Abstract
Cyclophilin A (CyPA) is a ubiquitously distributed peptidylprolyl cis-trans isomerase (PPIase) that possesses diverse biological functions. Extracellular CyPA is a potent chemokine, which can directly induce leukocyte chemotaxis and contribute to the pathogenesis of inflammation-mediated diseases. Although it has been identified that the chemotaxis activity of CyPA is mediated through its cell surface signaling receptor CD147, the role of CyPA PPIase activity in this process is disputable, and the underlying molecular mechanism is still poorly understood. In this study, we present the first evidence that CyPA induces leukocyte chemotaxis through a direct binding with the ectodomain of CD147 (CD147ECT), independent of its PPIase activity. Although NMR study indicates that the CD147ECT binding site on CyPA overlaps with the PPIase active site, the PPIase inactive mutant CyPAR55A exhibits similar CD147ECT binding ability and chemotaxis activity to those of CyPAWT. Furthermore, we have identified three key residues of CyPA involved in CD147ECT binding and found that mutations H70A, T107A, and R69A result in similar levels of reduction in CD147ECT binding ability and chemotaxis activity for CyPA, without affecting the PPIase activity. Our findings indicate that there exists a novel mechanism for CyPA to regulate cellular signaling processes, shedding new light on its applications in drug development and providing a new targeting site for drug design.
Keywords: Cell Surface Receptor, Chemotaxis, NMR, Prolyl Isomerase, Protein-Protein Interactions, CD147, cyclophilin
Introduction
Leukocyte trafficking and recruitment are critical processes and essential aspects in host immune surveillance as well as for inflammation-mediated pathology. Although chemokines are the main regulators of leukocyte trafficking, extracellular cyclophilins have been shown to have potent chemoattractant properties for human leukocytes (1). Cyclophilins are ubiquitously expressed enzymes that catalyze the isomerization of peptidyl-prolyl bonds and regulate a diverse array of protein functions (2). Cyclophilin A (CyPA),4 the most abundant cytoplasmic cyclophilin (3, 4), initially gained its recognition as the target of the immunosuppressive drug cyclosporine A (CsA) (5, 6), and the CyPA-CsA complex can inhibit the activity of calcineurin. It was later found that CyPA plays important roles in protein folding, trafficking, assembly, immunomodulation, and cell signaling.
Many structures of free CyPA and CyPA complexes with interacting molecules, such as CsA or CsA variants, substrate peptides, as well as the HIV-1 capsid protein, have been determined (7). The structure of CyPA is composed of eight strands of antiparallel β-sheets in a flattened β-barrel with two helices capping the top and bottom. A shallow pocket on the barrel surface, constituting residues Arg-55, Phe-60, Met-61, Gln-63, Phe-113, Trp-121, Leu-122, and His-126, is the PPIase active site, and Arg-55 was identified to be a crucial catalytic residue. CsA also binds the same area of CyPA through interacting with residues Arg-55, Phe-60, Met-61, Gln-63, Gly-72, Ala-101, Asn-102, Ala-103, Gln-111, Phe-113, Trp-121, Leu-122, and His-126 (8, 9). Furthermore, NMR studies have demonstrated that CyPA efficiently catalyzes prolyl cis-trans isomerization of interleukin-2 tyrosine kinase (10, 11), cell signaling adaptor protein Crk (12), and HIV-1 capsid protein (13, 14) and thus regulates their functions.
CD147, a type I transmembrane protein, was identified to be the cell surface signaling receptor for extracellular CyPA (1, 15). The chemotaxis activity of CyPA is dependent upon its interaction with CD147, and inhibition of the CyPA/CD147 interaction with anti-CD147 mAb or CsA greatly decreases migration of immune cells to the sites of inflammation in mouse models of diseases such as acute lung injury, asthma, and rheumatoid arthritis (1, 16–18). Moreover, CyPA/CD147 interaction also plays a role during the infection of many viruses, including HIV-1 (19), severe acute respiratory syndrome coronavirus (SARS-CoV) (20), vaccinia virus, vesicular stomatitis virus, and others (21, 22). In addition, the cell surface expression of CD147 was reported to be regulated by the cytosolic CyPA (23). Therefore, understanding the molecular mechanisms of CyPA/CD147 interaction may have profound therapeutic value with respect to therapies for inflammation and viral infection (24–26).
So far, the molecular mechanism of CyPA/CD147 interaction is still poorly understood. Two CyPA-responsive sites on CD147 have been identified. It was first reported that residues Pro-180 and Gly-181 in the ectodomain of CD147 are critical for the signaling and chemotaxis induced via CyPA. The PPIase activity of CyPA was suggested to be crucial for the CyPA/CD147 interaction responsible for chemotaxis as CyPAH126A and CyPAF60A mutants with less than 1% of the wild-type (WT) CyPA catalytic efficiency failed to initiate the signaling event of chemotaxis (15). It was later reported that the cell surface expression of CD147 correlates with the capacity of its transmembrane domain to interact with CyPA. Residue Pro-211 in the transmembrane domain of CD147, instead of residue Pro-180 in the ectodomain, was found to be critical for this interaction (23). Recently, it was reported that CyPA can catalyze the isomerization of Pro-211 but not Pro-180 in vitro (27). Nevertheless, the mechanism of the signal transduction attributable to extracellular interaction between CD147 and its ligand, secreted CyPA, remains unclear.
Here, we report that the ectodomain of CD147 (CD147ECT) can directly interact with CyPA, which should be responsible for the leukocyte chemotaxis induced by CyPA. We found that the CD147ECT binding site on CyPA overlaps with the PPIase active site and the CsA binding site, but the chemotaxis activity of CyPA is unrelated to its PPIase activity.
EXPERIMENTAL PROCEDURES
Construction of Plasmids
The cDNA fragment encoding human CyPA was amplified by PCR using primers 5′-CGGCATATGGTCAACCCCACCGTGTTCTTCGAC-3′ and 5′-CCGCCTCGAG TTCGAGTTGTCCACAGTCAGC-3′, in which NdeI and XhoI restriction enzyme sites (underlined) have been added, respectively. The PCR product was doubly digested with NdeI and XhoI and ligated into the expression vector pET-32a+, with a C-terminal His6 tag introduced. The cDNA fragment encoding CD147ECT (residues 22–205) was amplified via PCR with primers 5′-GCGGAATTCATATGGCTGCTGGTACCGTTTTCACCACCGTTGAAG ACCTG-3′ and 5′-CAATACTCGAGGTGCGTGCGCACGCGGAGCG-3′, in which NdeI and XhoI restriction enzyme sites (underlined) have been added, respectively. The PCR product was doubly digested with NdeI and XhoI and ligated into the expression vector pGEX, which produces CD147ECT with a GST tag at the N terminus. CyPA site-directed mutagenesis was carried out using a QuikChange kit from Qiagen. These DNA constructs were verified by DNA sequencing.
Protein Purification and Sample Preparation
For protein expression, all plasmids were transformed into Escherichia coli strain BL21(DE3), respectively. For wild-type CyPA and its mutants, the bacteria were cultured to in LB medium at 16 °C until A600 reached 1.0, and isopropyl-β-d-1-thiogalactopyranoside was then added to the culture to induce protein expression at a final concentration of 100 mg/liter. Cells were harvested 12 h later, resuspended in 50 mm phosphate-buffered saline (PBS) at pH 7.0, and homogenized by sonication. The cell lysate was centrifuged, and the supernatant was loaded directly onto a Ni2+-nitrilotriacetic acid agarose column (Qiagen) pre-equilibrated in PBS (pH 7.0) with 10 mm imidazole and 300 mm NaCl. The target protein was eluted using PBS (pH 7.0) with 200 mm imidazole and was further purified by gel filtration with a Superdex 75 size-exclusion column (GE Healthcare) equilibrated with 50 mm Tris buffer (pH 7.0) with 50 mm NaCl using an ÄKTApurifier 100 system. Uniformly 15N-labeled protein was prepared by growing bacteria in minimal medium with 15NH4Cl as the sole nitrogen.
For GST-fused CD147ECT, E. coli BL21(DE3) cells harboring the recombinant plasmid pGEX/CD147ECT were grown in LB medium at 18 °C until A600 reached 0.8, protein expression was induced with 100 mg/liter isopropyl-β-d-1-thiogalactopyranoside, and the cells was harvested after 20 h of growth. GST-CD147ECT was purified from the soluble fraction using a Sepharose 4B-glutathione column (Amersham Biosciences) and subsequent gel filtration with Superdex 75 column (Amersham Biosciences) in 20 mm HEPES buffer (pH 7.3). Expression and purification of untagged CD147ECT protein were as described previously (28).
Chemotaxis Assay
Chemotaxis assays (29) were conducted with the neutrophil-like cell line, HL-60, in a 48-well modified Boyden chamber (Neuro Probe), with the upper and lower compartments separated by a polyvinyl pyrrolidone-free polycarbonate filter of 5-μm pore size (Neuro Probe). HL-60 cells (1 × 106 cells/ml) in binding medium (serum-free RPMI 1640 medium (Invitrogen) supplemented with 2% bovine serum albumin) were added into the upper compartments, whereas the binding medium containing 10 nm CyPAWT or CyPA mutants or medium alone (for random migration) was added to the lower wells. For chemokinesis control, 10 nm CyPAR55A was added to both the upper and the lower chambers. Anti-CD147 antibody (50 μg/ml) (BD Biosciences) was added to the cells in the upper chambers to investigate its effect. The chambers were incubated at 37 °C with 5% CO2 for 90 min. The filter was then removed, fixed, and stained with Wright Giemsa (Sigma-Aldrich), and the number of HL-60 cells appearing on the bottom surface of the filter was counted under a microscope in at least four randomly selected fields for each well. Each experimental condition was assayed in triplicate wells. The chemotaxis cell number was calculated by deducting the average migrated cell number for the random migration control test from that for the CyPAWT or CyPA mutants. The relative chemotaxis ability of the CyPA mutant to CyPAWT was calculated by dividing the chemotaxis cell number of CyPA mutants by that of CyPAWT.
NMR Spectroscopy
All uniformly 15N-labeled CyPA NMR samples were in 50 mm Tris buffer (pH 7.5) with 50 mm NaCl and 0.006% 2,2-dimethyl-2-silapentanesulfonic acid in 90% H2O, 10% D2O plus Complete, an EDTA-free protease inhibitor mixture (Roche Applied Science). All NMR experiments were performed at 298 K on a Bruker Avance 800-MHz NMR spectrometer (with CryoProbe). Two-dimensional 1H-15N HSQC spectra were recorded for uniformly 15N-labeled CyPAWT or CyPA mutants (0.2 mm) with or without CD147ECT (0.6 mm). All NMR spectra were processed with NMRPipe and analyzed using NMRView.
GST Pulldown Assay
An equal amount of the purified GST or GST-CD147ECT (1 μg) was applied to a 1.5-ml Eppendorf tube containing 0.1 ml of Sepharose 4B-glutathione beads suspension (Amersham Biosciences). The beads were washed with 600 μl of washing buffer (20 mm HEPES, 50 mm NaCl, pH 7.3). Then, 1 μg of purified CyPAWT or CyPA mutants was added. After incubation for 2 h at 4 °C, the beads were washed again four times using the washing buffer with protease inhibitor mixture. The beads were then collected and resuspended in SDS-PAGE sample loading buffer for Western blot analysis. Two primary antibodies, anti-CD147 and anti-His tag, were used to detect the GST-CD147ECT and CyPAWT or its mutants, respectively. Protein bands were visualized using the ECL Plus detection system (Amersham Biosciences) with HRP-conjugated secondary goat anti-mouse antibody. The protein amount was taken as the grayscale of each visualized band. The binding ability of CyPAWT or CyPA mutants toward CD147ECT was calculated as the proportion of CyPA protein amount to the corresponding GST-CD147ECT protein amount for each pulldown test.
PPIase Assay
PPIase activity was determined in a coupled assay with chymotrypsin using the synthetic tetrapeptide succinyl-Ala-Ala-Pro-Phe-4-nitroanilide (Sigma) (30). 930 μl of 0.1 m Tris-HCl (pH 8.0) and 50 μl of 200 μm solution of α-chymotrypsin were mixed in the spectrometer cell and preincubated at 0 °C for 10 min. Then 10 μl of 1 μm CyPA or mutant solution to be assayed was added, and after 5 min, the assay was initiated by adding 10 μl of a 7.8 mm solution of the peptide dissolved in 0.47 m LiCl in trifluoroethanol. The cis-to-trans isomerization of the Ala-Pro peptide bond, coupled with the chymotryptic cleavage of the trans peptide, was followed by the increase in absorbance at 390 nm for 40 s in a spectrophotometer. The temperature was controlled at 0 °C during the assay. The uncatalyzed cis-trans isomerization in the absence of CyPA was performed as a blank control, and the CyPA PPIase activity on CD147 engagement was performed with the addition of 10 μl of 2 μm CD147ECT to the reaction system.
RESULTS
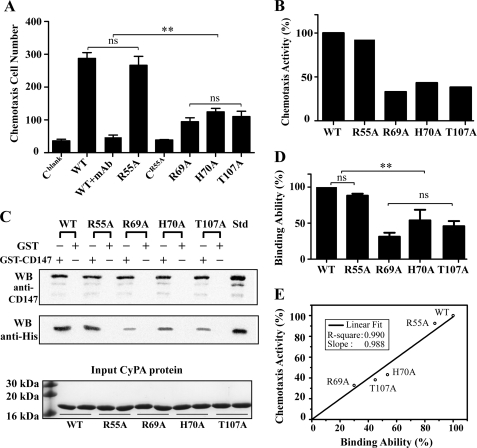
We first performed the in vitro chemotaxis assay to determine the chemotactic activity of wild-type CyPA (CyPAWT) and the inactive mutant CyPAR55A toward the neutrophil-like cell HL-60 using a Boyden chamber. Residue Arg-55 of CyPA is an essential catalytic residue, and the R55A mutation results in complete loss of the PPIase activity (31). The response of HL-60 cells to CyPAWT was dose-dependent and exhibited a typical bell-shaped curve with an optimal chemotactic dose of 10 nm (data not shown). When anti-CD147 antibody was added to HL-60 cells, the number of cells that migrated in response to CyPAWT decreased significantly (Fig. 1A), indicating that the chemotactic activity of CyPA is indeed due to the CD147 expression on HL-60 cells. Unexpectedly, we found that the CyPAR55A mutant displays similar chemotactic ability to CyPAWT, and the number of cells that migrated in response to CyPAR55A was about 90% of that for CyPAWT, at 10 nm. To control for chemokinesis, we conducted the experiment with CyPAR55A added into both the upper and the lower chambers at the same concentration, and the migrated cell number was about the same as that of the negative control group without CyPA (Fig. 1A). These results indicate that the PPIase function of CyPA is not critical to its chemotactic activity mediated by CD147.
FIGURE 1.
Chemotactic activity of CyPA and direct binding between CyPA and CD147 ectodomain. A, the average of migrated HL-60 cell numbers in response to CyPAWT (WT) and mutants CyPAR55A (R55A), CyPAR69A (R69A), CyPAH70A (H70A), and CyPAT107A (T107A) in the chemotaxis assay using the Boyden chamber. HL-60 cells are in the upper chambers at 1 × 106 cells/ml. All proteins are in the lower chambers at a concentration of 10 nm. Cblank stands for negative control with no CyPA in the lower chambers. CR55A indicates the chemokinesis assay with 10 nm CyPAR55A in both the upper and the lower chambers. WT+mAb represents negative control with anti-CD147ECT antibody in the upper chambers and CyPAWT protein in the lower chambers. Only cells that migrated to the bottom surface of the separation filter are counted. Data are presented as the mean of three independent wells with S.D. indicated. ns, not significant; **, p < 0.01. B, the relative chemotaxis activity of CyPA mutants in percentages. The relative chemotaxis activity is calculated by dividing the chemotaxis cell number of each CyPA test group by that of CyPAWT, whereas the average chemotaxis cell number is defined by subtracting the average migrated cell number of the negative control group from that of the CyPA test group. C, GST pulldown assay for the interaction between CD147ECT and CyPAWT or mutants CyPAR55A, CyPAR69A, CyPAH70A, and CyPAT107A. For each CyPA sample, GST-CD147ECT was used as bait, and GST alone was used as negative control. Anti-CD147 antibody was used for detecting GST-CD147ECT. All CyPA proteins have a C-terminal His6 tag, and anti-His antibody was used for detecting CyPA. At the bottom, an SDS-PAGE gel shows the amount of CyPA sample used in each pulldown test. Std designates standard CyPA-His or GST-CD147ECT as positive controls for anti-His and anti-CD147 antibodies in Western blot (WB) experiment. D, the relative CD147 binding ability of CyPA. For each pulldown test, the ratio between the grayscale intensity of the CyPA band and that of the GST-CD147ECT band was calculated, and the ratio was normalized to that of CyPAWT. Each bar represents the average of normalized ratios from three independent pulldown experiments, and error bars are indicated. **, p < 0.01. E, correlation between the relative chemotaxis activity and the relative CD147 binding ability for CyPA and its mutants. The line is from a linear fitting of the five data points, and fitting parameters are indicated.
Because CD147 has been identified as the cell surface receptor of CyPA and it is essential for cyclophilin-mediated signaling (1, 15), we next conducted the in vitro GST pulldown assay to examine whether the ectodomain of CD147 (CD147ECT, residues 22–205) is capable of directly interacting with CyPA. CyPAWT with a C-terminal His tag was incubated with GST-fused CD147ECT (GST-CD147ECT) bound to glutathione-Sepharose 4B beads. After washing extensively, proteins bound to the beads were analyzed with Western blot using anti-His tag and anti-CD147 monoclonal antibodies. As shown in Fig. 1C, CyPAWT does bind GST-CD147ECT, but not free GST as the negative control, indicating that there is a direct binding between CyPA and the ectodomain of CD147. We also performed GST pulldown assay to detect the binding between CD147ECT and CyPAR55A. The amount of CyPAR55A binding to GST-CD147ECT was about 90% of that for CyPAWT, suggesting that CyPAR55A also binds CD147ECT with a comparable affinity.
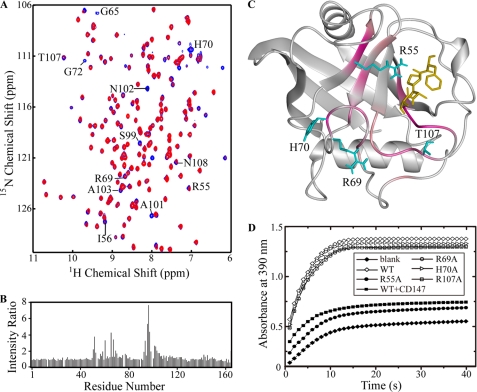
Next, we used NMR spectroscopy to probe the interaction between CyPAWT and CD147ECT. Because protein backbone NH signals (cross-peak) in the two-dimensional 1H-15N HSQC spectrum are sensitive to nearby local chemical environment change, the binding of another protein will cause NH signal intensity or chemical shift changes for residues at the binding site. Therefore, the CD147 binding site on CyPA can be identified from the comparison of the two-dimensional 1H-15N HSQC spectra of CyPA with and without CD147ECT. With the addition of unlabeled CD147ECT, some NH signals in the two-dimensional 1H-15N HSQC spectrum of 15N-labeled CyPAWT became weaker or even disappeared, accompanied with small chemical shift changes (Fig. 2A). The decrease in NH signal intensity is most likely due to the intermediate NMR time scale conformational exchange for CyPA and CD147ECT binding, which results in line broadening of NMR signals. We calculated the NH signal intensity ratio for each residue of 15N-labeled CyPAWT with and without 3-fold unlabeled CD147ECT (Fig. 2B). Residues with over 2.8-fold intensity reduction include Ile-56, Gly-65, His-70, Gly-72, Ser-99, Ala-101, Asn-102, Ala-103, Thr-107, and Asn-108 (labeled in Fig. 2A), and residues Arg-55, Ile-57, Met-61, Gln-63, Gly-74, Gly-75, Lys-82, Leu-98, Gly-104, Gly-109, Ser-110, and Gln-111 also show significant reduction (1.8–2.8-fold) in NH signal intensities. By mapping these residues onto the crystal structure of CyPA, it is obvious that they are spatially clustered and mainly located at the area constituting the PPIase active site and two nearby loops consisting of residues 65–75 and 102–110 (Fig. 2C). This area should be the CD147ECT binding site on CyPA, and it also overlaps with the CsA binding site on CyPA. We thus examined the effect of CD147ECT binding on the PPIase activity of CyPA and found that CyPA does display dramatically lowered PPIase activity in the presence of CD147ECT (Fig. 2D), confirming that CD147ECT has an inhibitory effect on the PPIase activity, as does CsA.
FIGURE 2.
CD147ECT binding site on CyPA. A, an overlay of two-dimensional 1H-15N HSQC spectra of free CyPAWT (blue) and CyPAWT/CD147ECT (ratio 1:3, red). Residues displaying significant NH signal intensity reduction (with NH signal intensity ratio over 2.8 in panel B) upon CD147ECT binding (mentioned under “Results”) are indicated by the one-letter amino acid code and residue number. B, plot of NH signal intensity ratio between CyPAWT/CD147ECT (ratio 1:3) and free CyPAWT. C, ribbon diagram of structure of CyPA in complex with substrate peptide suc-AAPF-NA (Protein Data Bank (PDB) code: 1RMH). Residues displaying significant NH signal intensity reduction upon CD147ECT binding are highlighted in pink. The bound substrate peptide is shown in golden. The side chain of CyPA residue Arg-55, Arg-69, His-70, and Thr-107 is shown in cyan. D, activity of wild-type and mutant CyPA in the protease-coupled prolyl isomerase assay. The time course of the cis-to-trans isomerization of the Ala-Pro peptide bond in the assay peptide succinyl-Ala-Ala-Pro-Phe-4-nitroanilide is measured by the increase in A390 after the coupled hydrolysis by chymotrypsin. The uncatalyzed cis-trans isomerization in the absence of CyPA is shown as a blank control, and isomerization catalyzed by 10 nm wild-type CyPA and mutants and wild-type CyPA preincubated with 10 nm CD147ECT are shown as indicated, respectively.
Analyzing structures of free CyPA and its complexes with peptide substrates or CsA revealed that most of the CyPA residues affected by CD147ECT binding are related to substrates and CsA binding or involved in structural packing, except residues His-70 and Thr-107. To determine whether this direct binding between CyPA and the ectodomain of CD147 is related to the chemotactic activity of CyPA, we mutated His-70, Thr-107, and Arg-69 (the residue preceding His-70) into alanine individually because the side chains of these three residues are known to be frequently involved in intermolecular interaction. Meanwhile, the side chains of the three residues are solvent-exposed and pointed away from the substrate and CsA binding sites on CyPA; thus mutations H70A, T107A, and R69A would not have a significant effect on the overall structure of CyPA. It has been reported that H70A mutation does not affect the PPIase activity of CyPA (32), and our enzymatic assay results also indicated that mutations R69A, H70A, and T107A have little effect on the PPIase activity of CyPA (Fig. 2D).
We again employed the GST pulldown assay to study the effect of these mutations on the binding affinity between CyPA and CD147ECT. Indeed, all three CyPA mutants showed significantly decreased affinity toward CD147ECT as compared with CyPAWT. As shown in Fig. 1D, CyPAR69A only retains 30% of the binding ability for CyPAWT to CD147ECT, whereas CyPAH70A and CyPAT107A possess 54 and 45% of the CD147ECT binding abilities, respectively. Two-dimensional 1H-15N HSQC spectra of these CyPA mutants confirmed that the three point mutations do not cause a large change to the structure of CyPA as the spectra of the mutants are quite similar to that of CyPAWT and only NH signals of residues close to the mutated residue are affected by the mutations (supplemental Fig. 1). These results suggest that the side chains of residues Arg-69, His-70, and Thr-107 are mainly responsible for binding CD147ECT.
We then carried out chemotaxis experiments for CyPAR69A, CyPAH70A, and CyPAT107A mutants and found that these mutants also displayed decreased chemotactic ability. At 10 nm, the chemotaxis cell number for CyPAR69A was 33% of that for CyPAWT, and the chemotactic abilities of CyPAH70A and CyPAT107A were reduced to 43 and 38% of that for CyPAWT, respectively (Fig. 1B). These results suggest that the residues Arg-69, His-70, and Thr-107 are also important for CyPA to function in the leukocyte chemotaxis, although they are not involved in the PPIase activity of CyPA, which confirms that the PPIase function of CyPA is unrelated to its chemotactic activity. It is interesting to note that the degrees of reduction in chemotactic ability for CyPAR55A, CyPAT107A, CyPAH70A, and CyPAR69A mutants correlate well with the degrees of reduction in the binding affinity with CD147ECT (Fig. 1E), strongly suggesting that the direct interaction between CyPA and the ectodomain of CD147 should be responsible for the chemotaxis activity of CyPA.
DISCUSSION
Although it is well documented that extracellular CyPA induces the migration of leukocyte through its receptor CD147, the underlying mechanism is still poorly understood. In this study, for the first time, we demonstrate that the PPIase activity of CyPA is not related to its chemotaxis function because on the one hand, a PPIase inactive mutant CyPAR55A has a comparable chemotactic ability for the neutrophil as CyPAWT. On the other hand, we have identified CyPA mutations H70A, T107A, and R69A at the CD147ECT binding site that significantly decrease the chemotactic ability of CyPA without affecting its PPIase activity.
So far, there are two well established mechanisms for CyPA to regulate the biological functions of its interacting proteins: (i) induce protein conformational change through catalyzing the cis-trans isomerization of a prolyl peptide bond, as in the case of interleukin-2 tyrosine kinase and Crk (10–12); and (ii) inhibit the phosphatase activity of calcineurin by forming complex together with CsA (33). Here, we demonstrate that CyPA can directly interact with the ectodomain of CD147, rather than interacting as an enzyme and a substrate, and our NMR data revealed that the CD147ECT binding site on CyPA overlaps with the PPIase active site and the CsA binding site of CyPA. The three mutations of the binding site residues, H70A, T107A, and R69A, reduced the binding affinity of CyPA for the ectodomain of CD147 and also decreased the chemotactic ability of CyPA to similar levels. Therefore, the PPIase activity-independent interaction between CyPA and the ectodomain of CD147 should be responsible for the leukocyte chemotaxis activity of CyPA.
Our results are not in conflict with the previous observation that mutants CyPAH126A and CyPAF60A fail to initiate the signaling event of chemotaxis, whereas they have less than 1% of the CyPAWT catalytic activity (15). In our NMR experimental data, NH signals of CyPA residues Phe-60 and His-126 are not significantly affected by CD147ECT binding. Analysis of CyPA structures suggests that the aromatic side chains of both residue Phe-60 and residue His-126 play a role in defining the conformation of the loop consisting of residues 118–125 (supplemental Fig. 2). Therefore, mutating either side chain into a methyl group may result in the loop collapse into the PPIase active site pocket, which thus possibly blocks the binding of peptide substrates or CD147. Two-dimensional 1H-15N HSQC spectra of 15N-labeled CyPAH126A and CyPAF60A are significantly different from that of CyPAWT with a large number of NH signals not overlapping (supplemental Fig. 2), strongly suggesting that mutations H126A and F60A have a much more broad impact on the structure of CyPA.
Our finding is consistent with the previous results that the ectodomain of CD147 does not exist in an inherent cis-trans equilibrium dictated by a proline residue, and no peptidylprolyl cis-trans isomerization catalyzed by CyPA for the ectodomain of CD147 was observed (27). Recently, more studies have started to reveal that CyPA can regulate biological processes independent of its PPIase activity (34, 35). It was found that intracellular CyPA regulates influenza A virus infectivity through direct interaction with the virus matrix protein (M1) in a PPIase-independent manner (34). Also, it was recently reported that CyPA is involved in the functional expression of Na+-Ca2+ exchanger (NCX), which can be inhibited by CsA, whereas the PPIase activity of CyPA is not mandatory (35). Taken together, we speculate that it may be a general property for CyPA to regulate cellular processes through a novel PPIase-independent mechanism.
Although CsA is a potent inhibitor of the PPIase activity of CyPs (36), CsA derives not from inhibition of isomerase activity but rather from interruption of signaling events in complex with CyPA (37). Our NMR data showed that the CD147ECT binding site on CyPA overlaps with both the PPIase active site and the CsA binding site (38), and it is expected that CsA inhibits CyPA-induced chemotaxis by blocking the interaction between CyPA and the ectodomain of CD147 because of the high binding affinity of CsA to CyPA (Kd = 10−8-10−9 m). Therefore, CsA can block the direct interaction between CyPA and CD147. In addition, it can also block any function of CyPA involving the area that CsA binds. As CsA is a widely used tool in research on CyPA and other cyclophilins, our results illustrate that it is a common misunderstanding that the inhibitory effect of CsA is simply viewed as the effect of blocking the PPIase activity of CyPA (39, 40).
In summary, our findings indicate that CyPA induces chemotaxis through a direct interaction with the ectodomain of CD147 in a PPIase-independent manner, suggesting that there exists a novel mechanism for CyPA to regulate cell signaling. Given its role in the pathogenesis of inflammatory diseases, the CyPA/CD147 interaction has been suggested to be an attractive target for therapeutic interventions. The fact that residues Arg-69, His-70, and Thr-107 of CyPA are critical for both CD147 binding and chemotaxis, but not for the PPIase activity, may hint at the possibility for developing novel anti-inflammatory drugs that selectively inhibit the chemotaxis activity without affecting the PPIase activity of CyPA. Further studies are needed to establish the roles of CD147 ectodomain in this cellular process and the detailed molecular mechanism for the function of extracellular CyPA, and thus to explore new therapeutic applications for CyPA.
Supplementary Material
This work was supported by Grant 30900234 from the National Natural Science Foundation of China and Grant 2009CB521700 from the 973 Program.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- CyPA
- cyclophilin A
- CsA
- cyclosporine A
- HSQC
- heteronuclear single quantum coherence
- PPIase
- peptidylprolyl cis-trans isomerase
- suc
- succinyl
- NA
- nitroanilide.
REFERENCES
- 1. Arora K., Gwinn W. M., Bower M. A., Watson A., Okwumabua I., MacDonald H. R., Bukrinsky M. I., Constant S. L. (2005) J. Immunol. 175, 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andreotti A. H. (2003) Biochemistry 42, 9515–9524 [DOI] [PubMed] [Google Scholar]
- 3. Fischer G., Wittmann-Liebold B., Lang K., Kiefhaber T., Schmid F. X. (1989) Nature 337, 476–478 [DOI] [PubMed] [Google Scholar]
- 4. Takahashi N., Hayano T., Suzuki M. (1989) Nature 337, 473–475 [DOI] [PubMed] [Google Scholar]
- 5. Handschumacher R. E., Harding M. W., Rice J., Drugge R. J., Speicher D. W. (1984) Science 226, 544–547 [DOI] [PubMed] [Google Scholar]
- 6. Galat A. (1993) Eur. J. Biochem. 216, 689–707 [DOI] [PubMed] [Google Scholar]
- 7. Ke H., Huai Q. (2004) Front Biosci. 9, 2285–2296 [DOI] [PubMed] [Google Scholar]
- 8. Hur S., Bruice T. C. (2002) J. Am. Chem. Soc. 124, 7303–7313 [DOI] [PubMed] [Google Scholar]
- 9. Li G., Cui Q. (2003) J. Am. Chem. Soc. 125, 15028–15038 [DOI] [PubMed] [Google Scholar]
- 10. Mallis R. J., Brazin K. N., Fulton D. B., Andreotti A. H. (2002) Nat. Struct. Biol. 9, 900–905 [DOI] [PubMed] [Google Scholar]
- 11. Brazin K. N., Mallis R. J., Fulton D. B., Andreotti A. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1899–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nicholson L. K., Lu K. P. (2007) Mol. Cell 25, 483–485 [DOI] [PubMed] [Google Scholar]
- 13. Bosco D. A., Eisenmesser E. Z., Pochapsky S., Sundquist W. I., Kern D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5247–5252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Howard B. R., Vajdos F. F., Li S., Sundquist W. I., Hill C. P. (2003) Nat. Struct. Biol. 10, 475–481 [DOI] [PubMed] [Google Scholar]
- 15. Yurchenko V., Zybarth G., O'Connor M., Dai W. W., Franchin G., Hao T., Guo H., Hung H. C., Toole B., Gallay P., Sherry B., Bukrinsky M. (2002) J. Biol. Chem. 277, 22959–22965 [DOI] [PubMed] [Google Scholar]
- 16. Gwinn W. M., Damsker J. M., Falahati R., Okwumabua I., Kelly-Welch A., Keegan A. D., Vanpouille C., Lee J. J., Dent L. A., Leitenberg D., Bukrinsky M. I., Constant S. L. (2006) J. Immunol. 177, 4870–4879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Damsker J. M., Bukrinsky M. I., Constant S. L. (2007) J. Leukoc. Biol. 82, 613–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sherry B., Yarlett N., Strupp A., Cerami A. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 3511–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pushkarsky T., Zybarth G., Dubrovsky L., Yurchenko V., Tang H., Guo H., Toole B., Sherry B., Bukrinsky M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6360–6365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen Z., Mi L., Xu J., Yu J., Wang X., Jiang J., Xing J., Shang P., Qian A., Li Y., Shaw P. X., Wang J., Duan S., Ding J., Fan C., Zhang Y., Yang Y., Yu X., Feng Q., Li B., Yao X., Zhang Z., Li L., Xue X., Zhu P. (2005) J. Infect. Dis. 191, 755–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang H., Chen J., Yang J., Qiao S., Zhao S., Yu L. (2007) Biochem. Biophys. Res. Commun. 361, 763–767 [DOI] [PubMed] [Google Scholar]
- 22. Li M., Zhai Q., Bharadwaj U., Wang H., Li F., Fisher W. E., Chen C., Yao Q. (2006) Cancer 106, 2284–2294 [DOI] [PubMed] [Google Scholar]
- 23. Yurchenko V., Pushkarsky T., Li J. H., Dai W. W., Sherry B., Bukrinsky M. (2005) J. Biol. Chem. 280, 17013–17019 [DOI] [PubMed] [Google Scholar]
- 24. Obchoei S., Wongkhan S., Wongkham C., Li M., Yao Q., Chen C. (2009) Med. Sci. Monit. 15, RA221–RA232 [PubMed] [Google Scholar]
- 25. Yurchenko V., Constant S., Bukrinsky M. (2006) Immunology 117, 301–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yurchenko V., Constant S., Eisenmesser E., Bukrinsky M. (2010) Clin. Exp. Immunol. 160, 305–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schlegel J., Redzic J. S., Porter C. C., Yurchenko V., Bukrinsky M., Labeikovsky W., Armstrong G. S., Zhang F., Isern N. G., DeGregori J., Hodges R., Eisenmesser E. Z. (2009) J. Mol. Biol. 391, 518–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu X. L., Hu T., Du J. M., Ding J. P., Yang X. M., Zhang J., Yang B., Shen X., Zhang Z., Zhong W. D., Wen N., Jiang H., Zhu P., Chen Z. N. (2008) J. Biol. Chem. 283, 18056–18065 [DOI] [PubMed] [Google Scholar]
- 29. Boyden S. (1962) J. Exp. Med. 115, 453–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moss M. L., Palmer R. E., Kuzmic P., Dunlap B. E., Henzel W., Kofron J. L., Mellon W. S., Royer C. A., Rich D. H. (1992) J. Biol. Chem. 267, 22054–22059 [PubMed] [Google Scholar]
- 31. Zydowsky L. D., Etzkorn F. A., Chang H. Y., Ferguson S. B., Stolz L. A., Ho S. I., Walsh C. T. (1992) Protein Sci. 1, 1092–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Eisenmesser E. Z., Millet O., Labeikovsky W., Korzhnev D. M., Wolf-Watz M., Bosco D. A., Skalicky J. J., Kay L. E., Kern D. (2005) Nature 438, 117–121 [DOI] [PubMed] [Google Scholar]
- 33. Clardy J. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu X., Sun L., Yu M., Wang Z., Xu C., Xue Q., Zhang K., Ye X., Kitamura Y., Liu W. (2009) Cell Microbiol. 11, 730–741 [DOI] [PubMed] [Google Scholar]
- 35. Elbaz B., Valitsky M., Davidov G., Rahamimoff H. (2010) Biochemistry 49, 7634–7642 [DOI] [PubMed] [Google Scholar]
- 36. Kofron J. L., Kuzmic P., Kishore V., Colón-Bonilla E., Rich D. H. (1991) Biochemistry 30, 6127–6134 [DOI] [PubMed] [Google Scholar]
- 37. Walsh C. T., Zydowsky L. D., McKeon F. D. (1992) J. Biol. Chem. 267, 13115–13118 [PubMed] [Google Scholar]
- 38. Spitzfaden C., Braun W., Wider G., Widmer H., Wüthrich K. (1994) J. Biomol. NMR 4, 463–482 [DOI] [PubMed] [Google Scholar]
- 39. Calhoun C. C., Lu Y. C., Song J., Chiu R. (2009) Mol. Cell Biochem. 320, 35–43 [DOI] [PubMed] [Google Scholar]
- 40. Zheng J., Koblinski J. E., Dutson L. V., Feeney Y. B., Clevenger C. V. (2008) Cancer Res. 68, 7769–7778 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.