Abstract
Huntington disease results from an expanded polyglutamine region in the N terminus of the huntingtin protein. HD pathology is characterized by neuronal degeneration and protein inclusions containing N-terminal fragments of mutant huntingtin. Structural information is minimal, though it is believed that mutant huntingtin polyglutamine adopts β structure upon conversion to a toxic form. To this end, we designed mammalian cell expression constructs encoding compact β variants of Htt exon 1 N-terminal fragment and tested their ability to aggregate and induce toxicity in cultured neuronal cells. In parallel, we performed molecular dynamics simulations, which indicate that constructs with expanded polyglutamine β-strands are stabilized by main-chain hydrogen bonding. Finally, we found a correlation between the reactivity to 3B5H10, an expanded polyglutamine antibody that recognizes a compact β rich hairpin structure, and the ability to induce cell toxicity. These data are consistent with an important role for a compact β structure in mutant huntingtin-induced cell toxicity.
Keywords: Amyloid, Huntington Disease, Molecular Dynamics, Polyglutamine Disease, Protein Conformation, huntingtin, Polyglutamine
Introduction
Huntington disease (HD)5 is a genetic neurodegenerative disorder caused by an expanded polyglutamine (polyQ) repeat in the huntingtin (Htt) protein (1–3). The function of Htt is poorly understood, but is thought to involve vesicular transport and recycling, as well as regulation of gene transcription (4). The Htt protein is more than 3,000 amino acids in length, with the polyQ stretch located at the extreme N terminus, and numerous regions predicted to form highly structured HEAT repeats (5). Normal length polyQ regions vary between 10 and 25 residues, while longer repeats of 36 glutamines or more can cause HD (6). Clinical studies in HD patients have shown that a longer repeat results in an earlier age of onset and a more rapid disease progression (7, 8). Based on more recent work, Htt is thought to undergo several proteolytic cleavage events, resulting in a number of Htt truncation products (9, 10). One cleavage product of particular interest is a short N-terminal fragment similar in size to the product of the first exon of the Htt gene. Htt exon-1 N-terminal fragment is comprised of the first 90 residues of the protein (numbering is based on 23 glutamine repeats as in the original published sequence; (3)) and is believed to be a mediator of toxicity in animal models of HD, as well as in human patients. The crystal structure of an N-terminal fragment of Htt exon-1 fused to the C terminus of maltose-binding protein shows that the initial 17 amino acids N-terminal to the polyQ stretch can form an α-helix that extends into the polyQ region (11). However, the structure of mutant Htt with an expanded polyQ repeat has not yet been determined.
A pathological hallmark of HD is the presence of intranuclear inclusions composed largely of aggregated N-terminal fragments of mutant Htt. While inclusions are pathognomonic for the disease, they are not most enriched in the striatum, the region of the brain most affected in HD, and are not always present in those neurons most likely to degenerate (12). Further, experiments carried out using neuronal cells in culture have demonstrated that the formation of inclusions and toxicity are separate events (13). At present, the formation of large inclusions is, in part, believed to be a cellular protective response to aggregated proteins (14–16).
While the aggregation pathway of mutant Htt is complex and remains poorly understood, it is likely to include a number of intermediates that may be on- or off-pathway to the formation of inclusions or to formation of the toxic species. The toxic species in HD has not been identified, but is believed to be an abnormally-folded monomer or a small soluble oligomer (17–19). Intramolecular interactions within the N-terminal 17-residue α-helical region of Htt may initiate the formation of oligomeric species (17, 20).
Whereas the conformation of the polyQ expansion in the toxic form of mutant Htt has not been determined, a variety of structures have been proposed (21, 22). In a previous study, we made alterations in the polyQ stretch to induce β-turn formation, and found that Htt proteins able to form a compact β structure comprised of alternating β-strands and β-turns were also toxic to cultured cells (23). Monoclonal expanded polyQ antibody 3B5H10 has been shown in a number of recent studies to detect soluble Htt in situ (24–26). Using computational methods and small angle x-ray scattering techniques, 3B5H10 was found to best recognize expanded polyQ in a compact, two-stranded, hairpin conformation with β-rich character.6 Thus, this antibody can be a useful tool in distinguishing compact versus extended β conformations of expanded polyQ.
In the present study, we have made a series of novel Htt exon-1 constructs that have varying propensities to form β-strands within the polyQ region and tested their ability to aggregate. In parallel, we have carried out molecular dynamics simulations, which indicate that constructs with expanded polyQ β-strands are stabilized by main-chain hydrogen bonding. Further, we have found a correlation between 3B5H10 reactivity to each Htt exon-1 construct and the ability to induce cell toxicity. Taken together, our data are consistent with an important role for a compact β structure in mutant Htt exon-1-mediated cell toxicity.
EXPERIMENTAL PROCEDURES
Plasmid Construction
Preparation of the Htt exon-1 76Q DNA for expression in Neuro2a (N2a) cells was carried out using a multi-step process as previously described (23). To generate all other DNA constructs, synthetic DNA fragments comprised of a mix of CAG/CAA sequence and encoding for Q3[PGQ9]6PGQ3 (PGQ9, see Fig. 1), Q3[PGQ4PQ4]4PGQ9[PGQ4PQ4]PGQ3 (PGQP), Q3[PGQ4WQ4]4PGQ9[PGQ4WQ4]PGQ3 (PGQW), Q3[EDQ9]6EDQ3 (EDQ9), and Q3[PGQ6PGQ12]3PGQ4 (PGQ6/12) were synthesized and cloned into a modified pUC vector by Blue Heron Biotechnology. These fragments were then subcloned using BseRI and BsgI restriction sites into a modified Htt exon-1 cDNA described previously (27). Finally, PGQ9, PGQP, PGQW, EDQ9, and PGQ6/12 Htt exon-1 constructs were subcloned into a modified pCMV-2B vector (Stratagene) via SalI and NotI restriction sites to allow for protein expression.
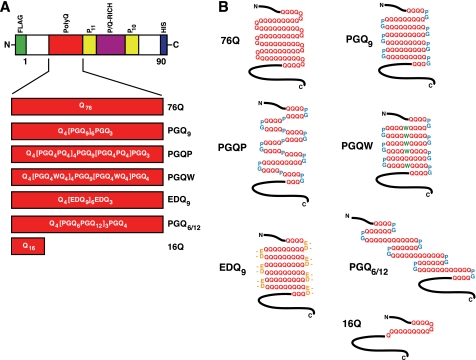
FIGURE 1.
Htt exon-1 polyQ and compact β polyQ proteins used for mammalian cell culture transfection experiments. A, domain structure of Htt exon-1 fragment, indicating polyQ region (red). Also shown are two Pro-repeat regions (yellow) and the P/Q-rich region (purple). An N-terminal FLAG tag (green) and a C-terminal His6 tag (blue) were engineered into each construct. The primary structure for each polyQ region is shown. B, predicted secondary structure of polyQ region in the expressed protein.
Antibodies
A mouse anti-FLAG monoclonal antibody was purchased from Sigma. MW7, a mouse monoclonal IgM specific for the polyproline-rich regions of Htt, was prepared as described previously (28). Anti-Htt exon-1, a goat polyclonal antibody, was generated against an Htt exon-1 internal deletion mutant as previously described (29). 3B5H10, a mouse monoclonal IgG selective for β-hairpin structure in expanded polyQ, was prepared as described (30).
Cell Culture and Transfection
Mouse N2a cells (American Type Culture Collection, ATCC) were maintained in N2a growth medium (50% high glucose Dulbecco's Modified Eagle Medium (DMEM; ATCC), 50% Opti-Minimal Essential Medium (-MEM; ATCC), 5% fetal bovine serum (FBS; Invitrogen), and 1% penicillin-streptomycin (Invitrogen). Mouse hippocampal-derived HT22 cells were grown in low glucose DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. Cells were grown on coverslips in 6-well plates (Corning Incorporated Life Sciences) to ∼50% confluency at the time of transfection. Transfections were carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Quantification of Aggregated Htt Exon-1 PolyQ and Compact β PolyQ Proteins in N2a Cells
Cells were fixed and permeabilized using 4% paraformaldehyde (Sigma) and 0.1% Triton X-100 (Sigma) in phosphate buffered saline (PBS), pH 7.4 at 48 h post-transfection. Fixed cells were stained with MW7 (1:2500) monoclonal antibody, followed by cy3-anti mouse IgM (Jackson ImmunoResearch), and labeled with Hoechst (Invitrogen/Molecular Probes) as described (23). Finally, coverslips were mounted with Vectashield mounting medium (Vector Laboratories). Indirect immunofluorescence (IIF) staining was viewed using a Zeiss Axiovert 200 m fluorescence microscope. For quantification, images were captured from 15 random selected fields for each coverslip (∼150 cell total). Duplicate coverslips for the transfection of each construct were examined in each individual experiment. Data presented are an average (± S.D.) of three separate experiments.
Quantification of 3B5H10 Reactivity in HT22 Cells
Cells were fixed and permeabilized as described above at 24 h post-transfection. Fixed cells were co-stained with MW7 (1:2500) and 3B5H10 (1:5000) monoclonal antibodies, followed by Cy3-anti mouse IgM (Jackson ImmunoResearch) and FITC-anti-mouse IgG (Chemicon International), and labeled with Hoechst (Invitrogen/Molecular Probes) as described (23). Images were collected on a Zeiss 510 Meta confocal microscope. A reference sample was used periodically throughout each experiment to confirm that image intensity remained consistent over time. For each experiment, one photomultiplier tube (PMT) detector was set to a single gain and offset setting for 3B5H10 fluorescence, one for MW7, and a third for Hoechst staining. PMT gain and offset settings were determined by the detection range necessary to image, with a single setting across all Htt exon-1 constructs and across all trials. Images were acquired using a 63× oil immersion lens, then imported into and analyzed by Volocity Quantitation software (Perkin Elmer). Cells with soluble staining were selected using MW7 staining, without regard for 3B5H10 reactivity. Htt that was included in the final quantification had to be within a lower and upper boundary of MW7 fluorescence. The lower boundary represents the minimum detectable soluble Htt staining, while the upper boundary prevented inclusion of aggregates in the calculations. For the Htt in each cell that was between the lower and upper boundaries of MW7 fluorescence, the corresponding 3B5H10 intensity was then quantified. These data were subsequently averaged for each Htt exon-1 compact β construct and normalized against Htt exon-1 76Q intensity per trial. The final data represent the average (± S.D.) of three independent trials.
Cell Lysis and Immunoblotting
Protein extracts from transfected cells were prepared in m-PER cell lysis buffer (Pierce) supplemented with a protease inhibitor mixture containing 4-(2-aminoethyl)benzenesulfonyl fluoride-hydrochloric acid, aprotinin, E-64, EDTA, and leupeptin hemisulfate (CalBiochem). Following a 10 min centrifugation of the cell lysates at 4 °C and 14,000 × g, detergent-soluble supernatant and detergent-insoluble pellet fractions were collected. Detergent soluble supernatants were prepared for SDS-PAGE as previously described (23). The insoluble fractions were solubilized by resuspension in m-PER supplemented with benzonase nuclease (Novagen), sonication, and centrifugation for 10 min at 4 °C and 4,000 × g. Protein determination was carried out on each lysate using a Bradford protein assay (Bio-Rad). For both soluble and insoluble fractions, identical amounts of total protein were analyzed by SDS-PAGE and Western blotting with an anti-Htt exon-1 antibody (1:5000) and anti-FLAG (1:2000) as previously described (23).
Determination of Cell Viability in N2A Cells using a pSIVA-based Cell Suspension Toxicity Assay
Cells maintained in growth medium were transfected as described above and treated with pSIVA, a membrane polarity-sensitive annexin-based green fluorescent probe (31), 72 h after transfection using a pSIVA concentration of 250 ng/ml. Following a 2 h staining period, cells were harvested and spun at 500 × g for 5 min at room temperature. Following successive cycles of pelleting and suspension, cells were fixed and permeabilized using 4% paraformaldehyde (Sigma) and 0.02% Digitonin (Sigma) in PBS with calcium and magnesium. Fixed cells were stained with MW7 (1:2500) followed by Cy3-anti mouse IgM (Jackson ImmunoResearch) and labeled with Hoechst (Invitrogen/Molecular Probes) as described (23). Following antibody and Hoechst staining, cells were resuspended in fluoromount (Sigma), added to a glass coverslip, and mounted. Cells were imaged at 10× using the MosaiX feature of Axiovision with a Zeiss Axiovert 200 m fluorescence microscope and motorized stage. The resulting images were imported into and analyzed with Volocity Quantitation software (Perkin Elmer). A lower threshold was defined for Htt-positive staining using MW7 fluorescence. Average pSIVA intensity was found for each Htt positive cell. Htt toxicity is reported as a ratio between healthy and unhealthy cells and normalized against Htt exon-1 16Q, the non-expanded polyQ nontoxic control. Data presented are an average (± S.D.) of three separate experiments.
Molecular Dynamics Simulations
All models were built manually with the programs QUANTA (Accelrys, Inc.) and O (32). PolyQ peptides are interrupted by three turns to form a four-strand antiparallel β-sheet structure. The turns were modeled as a β-turn conformation and all the Gln side chains were aligned to maximize the number of hydrogen bonds (H-bonds). In the PGQ9 (EDQ9) structures, the turn QQQQ was mutated to QPGQ (QEDQ); in PGQP (PGQW), residues 7, 18, 40 were mutated to Pro (Trp) in addition to the turn mutations. All-atom molecular dynamics (MD) simulations were performed with the program NAMD (33) using CHARMM 22/27 force field. Periodic boundary conditions were used in all simulations. The electrostatic interactions were calculated using Particle Mesh Ewald algorithm with 12 Å real space cutoff distance and grid widths of 0.89–0.95 Å. We used a 12 Å cutoff radius for the Lennard-Jones interactions. Calculations were performed in the NPT ensemble. Nose-Hoover Langevin piston pressure control was used to maintain the system at a constant pressure of 1 atm. A constant temperature of 300K was controlled by Langevin dynamics with a damping coefficient of 2.5/ps. The system consisted of 23000–25000 atoms, depending on the specific run and including ∼7800 water molecules. MD runs used an integration step of 2 fs with SHAKE algorithm. Before all MD runs, the manually built structures were subjected to 5000 steps of energy minimization with the conjugate gradients algorithm used by NAMD. The models were then solvated in a rectangular box with TIP3 water molecules. The whole system (polypeptide with solvent) was again minimized and then brought to 300K in 10,000 steps. Finally, MD simulations were performed to study the conformational stability of the proposed models for 10 ns (20 ns for PGQ6/12) in NPT ensemble. Model figures were generated with the program VMD (34).
RESULTS
A Compact β Model of Mutant Htt poly Q Structure
To investigate the relevance of the compact β model on mutant Htt polyQ aggregation and toxicity, five novel Htt exon-1 cDNAs (designed based on their propensity to form a compact β structure) were prepared for expression in mammalian cells (Fig. 1A). The first two constructs, Htt exon-1 PGQ9, a putative β-turn former (Fig. 1B) and PGQP, a putative β-strand breaker (Fig. 1B), were modified from constructs originally described in (23). Htt exon-1 PGQW (Fig. 1A), a PGQP variant, and Htt exon-1 EDQ9 (Fig. 1A), a PGQ9 variant, were designed to investigate the influence of different amino acid substitutions at the central putative β-strand residue or at the β-turn positions. The prediction was that both of these constructs would form a compact β structure (Fig. 1B) and, therefore, be prone to aggregate, albeit for different reasons. Compact β structure within Htt exon-1 PGQW could be stabilized by hydrophobic interactions between Trp side chains within the β-strand. Htt exon-1 EDQ9 was predicted to form a compact β structure because Glu/Asp is the second most probable β-turn-inducing pair (following Pro/Gly) at the i, i+1 β-turn positions within β-strands of known proteins (35). The final Htt exon-1 construct, PGQ6/12, was designed with alternating PGQ6/PGQ12 elements (Fig. 1, A and B) to determine the effect of altering polyQ length within putative β-strands of the same peptide. We reasoned that this design would prevent β-strands from aligning for maximum hydrogen bonding, rendering the putative compact β structure less stable and therefore less prone to aggregate.
Aggregation Propensity of Htt Exon-1 PolyQ and Compact β PolyQ Variants in N2a Cells
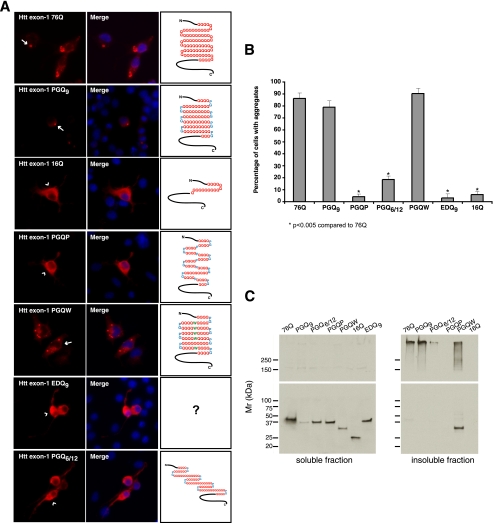
Previous studies demonstrated that placement of a single Pro residue, a known β-strand breaker, within the center of three out of four PGQ9 elements effectively blocked aggregation of Htt exon-1 in mammalian cells (23). This observation was confirmed in the present studies using a modified Htt exon-1 protein in which five out of six PGQ9 elements contained a central Pro (PGQP, Figs. 1, A and B, and 2A). To determine whether this effect is specific for Pro, or could be observed with a different amino acid not typically associated with β-strand breaks, the central Pro residue was replaced with a Trp residue (PGQW, Fig. 1, A and B). Following expression in N2a cells, Htt exon-1 PGQW formed numerous cellular aggregates that were indistinguishable from those formed by Htt exon-1 PGQ9 or 76Q proteins (Fig. 2A). Quantitative analysis of these data (Fig. 2B) demonstrated that 79% of Htt exon-1 PGQ9-expressing cells and 90% of PGQW-expressing cells contained visible aggregates, while only 4% of PGQP-expressing cells had detectable aggregates. Western blot analysis of N2a cell extracts showed a slightly faster migration rate for PGQW compared with other expanded polyQ and compact β polyQ Htt exon-1 mutants (Fig. 2C, left panel). This observation could result from a higher affinity of the hydrophobic Trp residue to SDS, rendering PGQW with a greater negative charge and faster mobility on SDS-PAGE. In addition, analysis of detergent-solubilized extracts prepared from Htt exon-1 76Q-, PGQ9-, and PGQW-expressing cells demonstrated that while 76Q and PGQ9-derived aggregates were completely stable in SDS, PGQW-derived aggregates were only partially resistant to denaturation in SDS (Fig. 2C, right panel). These data suggest that the PGQW polyQ region may be conformationally different compared with 76Q and PGQ9.
FIGURE 2.
Aggregation propensities of Htt exon-1 polyQ and compact β polyQ proteins in transiently transfected N2a cells. A, immunofluorescence images of transfected N2a cells showing MW7 staining (red, left panels) 48 h after transfection. Arrows indicate Htt exon-1 76Q, PGQ9, and PGQW cytoplasmic and perinuclear aggregates, while arrowheads show diffuse cytoplasmic staining in Htt exon-1 16Q, PGQP-, EDQ9-, and PGQ6/12-expressing cells. Nuclei in merged images were stained with Hoechst (blue, middle panels). B, quantification of aggregated Htt exon-1 by cell counting analysis. Data shown are the average (± S.D.) of three independent experiments. C, Western blot analysis of Htt exon-1 expression 48 h after transfection. Detergent-soluble supernatant (left panels) and insoluble pellet fraction (right panels) samples were prepared as described under “Experimental Procedures.” Htt exon-1 EDQ9 remained mostly soluble by IIF, as 16Q, and was not included in the insoluble fraction samples (right panels). Blots were stained with anti-FLAG (upper panels) and anti-Htt exon-1 (Ref. 29, lower panels) antibodies.
In the next set of experiments designed to investigate the effect of an amino acid pair other than Pro/Gly at the β-turn position, N2a cells were transfected with the Glu/Asp-containing-EDQ9 construct (Fig. 1, A and B). Following expression in N2a cells, Htt exon-1 EDQ9 remained completely soluble (Fig. 2A), with only 3% of transfected cells showing aggregates (Fig. 2B). This result was comparable to that of non-expanded Htt exon-1 16Q protein (6% of cells with aggregates, Fig. 2B), and was confirmed by Western blotting analysis (Fig. 2C). Taken together, these data demonstrated that in contrast to Htt exon-1 PGQ9 and despite similar levels of overexpressed protein (data not shown), Glu/Asp at the β-turn position prevented expanded Htt exon-1 polyQ from forming visible aggregates in cells.
To determine the effect of altering polyQ length within putative β-strands of the same peptide, N2a cells were transfected with Htt exon-1 PGQ6/12. In contrast to 76Q and PGQ9 proteins, Htt exon-1 PGQ6/12 formed aggregates in only 18% of transfected cells (Fig. 2, A and B). Western blot analysis confirmed this result (Fig. 2C), indicating that a polyQ region comprised of PGQ elements of different length diminished the ability of Htt exon-1 to form the visible aggregates observed for Htt exon-1 76Q, PGQ9, and PGQW proteins.
Monoclonal Antibody 3B5H10 Recognizes an Epitope in Htt Exon-1 PolyQ and Selective Compact β PolyQ Proteins
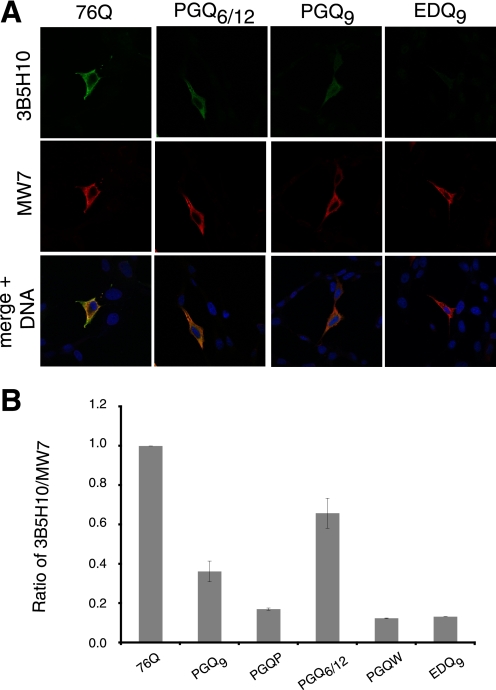
Recent studies have demonstrated that 3B5H10 monoclonal antibody recognizes a compact, two-stranded β-rich hairpin structure formed by polyQ.6 To probe for 3B5H10-reactive Htt species in the present studies, HT22 cells were transfected with Htt exon-1 polyQ and compact β polyQ constructs and processed for immunocytochemistry 24 h following transfection. As expected, 3B5H10 strongly stained cells containing diffuse Htt exon-1 76Q (Fig. 3A). In addition, Htt exon-1 PGQ6/12 and PGQ9 proteins were also recognized by 3B5H10 (Fig. 3A), suggesting that both of these Htt exon-1 mutants have compact β hairpin structure in their polyQ region. In contrast, 3B5H10 staining was not detected for Htt exon-1 PGQP, PGQW, or EDQ9 proteins, despite strong staining with monoclonal antibody MW7 (Fig. 3A), which recognizes a broad array of Htt species. The levels of 3B5H10-reactive Htt species were quantified, relative to MW7 staining, and are presented in Fig. 3B. Relative to Htt exon-1 76Q (set to a ratio of 1.0), the ratio of 3B5H10:MW7 reactivity was 0.65 for PGQ6/12 and 0.39 for PGQ9. Thus, the compact, hairpin structure of polyQ recognized by 3B5H10 is preferentially found in only a subset of our Htt constructs (Htt exon-1 76Q > PGQ6/12 > PGQ9 ⋙ PGQP, PGQW, EDQ9).
FIGURE 3.
Reactivity of Htt exon-1 polyQ and compact β polyQ proteins with 3B5H10, an antibody that recognizes compact, two-stranded hairpins of polyQ, and MW7, an antibody that recognizes a wide range of Htt species, in transiently-transfected mammalian cells. A, selected immunofluorescence images of transfected HT22 cells showing 3B5H10 (green, upper panels) and MW7 (red, middle panels) staining 48 h after transfection. Nuclei in merged images were stained with Hoechst (blue, bottom panels). B, quantification of 3B5H10 reactivity, relative to MW7 staining, for Htt exon-1 polyQ and compact β polyQ proteins. All proteins are compared with Htt exon-1 76Q. Data shown are the average (± S.D.) of three independent experiments.
Expanded Htt Exon-1 PolyQ and Select Compact β PolyQ Proteins Are Toxic in Mammalian Cells
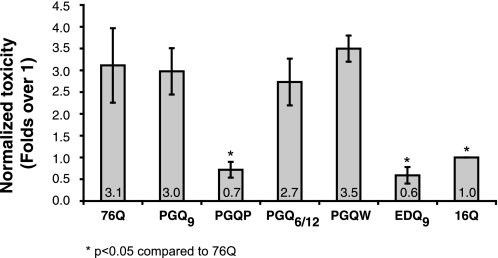
Previous studies carried out in N2a cells and in primary cortical neurons have demonstrated that the Htt exon-1 compact β construct PGQ9, a putative β-turn former, is as toxic as the uninterrupted 76Q protein, while Htt exon-1 PGQP, a putative β-strand breaker, has a toxicity comparable to the non-expanded Htt exon-1 16Q protein (23). To test for polyQ-mediated toxicity of all compact β Htt exon-1 constructs outlined in Fig. 1, we have developed a novel suspension-based fluorescent cell viability assay. This assay is based on co-staining of cells with the monoclonal antibody MW7, a monoclonal antibody specific for the proline-rich region within Htt exon-1 (28) and pSIVA, a membrane polarity-sensitive annexin-based green fluorescent probe that can be used to detect dying or dead cells (31). Cells were treated with pSIVA 72 h following transfection but prior to fixation. After pSIVA staining, cells were harvested, fixed, and stained with MW7 and Hoechst in suspension, and mounted for immunofluorescence imaging. As shown in Fig. 4, Htt exon-1 76Q and PGQ9 displayed similar levels of toxicity when expressed in N2a cells, with a 3-fold increase relative to non-expanded 16Q. In contrast, Htt exon-1 PGQP toxicity was comparable to 16Q. These data are consistent with previous observations (23), and suggest that disruption of compact β structure ameliorates cell toxicity of Htt exon-1. Substitution of a Trp residue in Htt exon-1 PGQW resulted in a dramatic 5-fold increase in toxicity as compared with PGQP (Fig. 4), while replacement of the Pro/Gly pair with a Glu/Asp pair in EDQ9 resulted in an equally dramatic 5-fold decrease in toxicity relative to PGQ9 (Fig. 4). Interestingly, altering polyQ length within putative β-strands of the same peptide in Htt exon-1 PGQ6/12 inhibited aggregation (Fig. 2), but led to a nearly 3-fold increase in toxicity as compared with non-expanded 16Q (Fig. 4). So, while Htt exon-1 PGQ6/12 contains a compact, hairpin polyQ structure (as detected by 3B5H10 staining (Fig. 3)), it remains soluble and toxic when expressed in N2a cells.
FIGURE 4.
Toxicity of Htt exon-1 polyQ and compact β polyQ proteins in N2a cells. pSIVA-based suspension assay demonstrates that Htt exon-1 76Q, PGQ9, PGQ6/12, and PGQW constructs are toxic, while Htt exon-1 PGQP and EDQ9 are not toxic. Cells were stained with pSIVA, a membrane polarity-sensitive annexin-based green fluorescent probe that can be used to detect dead or dying cells (31), 72 h after transfection. Cells were then fixed and stained with MW7 antibody and processed as described under “Experimental Procedures.” Toxicity values are reported relative to non-expanded Htt exon-1 16Q, which has been previously shown in vitro and in vivo using numerous assays to be soluble and non-toxic. Data shown are the average (± S.D.) of three independent experiments.
Molecular Dynamics Confirms the Stability of Certain Compact β PolyQ Structures
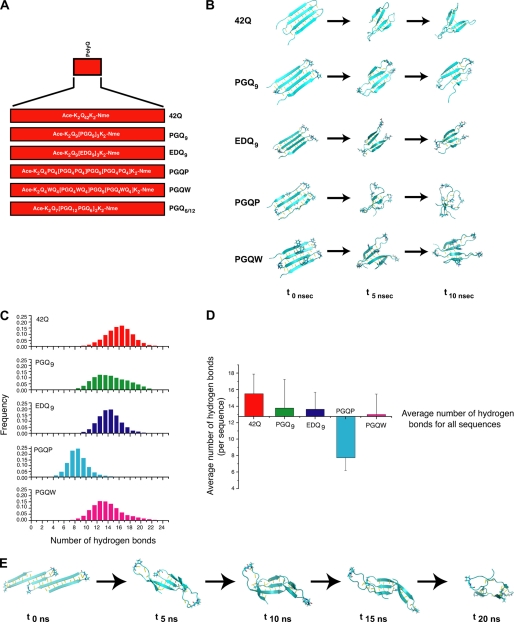
To confirm conformational preferences of Htt exon-1 polyQ and compact β polyQ monomers and the role of hydrogen bonding in stabilizing the compact β structure, we carried out MD simulations. These calculations are based on the hypothesis that mutant Htt forms an abnormal β structure and that this structure has a compact β conformation. Our working hypothesis is that while not all sequences that are able to form a compact β structure will be toxic, sequences that do not form a compact β structure will not be toxic. The relative ability of different sequences to form a stable, four-stranded compact β structure was investigated by analyzing the stability of pre-formed four-stranded compact β-hairpin structures during long MD simulations. Peptide sequences for the simulations are outlined in Fig. 5A and include an uninterrupted 42Q, along with comparable-length compact β sequences modeled after the Htt exon-1 cell expression constructs (Fig. 1A).
FIGURE 5.
Molecular dynamics simulation studies. A, schematic representation of polyQ and compact β polyQ sequences. These sequences are analogous to the constructs presented in Fig. 1A. B, snapshots for each sequence at the initial (t0 ns), midway (t5 ns), and final (t10 ns) time points of each 10 ns simulation. (C) Histograms of the number of main-chain hydrogen bonds over the 10-ns MD simulation for each sequence. D, average number of main-chain hydrogen bonds for each structure. Error bars represent root mean squared (rms) fluctuations in the number of hydrogen bonds with respect to the average. E, snapshots of PGQ6/12 at the initial (t0 ns), midway (t5 ns, t10 ns, t15 ns), and final (t20 ns) time points over the 20-ns molecular dynamics simulation.
Inter-strand Hydrogen Bonding Stabilizes the PolyQ Compact β Structure
To study the stability of the postulated antiparallel, β-strand/β-turn conformation illustrated in Fig. 1B, we followed the folded peptides in Fig. 5A through a 10 ns MD simulation and monitored preservation of the compact, β-strand/β-turn structure throughout the simulation period. Shown in Fig. 5B are representative “snapshot” images for each sequence taken at times 0, 5 ns, and 10 ns of the simulation. To quantitatively evaluate how different peptide sequences affect stability of the folded β hairpins, we monitored main chain hydrogen bond (H-bond) formation over the 10 ns simulation period. Assessment of main-chain hydrogen bonding demonstrated that 42Q, PGQ9, EDQ9, and PGQW maintained a similar number of H-bonds during the 10 ns simulation (Fig. 5C). With an average number of 15 H-bonds, 42Q was only slightly higher in main-chain H-bond formation than the compact β sequences (13–13.5 H-bonds; Fig. 5D). H-bond formation for PGQP was markedly reduced compared with the other sequences (Fig. 5C), with an average of 8 H-bonds present over the 10 ns simulation period. These data suggest that, based on main-chain H-bond formation, the PGQP monomer forms the least stable four-stranded structure of all sequences examined. In addition, these data suggest that stability of the EDQ9 monomer is comparable to PGQ9, and imply that EDQ9 is able to form a monomeric compact β structure in solution, despite the fact that the Htt exon-1 EDQ9 protein does not form visible aggregates in transfected cells (Fig. 2A).
Altering PolyQ Repeat Length Within Consecutive β-Strand Elements of the Same Peptide Results in Formation of an Early Stage β-Barrel Structure
To determine the effect of altering polyQ-repeat length within consecutive β-strands of the same peptide (thus, reducing the number of main-chain H-bonds) on compact β monomer stability, we built the PGQ6/12 sequence. For this analysis, a 20-ns MD simulation was carried out. Interestingly, over the 20-ns simulation period, the PGQ6/12 sequence loses the extended β-sheet conformation preserved in the other polyQ and compact β polyQ sequences (Fig. 5B), and adopts a structure that is reminiscent of an early-stage β-barrel (Fig. 5E, supplemental Fig. S1). These data suggest that an expanded polyQ sequence designed with alternating PGQ6/PGQ12 elements may form a globular conformation that would lack a suitable binding site for homomeric or heteromeric associations.
DISCUSSION
One attractive hypothesis that has been suggested for HD pathogenesis is the formation of a toxic conformation by which abnormally-folded mutant Htt can recruit additional Htt molecules, or other cellular binding partners to alter their function (14). In the present study, we have explored this hypothesis in the context of the compact β structural model of mutant Htt polyQ (23). To this end, we have designed mammalian cell expression constructs encoding compact β variants of Htt exon-1 to address the relationship between mutant polyQ structure and both aggregation and neurotoxicity. In parallel, we carried out an MD study, using similar expanded polyQ sequences to gain insight into mutant polyQ aggregation on a structural level. This MD study was based on the hypothesis that formation of a compact β is a pre-requisite for toxicity.
Using our cell expression constructs, we demonstrate that an Htt exon-1 protein with Trp interruptions within the polyQ region was able to aggregate and was toxic when expressed in cultured neuronal cells, even though Pro interruptions at the same positions abrogated aggregation and toxicity. In contrast, placement of Glu/Asp, a predicted β-turn former, at the β-turn position prevented aggregation and toxicity, even though placement of Pro/Gly at the β-turn positions preserved aggregation and toxicity. MD simulations using peptide sequences based on the polyQ region within these constructs suggest that main chain hydrogen bonding is involved in stabilizing the compact β polyQ structure. Secondly, we found that altering polyQ length within putative β-strands of the same mutant Htt exon-1 protein resulted in a dramatic decrease in the formation of visible aggregates in cells, despite the presence of β-hairpin structure. Interestingly, while this protein remained mostly soluble, it was as toxic as an uninterrupted polyQ Htt exon-1 protein. Taken together, these studies imply that the formation of a compact hairpin of polyQ within Htt exon-1 is necessary but insufficient for both aggregation and toxicity of Htt.
Recent studies demonstrate that the monoclonal antibody 3B5H10 recognizes expanded polyQ in a compact, two-stranded hairpin conformation,6 and that a minimum of 8 consecutive glutamines is required for binding.7 In the present studies, we show that 3B5H10 reacts in situ with Htt exon-1 76Q, the PGQ9 β-turn former, and the β-strand-alternating PGQ6/12 protein. 3B5H10 reactivity was slightly higher for the PGQ6/12 protein than for PGQ9 β-turn former. This difference may be explained by the longer β-strand segments present in the PGQ6/12 construct, as 3B5H10 reactivity is stronger with longer, expanded polyQ tracts.6 While MD studies suggest that PGQW and EDQ9 proteins both form a compact β monomer, neither protein was recognized by 3B5H10, and could be due to steric hindrance (PGQW) and electrostatic repulsion (EDQ9). The electrostatic repulsion that possibly prevents 3B5H10 binding to EDQ9 may also prevent homotypic association (oligomerization and aggregation) and heterotypic formation of toxic protein complexes.
Based on the original Htt exon-1 compact β cell model (23), a correlation was observed between Htt exon-1 proteins able to form a compact β structure and toxicity. The results of the present study are summarized in Table 1. While a correlation was observed between the formation of visible aggregates and toxicity, there was an exception to this finding. Specifically, the Htt exon-1 β-strand-alternating PGQ6/12 protein did not form visible aggregates in situ, yet was highly toxic in cultured cells. Interestingly, this protein displays 3B5H10 reactivity, demonstrating the presence of a compact two-stranded hairpin conformation of polyQ,6 which is believed to be associated with a toxic form of Htt.6 Molecular-simulating unfolding data suggests that this sequence may form a more globular structure (early stage β-barrel) with somewhat reduced surface area for self-associations, and may be less able to form larger oligomeric species. The present data cannot rule out the possibility that small oligomeric species are formed by this protein. Future in vitro studies with purified recombinant protein will be necessary to determine whether or not the Htt exon-1 PGQ6/12 protein is able to form oligomeric structures.
TABLE 1.
Summary of cell transfection and molecular dynamics data
ND, not determined.
| Construct transfected | Aggregation | Toxicity | 3B5H10 reactivity | Predicted compact β monomer by MD |
|---|---|---|---|---|
| 76Q | + | + | + | + |
| PGQ9 | + | + | + | + |
| EDQ9 | - | - | - | + |
| PGQP | - | - | - | - |
| PGQW | + | + | - | + |
| PGQ6/12 | - | + | + | + |
| 16Q | - | - | ND | ND |
In these studies, a combined cell biological, biochemical, and biophysical approach was used to investigate the relevance of the compact β model of mutant polyQ structure on Htt aggregation and toxicity. This approach does have some limitations, including the use of artificial polyQ constructs, the use of assumptions for MD studies, and the inability to detect soluble oligomeric species in cells. Nevertheless, our data support a compact β model for polyQ cell toxicity. These data suggest that the compact β conformation may be critical for both oligomerization and toxicity. Our data also suggest that compact β-mediated oligomerization can be decoupled from toxicity (PGQ6/12), suggesting that the compact β conformation could be toxic in both monomeric and oligomeric forms of Htt. Taken together, these studies may help elucidate the nature of the toxic species, and could provide targets for experimental therapeutic to treat Huntington disease.
Supplementary Material
Acknowledgments
We thank Paul Patterson and Jan Ko for providing MW7 antibody and for critical reading of the manuscript, Frances Davenport for providing N2a cells, Donald DeFranco and Marcia Lewis for providing mouse hippocampal HT22 cells, Xia Feng for assistance with initial aggregation studies, and Randy Lawrence, Eka Chighladze, and Otto Schoek for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01NS053679 (to M. A. P.), P01NS16375 (to C. A. R.), 2NS45091 (to S. F.), AG027936 (to R. L.), and DOE DE-FG02–04ER25626 (to M. L. A.), the National Institutes of Health-NIGMS UCSF Medical Scientist Training Program (to J. M.), and a fellowship from the University of California at San Francisco Hillblom Center for the Biology of Aging (to J. M.). This work was also supported by the Huntington Disease Society of America Coalition for the Cure and by CHDI, Inc.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
J. Miller and S. Finkbeiner, submitted for publication.
J. Miller and S. Finkbeiner, personal communication.
- HD
- Huntington disease
- polyQ
- polyglutamine
- Htt
- huntingtin
- N2a
- Neuro2a
- IIF
- indirect immunofluorescence
- PMT
- photomultiplier tube
- MD
- molecular dynamics
- H-bond
- hydrogen bond.
REFERENCES
- 1. Ross C. A., Margolis R. L., Rosenblatt A., Ranen N. G., Becher M. W., Aylward E. (1997) Medicine 76, 305–338 [DOI] [PubMed] [Google Scholar]
- 2. Zoghbi H. Y., Orr H. T. (2000) Annu. Rev. Neurosci. 23, 217–247 [DOI] [PubMed] [Google Scholar]
- 3. Huntington's Disease Collaborative Research Group (1993) Cell 72, 971–983 [DOI] [PubMed] [Google Scholar]
- 4. Ross C. A., Tabrizi S. J. (2011) Lancet 10, 83–98 [DOI] [PubMed] [Google Scholar]
- 5. Andrade M. A., Bork P. (1995) Nat. Genet. 11, 115–116 [DOI] [PubMed] [Google Scholar]
- 6. Duyao M., Ambrose C., Myers R., Novelletto A., Persichetti F., Frontali M., Folstein S., Ross C., Franz M., Abbott M., Gray J., Conneally P., Young A., Penney J., Hollingsworth Z., Shoulson I., Lazzarini A., Falek A., Koroshetz W., Sax D., Bird E., Vonsattel J., Bonilla E., Alvir J., Bickham Conde J., Cha J.-H., Dure L., Gomez F., Ramos M., Sanchez-Ramos J., Snodgrass S., de Young M., Wexler N., Moscowitz C., Penchaszadeh G., MacFarlane H., Anderson M., Jenkins B., Srinidhi J., Barnes G., Gusella J., MacDonald M. (1993) Nat. Genet. 4, 387–392 [DOI] [PubMed] [Google Scholar]
- 7. Rosenblatt A., Liang K. Y., Zhou H., Abbott M. H., Gourley L. M., Margolis R. L., Brandt J., Ross C. A. (2006) Neurology 66, 1016–1020 [DOI] [PubMed] [Google Scholar]
- 8. Ravina B., Romer M., Constantinescu R., Biglan K., Brocht A., Kieburtz K., Shoulson I., McDermott M. P. (2008) Mov. Disord. 23, 1223–1227 [DOI] [PubMed] [Google Scholar]
- 9. Kim Y. J., Yi Y., Sapp E., Wang Y., Cuiffo B., Kegel K. B., Qin Z. H., Aronin N., DiFiglia M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12784–12789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ratovitski T., Nakamura M., D'Ambola J., Chighladze E., Liang Y., Wang W., Graham R., Hayden M. R., Borchelt D. R., Hirschhorn R. R., Ross C. A. (2007) Cell Cycle 6, 2970–2981 [DOI] [PubMed] [Google Scholar]
- 11. Kim M. W., Chelliah Y., Kim S. W., Otwinowski Z., Bezprozvanny I. (2009) Structure 17, 1205–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kuemmerle S., Gutekunst C. A., Klein A. M., Li X. J., Li S. H., Beal M. F., Hersch S. M., Ferrante R. J. (1999) Ann. Neurol. 46, 842–849 [PubMed] [Google Scholar]
- 13. Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S. (2004) Nature 431, 805–810 [DOI] [PubMed] [Google Scholar]
- 14. Ross C. A., Poirier M. A. (2005) Nat. Rev. Mol. Cell Biol. 6, 891–898 [DOI] [PubMed] [Google Scholar]
- 15. Ravikumar B., Rubinsztein D. C. (2006) Mol. Aspects Med. 27, 520–527 [DOI] [PubMed] [Google Scholar]
- 16. Kopito R. R. (2000) Trends Cell Biol. 10, 524–530 [DOI] [PubMed] [Google Scholar]
- 17. Thakur A. K., Jayaraman M., Mishra R., Thakur M., Chellgren V. M., Byeon I. J., Anjum D. H., Kodali R., Creamer T. P., Conway J. F., Gronenborn A. M., Wetzel R. (2009) Nat. Struct. Mol. Biol. 16, 380–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wacker J. L., Zareie M. H., Fong H., Sarikaya M., Muchowski P. J. (2004) Nat. Struct. Mol. Biol. 11, 1215–1222 [DOI] [PubMed] [Google Scholar]
- 19. Legleiter J., Mitchell E., Lotz G. P., Sapp E., Ng C., DiFiglia M., Thompson L. M., Muchowski P. J. (2010) J. Biol. Chem. 285, 14777–14790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tam S., Spiess C., Auyeung W., Joachimiak L., Chen B., Poirier M. A., Frydman J. (2009) Nat. Struct. Mol. Biol. 16, 1279–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Perutz M. (1994) Protein Sci. 3, 1629–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ross C. A., Poirier M. A., Wanker E. E., Amzel M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Poirier M. A., Jiang H., Ross C. A. (2005) Hum. Mol. Genet. 14, 765–774 [DOI] [PubMed] [Google Scholar]
- 24. Miller J., Arrasate M., Shaby B. A., Mitra S., Masliah E., Finkbeiner S. (2010) J. Neurosci. 30, 10541–10550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Legleiter J., Lotz G. P., Miller J., Ko J., Ng C., Williams G. L., Finkbeiner S., Patterson P. H., Muchowski P. J. (2009) J. Biol. Chem. 284, 21647–21658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sathasivam K., Lane A., Legleiter J., Warley A., Woodman B., Finkbeiner S., Paganetti P., Muchowski P. J., Wilson S., Bates G. P. (2010) Hum. Mol. Genet. 19, 65–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Poirier M. A., Li H., Macosko J., Cai S., Amzel M., Ross C. A. (2002) J. Biol. Chem. 277, 41032–41037 [DOI] [PubMed] [Google Scholar]
- 28. Ko J., Ou S., Patterson P. H. (2001) Brain Res. Bull. 56, 319–329 [DOI] [PubMed] [Google Scholar]
- 29. Peters M. F., Ross C. A. (2001) J. Biol. Chem. 276, 3188–3194 [DOI] [PubMed] [Google Scholar]
- 30. Peters-Libeu C., Newhouse Y., Krishnan P., Cheung K., Brooks E., Weisgraber K., Finkbeiner S. (2005) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61, 1065–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim Y. E., Chen J., Chan J. R., Langen R. (2010) Nat. Methods 7, 67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 33. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graph. 14, 33–38, 27–38 [DOI] [PubMed] [Google Scholar]
- 35. Hutchinson E. G., Thornton J. M. (1994) Protein Sci. 3, 2207–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.