Abstract
P2X7 receptors are important in mediating the physiological functions of extracellular ATP, and altered receptor expression and function have a causative role in the disease pathogenesis. Here, we investigated the mechanisms determining the P2X7 receptor function by following two human single-nucleotide polymorphism (SNP) mutations that replace His-155 and Ala-348 in the human (h) P2X7 receptor with the corresponding residues, Tyr-155 and Thr-348, in the rat (r) P2X7 receptor. H155Y and A348T mutations in the hP2X7 receptor increased ATP-induced currents, whereas the reciprocal mutations, Y155H and T348A, in the rP2X7 receptor caused the opposite effects. Such a functional switch is a compelling indication that these residues are critical for P2X7 receptor function. Additional mutations of His-155 and Ala-348 in the hP2X7 receptor to residues with diverse side chains revealed a different dependence on the side chain properties, supporting the specificity of these two residues. Substitutions of the residues surrounding His-155 and Ala-348 in the hP2X7 receptor with the equivalent ones in the rP2X7 receptor also affected ATP-induced currents but were not fully reminiscent of the H155Y and A348T effects. Immunofluorescence imaging and biotin labeling assays showed that H155Y in the hP2X7 receptor increased and Y155H in the rP2X7 receptor decreased cell-surface expression. Such contrasting effects were not obvious with the reciprocal mutations of residue 348. Taken together, our results suggest that residues at positions 155 and 348 contribute to P2X7 receptor function via determining the surface expression and the single-channel function, respectively. Such interpretations are consistent with the locations of the residues in the structural model of the hP2X7 receptor.
Keywords: ATP, Biophysics, Immunochemistry, Ion Channels, Receptor Structure-Function, Immunofluorescence Confocal Imaging, P2X Receptor, Patch-Clamp Recording
Introduction
P2X7 receptors belong to the ionotropic purinergic P2X receptor family (1–5). The receptor expression is well documented in immune cells, glial cells in the brain, satellite cells in the peripheral nervous system, bone, and epithelial cells, where the receptor serves as the primary mediator for numerous physiological functions of extracellular ATP, including immune responses, inflammation, cell proliferation, neuron-glial cell interactions, nociception, bone remodeling, and saliva secretion (6–17). The receptor is thought to be a homotrimer (18) with three subunits intertwining together along a vertical ion-conducting pathway. Each subunit contains two transmembrane domains (TM1 and TM2) joined by a large ectodomain, and both N and C termini reside intracellularly. Functional characterizations of mammalian P2X7 receptors have identified several distinguishing features, including activation by submillimolar concentrations of ATP and a higher sensitivity to benzoylbenzoyl-ATP (BzATP),4 a synthetic ATP analog, than to ATP (19–21). In addition to operating as ATP-gated Ca2+-permeable cation channels that open within milliseconds upon agonist binding, P2X7 receptors induce formation of a large pore, activation of the inflammasome, and numerous other downstream signaling events in response to prolonged stimulation (1, 19–22).
The functional expression level of P2X7 receptor is crucial in its role in the disease pathogenesis. This has been highlighted by an increasing number of findings that aberrant or altered receptor expression and function have a causative role in chronic lymphocytic leukemia (8, 23, 24), reduced bacterial-killing ability of macrophages (26, 27), impaired release of cytokines (6, 28–30), inflammatory and neuropathic pain (11, 16), rheumatoid arthritis (7), neurodegenerative and neuroinflammatory disorders (13, 14, 31–36), mood-depressive disorders (37–40), and epithelial cancers (41–43). Studies of the mammalian P2X7 receptors, and particularly the human (h) and rat (r) P2X7 receptors, have revealed striking species differences in the receptor properties, including receptor-mediated functional responses and sensitivities to agonists, antagonists, and modulators (19–21, 44–52). However, the physiological implications and underlying mechanisms are still not well understood.
Our recent study characterizing the mutations resulting from non-synonymous SNPs in the human P2RX7 gene has shown that H155Y and A348T mutations increase ATP-induced currents (53). These two gain-of-function mutations substitute the residues in the hP2X7 receptor with the corresponding ones in the rP2X7 receptor. Here, we show evidence for a critical role of residues at positions 155 and 348 in determining the ATP-induced currents mediated by hP2X7 and rP2X7 receptors and the distinct mechanisms that underlie their contribution.
MATERIALS AND METHODS
Cell and Molecular Biology
Human embryonic kidney HEK293 cells were used to transiently express WT and mutant P2X7 receptors. Cell preparation and transfection using Lipofectamine 2000 reagent (Invitrogen) were performed according to the manufacturer's instructions. In brief, for patch-clamp recordings, HEK293 cells in a 35-mm Petri dish were cotransfected with 1 μg of P2X7 plasmid and 0.1 μg of enhanced GFP plasmid or otherwise as specifically indicated. Cells were dispersed on coverslips 12–24 h after transfection and used within 48 h. The transfection efficiency, estimated based on the GFP-positive cells, was >50%, and single cells with similar GFP expression levels were used. For biotin labeling and immunostaining, HEK293 cells were transfected in the same way except for omission of GFP plasmid for immunostaining experiments. The cDNAs encoding the rP2X7 (Swiss-Prot accession number Q64663) and hP2X7 (accession number Q99572) proteins with a C-terminal EYMPME (referred to as EE) epitope were inserted in the pcDNA3 or pcDNA3.1 vector (Invitrogen). Mutations were introduced by site-directed mutagenesis and confirmed by sequencing.
Patch-Clamp Recording
Whole-cell currents were recorded at room temperature as described previously (54) using an HEKA EPC 10 amplifier at a holding potential of −60 mV. Normal extracellular solution contained 147 mm NaCl, 2 mm KCl, 1 mm MgCl2, 2 mm CaCl2, 10 mm HEPES, and 13 mm glucose (pH 7.3). The maximal currents in cells expressing P2X7 receptors were not readily observed in the normal solution by applying 5–10 mm ATP (53), and therefore, most experiments were carried out in low divalent cation extracellular solution containing no MgCl2 and 0.3 mm CaCl2. Intracellular solution contained 145 mm NaCl, 10 mm EGTA, and 10 mm HEPES (pH 7.3). The currents were compared upon full facilitation (51) by repeated applications of submaximal concentrations of agonist every 1 min. Bisperoxo(1,10-phenanthroline)oxovanadate (V) (100 μm) was used to mitigate current decline in some experiments (55). The concentration-current response curves shown in the figures were constructed using the recordings that showed <10% decline in the currents induced by the same concentration of agonists used for facilitation before and after measuring the concentration-current responses. To examine each subset of mutations, only the data for the WT receptor obtained in parallel experiments were used for comparison. To test the KN-62 antagonist, currents induced by 1 mm ATP, when reproducible, were exposed to each accumulative concentration for 4 min. The ATP concentration-current responses were also measured before and 4 min after exposure to 100 nm KN-62. Agonists and antagonists were delivered using a Bio-Logic RSC160 system.
Structural Modeling
The structural model of the hP2X7 receptor was generated based on the crystal structure of the zebrafish P2X4 receptor (Protein Data Bank code 3H9V) (56). As we have described recently (53), the modeling was carried out using Modeler Version 8.2 (57). The non-conserved loop region between the β2 and β3 strands was modeled de novo using the ModLoop server (58, 59). Images were generated using UCSF Chimera Version 1.4 (60).
Immunofluorescence Confocal Imaging
These experiments were carried out as detailed previously (61, 62). To examine the subcellular distribution of the WT and mutant P2X7 receptor proteins, cells were stained with rabbit anti-EE primary antibody (1:500 or 1:000 dilution; Bethyl) for 2–3 h (or overnight at 4 °C) and Alexa Fluor 488-conjugated anti-rabbit IgG secondary antibody (1:4000 or 1:500; Molecular Probes) for 1–2 h. To examine the co-localization of hP2X7 proteins and calreticulin, an endoplasmic reticulum (ER) protein marker, cells transfected with hP2X7 plasmid were first stained with mouse anti-calreticulin primary antibody (1:250; Stressgen) overnight at 4 °C and FITC-conjugated goat anti-mouse IgG secondary antibody (1:500; Sigma) for 2 h. Cells were then stained with rabbit anti-EE primary antibody (1:500) for 3 h and Alexa Fluor 546-conjugated goat anti-rabbit IgG secondary antibody (1:500; Molecular Probes) for 2 h. To examine the co-localization of hP2X7 proteins and LAMP1 (lysosome-associated membrane protein 1) in the late endocytic organelles (late endosomes and lysosomes), cells were cotransfected with 1 μg of hP2X7 plasmid and 0.5 μg of plasmid encoding LAMP1 fused at its C terminus to GFP (GFP-LAMP1) and stained with rabbit anti-EE primary antibody (1:1000 or 1:2000) for 3 h and Cy3-conjugated goat anti-rabbit IgG secondary antibody (1:2000; Molecular Probes) for 2 h. To examine the co-localization of hP2X7 proteins and TGN46, a trans-Golgi network protein, cells transfected with hP2X7 plasmid were stained with sheep anti-TGN46 primary antibody (1:100) overnight at 4 °C and FITC-conjugated donkey anti-sheep IgG secondary antibody (1:100; Jackson ImmunoResearch Laboratories) for 2 h. Cells were then stained with rabbit anti-EE primary antibody (1:1000 or 1:2000) for 3 h and Cy3-conjugated goat anti-rabbit IgG secondary antibody (1:2000) for 2 h. The images were captured using a Zeiss Axiovert 200M confocal microscope and LSM 150 META software or a Nikon Eclipse TE2000-E confocal microscope and NIS-Elements AR software.
Biotin Labeling and Western Blotting
Biotin labeling was performed by modifying the protocols described previously (62, 63). In brief, cells transfected with P2X7 plasmid (or an empty vector) and GFP plasmid were labeled with sulfo-NHS-LC-biotin (Pierce) for 30 min at 4 °C. Total protein concentrations of the cell lysates were determined using the BCA assay (Thermo Scientific). The biotinylated proteins were purified from a total of 300 μg of lysates using EZview red streptavidin affinity beads (Sigma) overnight at 4 °C and eluted in 50 μl of electrophoresis sample buffer (6% SDS, 10% glycerol, 50 mm Tris-HCl (pH 6.8), 2 mm EDTA, 0.05% bromphenol blue, and 10% β-mercaptoethanol). Whole-cell lysate (5 or 10 μg) or biotin-labeled samples (20 μl) were separated on 12% SDS-polyacrylamide gels. Proteins were detected using rabbit anti-EE primary antibody (1:5000) or mouse anti-GFP antibody (1:2000; Santa Cruz Biotechnology) and horseradish peroxidase-conjugated anti-rabbit secondary antibody (1:5000; Santa Cruz Biotechnology) or anti-mouse IgG antibody (1:2000; Santa Cruz Biotechnology) and visualized using SuperSignal West Femto maximal sensitivity substrate (Thermo Scientific).
Data Analysis
All results, where appropriate, are presented as means ± S.E. All currents are presented as current density, i.e. the currents divided by the cell capacitance, to minimize the effects of the variation in cell size. Agonist EC50 values were determined by least-squares fitting of the data from individual cells to the Hill equation: I = Imax/(1 + (EC50/[A])n), where I is the peak current density evoked by given agonist concentrations ([A]), Imax is the maximal current density, and n is the Hill coefficient. Figures show the curves fitted to the mean of all experiments. Antagonist IC50 values were derived by fitting the data from individual cells to the Hill equation: I = 100/(1 + ([B]/IC50)n), where I is the ATP-induced currents after exposure to given concentrations of antagonist ([B]) expressed as a percentage of the control currents before antagonist application, and n is the Hill coefficient. Curve fits were performed using Origin software. Statistical analysis was carried out using Student's t test for two groups and one-way analysis of variance test for more than two groups, and the difference was considered to be significant at p < 0.05.
RESULTS
Initial Comparison of hP2X7 and rP2X7 Receptor-mediated ATP-induced Currents
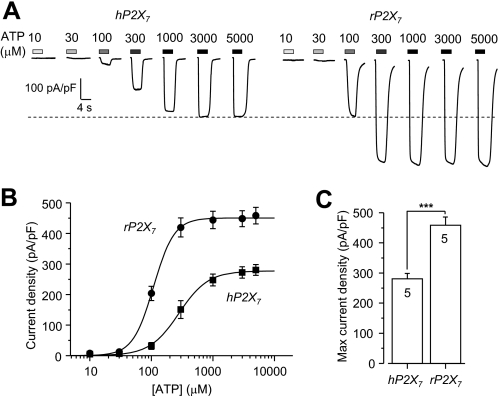
ATP is the physiological agonist for all P2X receptors, including the P2X7 receptor. Fig. 1A shows representative currents responses of cells expressing WT hP2X7 and rP2X7 receptors to the indicated concentrations of ATP recorded in low divalent cation extracellular solution (see “Materials and Methods”). Two differences are discernible. First, the hP2X7 receptor was less sensitive to ATP than the rP2X7 receptor, with EC50 values of 304 ± 49 μm (n = 5) and 110 ± 6 μm (n = 5; p < 0.01) for the human and rat receptors, respectively (Fig. 1B). Second, the amplitude of currents mediated by the hP2X7 receptor was lower than that mediated by the rP2X7 receptor. The average current density evoked by the maximal concentration of ATP (Imax) was 281 ± 18 pA/picofarads (pF; n = 5) for the hP2X7 receptor, corresponding to ∼60% of the value of 458 ± 27 pA/pF (n = 5; p < 0.001) obtained for the rP2X7 receptor (Fig. 1C). These results provide an initial indication of significant functional differences between these two receptors.
FIGURE 1.
ATP-induced current responses mediated by the hP2X7 and rP2X7 receptors. A, representative ATP-induced inward current densities in cells expressing the hP2X7 (left) or rP2X7 (right) receptor. The dashed line denotes the maximal current for the hP2X7 receptor. B, ATP concentration-current density curves from recordings shown in A. Lines represent the best fits to the Hill equation. C, mean maximal current density in parallel experiments. The cell number in each case for B and C is indicated in C. ***, p < 0.001 compared between the hP2X7 and rP2X7 receptors (t test).
Reciprocal Substitution of Residues at Positions 155 and 348 in the hP2X7 and rP2X7 Receptors
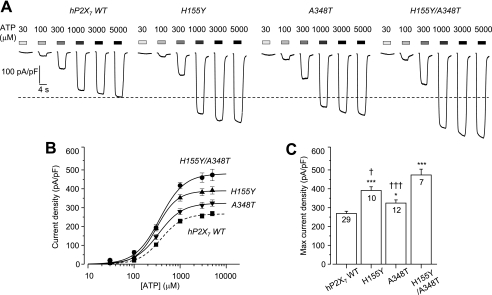
To test whether residues at positions 155 and 348 are involved in determining the P2X7 receptor function, and particularly the difference between human and rat receptors, we compared the ATP-induced currents mediated by hP2X7 and rP2X7 receptors bearing reciprocal mutations in low divalent cation extracellular solution. Fig. 2A shows representative ATP-induced currents in cells expressing the WT or mutant hP2X7 receptors. The A348T and H155Y mutations caused little or no change in the ATP sensitivity of the receptor (Fig. 2B). However, the ATP-induced Imax increased from 268 ± 12 pA/pF (n = 29) for the WT receptor to 391 ± 19 pA/pF (n = 10; p < 0.001) for the H155Y mutant and 324 ± 17 pA/pF (n = 12; p < 0.05) for the A348T mutant (Fig. 2C). These results confirmed our recent report that both the H155Y and A348T mutations increase hP2X7-mediated ATP-induced responses (53). The ATP-induced Imax was further increased by the double mutation H155Y/A348T (473 ± 31 pA/pF (n = 7); p < 0.001); the increase was significantly greater than that produced by the single mutations (Fig. 2C).
FIGURE 2.
Effects of H155Y and A348T mutations on ATP-induced currents mediated by the hP2X7 receptor. A, representative ATP-induced current densities in cells expressing the WT hP2X7 receptor or the H155Y, A348T, or H155Y/A348T mutant. The dashed line denotes the maximal current for the WT hP2X7 receptor. B, ATP concentration-current density curves from recordings shown in A. The lines represent the best fits to the Hill equation. The dashed line denotes the WT hP2X7 receptor. C, mean maximal current density in parallel experiments. The cell number in each case for B and C is indicated in C. ***, p < 0.001, and *, p < 0.05 compared with the WT receptor; †††, p < 0.001, and †, p < 0.05 compared with the H155Y/A348T double mutant receptor (t test).
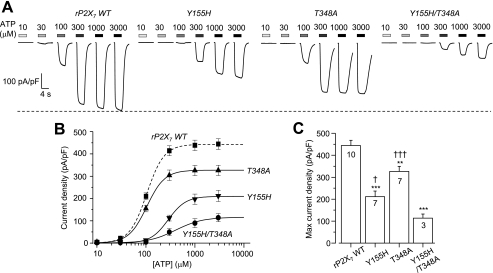
Fig. 3A shows representative ATP-induced currents under the same experimental conditions, resulting from expression of the WT rP2X7 receptor or the Y155H, T348A, or Y155H/T348A mutants. The T348A mutation did not alter the ATP sensitivity, whereas the Y155H and Y155H/T348A mutations caused small reductions (∼3- and 5-fold increases in the EC50 value, respectively) (Fig. 3B). The most prominent mutational effect was that the ATP-induced Imax was strongly attenuated from 444 ± 24 pA/pF (n = 10) for the WT receptor to 211 ± 25 pA/pF (n = 7; p < 0.001) for the Y155H mutant and 326 ± 23 pA/pF (n = 7; p < 0.01) for the T348A mutant (Fig. 3C). The ATP-induced Imax was further reduced by the double mutation Y155H/T348A (114 ± 18 pA/pF (n = 3); p < 0.001); the reduction was significantly greater than that yielded by the single mutations (Fig. 3C). We also examined ATP-induced currents mediated by the WT rP2X7 receptor and the Y155H and T348A mutants in a solution with normal extracellular divalent cation concentrations (see “Materials and Methods”). The reduction in the ATP-induced Imax caused by the Y155H and T348A mutations was also clear under these conditions (supplemental Fig. S1), in striking contrast with the increase in the ATP-induced Imax resulting from the H155Y and A348T mutations in the hP2X7 receptor under the same conditions, as reported in our recent study (53). These contrasting effects were also apparent in terms of BzATP-induced currents. The H155Y and A348T mutations in the hP2X7 receptor increased and the reciprocal Y155H and T348A mutations in the rP2X7 receptor reduced the BzATP-induced Imax (supplemental Fig. S2).
FIGURE 3.
Effects of Y155H and T348A mutations on ATP-induced currents mediated by the rP2X7 receptor. A, representative ATP-induced current densities in cells expressing the WT rP2X7 receptor or the Y155H, T348A, or Y155H/T348A mutant. The dashed line denotes the maximal current for the WT rP2X7 receptor. B, ATP concentration-current density curves from recordings shown in A. The lines represent the best fits to the Hill equation. The dashed line denotes the WT rP2X7 receptor. C, mean maximal current density in parallel experiments. The cell number in each case for B and C is indicated in C. ***, p < 0.001, and **, p < 0.01 compared with the WT receptor; †††, p < 0.001, and †, p < 0.05 compared with the Y155H/T348A double mutant receptor (t test).
All these results consistently indicated that reciprocal mutations of residues at positions 155 and 348 essentially switched the agonist-induced current responses mediated by the hP2X7 and rP2X7 receptors. Such findings strongly suggest a critical role for these two residues in determining the P2X7 receptor function and, in particular, the difference in the current responses between human and rat receptors.
Further Substitution of His-155 and Ala-348 in the hP2X7 Receptor
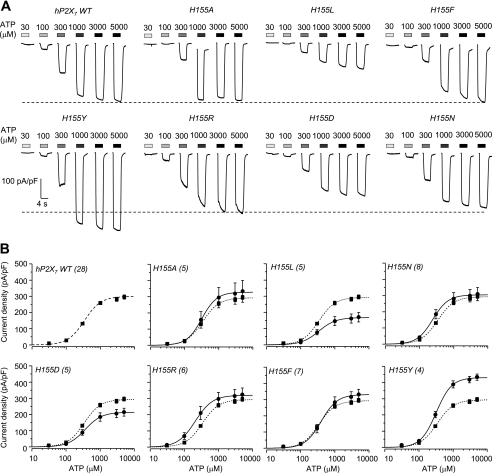
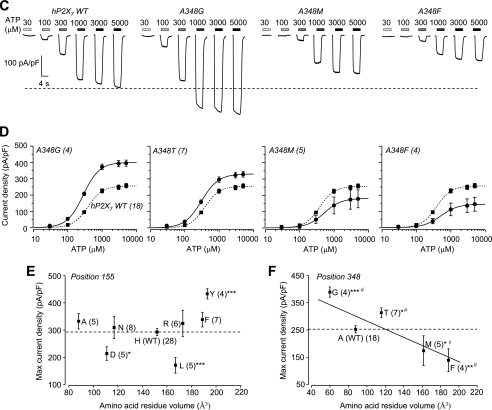
To gain more insight into the role of residues at positions 155 and 348 and to test the importance of the side chain, we mutated His-155 and Ala-348 in the hP2X7 receptor to residues with side chains that have diverse properties (small, lengthy, bulky, neutral, or negatively or positively charged). Fig. 4 shows representative ATP-induced currents and the concentration-response curves in cells expressing the hP2X7 receptors carrying mutations of His-155 (Fig. 4, A and B) and Ala-348 (Fig. 4, C and D), respectively. Overall, there was no or a small change in the ATP sensitivity for all mutants (Fig. 4, B and D). The ATP-induced Imax was reduced by the H155L and H155D mutations but was not changed by the H155A, H155N, H155R, and H155F mutations (Fig. 4E). In contrast, the ATP-induced Imax was significantly altered by each residue substituted for Ala-348 (Fig. 4F). Although the effects of mutations showed no clear relationship to the size of the residues introduced into His-155 (Fig. 4E), there was an apparent correlation between the effects of mutations and the volume of the residues used to replace Ala-348 (Fig. 4F): the one with the smallest side chain (A348G) conferred the greatest increase in the ATP-induced Imax, and the ones with the larger side chains (A348M and A348F) caused the greatest decrease. These results show a strong dependence of the effects of mutations on the side chains, suggesting that the side chain properties of residues at positions 155 and 348 are critical in determining the P2X7 receptor function.
FIGURE 4.
Mutational analysis of the roles of residues at positions 155 and 348 in the hP2X7 receptor. A, representative ATP-induced current densities in cells expressing the WT hP2X7 receptor and mutant receptors carrying mutations of His-155. B, ATP concentration-current density curves from recordings shown in A. C, representative ATP-induced current densities in cells expressing the WT hP2X7 receptor and mutant receptors carrying mutations of Ala-348. D, ATP concentration-current density curves from recordings shown in C. In A and C, the dashed lines denote the maximal currents for the WT hP2X7 receptor. In B and D, the lines represent the best fits to the Hill equation, and the dashed lines denote the WT hP2X7 receptor. The number of cells examined in each case is indicated in parentheses. E and F, the mean maximal current density from A–D is plotted against the volume of the amino acid residues (from Ref. 25) introduced into positions 155 (E) and 348 (F). The dashed lines denote the mean maximal currents for the WT hP2X7 receptor in parallel experiments. ***, p < 0.001, **, p < 0.01, and *, p < 0.05 compared with the WT hP2X7 receptor (t test). In F, # indicates that A348G differs from the WT receptor, A348M, and A348F; A348T differs from A348M and A348F; A348M differs from A348G and A348T; and A348F differs from A348G and A348T (one-way analysis of variance test, p < 0.05). The solid line shows a linear fit, and the R value is 0.7.
Substitutions of Residues Surrounding His-155 and Ala-348 in the hP2X7 Receptor
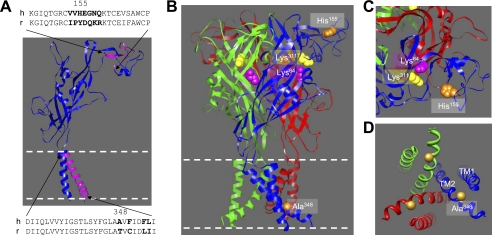
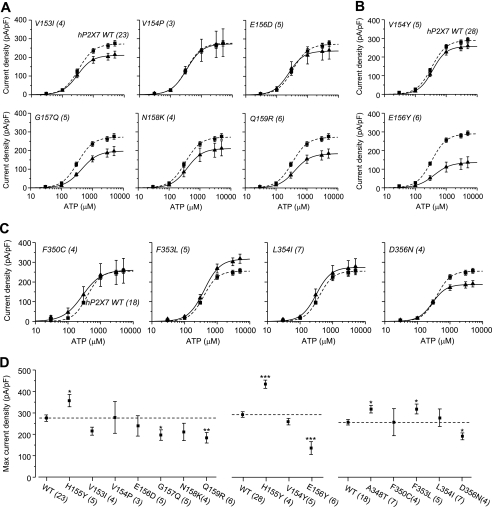
Several residues surrounding positions 155 and 348 are different between the hP2X7 and rP2X7 receptors (Fig. 5A). To investigate whether any of these residues have a similar role, we individually replaced the residues in the hP2X7 receptor with the corresponding ones in the rP2X7 receptor and examined the effects on the ATP-induced currents. Swapping residues in the region surrounding His-155 resulted in six mutants: V153I, V154P, E156D, G157Q, N158K, and Q159R. Fig. 6A shows their ATP concentration-response curves (see representative currents in supplemental Fig. S3A). The G157Q and Q159R mutations reduced the ATP-induced Imax, whereas the other mutations had no effect. We also introduced tyrosine into positions 154 and 156, flanking His-155. Fig. 6B shows the results (see representative currents in supplemental Fig. S3B). Unlike H155Y, the V154Y mutation had no effect on the ATP-induced Imax, whereas the E156Y mutation strongly decreased it (Fig. 6D). These results indicate that none of these mutations mimics H155Y or that none of these residues has a role similar to that of His-155. Fig. 6C summarizes the results from swapping the four residues in the region surrounding Ala-348 (see representative currents in supplemental Fig. S3C). The F350C and L354I mutations were without effect. However, the D356N mutation reduced and the F353L mutation increased the ATP-induced Imax (Fig. 6D). However, introduction of the reciprocal L353F mutation into the rP2X7 receptor had no significant effect (data not shown). Taken together, the results from these experiments suggest that His-155 and Ala-348 and also some surrounding residues, including Glu-156, Gly-157, Gln-159, Phe-353, and Asp-356, are involved in determining the hP2X7-mediated ATP-induced current responses. However, only residues at positions 155 and 348 are critical for the difference between hP2X7 and rP2X7 receptors.
FIGURE 5.
Structural model of hP2X7 receptors and locations of His-155 and Ala-348. A, the dolphin-shaped structural model of a single hP2X7 subunit, with the residues surrounding positions 155 and 348 highlighted in purple and those under investigation denoted in boldface in the sequences. B, the trimeric complex of the hP2X7 receptor, with three different subunits in blue, green, and red, respectively. His-155 and Ala-348 are shown in orange. Lys-64 (in purple) and Lys-311 (in yellow) represent the key residues in the intersubunit ATP-binding site. The structures in A and B are viewed parallel to the membrane plane. C, a snapshot of the structure surrounding His-155, viewed from along the axis of 3-fold symmetry from the extracellular side. His-155 is distant from the ATP-binding site. D, a snapshot of the structure surrounding Ala-348, viewed from along the axis of 3-fold symmetry from the intracellular side. TM2 contributes the ion-conducting pathway, and TM1 is largely peripheral. Ala-348 is positioned immediately intracellularly to the narrowest part (the gate) of the ion-conducting pathway.
FIGURE 6.
Mutational analysis of residues surrounding His-155 and Ala-348 in the hP2X7 receptor. A and B, ATP concentration-current density curves for the hP2X7 receptors carrying residue substitutions surrounding His-155. C, ATP concentration-current density curves for the hP2X7 receptors carrying residue substitutions surrounding Ala-348. The lines represent the best fits to the Hill equation. The dashed lines denote the WT hP2X7 receptor. The cell number in each case for A–C is indicated in parentheses. D, the mean maximal current density from A–C. The dashed lines denote the mean maximal currents for the WT hP2X7 receptor in parallel experiments. ***, p < 0.001, **, p < 0.01, and *, p < 0.05 compared with the WT receptor (t test).
Effects of Mutating Residues at Positions 155 and 348 on Cell-surface Expression of the P2X7 Receptors
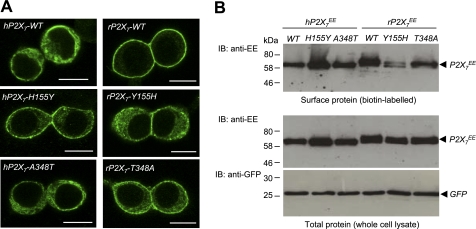
Whole-cell ionic currents are governed by two distinctive factors: the number of functional channels at the cell surface (surface expression) and the single-channel function (conductance and opening probability) (65). We therefore investigated whether residues at positions 155 and 348 are important in determining the surface expression of the hP2X7 and rP2X7 receptors. Using immunofluorescence confocal imaging, we compared the subcellular distribution of the WT and mutant human and rat receptors containing reciprocal mutations of residues at these two positions. Fig. 7A shows representative results. The immunoreactivity for the WT hP2X7 proteins was highly diffused. This diffuse distribution was largely retained for the A348T mutant. In contrast, the immunoreactivity for the H155Y mutant was highly enriched on or near the cell surface (Fig. 7A, left panels). On the other hand, the immunoreactivity for the WT rP2X7 proteins was located principally within or in close vicinity of the plasma membrane, as we reported previously (54, 66). Such membrane-restricted immunostaining was strongly disrupted by the Y155H mutation and only slightly reduced by the T348A mutation (Fig. 7A, right panels).
FIGURE 7.
Effect of reciprocal mutations of residues at positions 155 and 348 on subcellular distribution and the cell-surface and total expression of human and rat P2X7 receptor proteins. A, representative immunofluorescence confocal images of cells expressing the WT hP2X7 receptor or the H155Y or A348T mutant (left panels) and the WT rP2X7 receptor or the Y155H or T348A mutant (right panels). Scale bars = 10 μm. Similar results were observed in three independent experiments. B, representative Western blots showing cell-surface expression of the indicated P2X7 proteins (biotin-labeled; upper panel) and total expression of the indicated P2X7 and GFP proteins (whole-cell lysate; lower panel). Similar results were observed in two to five independent experiments. IB, immunoblot.
To further examine the effects of mutations on cell-surface expression, we used biotin labeling and Western blotting to analyze surface and total protein expression of the WT and mutant hP2X7 and rP2X7 receptors. Fig. 7B shows representative results. The H155Y mutation increased the surface protein expression (upper panel) and also increased the total protein expression (lower panel) of the hP2X7 receptor. However, there was no change in the control GFP protein expression (lower panel). We also examined the H155N mutation, which did not change the ATP-induced Imax (Fig. 4E), and found that unlike the H155Y mutation, the H155N mutation had no effect on the surface and total protein expression (supplemental Fig. S4). In contrast with the H155Y mutation in the hP2X7 receptor, the reciprocal Y155H mutation strongly reduced the surface protein expression and, to a lesser degree, the total protein expression of the rP2X7 receptor (Fig. 7B). However, such contrasting changes in the surface and total protein expression were not observed for the A348T mutation in the hP2X7 receptor and the T348A mutation in the rP2X7 receptor (Fig. 7B), consistent with the immunostaining results (Fig. 7A).
Therefore, our results show that the H155Y mutation in the hP2X7 receptor enhanced the surface expression of the protein and conferred a subcellular distribution close to that of the WT rP2X7 receptor. Conversely, the reciprocal mutation Y155H in the rP2X7 receptor decreased the surface expression and resulted in a subcellular distribution that was similar to that of the WT hP2X7 receptor. There was no such effect on both subcellular distribution and surface expression by the reciprocal mutations of residue 348. These results, together with the effects of mutations on the ATP-induced Imax, imply an important role for residue 348 in determining the single-channel function and provide direct evidence to support a critical role for residue 155 in surface expression. As discussed below, such interpretations are consistent with the structural model of the hP2X7 receptor based on the recently published crystal structure of the zebrafish P2X4 receptor (55).
Subcellular Localization of Intracellular hP2X7 Receptor Proteins
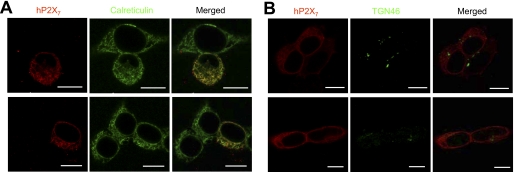
Immunofluorescence confocal imaging showed extensive intracellular location of the hP2X7 receptor proteins (Fig. 7A). To investigate in which subcellular compartments they might be located, we performed double immunostaining of hP2X7 proteins and intracellular organelle marker proteins. Fig. 8 shows representative confocal images. There was strong co-localization of the hP2X7 proteins with calreticulin, an ER protein marker (Fig. 8A), but not with TGN46, a trans-Golgi network protein marker (Fig. 8B). In addition, the immunoreactivity for the hP2X7 proteins exhibited no apparent co-localization with the coexpressed GFP-LAMP1, a protein localized to the late endocytic compartments and lysosomes (supplemental Fig. S6). These results indicate that the intracellular hP2X7 proteins reside mainly within the ER.
FIGURE 8.
Localization of intracellular hP2X7 proteins within the ER compartment. Shown are representative immunofluorescence confocal images of double staining for WT hP2X7 proteins (red) and calreticulin (green), an ER protein marker (A), and double staining for WT hP2X7 proteins (red) and TGN46 (green), a trans-Golgi network protein marker (B). Scale bars = 10 μm.
DISCUSSION
By combining site-directed mutagenesis with whole-cell current patch-clamp recording and protein expression assays, we have shown that the residues at positions 155 and 348 are critical in determining ATP-induced currents mediated by P2X7 receptors, as well as the difference between the human and rat receptors. We have provided further evidence to suggest that distinct molecular mechanisms underlie their contribution.
Importance of Residues at Positions 155 and 348 in Determining the P2X7 Receptor Function
The gene encoding the hP2X7 receptor has extensive SNPs (650 in total as of today), including many non-synonymous ones, several of which have been shown to alter the receptor functions (23, 24, 26–29, 53–55, 61, 64, 67–78). H155Y and A348T are unique in that both mutations change residues in the hP2X7 receptor to the corresponding ones in the rP2X7 receptor (Fig. 5A). Interestingly, these two mutations confer gain of function (Fig. 2) (53, 67), hinting that Tyr-155 and Thr-348 may contribute to the greater currents mediated by the rP2X7 receptor than those by the hP2X7 receptor (Fig. 1). The results from studying the hP2X7 and rP2X7 receptors bearing the reciprocal mutations show that such swapping of residues at positions 155 and 348 results in opposite effects on the ATP-induced Imax (Figs. 2 and 3), providing compelling evidence to indicate that the residues at positions 155 and 348 are important in determining the hP2X7 and rP2X7 receptor functions.
The effects of mutating His-155 on the ATP-induced Imax clearly depend on the introduced residues, but there was no clear correlation between the side chain properties of the residues and the functional phenotypes (Fig. 4E). For example, phenylalanine also contains a bulky side chain but failed to mimic tyrosine (Fig. 4E). In contrast, the effects of mutating Ala-348 on the ATP-induced Imax showed a clear correlation with the side chain size of the introduced residues (Fig. 4F). Swapping residues surrounding His-155 and Ala-348 identified several other residues, including Glu-156, Gly-157, Gln-159, Phe-353, and Asp-356, that influence the hP2X7-mediated ATP-induced currents, but none of these mutations were fully reminiscent of H155Y and A348T (Fig. 6D). These results show that the unique location and properties of the residues at positions 155 and 348 are critical in determining the P2X7 receptor function.
Distinct Mechanisms Underlying the Contribution of Residues at Positions 155 and 348
The identification of residues 155 and 348 adds to the growing list of residues that contribute to determination of the P2X7 receptor functional properties, but only in a few cases is the underlying mechanism clearly understood (23, 26–29, 53–55, 61, 64, 67–78). As mentioned above, the maximal agonist-induced whole-cell currents, which are widely used to indicate the functional expression of an ion channel, are determined by both the surface expression (channel number) and the single-channel function (conductance and opening probability) (64). Structural information proves powerful in helping to infer a mechanistic understanding of the P2X receptor functions from determined mutational effects, as we have reviewed recently (5). According to the structural model of the hP2X7 receptor based on the crystal structure of the zebrafish P2X4 receptor (Fig. 5) (56), His-155 is in the extracellular domain and is positioned at the periphery of the receptor complex and well away from the intersubunit ATP-binding site that contains the key residues Lys-64 and Lys-311 (Fig. 5, B and C) (5, 68). Consistent with this, mutating His-155 to residues with diverse side chain properties, despite significant alterations in the ATP-induced Imax, had no or a small effect on the ATP sensitivity (Fig. 4B). In addition, the H155Y mutant receptor exhibited virtually the same sensitivity to the hP2X7-specific antagonist KN-62 (44) as the WT receptor (IC50 = 127 ± 38 nm (n = 3) for the WT receptor and 129 ± 20 nm (n = 3) for the H155Y mutant; p > 0.1). Furthermore, KN-62 at 100 nm resulted in almost identical inhibition of the ATP-induced currents mediated by the WT and H155Y mutant receptors (supplemental Fig. S5). These results strongly support the conclusion that the H155Y mutation causes no major changes in the global conformation (and particularly, the ligand-binding sites) of the receptor. Moreover, the location of His-155 is distant from the ion- permeating pathway (Fig. 5B). Thus, the functional results and structural considerations consistently disfavor significant involvement of residue 155 in determining the single-channel function and instead point to a role in the cell-surface expression of the receptor. Indeed, despite no indication of a change in the surface expression in a previous study using immunofluorescence flow cytometry (67), our immunofluorescence confocal imaging and biotin labeling studies clearly show that the H155Y mutation increases the surface expression of the hP2X7 receptor, whereas the reciprocal mutation, Y155H, reduces the surface expression of the rP2X7 receptor (Fig. 7). These results provide compelling evidence to support a role for residue 155 in determining the surface expression of the P2X7 receptors. Residue 155 is expected to be located in the lumen of the ER and the trans-Golgi network before the receptor is inserted into the plasma membrane or inside the endosomes and lysosomes after retrieval from the cell surface. Our immunostaining results indicate that the intracellular hP2X7 proteins reside mainly in the ER (Fig. 8). Thus, one simple and consistent interpretation is that residue 155 is critical in the P2X7 protein folding in the ER, resulting in P2X7 receptors that are either transported from the ER to the plasma membrane or retained in the ER and degraded. This can also explain that mutation of residue 155 altered the total protein expression level (Fig. 7). However, we cannot exclude other possibilities such as altered protein synthesis or mRNA stability.
On the other hand, the model predicts Ala-348 to be part of the intracellular pore and close to the physical gate that occludes the ion-permeating pathway in the closed state (Fig. 5D) (5, 56), a position well purported for Ala-348 to have a role in channel gating and conductance. A recent study has proposed that a significant narrowing of the intracellular pore occurs during the P2X receptor channel opening (79). This notion is concordant with the strong correlation of the effects of mutating Ala-348 on the ATP-induced Imax with the side chain volume or size of the introduced residues (Fig. 4F). The immunofluorescence confocal imaging and biotin labeling results (Fig. 7) largely rule out a significant contribution of residue 348 to cell-surface expression and indirectly support a more important role in determining the single-channel properties.
Taken together, our results suggest distinct mechanisms underlying the contribution of residues at positions 155 and 348 to the P2X7 receptor function: an important role for residue 155 in the surface expression and a predominant role for residue 348 in the single-channel function. Further studies are required to better understand these specific roles.
Physiological and Pathological Implications
The expression and function of the P2X7 receptor are vital to its role in mediating the functions of extracellular ATP (see the Introduction). As mentioned above, the H155Y and A348T mutations result from non-synonymous SNPs in the human P2RX7 gene. A very recent study shows that monocytes from individuals with haplotypes of the hP2X7 receptors carrying A348T exhibit increased ATP-induced secretion of interleukin-1β (78), a pro-inflammatory cytokine that is important in the pathological roles of the P2X7 receptors (13, 14, 80, 81). As demonstrated in our study, the strong difference in hP2X7 receptor function (Fig. 3) depending on which residues at positions 155 and 348 individuals carry is expected to confer differential susceptibilities to diseases. Genetic linkage evidence for disease association of these two non-synonymous SNPs has not yet come forward. However, a previous study reported a trend for linkage of the A348T SNP with the severity of the anxiety disorders (39). Clearly, the results reported and the concepts formulated here should facilitate future genetic linkage studies and therapeutic exploitations (82–84). As far as we are aware, this study provides the first insight into the molecular mechanisms controlling the species difference in the P2X7 receptor functions. Such and other differences in the receptor properties (see the Introduction) may call for cautions to be exercised when the functional roles of the hP2X7 receptor are inferred from studies using laboratory animals.
In conclusion, we have shown that residues at positions 155 and 348 are important in determining the P2X7 receptor function and contribute to the difference in ATP-induced responses between the human and rat receptors. We have provided evidence suggesting that distinct mechanisms underlie the contribution of these two residues. Such insight into mechanisms controlling the P2X7 receptor functions will help to evolve a better understanding of the important and diverse roles for this receptor in mediating an increasing number of physiological and pathological functions of extracellular ATP.
Supplementary Material
Acknowledgments
We are grateful to P. Boquet (INSERM452, Nice, France) for the GFP-LAMP1 construct, Drs. C. Cockcroft and R. Karnik for expert assistance in immunostaining, Dr. M. Young (University of Cardiff) for biotin labeling, and Dr. N. Gamper for confocal imaging. We thank Prof. D. J. Beech and Dr. J. Lippiat for constructive discussions during this project.
This work was supported in part by the Biotechnology and Biological Science Research Council.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- BzATP
- benzoylbenzoyl-ATP
- h
- human
- r
- rat
- ER
- endoplasmic reticulum
- pF
- picofarads.
REFERENCES
- 1. North R. A. (2002) Physiol. Rev. 82, 1013–1067 [DOI] [PubMed] [Google Scholar]
- 2. Vial C., Roberts J. A., Evans R. J. (2004) Trends Pharmacol. Sci. 25, 487–493 [DOI] [PubMed] [Google Scholar]
- 3. Khakh B. S., North R. A. (2006) Nature 442, 527–532 [DOI] [PubMed] [Google Scholar]
- 4. Surprenant A., North R. A. (2009) Annu. Rev. Physiol. 71, 333–359 [DOI] [PubMed] [Google Scholar]
- 5. Browne L. E., Jiang L. H., North R. A. (2010) Trends Pharmacol. Sci. 31, 229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Solle M., Labasi J., Perregaux D. G., Stam E., Petrushova N., Koller B. H., Griffiths R. J., Gabel C. A. (2001) J. Biol. Chem. 276, 125–132 [DOI] [PubMed] [Google Scholar]
- 7. Labasi J. M., Petrushova N., Donovan C., McCurdy S., Lira P., Payette M. M., Brissette W., Wicks J. R., Audoly L., Gabel C. A. (2002) J. Immunol. 168, 6436–6445 [DOI] [PubMed] [Google Scholar]
- 8. Adinolfi E., Melchiorri L., Falzoni S., Chiozzi P., Morelli A., Tieghi A., Cuneo A., Castoldi G., Di Virgilio F., Baricordi O. R. (2002) Blood 99, 706–708 [DOI] [PubMed] [Google Scholar]
- 9. Ke H. Z., Qi H., Weidema A. F., Zhang Q., Panupinthu N., Crawford D. T., Grasser W. A., Paralkar V. M., Li M., Audoly L. P., Gabel C. A., Jee W. S., Dixon S. J., Sims S. M., Thompson D. D. (2003) Mol. Endocrinol. 17, 1356–1367 [DOI] [PubMed] [Google Scholar]
- 10. Duan S., Anderson C. M., Keung E. C., Chen Y., Chen Y., Swanson R. A. (2003) J. Neurosci. 23, 1320–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chessell I. P., Hatcher J. P., Bountra C., Michel A. D., Hughes J. P., Green P., Egerton J., Murfin M., Richardson J., Peck W. L., Grahames C. B., Casula M. A., Yiangou Y., Birch R., Anand P., Buell G. N. (2005) Pain 114, 386–396 [DOI] [PubMed] [Google Scholar]
- 12. Adinolfi E., Callegari M. G., Ferrari D., Bolognesi C., Minelli M., Wieckowski M. R., Pinton P., Rizzuto R., Di Virgilio F. (2005) Mol. Biol. Cell 16, 3260–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matute C., Torre I., Pérez-Cerdá F., Pérez-Samartín A., Alberdi E., Etxebarria E., Arranz A. M., Ravid R., Rodríguez-Antigüedad A., Sánchez-Gómez M., Domercq M. (2007) J. Neurosci. 27, 9525–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choi H. B., Ryu J. K., Kim S. U., McLarnon J. G. (2007) J. Neurosci. 27, 4957–4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang X., Chen Y., Wang C., Huang L. Y. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9864–9869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen Y., Zhang X., Wang C., Li G., Gu Y., Huang L. Y. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16773–16778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakamoto T., Brown D. A., Catalán M. A., Gonzalez-Begne M., Romanenko V. G., Melvin J. E. (2009) J. Biol. Chem. 284, 4815–4822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nicke A. (2008) Biochem. Biophys. Res. Commun. 377, 803–808 [DOI] [PubMed] [Google Scholar]
- 19. Surprenant A., Rassendren F., Kawashima E., North R. A., Buell G. (1996) Science 272, 735–738 [DOI] [PubMed] [Google Scholar]
- 20. Rassendren F., Buell G. N., Virginio C., Collo G., North R. A., Surprenant A. (1997) J. Biol. Chem. 272, 5482–5486 [DOI] [PubMed] [Google Scholar]
- 21. Chessell I. P., Simon J., Hibell A. D., Michel A. D., Barnard E. A., Humphrey P. P. (1998) FEBS Lett. 439, 26–30 [DOI] [PubMed] [Google Scholar]
- 22. Di Virgilio F. (2007) Trends Pharmacol. Sci. 28, 465–472 [DOI] [PubMed] [Google Scholar]
- 23. Wiley J. S., Dao-Ung L. P., Gu B. J., Sluyter R., Shemon A. N., Li C., Taper J., Gallo J., Manoharan A. (2002) Lancet 359, 1114–1119 [DOI] [PubMed] [Google Scholar]
- 24. Thunberg U., Tobin G., Johnson A., Söderberg O., Padyukov L., Hultdin M., Klareskog L., Enblad G., Sundström C., Roos G., Rosenquist R. (2002) Lancet 360, 1935–1939 [DOI] [PubMed] [Google Scholar]
- 25. Zamyatnin A. A. (1972) Prog. Biophys. Mol. Biol. 24, 107–123 [DOI] [PubMed] [Google Scholar]
- 26. Saunders B. M., Fernando S. L., Sluyter R., Britton W. J., Wiley J. S. (2003) J. Immunol. 171, 5442–5446 [DOI] [PubMed] [Google Scholar]
- 27. Shemon A. N., Sluyter R., Fernando S. L., Clarke A. L., Dao-Ung L. P., Skarratt K. K., Saunders B. M., Tan K. S., Gu B. J., Fuller S. J., Britton W. J., Petrou S., Wiley J. S. (2006) J. Biol. Chem. 281, 2079–2086 [DOI] [PubMed] [Google Scholar]
- 28. Sluyter R., Shemon A. N., Wiley J. S. (2004) J. Immunol. 172, 3399–3405 [DOI] [PubMed] [Google Scholar]
- 29. Sluyter R., Dalitz J. G., Wiley J. S. (2004) Genes Immun. 5, 588–591 [DOI] [PubMed] [Google Scholar]
- 30. Glas R., Sauter N. S., Schulthess F. T., Shu L., Oberholzer J., Maedler K. (2009) Diabetologia 52, 1579–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parvathenani L. K., Tertyshnikova S., Greco C. R., Roberts S. B., Robertson B., Posmantur R. (2003) J. Biol. Chem. 278, 13309–13317 [DOI] [PubMed] [Google Scholar]
- 32. McLarnon J. G., Ryu J. K., Walker D. G., Choi H. B. (2006) J. Neuropathol. Exp. Neurol. 65, 1090–1097 [DOI] [PubMed] [Google Scholar]
- 33. Sanz J. M., Chiozzi P., Ferrari D., Colaianna M., Idzko M., Falzoni S., Fellin R., Trabace L., Di Virgilio F. (2009) J. Immunol. 182, 4378–4385 [DOI] [PubMed] [Google Scholar]
- 34. Díaz-Hernández M., Díez-Zaera M., Sánchez-Nogueiro J., Gómez-Villafuertes R., Canals J. M., Alberch J., Miras-Portugal M. T., Lucas J. J. (2009) FASEB J. 23, 1893–1906 [DOI] [PubMed] [Google Scholar]
- 35. Yiangou Y., Facer P., Durrenberger P., Chessell I. P., Naylor A., Bountra C., Banati R. R., Anand P. (2006) BMC Neurol. 6, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brough D., Le Feuvre R. A., Iwakura Y., Rothwell N. J. (2002) Mol. Cell. Neurosci. 19, 272–280 [DOI] [PubMed] [Google Scholar]
- 37. Barden N., Harvey M., Gagne B., Shink E., Tremblay M., Raymond C., Labbe M., Villeneuve A., Rochette D., Bordeleau L., Stadler H., Holsboer F., Muller-Myhsok B. (2006) Am. J. Med. Genet. B Neuropsychiatr. Genet. 141, 374–382 [DOI] [PubMed] [Google Scholar]
- 38. Lucae S., Salyakina D., Barden N., Harvey M., Gagné B., Labbé M., Binder E. B., Uhr M., Paez-Pereda M., Sillaber I., Ising M., Brückl T., Lieb R., Holsboer F., Müller-Myhsok B. (2006) Hum. Mol. Genet. 15, 2438–2445 [DOI] [PubMed] [Google Scholar]
- 39. Erhardt A., Lucae S., Unschuld P. G., Ising M., Kern N., Salyakina D., Lieb R., Uhr M., Binder E. B., Keck M. E., Müller-Myhsok B., Holsboer F. (2007) J. Affect. Disord. 101, 159–168 [DOI] [PubMed] [Google Scholar]
- 40. Basso A. M., Bratcher N. A., Harris R. R., Jarvis M. F., Decker M. W., Rueter L. E. (2009) Behav. Brain Res. 198, 83–90 [DOI] [PubMed] [Google Scholar]
- 41. Zhou L., Qi X., Potashkin J. A., Abdul-Karim F. W., Gorodeski G. I. (2008) J. Biol. Chem. 283, 28274–28286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Solini A., Cuccato S., Ferrari D., Santini E., Gulinelli S., Callegari M. G., Dardano A., Faviana P., Madec S., Di Virgilio F., Monzani F. (2008) Endocrinology 149, 389–396 [DOI] [PubMed] [Google Scholar]
- 43. Feng Y. H., Li X., Wang L., Zhou L., Gorodeski G. I. (2006) J. Biol. Chem. 281, 17228–17237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Humphreys B. D., Virginio C., Surprenant A., Rice J., Dubyak G. R. (1998) Mol. Pharmacol. 54, 22–32 [DOI] [PubMed] [Google Scholar]
- 45. Hibell A. D., Kidd E. J., Chessell I. P., Humphrey P. P., Michel A. D. (2000) Br. J. Pharmacol. 130, 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jiang L. H., Mackenzie A. B., North R. A., Surprenant A. (2000) Mol. Pharmacol. 58, 82–88 [PubMed] [Google Scholar]
- 47. Stokes L., Jiang L. H., Alcaraz L., Bent J., Bowers K., Fagura M., Furber M., Mortimore M., Lawson M., Theaker J., Laurent C., Braddock M., Surprenant A. (2006) Br. J. Pharmacol. 149, 880–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Young M. T., Pelegrin P., Surprenant A. (2007) Mol. Pharmacol. 71, 92–100 [DOI] [PubMed] [Google Scholar]
- 49. Sluyter R., Shemon A. N., Hughes W. E., Stevenson R. O., Georgiou J. G., Eslick G. D., Taylor R. M., Wiley J. S. (2007) Am. J. Physiol. Regul. Integr. Comp. Physiol. 293, R2090–R2098 [DOI] [PubMed] [Google Scholar]
- 50. Moore S. F., Mackenzie A. B. (2008) Biochem. Pharmacol. 76, 1740–1747 [DOI] [PubMed] [Google Scholar]
- 51. Roger S., Pelegrin P., Surprenant A. (2008) J. Neurosci. 28, 6393–6401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Donnelly-Roberts D. L., Namovic M. T., Han P., Jarvis M. F. (2009) Br. J. Pharmacol. 157, 1203–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Roger S., Mei Z. Z., Baldwin J. M., Dong L., Bradley H., Baldwin S. A., Surprenant A., Jiang L. H. (2010) J. Psychiatr. Res. 44, 347–355 [DOI] [PubMed] [Google Scholar]
- 54. Liu X., Ma W., Surprenant A., Jiang L. H. (2009) Br. J. Pharmacol. 156, 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim M., Jiang L. H., Wilson H. L., North R. A., Surprenant A. (2001) EMBO J. 20, 6347–6358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kawate T., Michel J. C., Birdsong W. T., Gouaux E. (2009) Nature 460, 592–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fiser A., Sali A. (2003) Bioinformatics 19, 2500–2501 [DOI] [PubMed] [Google Scholar]
- 59. Fiser A., Sali A. (2003) Methods Enzymol. 374, 461–491 [DOI] [PubMed] [Google Scholar]
- 60. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 61. Liu X., Surprenant A., Mao H. J., Roger S., Xia R., Bradley H., Jiang L. H. (2008) Mol. Pharmacol. 73, 252–259 [DOI] [PubMed] [Google Scholar]
- 62. Mei Z. Z., Xia R., Beech D. J., Jiang L. H. (2006) J. Biol. Chem. 281, 38748–38756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xia R., Mei Z. Z., Mao H. J., Yang W., Dong L., Bradley H., Beech D. J., Jiang L. H. (2008) J. Biol. Chem. 283, 27426–27432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sun C., Chu J., Singh S., Salter R. D. (2010) Purinergic Signal. 6, 31–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hille B. (1992) Ionic Channels in Excitable Membranes, 2nd Ed., Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- 66. Jiang L. H., Rassendren F., Mackenzie A., Zhang Y. H., Surprenant A., North R. A. (2005) Am. J. Physiol. Cell Physiol. 289, C1295–C1302 [DOI] [PubMed] [Google Scholar]
- 67. Cabrini G., Falzoni S., Forchap S. L., Pellegatti P., Balboni A., Agostini P., Cuneo A., Castoldi G., Baricordi O. R., Di Virgilio F. (2005) J. Immunol. 175, 82–89 [DOI] [PubMed] [Google Scholar]
- 68. Wilkinson W. J., Jiang L. H., Surprenant A., North R. A. (2006) Mol. Pharmacol. 70, 1159–1163 [DOI] [PubMed] [Google Scholar]
- 69. Worthington R. A., Smart M. L., Gu B. J., Williams D. A., Petrou S., Wiley J. S., Barden J. A. (2002) FEBS Lett. 512, 43–46 [DOI] [PubMed] [Google Scholar]
- 70. Gu B. J., Zhang W., Worthington R. A., Sluyter R., Dao-Ung P., Petrou S., Barden J. A., Wiley J. S. (2001) J. Biol. Chem. 276, 11135–11142 [DOI] [PubMed] [Google Scholar]
- 71. Adriouch S., Dox C., Welge V., Seman M., Koch-Nolte F., Haag F. (2002) J. Immunol. 169, 4108–4112 [DOI] [PubMed] [Google Scholar]
- 72. Wiley J. S., Dao-Ung L. P., Li C., Shemon A. N., Gu B. J., Smart M. L., Fuller S. J., Barden J. A., Petrou S., Sluyter R. (2003) J. Biol. Chem. 278, 17108–17113 [DOI] [PubMed] [Google Scholar]
- 73. Gu B. J., Sluyter R., Skarratt K. K., Shemon A. N., Dao-Ung L. P., Fuller S. J., Barden J. A., Clarke A. L., Petrou S., Wiley J. S. (2004) J. Biol. Chem. 279, 31287–31295 [DOI] [PubMed] [Google Scholar]
- 74. Le Stunff H., Auger R., Kanellopoulos J., Raymond M. N. (2004) J. Biol. Chem. 279, 16918–16926 [DOI] [PubMed] [Google Scholar]
- 75. Denlinger L. C., Sommer J. A., Parker K., Gudipaty L., Fisette P. L., Watters J. W., Proctor R. A., Dubyak G. R., Bertics P. J. (2003) J. Immunol. 171, 1304–1311 [DOI] [PubMed] [Google Scholar]
- 76. Adriouch S., Scheuplein F., Bähring R., Seman M., Boyer O., Koch-Nolte F., Haag F. (2009) Purinergic Signal. 5, 151–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Young M. T., Pelegrin P., Surprenant A. (2006) Br. J. Pharmacol. 149, 261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Stokes L., Fuller S. J., Sluyter R., Skarratt K. K., Gu B. J., Wiley J. S. (2010) FASEB J. 24, 2916–2927 [DOI] [PubMed] [Google Scholar]
- 79. Li M., Kawate T., Silberberg S. D., Swartz K. J. (2010) Nat. Commun. 1, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Monif M., Reid C. A., Powell K. L., Smart M. L., Williams D. A. (2009) J. Neurosci. 29, 3781–9391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Skaper S. D., Debetto P., Giusti P. (2010) FASEB J. 24, 337–345 [DOI] [PubMed] [Google Scholar]
- 82. Donnelly-Roberts D. L., Jarvis M. F. (2007) Br. J. Pharmacol. 151, 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gorodeski G. I. (2009) Expert Opin. Ther. Targets 13, 1313–1332 [DOI] [PubMed] [Google Scholar]
- 84. Gunosewoyo H., Kassiou M. (2010) Expert Opin. Ther. Patents 20, 625–646 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.