypeptide (p36), which was detected using the anti–HhH-PVD pretreated on TrpE–Sepharose but not on TrpE-HhH-PVD–Sepharose (Figure 4A, bottom panels). On SDS-PAGE, p36 in the partially purified fraction showed the same migration as the recombinant OGG1–1a but not OGG1–1b, as detected using anti–HhH-PVD but not by using anti–2a-CT (Figure 4B). These results demonstrate that the nicking activity in nuclear extract from Jurkat cells is attributed to the authentic OGG1–1a (p36).
Table 1.
Purification of human 8-oxoguanine DNA glycosylase from Jurkat cells
| Fraction | Protein (mg) | Specific activity (× 103 U/mg) | Fold purification |
|---|---|---|---|
| Crude nuclear extract | 506 | 0.059 | 1 |
| Phosphocellulose (P11) | 25.5 | 4.789 | 81 |
| HiLoad 26/60 Superdex 75 pg | 0.745 | 579.7 | 9892 |
| RESOURCE Q | 0.630 | 660.8 | 11276 |
| RESOURCE S | 0.056 | 2135 | 36435 |
Figure 4.
Chromatographic identification of nuclear OGG1 protein in human cells. (A) RESOURCE S column chromatography of 8-oxoG DNA glycosylase and OGG1 protein present in nuclear extracts prepared from Jurkat cells. The extracts were applied to successive column chromatographies, as described in MATERIALS AND METHODS. The flow-through fraction from the RESOURCE Q column that contains activity introducing a nick adjacent to 8-oxoG paired with cytosine in double-stranded oligonucleotides was loaded onto a RESOURCE S column. The active fractions were eluted at 225–350 mM KCl (top). Fractions (70 μl each) eluted from the column were analyzed by Western blotting, using anti–HhH-PVD, which was preincubated with TrpE–Sepharose (middle) or TrpE-HhH-PVD–Sepharose (bottom). (B) Identification of human OGG1–1a in a partially purified preparation of the nuclear 8-oxoG DNA glycosylase. Fraction 6 (70 μl) eluted from RESOURCE S column and recombinant OGG1 proteins expressed in E. coli (OGG1–1a, -1b, and -2a) were subjected to Western blotting, using anti–HhH-PVD and anti–2a-CT.
Differential Intracellular Localization of Recombinant Human OGG1 Proteins
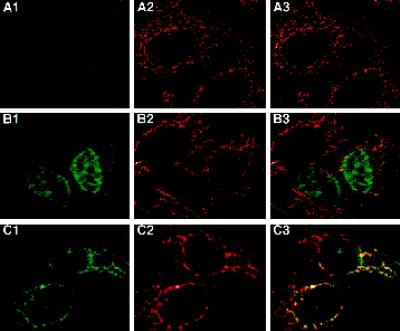
The visualization of OGG1 proteins exogenously expressed in human cells provides supporting evidence for the differential intracellular localization of the authentic human OGG1 proteins (OGG1–1a and -2a). We transfected HeLa MR cells with expression plasmids encoding HA epitope-tagged OGG1–1a or OGG1–2a, and cells were labeled for mitochondria with MitoTracker and stained for the HA epitope. Cells transiently expressing OGG1–1a:HA showed strong immunoreactivity exclusively in their nuclei (Figure 5, B1) and were not colocalized with signals for MitoTracker (Figure 5, B2 and B3). On the other hand, OGG1–2a:HA was seen in the cytosol on punctuate structures distributed around the nucleus (Figure 5, C1), and the structures were also labeled with MitoTracker (Figure 5, C2 and C3). These results support the notion that OGG1–1a is preferentially localized in the nucleus, whereas OGG1–2a is mainly localized in mitochondria in human cells.
Figure 5.
Different intracellular localizations of exogenous OGG1–1a and -2a transiently expressed in HeLa MR cells. The cells were transfected with pcDEBΔ (A), pcDEBΔ::OGG1–1a:HA (B), or pcDEBΔ::OGG1–2a:HA (C) and incubated in the presence of MitoTracker. After being fixed, the cells were subjected to immunostaining with the combination of anti-HA (12CA5) and Cy5-labeled anti-mouse IgG. Fluorescence of Cy5 (A1–C1) and MitoTracker (A2–C2) are shown in green and red, respectively. In merged images (A3–C3), colocalized signals for Cy5 and MitoTracker are in yellow.
Mitochondrial Localization of Human OGG1 Protein and Its Mitochondrial Targeting Signal
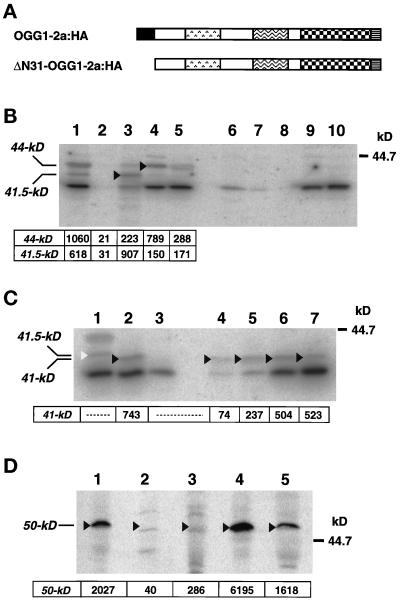
We established a cell line MR-2a:HA stably expressing OGG1–2a:HA. On Western blot analysis with anti–2a-CT, 44-kDa (p44) and 41.5-kDa (p41.5) fusion polypeptides were detected in whole-cell extracts from MR-2a:HA cells (Figure 6B, lane 1). No specific band was detected in MR-V (Figure 6B, lane 6) because the original cell line produces a much lower level of the authentic p40, which is under the limit of detection by anti–2a-CT.
Figure 6.
Intracellular localization of OGG1–2a:HA and ΔN31-OGG1–2a:HA in HeLa MR cells. (A) Structures of OGG1–2a:HA and ΔN31-OGG1–2a:HA. The latter lacks the N-terminal 30 amino acid residues (aa 2–31) of the former. To each C-terminal end, the HA epitope was attached. HeLa MR cells expressing each protein stably were established and designated MR-2a:HA (OGG1–2a:HA) and MR-ΔN31:HA (ΔN31-OGG1–2a:HA), respectively. (B) Subcellular distribution of OGG1–2a:HA. Proteins of the whole-cell extracts (lanes 1 and 6: 40 μg), nuclear fractions (lanes 2 and 7: 20 μg), mitochondrial fractions (lanes 3 and 8: 20 μg), microsomal fractions (lanes 4 and 9: 20 μg), and cytosol fractions (lanes 5 and 10: 20 μg) prepared from MR-2a:HA (lanes 1–5) or MR-V cells (lanes 6–10) to which pcDEBΔ was introduced were separated on 12.5% SDS-PAGE and subjected to Western blotting with anti–2a-CT. Arrows and arrowheads indicate 44- and 41.5-kDa OGG1–2a:HA polypeptides. Radioactivities of bands corresponding to these polypeptides are shown. (C) Subcellular distribution of ΔN31-OGG1–2a:HA. Proteins of the whole-cell extracts (lanes 1–3: 40 μg), nuclear fractions (lane 4: 20 μg), mitochondrial fraction (lane 5: 20 μg), microsomal fraction (lane 6: 20 μg), and cytosol fraction (lane 7: 20 μg) prepared from MR-2a:HA (lane 1), MR-V cells (lane 3), and MR-ΔN31:HA cells (lanes 2, 4–7) were separated on 12.5% SDS-PAGE and subjected to Western blotting with anti–2a-CT. An open arrowhead indicates the 41.5-kDa OGG1–2a:HA polypeptide, and closed arrowheads indicate 41-kDa ΔN31-OGG1–2a:HA polypeptides. Radioactivities of bands corresponding to the latter are shown. (D) Distribution of anti-HA–reacting p50 in subcellular fractions. To estimate contamination of microsomes into other fractions, amounts of p50 in each subcellular fraction (100 μg protein) prepared from MR-2a:HA cells were determined by Western blotting. Lane 1: the whole-cell extract; lane 2: nuclear fraction; lane 3: mitochondrial fraction; lane 4: microsomal fraction; and lane 5: cytosol fraction. Radioactivities for p50 are shown (means from 4 independent experiments).
To examine the intracellular localization of the OGG1–2a:HA more precisely, subcellular fractionation of MR-2a:HA cells was performed, and we estimated the amount of OGG1–2a:HA in each fraction by Western blotting using anti–2a-CT (Figure 6B). In whole-cell extracts, the relative ratio of the amount of p44 to that of p41.5 was 5:3 (Figure 6B, lane 1). On the other hand, the ratio in mitochondrial fraction was 2:9 (Figure 6B, lane 3), indicating that p41.5 accumulates preferentially in mitochondria. The amount of OGG1–2a:HA in mitochondria was estimated to be 20–30 times higher than that of the authentic p40 (our unpublished results). On the contrary, neither p44 nor p41.5 was detected in nuclear fractions (Figure 6B, lane 2). It is noteworthy that a substantial amount of p44 was detected in the microsomal fraction (Figure 6B, lane 4).
By measuring activities of the mitochondrial succinate–cytochrome c reductase and cytosolic lactate dehydrogenase and the amount of the microsomal protein (p50) reactive with anti-HA (Figure 6D) in each subcellular fraction (Table 2), we confirmed that the mutual contamination of nuclear, mitochondrial, or microsomal proteins into other fractions was only a few percentage points or less.
Table 2.
Relative specific content of marker proteins in subcellular fractions
| Marker | Whole | Nuclei | Mitochondria | Microsomes | Cytosol |
|---|---|---|---|---|---|
| Succinate–cytochrome c reductase | 5.6 | ND | 100a | 0.2 | ND |
| Lactate dehydrogenase | 25.3 | 0.3 | 2.3 | 5.1 | 100b |
| Anti-HA–reacting protein (p50) | 32.7 | 0.1 | 4.6 | 100c | 26.1 |
All the fractions are derived from MR-2a:HA cells. Each value is a mean of four independent experiments. ND, not detected.
172.8 μmol · min−1 · mg−1 of protein.
1166.0 μmol · min−1 · mg−1 of protein.
1.9 photo-stimulated luminescence (PSL)/μg of protein.
To examine the role of N-terminal region of OGG1–2a, we established MR-ΔN31:HA cells stably expressing ΔN31-OGG1–2a:HA that lacks the N-terminal region (aa from Pro2 to Ser31) from OGG1–2a:HA (Figure 6A). Only the 41-kDa polypeptide (p41) was detected in whole-cell extract by Western blotting using anti–2a-CT (Figure 6C, lane 2), and also in all subcellular fractions, even in the nuclear fraction (Figure 6C, lanes 4–7). Thus, ΔN31-OGG1–2a:HA lost the potential of preferential targeting into mitochondria, indicating that the N-terminal region indeed functions as MTS.
Because on SDS-PAGE the ΔN31-OGG1–2a:HA (p41) migrated faster than did p41.5 detected in mitochondrial fraction from MR-2a:HA cells (Figure 6C, lanes 1 and 2), it seems likely that OGG1–2a, after being translocated into mitochondria, is processed at a residue closer to the N terminus than to the residue Pro31.
Inner Membrane–associated Localization of Human OGG1 Protein in Mitochondria
Isolated mitochondria from MR-2a:HA cells were sonicated and separated into the supernatant and pellet by centrifugation, the former representing the soluble fraction and the latter representing the insoluble membrane fraction, respectively. Recovery of the mitochondrial p41.5 into each fraction was examined by Western blotting with anti–2a-CT (Table 3). The relative specific content of p41.5 in each fraction was 74.8 and 43.4% of that in the total homogenate of mitochondria, respectively. With the fractionation procedure applied here, the matrix protein MTH1 was mostly found in the supernatant and the specific content in the fraction was more than four times higher than that in the total homogenate, whereas the succinate–cytochrome c reductase, tightly associated with the inner membrane, was found exclusively in the pellet with the relative specific content of 134.7%.
Table 3.
Membrane-associated localization of human OGG1 protein in mitochondria
| Protein recovered (% specific content)
|
|||
|---|---|---|---|
| Whole mitochondria | Soluble fraction | Membrane fraction | |
| Mitochondrial OGG1-2a:HA | 100a | 74.8 | 43.4 |
| Bcl2 | 100b | 22.5 | 53.1 |
| MTH1 | 100c | 413.8 | 18.6 |
| Succinate–cytochrome c reductase | 100d | 7.2 | 134.7 |
17.5 PSL/μg of protein.
204.4 PSL/μg of protein.
94.5 PSL/μg of protein.
172.8 μmol · min−1 · mg−1 of protein.
We assumed that the succinate–cytochrome c reductase detected in the insoluble pellet represents the net recovery of the membrane fraction, and that MTH1 in the supernatant represents the net recovery of the soluble fraction (Kang et al., 1995). On the basis of this assumption, we estimated that 32% of the total amount of p41.5 was fractionated into the membrane fraction, and 18% of that was fractionated into the soluble fraction. In the case of Bcl2, which is reportedly a membrane-anchored protein (Nguyen et al., 1993), 40% of the protein was fractionated into the membrane fraction, as compared with p41.5, whereas 5% of that went into the soluble fraction. These results suggest that the p41.5 present in mitochondria may be in its matrix, with weak interaction with the inner membrane.
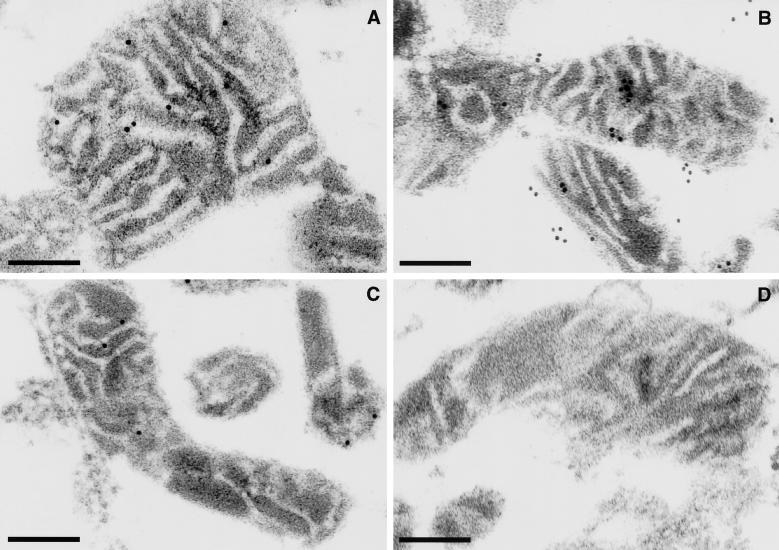
To obtain direct evidence for the membrane-associated localization of p41.5 in mitochondria, we prepared thin sections from fixed mitochondrial pellets prepared from MR-2a:HA, MR-ΔN31:HA, and MR-V cells and examined the localization of p41.5 by electron microscopic immunocytochemistry, using anti–2a-CT in combination with protein A–gold (Figure 7). In all mitochondrial sections from MR-2a:HA cells, more than 10 specific signals were observed in low electron-dense areas (Figure 7A), which correspond to the inner membrane of mitochondria. Although a similar number of specific signals were observed in each section from MR-ΔN31:HA cells, approximately half were localized in electron-dense areas corresponding to the matrix, and the other half were seen to be associated with the outside of each mitochondrion (Figure 7B), thus confirming the results obtained in the case of subcellular fractionation (Figure 6C).
Figure 7.
Submitochondrial localization of human OGG1 proteins, determined by electron microscopic immunocytochemistry. After each mitochondria was isolated from MR-2a:HA (A), MR-ΔN31:HA (B), and MR-V cells (C), thin sections (∼0.1 μm) were prepared for electron microscopic immunocytochemistry with anti–2a-CT in combination with protein A–gold. Anti–2a-CT preabsorbed with TrpE-2a-CT–Sepharose was applied to mitochondria from MR-V cells (D). Bars, 0.2 μm.
In one of seven mitochondrial sections prepared from MR-V cells carrying a control plasmid, we detected a few signals in low electron-dense areas (Figure 7C). Thus the increased number of signals detected in each section from MR-2a:HA or MR-ΔN31:HA demonstrated that these signals represent exogenously expressed OGG1–2a derivatives. When anti–2a-CT preabsorbed by TrpE-2a-CT–Sepharose was applied to sections from MR-V cells, no signal was observed (Figure 7D), indicating that a few signals in mitochondria from MR-V cells represent the authentic p40 polypeptide identified by Western blotting using the same antibody.
These results taken together suggest that OGG1–2a is preferentially localized in mitochondria and only weakly associating with the inner membrane and that the MTS at its N-terminal region is essential for proper mitochondrial localization.
Nuclear Localization Signal in Human OGG1 Protein Suppresses Mitochondrial Targeting of the Protein
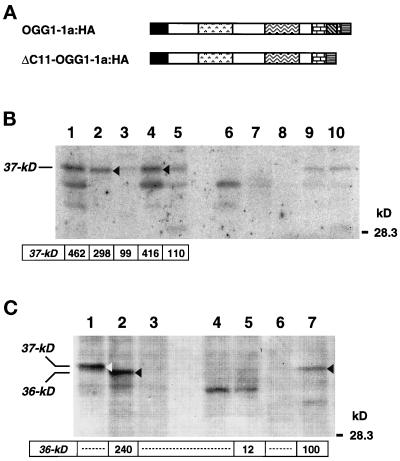
We established two more cell lines, one expressing OGG1–1a:HA (MR-1a:HA) and the other expressing ΔC11-OGG1–1a:HA lacking the putative C-terminal NLS (MR-ΔC11:HA) (Figure 8A). Subcellular fractionation of these cell lines was performed in combination with Western blotting of each protein. A 37-kDa polypeptide (p37) was detected in whole-cell extracts from MR-1a:HA cells, and the majority of p37 was fractionated into nuclear and microsomal fractions (Figure 8B, lanes 2 and 4). A small amount of p37 was also detected in mitochondrial fractions; however, a processed form of p37, expected to be 35.5 kDa, was not detected (Figure 8B, lane 3). As expected, a 36-kDa polypeptide (p36) was detected in the whole-cell extract from MR-ΔC11:HA cells (Figure 8C, lane 1), and the majority of p36 was detected in mitochondrial but not in nuclear fractions (Figure 8C, lanes 5 and 7). Again, a processed form of p36, expected to be 34.5 kDa, was not detected. Therefore, the 11 aa residues at the C-terminal end of OGG1–1a function as NLS and also suppress efficient mitochondrial targeting of the protein.
Figure 8.
Intracellular localization of OGG1–1a:HA and ΔC11-OGG1–1a:HA in HeLa MR cells. (A) Structures of OGG1–1a:HA and ΔC11-OGG1–1a:HA. The latter lacks the last 11 amino acids of OGG1–1a corresponding to the NLS. To each C-terminal end, the HA epitope was attached. HeLa MR cells expressing each protein stably were established and designated MR-1a:HA (OGG1–1a:HA) and MR-ΔC11:HA (ΔC11-OGG1–1a:HA), respectively. (B) Subcellular distribution of OGG1–1a:HA. Proteins of the whole-cell extracts (lanes 1 and 6: 40 μg), nuclear fractions (lanes 2 and 7: 20 μg), mitochondrial fractions (lanes 3 and 8: 20 μg), microsomal fractions (lanes 4 and 9: 20 μg), and cytosol fractions (lanes 5 and 10: 20 μg) prepared from MR-1a:HA (lanes 1–5) or MR-V cells (lanes 6–10) were separated on 12.5% SDS-PAGE and subjected to Western blotting with anti–HhH-PVD. An arrow and arrowheads indicate 37-kDa OGG1–1a:HA polypeptides. Radioactivities for bands corresponding to these polypeptides are shown. (C) Subcellular distribution of ΔC11-OGG1–1a:HA. Proteins of the whole-cell extracts (lanes 1–3: 40 μg), nuclear fractions (lanes 4 and 5: 20 μg), and mitochondrial fraction (lanes 6 and 7: 20 μg) prepared from MR-1a:HA (lane 1), MR-V (lanes 3, 4, and 6), and MR-ΔC11:HA cells (lanes 2, 5, and 7) were separated on 12.5% SDS-PAGE and subjected to Western blotting with anti–HhH-PVD, in combination with alkaline phosphatase-conjugated mouse anti-rabbit IgG. Bound IgG was detected with 5-bromo-4-chloro-3-indolyl-1-phosphate and nitro blue tetrazolium. An open arrowhead indicates the 37-kDa OGG1–1a:HA polypeptide, and closed arrowheads indicate 36-kDa ΔC11-OGG1–1a:HA polypeptides. Shown are intensities of bands corresponding to the ΔC11-OGG1–1a:HA, measured using NIH image software.
DISCUSSION
Our main conclusions are that at least two isoforms of human OGG1 proteins (OGG1–1a and OGG1–2a) encoded by alternatively spliced mRNAs differentially contribute to the base excision repair pathways for 8-oxoG generated in both nuclear and mitochondrial DNAs, and that the NLS at the C-terminal end of OGG1–1a dominates the MTS found at the N-terminal ends of all isoforms. The C-terminal region in OGG1–2a as well as the MTS seems to be required for appropriate localization and processing in mitochondria.
We identified seven different forms of human OGG1 cDNAs, with four newly isolated ones, and further demonstrated that in most human tissues, including brain, both type 1a and 2a are major OGG1 mRNAs, whereas all others with a few unidentified ones are also expressed, albeit at much lower levels. Each OGG1 mRNA is expected to encode a polypeptide sharing the first 190 aa in common and a unique C-terminal region. Among them, type 1a, 1b, 1c, and 2a proteins have been shown to possess activities of 8-oxoG DNA glycosylase and AP lyase, which catalyzes β-elimination to introduce a nick at the 3′ side of AP site, and/or to suppress the mutator phenotype of mutM− mutY− strain of E. coli (Aburatani et al., 1997; Arai et al., 1997; Bjoras et al., 1997; Radicella et al., 1997; Roldán-Arjona et al., 1997). These results indicate that the conserved region (aa 1–316) among the four isoforms, which includes the HhH-PVD motif, is important for repair functions, although there is no biochemical evidence that type 1c protein indeed possesses the catalytic activity (Lu et al. 1997). Because both OGG1–2d and -2e retain this region, they may have repair activities, whereas OGG1–2b and -2c, lacking a part or an entire region of the HhH-PVD motif, may not participate in the usual repair pathway for 8-oxoG. The biochemical activities and biological significance of these proteins need to be elucidated.
Although DNA glycosylase activity excising 8-oxoG in DNA or a DNA endonuclease activity incising DNA containing 8-oxoG have been detected in nuclear extracts from human cells (Bessho et al., 1993; Nagashima et al., 1997), there was no evidence that these enzymes are indeed encoded by the human OGG1 gene, and none of the OGG1 isoforms were seen to exist in human cells. Using antibodies that recognize the C-terminal region of OGG1–2a (anti–2a-CT), we detected for the first time a 40-kDa polypeptide (p40) in mitochondrial but not in nuclear fractions of cultured human cells. Among seven isoforms of OGG1 proteins, OGG1–2a and -2b have the same epitope to which the anti–2a-CT reacts, and their predicted molecular masses are 47,206 and 39,684, respectively. Judging from the facts that the recombinant OGG1–2a expressed in E. coli migrates as a 43-kDa polypeptide on SDS-PAGE and that HA epitope-tagged OGG1–2a (p44) expressed in HeLa cells was processed into p41.5 in mitochondria, we conclude that the p40 is the final form of OGG1–2a in mitochondria. The finding on RT-PCR analysis that type 2a mRNA is major among type 2 mRNAs in Jurkat cells supports this conclusion.
The MitoProt II (Claros and Vincens, 1996) can predict an amphipathic helical structure for a given MTS and its cleavage site in mitochondria. Using this program, we analyzed the N terminus of OGG1 protein and obtained predictions that the region from the 9th to 26th residues can form an amphipathic helical structure with values of 6.57 for hydrophobic moment and 4.33 for maximum hydrophobicity, indicating a relatively poor MTS, and that the MTS can be cleaved at the 23rd tryptophan residue after being translocated into mitochondria. A deletion of the aa 2–31 from the N-terminal end of OGG1–2a abolished both its preferential accumulation in mitochondria and processing, thus providing support for these predictions.
A part of ΔN31-OGG1–2a:HA lacking the MTS was translocated into mitochondrial matrix, and the rest remained outside of mitochondria in association with the outer membrane, suggesting that the deletion mutant still has some degree of the affinity for mitochondria. Interestingly, ΔC11-OGG1–1a:HA, lacking the NLS but with the MTS, lost its capability of being translocated into nuclei yet gained an ability to target itself specifically to mitochondria, although the mutant polypeptide was not processed in mitochondria. Because ΔC11-OGG1–1a:HA can be considered as a mutant form of OGG1–2a:HA that lacks the unique C-terminal region, we concluded that the C-terminal region of OGG1–2a is essential for proper localization and processing in mitochondria as well as the MTS essential for its proper targeting into mitochondria. Furthermore, it must be noted that we could not establish HeLa MR cells expressing a truncated form of OGG1 protein (aa 1–316), corresponding to the common region between OGG1–1a and -2a, thus indicating that parts of the C-terminal regions in the two OGG1 isoforms are essential for stable expression as well as for intracellular localization (our unpublished results).
It has been shown that a precursor polypeptide for uracil DNA glycosylase (UNG), with an MTS at the N-terminal region, is translocated into mitochondria and processed. With deletion of its MTS the protein was translocated into nuclei (Slupphaug et al., 1993), indicating that the potential to be translocated into nuclei is suppressed by MTS. As pointed out by Slupphaug et al. (1993), the most N-terminal signal in polypeptides with multiple signals for organella targeting usually dominates; however, such is not the case for OGG1 proteins. Analysis of the MTS of human UNG by the MitoProt II revealed that the MTS forms an amphipathic helix with values of 11.18 for hydrophobic moment and 4.64 for maximum hydrophobicity, indicating that the MTS in OGG1 protein is much less functional than that for UNG. Thus, the MTS of OGG1–1a cannot dominate the NLS at its C-terminal end and an associated function of the C-terminal region of OGG1–2a is required for proper mitochondrial localization.
Electron microscopic immunocytochemistry revealed that OGG1–2a resides on the inner membrane of mitochondria, and submitochondrial fractionation indicated that the OGG1–2a in mitochondria weakly associates with a membrane. Because the unique C-terminal region of OGG1–2a consists of two distinct regions—one is the N-terminal–sided acidic region (aa from Ile345 to Asp381) and the other is the C-terminal–sided hydrophobic one (the last 20 residues)—we speculate that the latter mediates a hydrophobic interaction between the OGG1–2a and the inner membrane of mitochondria. This affinity may be comparable to that in the association of Bcl2 with the mitochondrial membrane (Nguyen et al., 1993). Recombinant OGG1–2a, but not OGG1–1a, expressed in E. coli, was mostly insoluble, indicating that this C-terminal region contributes to the insolubility as a result of its hydrophobicity. On the other hand, the acidic region with a value of 3.63 for a local isoelectric pH may possibly provide a protein–protein interacting surface for another component(s), which could then be involved in its stabilization and/or the base excision repair pathway in mitochondria, as discussed below.
Because it has been shown that mitochondrial DNAs exist in association with the inner membrane (Albring et al., 1977), it seems likely that some OGG1–2a molecules in mitochondria colocalize with the DNAs. Indeed, the number of authentic OGG1–2a molecules in mitochondria is comparable to findings in the mitochondrial DNA: a few thousand molecules per cell. Recently, activities of an AP endonuclease, DNA ligase III, and DNA polymerase γ were fractionated from mitochondria of Xenopus oocytes (Pinz and Bogenhagen, 1998). It is likely that there is a repair complex containing the entire machinery essential for base excision repair and that it associates with the inner membrane and/or mitochondrial DNAs as expected for OGG1–2a; thus efficient repair by the machinery would contribute to maintaining the functions and integrity of mitochondrial DNAs, even if threatened by attacks of ROS produced during oxidative phosphorylation.
Because we did not detect OGG1 protein(s) in nuclear extracts by simple Western blotting, we partially purified an enzyme activity that specifically cleaves phosphodiester bonds adjacent to 8-oxoG paired with cytosine in double-stranded oligonucleotides from nuclear extract of Jurkat cells. The enzyme preparation from Jurkat cells also contained an additional activity that directly cleaves phosphodiester bonds at AP sites, namely AP lyase activity. Both activities preferred cytosine to thymine as an opposite base to each damage, but both showed little activity toward each damage opposite to guanine or adenine (our unpublished results). Substrate specificities as well as chromatographic behaviors of the two activities indicate that the Jurkat enzyme shares its biochemical properties with the 8-oxoG DNA glycosylase partially purified from other human cell lines (Bessho et al., 1993; Nagashima et al., 1997) or with the recombinant OGG1–1a (Aburatani et al., 1997; Bjoras et al., 1997; Lu et al., 1997; Radicella et al., 1997; Roldán-Arjona et al., 1997). With Western blotting, using the anti–HhH-PVD, it was clear that a single 36-kDa polypeptide (p36) reactive to the antibody is copurified with the activities. Because the p36 showed the same migration as the recombinant OGG1–1a and is not reactive to the anti–2a-CT, this is the first detection of authentic OGG1–1a, which is responsible for the major 8-oxoG DNA glycosylase activity in nuclei of human cells. At least two isoforms of OGG1 protein differentially participate in the repair pathway of 8-oxoG in nuclear and mitochondrial genomes in human cells.
Because the mitochondrial function of generating energy is crucial for maintaining life, the integrity of mitochondrial DNA has to be maintained. Accumulation of DNA damage in mitochondrial DNA is considered to cause mitochondrial dysfunction and further enhances production of ROS, thus finally triggering a programmed cell death (for a review see Petit and Kroemer, 1998). This cell death induced by oxidative stress has been implicated as a cause of neuronal or renal degeneration (for reviews see Lin and Flint Beal, 1998; Moraes, 1998). The high level of expression of the OGG1 gene in brain or kidney may well contribute to protection of these cells from such oxidative stress.
ACKNOWLEDGMENTS
We extend special thanks to Drs. M. Furuichi, K. Sakumi, Y. Tominaga, T. Muta, T. Tsuzuki, and M. Sekiguchi for helpful discussion, and to M. Ohara for useful comments on this manuscript. This work was supported by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan, and Showa Shell Sekiyu Foundation for Promotion of Environmental Research.
Abbreviations used:
- aa
amino acid
- AP
apurinic/apyrimidinic
- FAM
N-(3-fluoranthyl)maleimide
- HA
hemagglutinin
- HhH
helix-hairpin-helix
- MTS
mitochondrial targeting signal
- 8-oxoG
8-oxoguanine
- ROS
reactive oxygen species
- UNG
uracil DNA glycosylase
REFERENCES
- Aburatani H, et al. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 1997;57:2151–2156. [PubMed] [Google Scholar]
- Albring M, Griffith J, Attardi G. Association of a protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replication. Proc Natl Acad Sci USA. 1977;74:1348–1352. doi: 10.1073/pnas.74.4.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants, and the degenerative diseases of aging. Proc Natl Acad Sci USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai K, Morishita K, Shinmura K, Kohno T, Kim SR, Nohmi T, Taniwaki M, Ohwada S, Yokota J. Cloning of a human homolog of the yeast OGG1 gene that is involved in the repair of oxidative DNA damage. Oncogene. 1997;14:2857–2861. doi: 10.1038/sj.onc.1201139. [DOI] [PubMed] [Google Scholar]
- Au KG, Clark S, Miller JH, Modrich P. Escherichia coli mutY gene encodes an adenine glycosylase active on G-A mispairs. Proc Natl Acad Sci USA. 1989;86:8877–8881. doi: 10.1073/pnas.86.22.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly V, Verly WG, O’Connor T, Laval J. Mechanism of DNA strand nicking at apurinic/apyrimidinic sites by Escherichia coli [formamidopyrimidine] DNA glycosylase. Biochem J. 1989;262:581–589. doi: 10.1042/bj2620581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmeyer HU, Bernt E, Hess B. Methods of Enzymatic Analysis. H.U. Bergmeyer, New York: Academic Press; 1965. Lactic dehydrogenase; pp. 736–741. [Google Scholar]
- Bessho T, Tano K, Kasai H, Ohtsuka E, Nishimura S. Evidence for two DNA repair enzymes for 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) in human cells. J Biol Chem. 1993;268:19416–19421. [PubMed] [Google Scholar]
- Bjoras M, Luna L, Johnsen B, Hoff E, Haug T, Rognes T, Seeberg E. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J. 1997;16:6314–6322. doi: 10.1093/emboj/16.20.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera M, Nghiem Y, Miller JH. mutM, a second mutator locus in Escherichia coli that generates G·C→T·A transversions. J Bacteriol. 1988;170:5405–5407. doi: 10.1128/jb.170.11.5405-5407.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai JP, Kawate H, Ihara K, Yakushiji H, Nakabeppu Y, Tsuzuki T, Sekiguchi M. Significance of the conserved amino acid sequence for human MTH1 protein with antimutator activity. Nucleic Acids Res. 1997;25:1170–1176. doi: 10.1093/nar/25.6.1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-hydroxyguanine, an abundant form of oxidative DNA damage, causes G → T and A → C substitutions. J Biol Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active RNA from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W. DNA Repair and Mutagenesis. Washington, DC: American Society for Microbiology; 1995. [Google Scholar]
- Furuichi M, Yoshida MC, Oda H, Tajiri T, Nakabeppu Y, Tsuzuki T, Sekiguchi M. Genomic structure and chromosome location of the human mutT homologue gene MTH1 encoding 8-oxo-dGTPase for prevention of A:T to C:G transversion. Genomics. 1994;24:485–490. doi: 10.1006/geno.1994.1657. [DOI] [PubMed] [Google Scholar]
- Girard PM, Guibourt N, Boiteux S. The Ogg1 protein of Saccharomyces cerevisiae: a 7,8-dihydro-8-oxoguanine DNA glycosylase/AP lyase whose lysine 241 is a critical residue for catalytic activity. Nucleic Acids Res. 1997;25:3204–3211. doi: 10.1093/nar/25.16.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa M, Sugiyama S, Hattori K, Takasawa M, Ozawa T. Age-associated damage in mitochondrial DNA in human hearts. Mol Cell Biochem. 1993;119:95–103. doi: 10.1007/BF00926859. [DOI] [PubMed] [Google Scholar]
- Igarashi H, Tsuzuki T, Kakuma T, Tominaga Y, Sekiguchi M. Organization and expression of the mouse MTH1 gene for preventing transversion mutation. J Biol Chem. 1997;272:3766–3772. doi: 10.1074/jbc.272.6.3766. [DOI] [PubMed] [Google Scholar]
- Ishibashi T, Nakabeppu Y, Sekiguchi M. Artificial control of nuclear translocation of DNA repair methyltransferase. J Biol Chem. 1994;269:7645–7650. [PubMed] [Google Scholar]
- Kakuma T, Nishida J, Tsuzuki T, Sekiguchi M. Mouse MTH1 protein with 8-oxo-7,8-dihydro-2′-deoxyguanosine 5′-triphosphatase activity that prevents transversion mutation: cDNA cloning and tissue distribution. J Biol Chem. 1995;270:25942–25948. doi: 10.1074/jbc.270.43.25942. [DOI] [PubMed] [Google Scholar]
- Kang D, Nishida J, Iyama A, Nakabeppu Y, Furuichi M, Fujiwara T, Sekiguchi M, Takeshige K. Intracellular localization of 8-oxo-dGTPase in human cells, with special reference to the role of the enzyme in mitochondria. J Biol Chem. 1995;270:14659–14665. doi: 10.1074/jbc.270.24.14659. [DOI] [PubMed] [Google Scholar]
- Kang D, Takeshige K, Sekiguchi M, Singh KK. Mitochondrial DNA Mutations in Aging, Disease and Cancer. K.K. Singh, New York: Springer/Verlag; 1998. Introduction; pp. 1–15. [Google Scholar]
- Kasai H, Nishimura S. Hydroxylation of deoxyguanosine at the C-8 position by ascorbic acid and other reducing agents. Nucleic Acids Res. 1984;12:2137–2145. doi: 10.1093/nar/12.4.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebovitz RM, Zhang H, Vogel H, Cartwright J, Jr, Dionne L, Lu N, Huang S, Matzuk MM. Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase-deficient mice. Proc Natl Acad Sci USA. 1996;93:9782–9787. doi: 10.1073/pnas.93.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, et al. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- Lin M, Flint Beal M. Mitochondrial DNA Mutations in Aging, Disease and Cancer. K.K. Singh, New York: Springer/Verlag; 1998. Mitochondrial dysfunction and neurodegenerative diseases; pp. 265–296. [Google Scholar]
- Lu R, Nash HM, Verdine GL. A mammalian DNA repair enzyme that excises oxidatively damaged guanines maps to a locus frequently lost in lung cancer. Curr Biol. 1997;7:397–407. doi: 10.1016/s0960-9822(06)00187-4. [DOI] [PubMed] [Google Scholar]
- Maki H, Sekiguchi M. MutT protein specifically hydrolyzes a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- Michaels ML, Cruz C, Grollman AP, Miller JH. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci USA. 1992;89:7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes CT. Mitochondrial DNA Mutations in Aging, Disease and Cancer. K.K. Singh, New York: Springer/Verlag; 1998. Characteristics of mitochondrial DNA diseases; pp. 167–184. [Google Scholar]
- Nagashima M, Sasaki A, Morishita K, Takenoshita S, Nagamachi Y, Kasai H, Yokota J. Presence of human cellular protein(s) that specifically binds and cleaves 8-hydroxyguanine containing DNA. Mutat Res. 1997;383:49–59. doi: 10.1016/s0921-8777(96)00045-6. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y, Nathans D. A naturally occurring truncated form of FosB that inhibits Fos/Jun transcriptional activity. Cell. 1991;64:751–759. doi: 10.1016/0092-8674(91)90504-r. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y, Oda S, Sekiguchi M. Proliferative activation of quiescent Rat-1A cells by ΔFosB. Mol Cell Biol. 1993;13:4157–4166. doi: 10.1128/mcb.13.7.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash HM, Bruner SD, Schärer OD, Kawate T, Addona TA, Spooner E, Lane WS, Verdine GL. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr Biol. 1996;6:968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- Nash HM, Lu R, Lane WS, Verdine GL. The critical active-site amine of the human 8-oxoguanine DNA glycosylase, hOgg1: direct identification, ablation and chemical reconstitution. Chem Biol. 1997;4:693–702. doi: 10.1016/s1074-5521(97)90225-8. [DOI] [PubMed] [Google Scholar]
- Nghiem Y, Cabrera M, Cupples CG, Miller JH. The mutY gene: a mutator locus in Escherichia coli that generates G·C → T·A transversions. Proc Natl Acad Sci USA. 1988;85:2709–2713. doi: 10.1073/pnas.85.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M, Millar DG, Yong VW, Korsmeyer SJ, Shore GC. Targeting of Bcl-2 to the mitochondrial outer membrane by a COOH-terminal signal anchor sequence. J Biol Chem. 1993;268:25265–25268. [PubMed] [Google Scholar]
- Oda H, Nakabeppu Y, Furuichi M, Sekiguchi M. Regulation of expression of the human MTH1 gene encoding 8-oxo-dGTPase: alternative splicing of transcription products. J Biol Chem. 1997;272:17843–17850. doi: 10.1074/jbc.272.28.17843. [DOI] [PubMed] [Google Scholar]
- Ohtsubo T, Matsuda O, Iba K, Terashima I, Sekiguchi M, Nakabeppu Y. Cloning of AtMMH, an Arabidopsis thaliana ortholog of Escherichia coli mutM gene and characterization its products. Mol Gen Genet. 1998;259:577–590. doi: 10.1007/s004380050851. [DOI] [PubMed] [Google Scholar]
- Petit PX, Kroemer G. Mitochondrial DNA Mutations in Aging, Disease and Cancer. K.K. Singh, New York: Springer/Verlag; 1998. Mitochondrial regulation of apoptosis; pp. 147–165. [Google Scholar]
- Pinz KG, Bogenhagen DF. Efficient repair of abasic sites in DNA by mitochondrial enzymes. Mol Cell Biol. 1998;18:1257–1265. doi: 10.1128/mcb.18.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radicella JP, Dherin C, Desmaze C, Fox MS, Boiteux S. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldán-Arjona T, Wei YF, Carter KC, Klungland A, Anselmino C, Wang RP, Augustus M, Lindahl T. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc Natl Acad Sci USA. 1997;94:8016–8020. doi: 10.1073/pnas.94.15.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenquist TA, Zharkov DO, Grollman AP. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc Natl Acad Sci USA. 1997;94:7429–7434. doi: 10.1073/pnas.94.14.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakumi K, Furuichi M, Tsuzuki T, Kakuma T, Kawabata S, Maki H, Sekiguchi M. Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J Biol Chem. 1993;268:23524–23530. [PubMed] [Google Scholar]
- Schnaitman C, Greenawalt JW. Enzymatic properties of the inner and outer membranes of rat liver mitochondria. J Cell Biol. 1968;38:158–175. doi: 10.1083/jcb.38.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi M. MutT-related error avoidance mechanism for DNA synthesis. Genes Cells. 1996;1:139–145. doi: 10.1046/j.1365-2443.1996.d01-232.x. [DOI] [PubMed] [Google Scholar]
- Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- Shigenaga MK, Hagen TM, Ames BN. Oxidative damage and mitochondrial decay in aging. Proc Natl Acad Sci USA. 1994;91:10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupphaug G, Markussen F-H, Olsen LC, Aasland R, Aarsaether N, Bakke O, Krokan HE, Helland DE. Nuclear and mitochondrial forms of human uracil-DNA glycosylase are encoded by the same gene. Nucleic Acids Res. 1993;21:2579–2584. doi: 10.1093/nar/21.11.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupska MM, Baikalov C, Luther WM, Chiang JH, Wei YF, Miller JH. Cloning and sequencing a human homolog (hMYH) of the Escherichia coli mutY gene whose function is required for the repair of oxidative DNA damage. J Bacteriol. 1996;178:3885–3892. doi: 10.1128/jb.178.13.3885-3892.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tajiri T, Maki H, Sekiguchi M. Functional cooperation of MutT, MutM and MutY proteins in preventing mutations caused by spontaneous oxidation of guanine nucleotide in Escherichia coli. Mutat Res. 1995;336:257–267. doi: 10.1016/0921-8777(94)00062-b. [DOI] [PubMed] [Google Scholar]
- Takao M, Aburatani H, Kobayashi K, Yasui A. Mitochondrial targeting of human DNA glycosylases for repair of oxidative DNA damage. Nucleic Acids Res. 1998;26:2917–2922. doi: 10.1093/nar/26.12.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani M, et al. Genomic structure and chromosomal localization of the mouse Ogg1 gene that is involved in the repair of 8-hydroxyguanine in DNA damage. Mamm Genome. 1998;9:32–37. doi: 10.1007/s003359900675. [DOI] [PubMed] [Google Scholar]
- van der Kemp PA, Thomas D, Barbey R, de Oliveira R, Boiteux S. Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine. Proc Natl Acad Sci USA. 1996;93:5197–5202. doi: 10.1073/pnas.93.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh YC, Chang DY, Masin J, Lu AL. Two nicking enzyme systems specific for mismatch-containing DNA in nuclear extracts from human cells. J Biol Chem. 1991;266:6480–6484. [PubMed] [Google Scholar]