Abstract
BAK is a key effector of mitochondrial outer membrane permeabilization (MOMP) whose molecular mechanism of action remains to be fully dissected in intact cells, mainly due to the inherent complexity of the intracellular apoptotic machinery. Here we show that the core features of the BAK-driven MOMP pathway can be reproduced in a highly simplified in vitro system consisting of recombinant human BAK lacking the carboxyl-terminal 21 residues (BAKΔC) and tBID in combination with liposomes bearing an appropriate lipid environment. Using this minimalist reconstituted system we established that tBID suffices to trigger BAKΔC membrane insertion, oligomerization, and pore formation. Furthermore, we demonstrate that tBID-activated BAKΔC permeabilizes the membrane by forming structurally dynamic pores rather than a large proteinaceous channel of fixed size. We also identified two distinct roles played by mitochondrial lipids along the molecular pathway of BAKΔC-induced membrane permeabilization. First, using several independent approaches, we showed that cardiolipin directly interacts with BAKΔC, leading to a localized structural rearrangement in the protein that “primes” BAKΔC for interaction with tBID. Second, we provide evidence that selected curvature-inducing lipids present in mitochondrial membranes specifically modulate the energetic expenditure required to create the BAKΔC pore. Collectively, our results support the notion that BAK functions as a direct effector of MOMP akin to BAX and also adds significantly to the growing evidence indicating that mitochondrial membrane lipids are actively implicated in BCL-2 protein family function.
Keywords: Liposomes, Membrane Biophysics, Membrane Lipids, Membrane Reconstitution, Mitochondrial Apoptosis
Introduction
MOMP5 and the ensuing release of apoptogenic factors such as cytochrome c constitutes a pivotal step in commitment to apoptosis, which is tightly regulated by BCL-2 family proteins (1, 2). The BCL-2 family is composed of both anti- and pro-apoptotic proteins (1–3). The anti apoptotic members (BCL-2, BCL-XL, BCL-W, MCL-1, BFL-1/A1) prevent apoptosis primarily by inhibiting MOMP. Proapoptotic members of this family are traditionally subdivided into direct effectors of MOMP (BAX and BAK), which adopt an inactive form in healthy cells, and BH3-only proteins (BID, BIM, BAD, BIK, HRK, PUMA, NOXA, and BMF), which trigger functional activation of BAX/BAK and thereby promote MOMP.
Understanding the precise proapoptotic mode of BAK/BAX action is crucial to understanding how a cell makes the decision to undergo apoptosis (4). However, despite intense research effort, the exact molecular pathway leading to functional BAK/BAX activation during apoptosis has not yet reached consensus, and several possible models have been proposed (5–13). In addition, the architecture of the BAK/BAX pore remains a key and controversial issue in the field of cell death (1–4, 14–20). On top of this, increasing evidence indicates that the BCL-2-regulated MOMP pathway is a lipid-assisted process (16, 18, 20–27), but whether and how mitochondrial lipids participate in BAK proapoptotic function remains largely unexplored.
One of the major hurdles for gaining a molecular-level understanding of the proapoptotic mode of BAK action working with intact cells is the complexity of the network of intermolecular interactions that controls the BCL-2-regulated MOMP pathway at compositional, structural, and dynamic levels (28). In this regard, compositionally defined in vitro reconstituted systems proved to be an invaluable tool complementing in vivo cell biology approaches to reveal fundamental information concerning the mechanism of action of individual BCL-2 family members and their regulatory factors (6, 9, 11, 14–31).
Here, we demonstrate that the main features of the BAK-driven MOMP pathway can be reproduced in vitro using recombinant forms of BAK and tBID together with synthetic vesicles containing a defined lipid composition. Our studies with this novel in vitro reconstituted system shed new mechanistic insight on how tBID triggers functional BAK activation as well as important information regarding the actual molecular mechanism of pore formation by BAK. Additionally, we also identified two distinct roles played by mitochondrial lipids at different steps along the membrane permeabilization process driven by BAK.
EXPERIMENTAL PROCEDURES
Materials
Egg phosphatidylcholine (PC), egg phosphatidylethanolamine (PE), liver phosphatidylinositol (PI), heart cardiolipin (CL), myristoylated CL (mCL), monolysocardiolipin (LCL), egg phosphatidylglycerol (PG), egg phosphatidic acid (PA), brain phosphatidylserine (PS), liver phosphatidylinositol (PI), cholesterol (CHOL), 1,2-di-(9,10-dibromooctadecanoyl)-sn-glycero-3-phosphocholine (BrPC), 1-palmitoyl-2-stearoyl-(5-doxyl)-sn-glycero-3-phosphocholine (5-doxyl-PC), 1-palmitoyl-2-stearoyl-(10-doxyl)-sn-glycero-3-phosphocholine (10-doxyl-PC), 1-palmitoyl-2-stearoyl-(16-doxyl)-sn-glycero-3-phosphocholine (16-doxyl-PC), egg lysophosphatidylcholine (LPC), egg diacylglycerol (DAG), and CHOL were purchased from Avanti Polar Lipids (Alabaster, AL). KCl, HEPES, EDTA, dodecyl octaethylene glycol monoether (C12E8), TX-100, FD-20, FD-250, histone 1.2, gliotoxin, tetanolysin, melitin, cupper II sulfate (CuSO4), and phenanthroline were obtained from Sigma. P53 was obtained from Calbiochem. 1,3,6-Aminonaphtalene-trisulfonate (ANTS) and p-xilene-bisdipicolinic acid (DPX) were purchased from Molecular Probes (Eugene, OR). Anti-BAK G23 polyclonal antibody was from Santa Cruz (Santa Cruz, CA), and anti-BAK Ab-1 polyclonal antibody was from Calbiochem.
Recombinant Proteins and Synthetic Peptides
Recombinant human BAK lacking the carboxyl-terminal 21 amino acids (BAKΔC) and its mutant form G126S (BAKΔCG126S) (32), recombinant full-length murine BID with an amino-terminal His6 tag (BID) (33), recombinant caspase 8-cleaved murine BID with an amino-terminal His6 tag (tBID) and its mutant form tBID (93–96)A (33), the active carboxyl-terminal fragment of tBID (amino acids 60–195) (tcBID) (33), recombinant BIML lacking the carboxyl-terminal 23 amino acids and with an amino-terminal His6 tag (BIMLΔC) (9), recombinant human Bcl-XL lacking the carboxyl-terminal 24 amino acids and with an amino-terminal His6 tag (BCL-XLΔC) (9), recombinant mouse MCL-1 lacking the carboxyl-terminal 27 amino acids (MCL-1ΔC) (9), and recombinant human BCL-2 lacking the carboxyl-terminal 24 amino acids and with an amino-terminal His6 tag (BCL-2ΔC) (9) were obtained as previously described. All proteins were purified from soluble fractions of bacterial extracts and were >90% pure as evaluated by Coomassie-stained SDS-PAGE. Except for tcBID, all proteins were purified in the complete absence of detergents during the purification procedure. HPLC-purified peptides were obtained from Abgent. Peptide sequences were: 21BIDBH3, IRNIARHLAQVGDSMDRSIPP; F-21BIDBH3, FITC-IRNIARHLAQVGDSMDRSIPP; 29BIDBH3, SESQEDIIRNIARHLAQVGDSMDRSIPPG.
Cytochrome c Release Assays
Mitochondria were isolated from livers of male Harlan Sprague-Dawley rats as described previously (18). Isolated mitochondria (500 μg of protein/ml) were incubated with recombinant proteins in 125 mm KCl, 5 mm KH2PO4, 2 mm MgCl2, 1 mm DTT, 25 μm EGTA, 5 mm succinate, 5 μm rotenone, and 10 mm HEPES-KOH, pH 7.2, for 20 min at 30 °C under constant stirring, using an Eppendorf Thermomixer (Vfinal = 100 μl). Reaction mixtures were centrifuged at 14,000 × g for 10 min. Supernatant fractions and pellet fractions were subjected to 4–20% SDS-PAGE followed by Western blotting using anti-cytochrome c 7H8.2C-12 antibody (Pharmingen). Mitochondria were kept on ice and used within 3 h of preparation (R&D Systems, Minneapolis, MN).
Liposome Preparation
Unless otherwise indicated, liposomes were prepared with a lipid composition resembling that of MOM contact sites ((40 PC/35 PE/10 PI/15 CL (mol/mol)) (MOM-like liposomes) (18). Lipid mixtures at the indicated ratios were co-dissolved in chloroform/methanol (2:1), and organic solvents were removed by evaporation under an argon stream followed by incubation under vacuum for 2 h. For membrane permeabilization assays, dry lipid films were resuspended in 20 mm KCl, 10 mm Hepes, pH 7.0, 0.1 mm EDTA, 12.5 mm ANTS, and 45 mm DPX, except in experiments corresponding to Fig. 8 where 100 mm KCl, 10 mm Hepes, 0.1 mm EDTA (KHE buffer) supplemented with 10 mg/ml FD-20 and FD-250 was used. Unless otherwise specified, for other assays lipid films were resuspended in KHE buffer. Multilamellar vesicles were next subjected to 5 freeze/thaw cycles. Unless otherwise specified, frozen/thawed liposomes were then extruded 10 times through two polycarbonate membranes of 0.2-μm pore size (Nucleopore, San Diego, CA) to obtain large unilamellar vesicles (LUVs). Untrapped ANTS/DPX and FD-20/FD-250 were removed by gel filtration in Sephadex G-75 and CL2B-Sepharose columns, respectively, with KHE running as elution buffer.
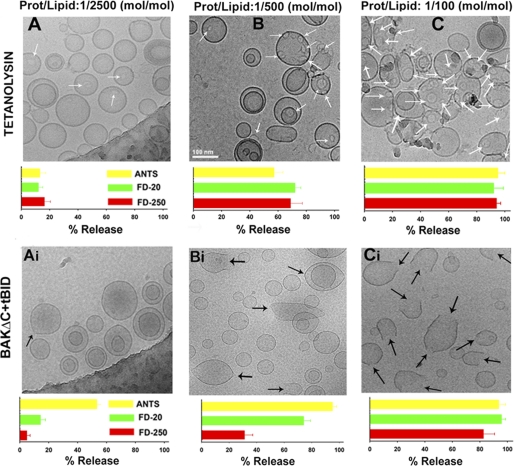
FIGURE 8.
tBID-activated BAKΔC and tetanolysin-permeabilized LUVs via different mechanisms as assessed by cryo-EM and vesicular marker release assays. A–C, top, typical cryo-EM images of 50PC/50CHOL LUV were treated with tetanolysin at the indicated protein/lipid ratios. White arrows indicate ring-shaped structures corresponding to the oligomeric tetanolysin channel. The same scale indicated in the panel B was used in all images. Bottom, release of liposome encapsulated fluorophores of various sizes was assessed by gel filtration chromatography on CL2B-Sepharose column under the conditions corresponding to the images on the top. Data correspond to the mean and S.E. of 3 independent measurements (except protein/lipid 1:100 mol/mol, n = 2). Ai–Ci, top, typical cryo-EM images of 25PC/35PE/10PI/30CL LUV treated with BAKΔC plus tBID (1 μm) at the indicated BAKΔC/lipid ratios. Black arrows indicate LUV-containing distortions in the membrane. The same scale indicated in panel B was used in all images. Bottom, release of liposome encapsulated fluorophores of various sizes was assessed by gel filtration chromatography on CL2B-Sepharose column under conditions corresponding to the images on the top. Data correspond to the mean and S.E. of 3–5 independent samples. In all cases, liposome concentration was 1 mm.
Fluorimetric Measurements of Vesicular Contents Release
Release of LUV-encapsulated ANTS/DPX was monitored in an 8100 Aminco-Bowman luminescence spectrometer (Spectronic Instruments, Rochester, NY) in a thermostatically controlled 1-cm path length cuvette with constant stirring, at 37 °C (Vfinal = 750 μl). The ANTS emission was monitored at 520 nm with the excitation wavelength set at 355 nm (slits, 4 nm). The FD-20 and FD-250 emission was monitored at 490 nm with the excitation wavelength set at 530 nm (slits, 4 nm). The extent of marker release was quantified on a percentage basis according to the equation (Fi − F0/F100 − F0) = 100, where Fi is the measured fluorescence of protein-treated LUV at time t, F0 is the initial fluorescence of the LUV suspension before protein addition, and F100 is the fluorescence value after complete disruption of LUV by the addition of C12E8 (final concentration, 0.5 mm). Unless otherwise specified, lipid concentration was 100 μm.
Binding of Proteins to MOM-like Liposomes
To assess the liposome binding capacity of BAKΔC alone or combined with tBID, a low speed vesicle sedimentation approach was used followed by detection of the protein by immunoblotting. Brominated PC was employed to allow efficient pelleting of liposomes by centrifugation. BAKΔC (200 nm) with or without tBID (100 nm) was incubated with freeze/thaw vesicles (1 mm) for 20 min at 37 °C under constant stirring using an Eppenford Thermomixer (Vfinal = 100 μl). Samples were centrifuged for another 10 min at 18,000 × g, and equivalent aliquots (10 μl) were taken from supernatant (corresponding to free protein) and pellet fractions (corresponding to liposome-bound protein). Half of the volume of both fractions was then subjected to reducing SDS-PAGE on 15% Tris-glycine gels followed by protein visualization by immunoblotting using anti-BAK G23 polyclonal antibody. Western blot signals were digitalized using a Bio-Rad Fluor-S-Max Imager, and integrated densities were measured using Quantity One program (Fujifilm USA, Valhalla, NY). Linearity of the response of the film-based system under conditions used was established using a serial dilution of recombinant purified BAKΔC protein.
Protein-Lipid Overlay Assay
Membrane lipid strips were purchased from Echelon Biosciences (Salt Lake City, UT). The strips were first blocked with 1% fat-free milk in PBS for 1 h and incubated for 1 more hour with BAKΔC in PBS at 37 °C. The strips were washed 3 times, 10 min each with PBS, and soaked in 1% fat-free milk in PBS with an anti-BAK polyclonal G23 antibody at 1:2000 dilution for 1 h at room temperature. The strips were washed twice with PBS and soaked in 1% fat free milk in PBS with horseradish peroxidase-conjugated anti mouse antibody (Bio-Rad) at a 1:20,000 dilution for 1 h at room temperature. After washing with PBS 3 × 10 min at room temperature, the protein was detected by chemiluminescence. Alternatively, stock solutions of different anionic lipids were solubilized in 2:1:0.8 MeOH:CHCl3:H20, spotted onto Hybond C nitrocellulose, and allowed to dry. The nitrocellulose was then subjected to the same procedure described above to detect BAKΔC by immunoblotting, protein bands were digitalized, and integrated densities were measured using Quantity One software. No detergent was added in any of the steps described for protein-lipid overlay assays to avoid any possible detergent-induced artifactual changes in BAKΔC conformation.
Gel Filtration Assays of BAKΔC Oligomerization
Gel filtration chromatography was performed in an AKTA system using a Superdex 200 HR 10/30 column from Amersham Biosciences equilibrated in KHE supplemented with 2% (v/v) CHAPS. The sample was applied to the column in a total volume of 0.25 ml at a flow rate of 0.6 ml/min, and 1-ml fractions were collected and analyzed by immunoblotting using anti-BAK G23 antibody.
SPR Analysis
SPR was measured with a BIAcoreTM 3000 instrument using an L1 sensor chip (BIAcore). To prepare the L1 chip with liposomes, 40 mm octyl β-d-glucopyranoside (Sigma) was injected over the surface at a flow rate of 100 μl/min for 30 s. Liposomes (2 mm final lipid concentration) were then injected at a flow rate of 2 μl/min for 3 min and 30 s. Binding analyses were performed at 25 °C in KHE (running buffer). Protein stocks were dialyzed against the running buffer and then diluted in the same buffer. Protein samples were injected over the liposome-coated chip at a flow rate of 10 μl/min. The resonance units change due to buffer was subtracted from the sample signal. The L1 chip was regenerated and stripped of liposomes by injecting a 2-min pulse of 40 mm octyl β-d-glucopyranoside followed by a 2-min pulse of running buffer.
BAKΔC Cross-linking Assays Using Copper (II) 1,10-Phenanthroline (CuPhe)
BAKΔC (200 nm) was first incubated with or without LUV of the indicated lipid compositions (1 mm) for 20 min at 37 °C under constant stirring using an Eppendorf Thermomixer (Vfinal = 100 μl). Then, samples were incubated with the redox catalyst CuPhe on ice for 30 min. The CuPhe stock was 30 mm CuSO4 and 100 mm 1,10-phenanthroline in 4:1 water/ethanol and diluted 50-fold into the sample. The oxidation was quenched by adding 20 mm EDTA to chelate copper and 20 mm N-ethylmaleimide to block unreacted cysteine residues, and the samples were analyzed by nonreducing SDS-PAGE and immunoblotting with anti-BAK G23 antibody.
Immunoprecipitations
Immunoprecipitations with the conformation-specific antibody Ab-1 were performed in 100-μl reactions in KHE buffer supplemented with 2% CHAPS (w/v) at 4 °C unless otherwise specified. BAKΔC and liposomes were added at concentrations of 400 nm and 1 mm, respectively. Immunoprecipitation was allowed to occur overnight on a rotator in KHE buffer. In the absence of liposomes, Triton X-100 or CHAPS were added at 0.5% (w/v) as positive and negative controls, respectively. Immunoprecipitates were collected by incubating with protein G-Sepharose for 2 h followed by centrifugation for 1 min. The pellets were washed 2 times with KHE supplemented with 0.5% CHAPS then an additional 3 times with KHE supplemented with 2% CHAPS. Immunoprecipitates were released from the beads in SDS loading buffer and analyzed by Western blotting with the BAK G23 antibody.
Far-UV Circular Dichroism (CD) Measurements
Far-UV CD spectra were recorded at 37 °C on a Jasco J-810 spectrapolarimeter (Jasco Spectroscopic Co. Ltd., Hachioji City, Japan) equipped with a JASCO PTC-423S temperature control unit using 1-mm path length cell. Data were collected every 0.2 nm at 50 nm/min from 250 to 200 nm with a bandwidth of 2 nm, and results were averaged from 20 scans. All samples were allowed to equilibrate for 10 min before CD analysis. Each spectrum represents the average of three distinct spectral recordings. The contribution of buffer with/or without liposomes to the measured ellipticity was subtracted as the blank. All samples were allowed to equilibrate for 10 min before CD analysis. Each spectrum represents the average of three distinct spectral recordings. The contribution of buffer with/or without liposomes to the measured ellipticity was subtracted as the blank. Molar ellipticity values were calculated using the expression φ = ϵ/10cnl, where ϵ is the ellipticity (millidegrees), c is the protein concentration (mol/liter), l is the cuvette path length, and n is the number of amino acid residues in the protein. To assess the thermal stability of the BAKΔC sample, components were incubated for 1 h in KHE buffer at 37 °C before the beginning of measurements. The midpoint of the melting transition (t½) was estimated using the first derivative converter available in the software of the CD instrument with a window of nine data points and a second-degree polynomial smoothing.
Steady-state Fluorescence Spectroscopy
BAKΔC intrinsic fluorescence was monitored by observing the protein emission spectrum from tryptophan fluorescence. Sample components were incubated for 10 min before beginning measurements. Fluorescence spectra (averaging three spectra) were recorded between 300 and 400 nm (5-nm bandpass) at a scan rate of 1 nm/s using an excitation wavelength of 295 nm (5 nm bandpass). The slit widths for excitation and emission were kept at 4 nm, with the experiment conducted at 37 °C. The contribution of buffer or liposomes alone to sample fluorescence was subtracted as blank.
Fluorescence Polarization Assays
BAKΔC or BCL-XLΔC (400 nm) were incubated for 2 h with F-BID BH3 peptide (5 nm). EC50 values were determined by fitting the experimental data using a sigmoidal dose-response nonlinear regression model with Sigma Plot software.
Isothermal Titration Calorimetry (ITC) Assays
ITC studies were performed using a VP-ITC microcalorimeter (Microcal Inc.). Titrations were performed in KHE buffer at 25 °C. Protein stocks were dialyzed against the same buffer used for preparing samples for ITC analysis. Titrations consisted of an initial injection of 2 μl followed by 29 × 10-μl injections of peptide at 40 μm into 1.4 ml of protein (5 μm). The data were analyzed to fit to a one-site model using the Origin software (OriginLab Corp.) provided by the manufacturer.
Electrophysiological Studies with Planar Lipid Bilayer Membranes
Experiments were performed in planar lipid bilayer membranes (BLMs) formed by the Mueller-Rudin technique across a 250-μm diameter hole. BLM composition was 50PC/25PE/25CL, and the solution bathing the membrane was KHE. Membrane thinning was monitored visually and by capacitance measurements. After planar membrane formation, a glass micropipette filled with a solution containing the protein was brought into close proximity to BLMs (∼2 μm). The whole BLM conductance was measured at voltage-clamp mode-holding potential from the site of protein addition. The resulting current was filtered (500 Hz corner frequency, H900 active filter, Frequency Devices, Haverhill, MA) and stored (100-μs sampling rate).
Cryoelectron Microscopy (Cryo-EM)
For the cryo-EM studies, tBID-activated BAKΔC or tetanolysin were incubated with LUVs at different protein/lipid ratios for at least 15 min. Grids were prepared following standard procedures and observed at liquid nitrogen temperatures in a JEM-2200FS/CR transmission electron microscope (JEOL Europe, Croissy-sur-Seine, France) operated at 200 kV. An in-column omega energy filter helped to record images with improved signal to noise ratio by zero-loss filtering. The energy selecting slit width was set at 9 eV. Digital images were recorded on UltraScan4000 CCD camera under low-dose conditions at a magnification of 55,058 obtaining a final pixel size of 2.7 Å/pixel.
RESULTS AND DISCUSSION
Functional Reconstitution of BAKΔC in CL-enriched MOM-like Liposomes
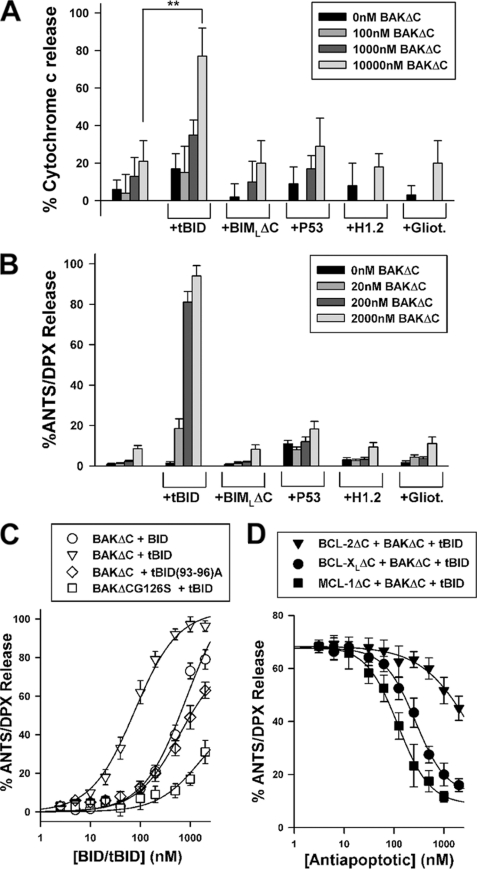
Our initial attempts to express and purify recombinant full-length human BAK in Escherichia coli using pRSETHisLipoTEV vector were hampered due to toxicity and insolubility problems associated with this construct (32). Thus, we decided to examine whether a recombinant purified form of human BAK lacking a 22-amino acid-long carboxyl-terminal segment containing a putative transmembrane region (termed BAKΔC hereafter) possesses an intrinsic membrane permeabilizing function (32). To this aim, isolated rat liver mitochondria and MOM-like LUVs loaded with the fluorophore/quencher pair ANTS/DPX were incubated in parallel with increasing concentrations of BAKΔC either alone or in combination with a number of putative BAK activators: caspase-8-cleaved BID (tBID), BIMLΔC, p53, histone 1.2, and gliotoxin (13, 31, 33–36). As shown in Fig. 1, BAKΔC by itself or in combination with BIMLΔC, p53, histone 1.2, or gliotoxin produced little release of mitochondrial cytochrome c or vesicular ANTS/DPX, indicating that BAKΔC is functionally inactive under these conditions. By contrast, the combination of BAKΔC with tBID produced near-to-complete release of both mitochondrial cytochrome c and vesicular ANTS/DPX. However, BAKΔC released ANTS/DPX from LUV with at least 20-fold higher potency than cytochrome c from mitochondria (Fig. 1B). Although the exact origin of this discrepancy remains to be determined, a likely possibility is that the two experimental systems present important differences regarding their content and availability of crucial modulators of BAK, including antiapoptotic BCL-2-type proteins, certain VDAC (voltage-dependent anion channel protein) isoforms, and/or CL (see below) (4, 37).
FIGURE 1.
Reconstitution of tBID-triggered BAKΔC permeabilizing function in MOM-like liposomes. A, rat liver mitochondria were incubated with the indicated recombinant apoptotic proteins for 20 min followed by enzyme-linked immunosorbent assay-based analysis of cytochrome c release. tBID, BIMLΔC, p53, histone 1.2 (H1.2), and gliotoxin (Gliot.) were added at 200 nm. The double asterisk denotes significant differences between experiments, as revealed by Student's t test (p < 0.01). All data points represent mean values and S.E. of three independent experiments (except p53, n = 2). B, ANTS/DPX-loaded MOM-like LUVs (100 μm) were treated with the indicated recombinant apoptotic proteins for 20 min followed by determination of the extents of vesicular contents release by spectrofluorimetry. tBID, BIMLΔC, p53, histone 1.2, and gliotoxin were added at 200 nm. Data represent the mean values and S.E. from 3–6 independent experiments. C, shown is the extent of ANTS/DPX release from MOM-like LUVs (100 μm) elicited by BAKΔC/BAKΔCG126S (200 nm) incubated for 20 min with increasing concentrations of indicated forms of tBID for 20 min. Data represent the mean values and S.E. of three independent experiments (except 3000 nm, n = 2). Experimental data were fitted with a sigmoidal dose-response nonlinear regression model. D, shown is concentration dependence of the inhibitory effect elicited by BCL-2ΔC, BCL-XLΔC, and MCL-1ΔC on the vesicular ANTS/DPX release induced by 200 nm BAKΔC plus 100 nm tBID. LUVs (100 μm) were first treated with antiapoptotic proteins for 5 min followed by incubation of the mixture with proapoptotic proteins for 20 min, and finally, extents of vesicular contents release were determined. When indicated, experimental data were fitted with a sigmoidal dose-response nonlinear regression model. Data represent mean values and S.E. from at least two independent experiments.
tBID was ≈13-fold more potent than BID activating BAKΔC-permeabilizing function in the liposomal system as determined by comparing the protein concentrations, giving half-maximal release (EC50 value), which is similar to the degrees of activation of cytochrome c release on BID cleavage reported in mitochondria isolated from a variety of cell types (typically 10–20-fold) (33, 38, 39) (Fig. 1C and supplemental Table I). The physiological relevance of the BAKΔC permeabilizing function triggered by tBID in MOM-like LUV was further supported by the finding that mutations known to inhibit the apoptogenic capacity of tBID/BAK in cells also diminished the ability of the tBID/BAKΔC mixture for releasing vesicular ANTS/DPX (Fig. 1C) (33, 40). Moreover, recombinant antiapoptotic BCL-XLΔC, MCL-1ΔC, and BCL-2ΔC proteins decreased tBID/BAKΔC-mediated liposome permeabilization in a dose-dependent manner with the inhibitory potency being higher for BCL-XLΔC/MCL-1ΔC than for BCL-2ΔC, in agreement with the higher binding affinity for BAK/tBID exhibited by the former antiapoptotic proteins relative to the latter (Fig. 1D) (41, 42).
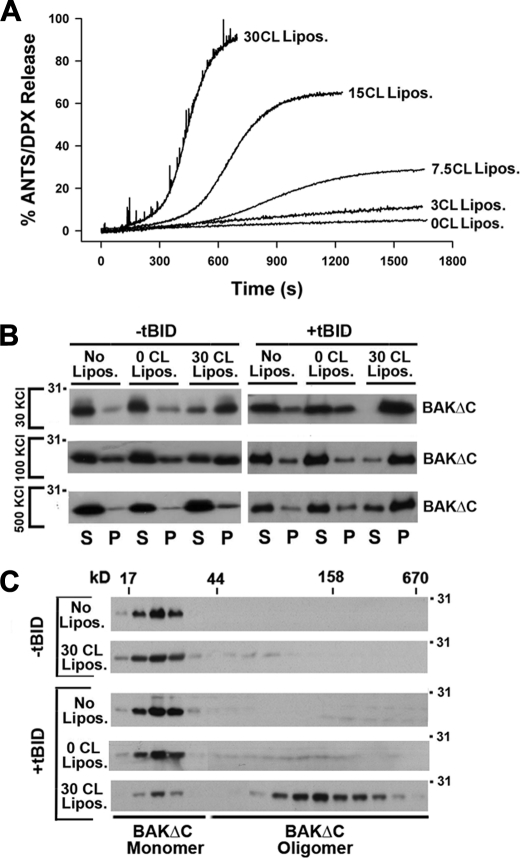
Previous studies by different groups revealed that CL plays an important role in the BAX-driven membrane permeabilization pathway triggered by tBID (16–18, 20, 23, 26, 27). Thus, we next decided to analyze the possible implication of CL in BAKΔC-induced liposome permeabilization. LUVs loaded with ANTS/DPX were prepared containing different amounts of CL followed by the addition of BAKΔC/tBID and monitoring of vesicular contents release by spectrofluorimetry. Increasing the concentration of CL progressively enhanced the extent of vesicular contents release induced by BAKΔC/tBID and also led to faster kinetics of liposome permeabilization, reaching a saturating response at ≈30 mol% CL (Fig. 2A, and supplemental Fig. 1). This effect was specific for the BAKΔC/tBID mixture, as incorporation of CL in LUV did not substantially change the permeabilizing activity of tetanolysin, a cholesterol binding pore-forming toxin, and even inhibited the release of vesicular ANTS/DPX induced by the antimicrobial peptide melittin (supplemental Fig. 1).
FIGURE 2.
Implication of CL in tBID-triggered functional activation of BAK. A, representative kinetics of ANTS/DPX release induced by BAKΔC (200 nm) plus tBID (100 nm) in LUVs (100 μm) containing lipid mixtures at the indicated the mol/mol ratios: 55PC/35PE/10PI (0CL Lipos.), 52PC/35PE/10PI/3CL (3CL Lipos.), 42.5PC/35 PE/10PI/7.5CL (7.5CL Lipos.), 40PC/35PE/10PI/15CL (15CL Lipos.), and 25PC/35PE/10PI/30CL (30CL Lipos.). B, lipid co-sedimentation assays are shown. BAKΔC (200 nm) with or without tBID (100 nm) was incubated for 20 min with freeze/thaw liposomes composed of 30BrPC/30PC/30PE/10PI (0CL Lipos.) or 30BrPC/30PE/10PI/30CL (30CL Lipos.) followed by sedimentation of the liposomes by low speed centrifugation, and immunoblot analysis of BAKΔC contents in the liposome-containing (P) and liposome-free (S) fractions using anti-BAK G23 polyclonal antibody is shown. Representative gels from at least three independent experiments are shown. No Lipos. corresponds to samples where proteins were incubated without liposomes. C, gel-filtration analysis of BAKΔC on Superdex 200 is shown. The indicated samples were treated with 2% CHAPS (w/v) applied to the column equilibrated with KHE + 2% CHAPS (w/v), and every second elution fraction was analyzed by SDS/PAGE and immunoblotting using anti-BAK G23 polyclonal antibody. Elution profiles of the molecular weight markers are indicated on the top.
Recent studies with an in vitro reconstituted system similar to ours identified two major consecutive steps in the BAX-driven membrane permeabilization process triggered by tBID; 1) penetration of otherwise cytosolic BAX segments into the lipid bilayer and 2) assembly of membrane-integrated BAX monomers into oligomeric species (11). As a first approach to assess the applicability of these postulates for tBID-triggered BAKΔC-permeabilizing function reconstituted in MOM-like liposomes, we examined the capacity of BAKΔC to interact with vesicles lacking or enriched in CL in the presence or absence of tBID. To try discriminating between adsorption of BAKΔC onto the liposome surface and penetration of BAKΔC segments into the bilayer hydrophobic core, liposomes were prepared and tested in buffers with different ionic strengths (i.e. 30 mm KCl, 100 mm KCl, and 500 mm KCl). Then, lipid-containing and lipid-free fractions were separated by centrifugation, and the amount of liposome-bound BAKΔC (pellet) and free BAKΔC (supernatant) were analyzed by SDS-PAGE/immunoblotting and quantitative densitometry. To avoid misinterpretation of the data, care was taken to stay within the linear range of the immunoblot assay (supplemental Fig. 2A). We found little BAKΔC bound to CL-lacking MOM-like liposomes under all conditions examined, whereas the majority of BAKΔC appeared associated to CL-enriched MOM-like liposomes in the presence of tBID irrespective of the ionic strength of the incubation medium (Fig. 2B and supplemental Fig. 3B). Remarkably, in the absence of tBID, a substantial amount of BAKΔC remained bound to CL-enriched MOM-like liposomes, and in this case increasing the ionic strength of the medium strongly decreased BAKΔC:liposome association.
We next studied the oligomeric status of BAKΔC by size-exclusion chromatography assays on Superdex 200. All samples were treated with 2% (w/v) CHAPS to allow for membrane solubilization. In this experimental setting, BAKΔC alone eluted close to its calculated monomeric mass (Fig. 2C and supplemental Fig. 2C). BAKΔC elution profile did not change when the protein was treated with tBID alone or in combination with MOM-like LUVs lacking CL. However, BAKΔC incubation with tBID plus CL-enriched MOM-like LUVs shifted the elution profile of the majority of the protein to high molecular weight fractions, indicating that BAKΔC forms oligomeric complexes under these conditions as observed during apoptosis with endogenous mitochondrial BAK. Of note, no shift in BAKΔC migration pattern was observed when the protein was incubated with CL-enriched liposomes in the absence of tBID.
To sum up, these findings led us to the following conclusions. First, basic aspects of the BAK-driven MOM permeabilization pathway can be reconstituted in vitro using purified recombinant BAKΔC/tBID proteins and MOM-like liposomes enriched in the mitochondrion-specific lipid CL. Second, in this minimalist reconstituted system, tBID-mediated activation of BAKΔC-permeabilizing function is correlated with membrane insertion and oligomerization of BAKΔC. Third, CL is implicated not only in functional BAKΔC activation by tBID but also in an interaction of BAKΔC itself with the membrane, which is not followed by intramembranous oligomerization of the protein and membrane damage.
BAKΔC Directly and Specifically Interacts with CL Leading to a Change in BAKΔC Conformation
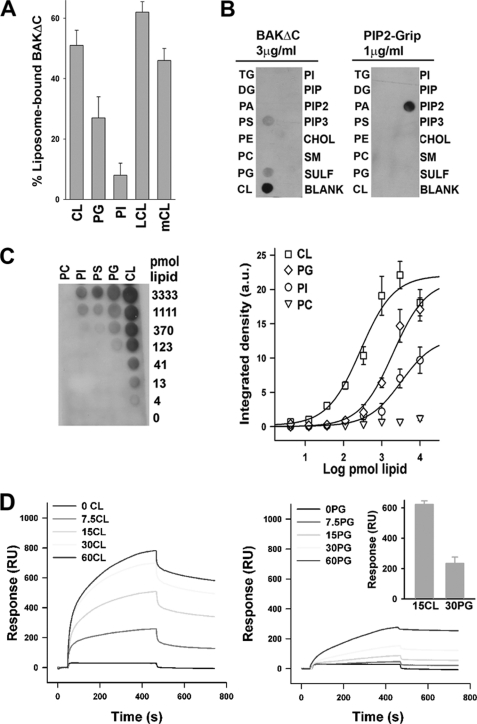
Next we wished to explore in more detail the mechanism underlying the interaction of BAKΔC itself with CL-enriched MOM-like LUVs. In principle, the effect of CL on BAKΔC-LUV binding may arise from the following changes induced by CL in bulk physical properties of the liposomal membrane; (i) increased net negative electric charge, (ii) generation of negative membrane monolayer curvature stress, or (iii) lateral segregation of membrane lipids into domains (43). To try discriminating between these possibilities, we probed for the impact of CL replacement by (i) two other anionic lipids (PG, PI) while keeping the net charge of the vesicle equal to that existing in CL-containing liposomes, (ii) LCL, which does not share with the parent lipid the potential to adopt a negatively curved disposition (43), and (iii) mCL, which unlike CL, segregated into distinct lipid domains in MOM-like liposomes (27). Substituting PG or PI for CL diminished but did not completely eliminate binding of BAKΔC to the liposomes, the reducing effect being more accentuated for PI than for PG (Fig. 3A). Of note, PG is a precursor of CL, which shares defined structural features with the parent molecule. These data imply that a net negative charge alone is not the sole defining feature that underlies BAKΔC binding to CL-enriched MOM-like liposomes. On the other hand, the amount of BAKΔC bound to liposomes did not change substantially when LCL or mCL was substituted for CL. Thus, the ability of CL to generate curvature or to form segregated domains does not appear to be the underlying cause for BAKΔC binding to CL-enriched MOM-like liposomes.
FIGURE 3.
BAKΔC binds CL directly and preferentially over other anionic lipids. A, BAKΔC was incubated with freeze/thaw liposomes composed of 30BrPC/30PE/10PI/30CL (CL), 30BrPC/10PI/60PG (PG), 30BrPC/70PI (PI), 30BrCL/30 PE/10 PI/30 (LCL), or 30 BrPC/30PE/10PI/30mCL (mCL) followed by sedimentation of the liposomes by centrifugation and quantitative assessment of liposome-bound BAKΔC fraction by immunoblotting and densitometry. Each bar represents the average and S.E. of at least three independent experiments (except LCL, n = 2). B, BAKΔC immunoreactivity is shown from a representative protein:lipid dot-blot experiment in which BAKΔC was overlaid onto commercially available nitrocellulose membranes spotted with some of the most common glycerolipids and sphingolipids present in mammalian cells. TG, triacylglycerol; PIP, phosphatidylinositol phosphate; PIP2, phosphatidylinositol bisphosphate; PIP3, phosphatidylinositol triphosphate; SM, sphingomyelin; SULF, sulfatide. PIP2-grip represents an internal control protein that binds PIP2 with high affinity provided by the producer. C, quantitative data on BAKΔC binding potency for different anionic lipids was determined using protein-lipid overlays in which increasing amounts of indicated lipids were spotted on nitrocellulose. Integrated BAKΔC immunoreactivity was measured at each spot and normalized for each lipid, and the data were fitted with a sigmoidal dose-response nonlinear regression model. EC50 values for CL, PG, and PI were 411.3 ± 51.2, 2232.6 ± 87.0, and 5674.2 ± 123.1 pmol, respectively. D, representative sensorgrams depict the interaction of BAKΔC with LUVs containing the indicated lipids immobilized on the surface of L1 sensor chip. Liposome lipid compositions were as follows: MOM, 55PC/35PE/10PI; 7.5CL, 47.5PC/35PE/10PI/7.5CL; 15CL, 40PC/35PE/10PI/15CL; 30CL, 25PC/35PE/10PI/25CL; 60CL, 20PC/10PE/10PI/60CL; 7.5PG, 47.5PC/35PE/10PI/7.5PG; 15PG, 40PC/35PE/10PI/15PG; 30PG, 25PC/35PE/10PI/30PG; 60PG, 20PC/10PE/10PI/60PG. Inset, summary of association binding responses obtained in four independent SPR experiments corresponding to BAKΔC:15CL LUV and BAKΔC:30PG LUV samples. RU, resonance units.
The above results raised the possibility that BAKΔC recognizes CL through a direct interaction rather than via a bilayer-mediated effect. To test this possibility, we performed a lipid dot-blot overlay assay in which a wide variety of mammalian zwitterionic and (poly)anionic phospholipids as well as other lipid species were presented to BAKΔC immobilized on top of a nitrocellulose membrane rather than in the form of free-standing bilayer vesicles. Among all lipid species examined, BAKΔC only recognized CL with high affinity in the lipid overlay assay (Fig. 3B). Nevertheless, similar to what is observed in liposome binding experiments, BAKΔC also recognized anionic phospholipid species other than CL by lipid overlay assay, albeit apparently with markedly lower affinity. To obtain more quantitative data on the relative affinity of BAKΔC for different anionic lipids, BAKΔC lipid binding capacity was tested using nitrocellulose blots on which representative anionic lipids were spotted in increasing amounts. In this experimental setup, the binding of BAKΔC was ≈5- and ≈12-fold stronger for CL than for PG and PI, respectively, whereas BAKΔC did not bind appreciably to PC (Fig. 3C and supplemental Table II).
As a further test of CL recognition by BAKΔC, surface plasmon resonance (SPR) spectroscopy was used. To this aim, liposomes containing increasing amounts of CL or PG were captured in the surface of the L1 sensor chip and used as ligands to probe for BAKΔC-membrane binding interactions in real time. Representative SPR response curves (sensorgrams) are presented in Fig. 3D. The reported sensorgrams showed that BAKΔC binding to MOM-like LUVs lacking CL was negligible, although increasing the concentration of CL progressively augmented BAKΔC binding to liposomes. BAKΔC showed lower binding responses with liposomes containing PG than with CL even when the two anionic lipids were compared at the same net charge density (Fig. 3D, inset). Unfortunately, kinetic rate constants could not be obtained by fitting these sensorgrams to a pseudo first-order binding model. There are precedents for complex sensorgrams obtained in other toxin-membrane interactions analyzed by SPR (44, 45). Irrespective of the origin of the sensorgram complexity (46), the SPR results obtained for BAKΔC-membrane interactions qualitatively agree with lipid-protein overlay and liposome association assays showing that BAKΔC recognizes CL directly and preferentially over other anionic lipids.
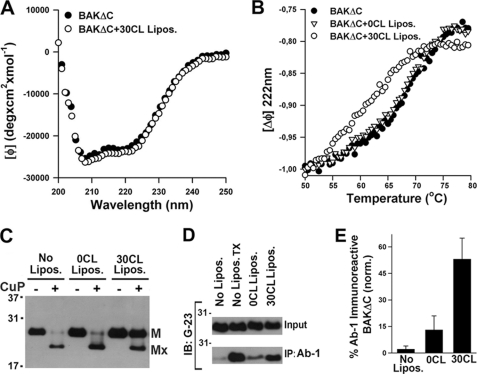
We next investigated whether interaction with CL-enriched MOM-like vesicles produces structural changes in BAKΔC. First, we examined BAKΔC secondary structure utilizing far-UV CD spectroscopy. As expected, the far-UV CD spectrum of BAKΔC revealed absorption minima at 208 and 222 nm characteristic of predominantly α-helical proteins (Fig. 4A). Upon incubation with CL-enriched MOM-like LUVs, the overall CD spectrum of BAKΔC was essentially unchanged. To explore whether interaction with CL-enriched membranes produces changes on BAKΔC tertiary structure, the conformational stability of the protein was analyzed by thermal denaturation assays. BAKΔC unfolding was monitored by the change in ellipticity at 222 nm as a function of temperature (Fig. 4B). The thermal unfolding curve of BAKΔC in the absence of liposomes showed an unfolding transition with partial loss of helicity occurring between ≈60 and ≈75 °C, with the midpoint transition for the protein melting curve being 68.1 °C. Importantly, when BAKΔC was incubated with CL-enriched, but not CL-lacking MOM-like LUV, the protein melting curve shifted ≈6 °C toward lower temperatures (Fig. 4B). Thus, interaction of BAKΔC with CL-enriched MOM-like LUVs significantly destabilizes the solution fold of the protein.
FIGURE 4.
Interaction with CL destabilizes BAKΔC solution fold and exposes amino-terminal regions of the protein. A, the CD spectrum of BAKΔC in the absence (BAKΔC) or presence of LUV composed of 25PC/35PE/10PI/30CL (BAKΔC+30CL Lipos.) is shown. B, thermal denaturation curves for BAKΔC incubated in the absence of LUV (BAKΔC) or in the presence of LUV composed of 55PC/35PE/10PI (BAKΔC+0CL Lipos.) or 25PC/35PE/10PI/30CL (BAKΔC+30CL Lipos.) is shown. Thermal denaturation curves were obtained by recording the temperature dependence of BAKΔC ellipticity at 222 nm. The molar ellipticity at 50 °C was set to −1.0 to compare BAKΔC thermal denaturation rates in the absence and presence of vesicles. BAKΔC and LUV concentrations were 5 μm and 2 mm, respectively. C, BAKΔC (200 nm) was incubated in the absence of liposomes (No Lipos.) or with LUV of the indicated compositions (0CL Lipos. and 30CL Lipos.) followed by treatment with CuPhe or with DMSO vehicle, SDS/PAGE under non-reducing conditions, and immunoblot analysis of BAKΔC content using anti-BAK G23 polyclonal antibody. The positions of monomeric BAKΔC (M) and monomeric BAKΔC harboring an intramolecular cross-link (Mx) are indicated. D, shown is a Western blot of BAKΔC immunoprecipitates (IP) after incubating the protein with 2% CHAPS (No Lipos., negative control) or with 0.2% TX-100 (No Lipos.TX, positive control) or with LUV of the indicated lipid compositions plus 2% CHAPS. Samples were subjected to immunoprecipitation with conformation-specific anti-BAK Ab-1 antibody conjugated to protein A-agarose beads followed by SDS-PAGE/immunoblot (IB) analysis using anti-BAK G23 antibody. Input represents an equivalent amount of BAKΔC loaded in each sample. Concentrations of BAKΔC and LUV were 400 nm and 1 mm, respectively. E, quantitation of BAKΔC immunoreactivity to Ab-1 was assessed by immunoprecipitation and densitometric analysis of immunoblots obtained from three independent experiments as described in panel D.
Early steps in BAK activation during apoptosis have been associated to conformational changes in the protein leading to exposure of amino-terminal regions that are otherwise hidden in the functionally inactive state of BAK (4, 13, 40, 47). However, exactly how this early conformational change in BAK is triggered remains obscure. In cellular studies the redox catalyst CuPhe has been successfully used to distinguish between inactive and “partially activated” BAK conformers (13, 40). In the inactive state of the protein, the proximity of amino-terminal (Cys-14) and carboxyl-terminal (Cys-166) cysteines allows intramolecular cross-linking of BAK by CuPhe. In contrast, in the partially activated state, the amino-terminal loop (containing Cys-14) of BAKΔC is disengaged from the rest of the protein, thereby abrogating intramolecular cross-linking of BAK by CuPhe. Inactive and partially activated BAKΔC conformers can then be detected by non-reducing SDS-PAGE and immunoblot analysis due to the faster migration pattern exhibited by intramolecular-cross-linked BAKΔC species.
As shown in Fig. 4C, treatment of isolated BAKΔC with CuPhe in our reconstituted system generated a faster migrating form of the protein (Mx), consistent with the protein adopting an intramolecular-cross-linked conformation as seen with endogenous inactive BAK. Incubation of BAKΔC with MOM-like LUV lacking CL followed by CuPhe treatment did not produce substantial changes in BAKΔC migration pattern. Remarkably, however, when BAKΔC was incubated with CL-enriched MOM-like liposomes followed by treatment with CuPhe, the majority of the Mx conformer was lost, suggesting interaction with CL results in disengagement of BAKΔC amino-terminal loop from the rest of the protein. Thus, BAKΔC bound to CL-enriched MOM-like LUV resembles the partially activated conformer detected in endogenous BAK early upon apoptotic stimulation (40).
The conformation-specific monoclonal antibody Ab-1 is also a commonly used marker in cellular assays to detect early steps in BAK activation pathway associated with exposure of a hidden epitope in its amino-terminal region (4, 47). Based on this information, we decided next to examine the pattern of Ab-1 recognition by BAKΔC in our reconstituted system. As seen with “inactive” BAK in cellular extracts, BAKΔC did not immunoprecipitate with Ab-1 in buffer containing the mild detergent CHAPS, but the protein could be artificially induced to adopt the Ab-1-reactive conformation by incubation in buffer containing Triton X-100 (Fig. 4D). However, when BAKΔC was incubated with CL-enriched MOM-like LUVs followed by treatment with CHAPS, a substantial part of the protein immunoprecipitated with Ab-1. Taking out CL from the lipid mixture potently reduced (but did not completely eliminate) BAKΔC immunoreactivity toward Ab-1 (Fig. 4, D and E).
Altogether these collective results led us to the following conclusions. First, BAKΔC recognizes CL through a direct interaction and with remarkable, albeit not absolute, specificity relative to other anionic lipids. Second, interaction with CL produces changes in the BAKΔC amino terminus that are like those observed in endogenous mitochondrial BAK early during apoptosis.
tBID Induces a Conformational Change in BAKΔC Distinct from That Triggered by CL Alone
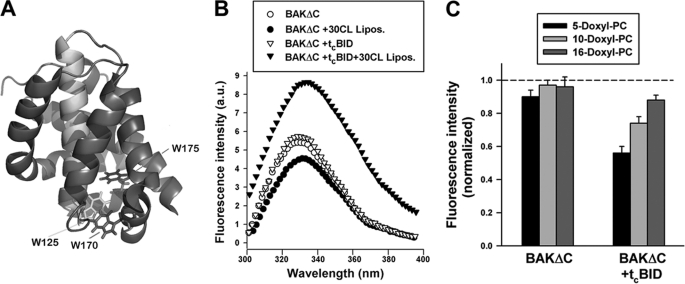
To further examine the effect of CL on BAKΔC structure and to try gaining more information on the mechanism of functional BAKΔC activation triggered by tBID, we measured changes in the tryptophan emission spectrum of the protein in the absence or presence of different lipid vesicles with or without tcBID, the carboxyl-terminal active fragment of tBID that does not interfere with BAKΔC fluorescence spectrum because it does not contain any tryptophan residues. In the absence of liposomes, the fluorescence emission of BAKΔC displayed a maximum value (λmax) of 329 nm, consistent with x-ray crystallography data showing that BAKΔC tryptophan residues are substantially buried in the interior of the soluble form of the protein (Fig. 5, A and B) (48). The addition of CL-enriched MOM-like LUVs gave rise to a small but reproducible change in BAKΔC fluorescence spectrum consisting in am ≈3-nm red shift in λmax accompanied by an ≈20% reduction in protein fluorescence intensity, suggesting under these conditions BAKΔC tryptophan residues shifted to a more polar environment relative to when the protein is free in solution (Fig. 5B). In contrast, the BAKΔC fluorescence spectrum remained virtually unaltered with MOM-like LUVs lacking CL. These results provide further evidence that interaction with CL produces a localized structural rearrangement in BAKΔC.
FIGURE 5.
Tryptophan fluorescence analysis of BAKΔC conformation. A, shown is the crystal structure of BAKΔC displaying the tryptophan residues that serve as spectroscopic probes of protein conformational changes in the presence of liposomes. The molecule is colored to highlight the location of W125 in helix 5 (green), W170 in helix 7 (red), and W175 in helix 8 (blue). B, shown are representative tryptophan fluorescence spectra of BAKΔC incubated for 20 min (i) in the absence of LUV (BAKΔC), (ii) in the presence of 25PC/35PE/10PI/30CL LUV (BAKΔC+30CL Lipos.), (iii) in the absence of LUV but with the carboxyl-terminal fragment of caspase-8-cleaved BID (BAKΔC+tcBID), or (iv) in the presence of tcBID and 25PC/35PE/10PI/30CL LUV (BAKΔC+tcBID+30 CL Lipos.). BAKΔC, tcBID, and LUV concentrations were 0.4 μm, 200 nm, and 200 μm, respectively. a.u., arbitrary units. C, quenching of tryptophan fluorescence of BAKΔC incubated with or without tcBID in the presence of LUV containing nitroxide moieties located at different depths in the membrane bilayer is shown. Liposome lipid composition was 25doxylPC/35 PE/10PI/30CL. Data from fluorescence quenching experiments were normalized to the fluorescence emission from protein in 25PC/35PE/10PI/30CL LUV, i.e. lacking nitroxylated lipids. Data represent the average and S.E. of 3–4 independent experiments.
Interestingly, despite that cBID did not produce significant changes in the fluorescence spectrum of BAKΔC in solution or in the presence of MOM-like LUVs lacking CL, incubation of BAKΔC/tcBID with CL-enriched vesicles led to an ≈65% increase in BAKΔC tryptophan fluorescence emission intensity, whereas the λmax for the fluorescence emission was shifted ≈4 nm to the red (Fig. 5B). Therefore, tcBID changed the environment sensed by BAKΔC tryptophan residues in the presence of CL-enriched MOM-like LUVs. Although the origin of the increased BAKΔC intrinsic fluorescence could be manifold, one possibility is that tcBID induces insertion of BAKΔC tryptophan residues into the liposomal membrane.
To more closely examine the position of BAKΔC tryptophan residues in the membrane, we examined quenching of tryptophan fluorescence by nitroxide groups attached at different positions along the phospholipid acyl chain (5-doxyl-PC, 10-doxyl-PC, and 16-doxyl PC). Upon incubating BAKΔC with CL-enriched MOM-like LUV alone, the extent of tryptophan fluorescence quenching by nitroxide-labeled lipids was either very small (5-doxyl-PC) or inexistent (10-doxyl-PC and 16-doxyl-PC) (Fig. 5C). On the other hand, when tested upon incubation with MOM-like LUV plus tcBID, the extent of BAKΔC fluorescence quenching increased with the profile: 5-doxyl-PC > 10-doxyl-PC > 16-doxyl-PC. These results suggest that in the absence of tcBID, BAKΔC tryptophan residues do not penetrate substantially into the acyl chain region of the bilayer, whereas in the presence of tcBID, BAKΔC tryptophan residues adopt on the average a shallow disposition within the hydrophobic core of the membrane.
Based on these data, we propose the following sequence of events in the BAKΔC-driven membrane permeabilization pathway reconstituted in CL-enriched MOM-like liposomes. First, direct interactions with CL lead to adsorption of BAKΔC onto the membrane surface accompanied by a localized structural rearrangement of the protein. Second, tBID triggers further structural changes in this intermediate membrane-associated state of BAKΔC, leading to penetration of distinct segments of BAKΔC into the lipid bilayer hydrophobic core, assembly of BAKΔC oligomers, and membrane poration.
An important question in the above-described hypothetical scheme is exactly how the change in BAKΔC conformation produced by CL might be connected to activation of BAKΔC permeabilizing function by tBID. One attractive possibility is that the change in BAKΔC conformation triggered by CL is a prerequisite for BAKΔC-tBID interaction. Of note, a similar but not identical mechanism was proposed previously for BAX based on data gathered in a reconstituted system alike ours (49). Because structural studies identified a putative BH3 binding groove in BAKΔC that adopts an “occluded” configuration in the soluble and functionally inactive form of the protein (48), we reasoned that interaction with CL could “open” the BAKΔC groove to allow insertion of tBID BH3 domain therein. In fact, the reduced potency of the tBID (93–96)A BH3 mutant for activating BAKΔC permeabilizing function is consistent with this view (Fig. 1C). Arguing along these lines, synthetic peptides representing the BID BH3 domain have been shown to reproduce tBID ability for activating BAX-permeabilizing function both in isolated mitochondria and in purified lipid vesicles (6, 9, 10, 29, 50). Thus, as a means to further test the above model, we decided to examine the impact of peptides representing the BID BH3 domain on BAKΔC.
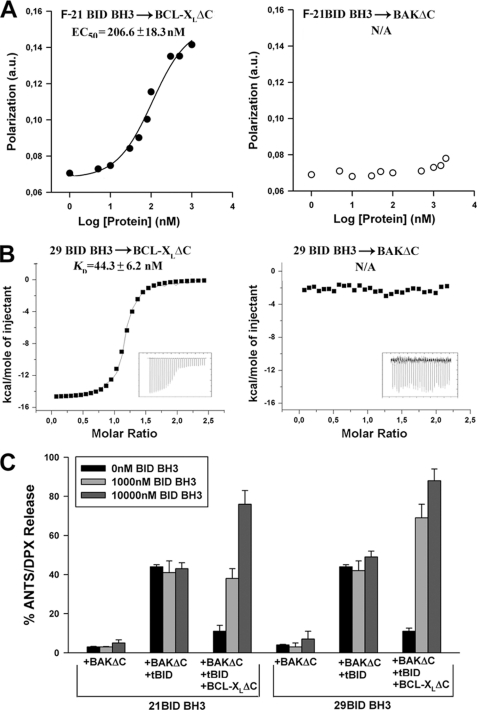
We began by performing fluorescence polarization studies to analyze whether an FITC-conjugated 21-mer peptide encompassing the BH3 domain of BID (F-21BID BH3) can interact with BAKΔC in solution. As shown in Fig. 6A, no evidence for F-21BID BH3 binding to BAKΔC was found even at the highest peptide concentrations tested (10 μm), whereas this peptide bound BCL-XLΔC with nanomolar affinity consistent with previous solution-based binding studies (50). Isothermal titration calorimetry studies using an unconjugated 29-mer BID BH3 peptide corroborated that the isolated BID BH3 domain bound BCL-XLΔC with nanomolar affinity but had negligible affinity for BAKΔC at least up to midmicromolar concentrations (Fig. 6B and supplemental Table III). As explained above, these results could be interpreted in light of the occluded BH3 binding groove present in the soluble form of BAKΔC according to x-ray crystallography (48).
FIGURE 6.
BID BH3 peptide neither interacts with BAKΔC in solution nor reproduces tBID-mediated activation of BAKΔC permeabilizing function. A, fluorescence polarization analysis of 21-mer FITC-labeled BID BH3 peptide (21 F-BID BH3) binding to BCL-XLΔC and BAKΔC is shown. Various concentrations of apoptotic proteins were incubated with 5 nm F-BID BH3 for 30 min followed by determination of fluorescence polarization. Data represent the mean (n = 3), with S.E. = 15% in all cases. B, ITC analysis of 29-mer BID BH3 peptide binding to BCL-XLΔC and BAKΔC. C, CL-enriched MOM-like LUV containing ANTS/DPX were treated for 20 min with the indicated protein/peptide combinations followed by determination of vesicular contents release. BAKΔC and BCL-XLΔC concentration were 200 and 1000 nm, respectively. tBID concentrations were 20 and 1000 nm in BAKΔC+tBID and BAKΔC+tBID+BCLXLΔC samples, respectively. Data represent the mean values and S.E. of at 3–5 independent experiments.
Next, we tested whether BID BH3 peptides trigger functional activation of BAKΔC in the presence of CL-enriched MOM-like liposomes. Contrary to such expectation, however, incubation of BAKΔC with unconjugated 21- or 29-mer BID BH3 peptides did not produce the robust vesicular ANTS/DPX release observed with tBID (Fig. 6C). Interestingly, 21- and 29-mer BID BH3 peptides successfully reversed the inhibition exerted by BCL-XLΔC on BAKΔC-permeabilizing function, indicating the peptides not only can bind to but can also functionally inactivate BCL-XLΔC (Fig. 6C). Thus, we concluded that the BH3 domain of BID in isolation does not recapitulate the ability of the entire tBID protein for activating the proapoptotic function of BAKΔC in our experimental setting.
A plausible explanation for these observations is that free BID BH3 peptides do not exactly reproduce the structure and/or orientation adopted by the BID BH3 domain in the context of membrane-associated tBID, which is required for functional activation of BAKΔC. Alternatively, there may be more in the mechanism of BAKΔC activation by tBID than insertion of BID BH3 domain into BAKΔC groove. For example, the interaction site between tBID and BAKΔC could include regions other than the BID BH3 domain and the BAKΔC grove, and/or lipid-mediated protein-protein interactions could also play a role in the process of functional BAKΔC activation, as previously proposed for BAX (9, 18, 26, 27). Clearly, further investigation is required to resolve this issue.
tBID-activated BAKΔC Forms Pores That Grow in Size and Are Susceptible to Modulation by Curvature-inducing Lipids
The nature of the apoptotic pore formed by BAK is another critical question under debate (1, 13–20, 27). Specifically, it is unclear whether BAK forms large proteinaceous channels with a fixed structure akin to certain pore-forming toxins or whether BAK forms structurally more dynamic pores composed at least in part by lipid molecules (proteolipidic pores).
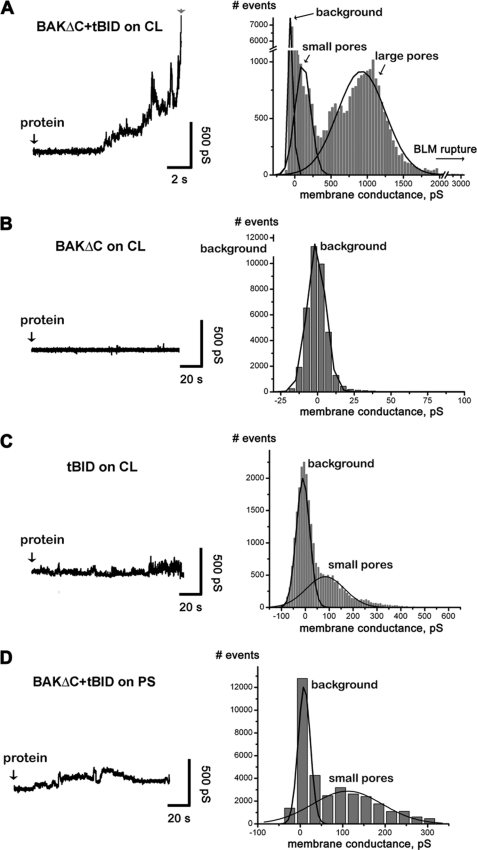
To improve our understanding of the mechanism of pore formation by BAK, we first conducted electrophysiological experiments with planar lipid BLMs. In a typical experiment, soon after the addition of BAKΔC and tBID to the aqueous solution, bathing CL-containing BLM-increased ion conductance events were detected (Fig. 7A, left). We consistently observed a lack of abrupt step-like fluctuations typical of ionic channels. The predominant pattern was that of brief, irregular current “spikes” in which amplitude increased with time until the membrane ruptured within minutes after protein addition. Lowering protein concentrations caused similar but delayed electrical responses (data not shown). Consistent with results obtained in liposome permeabilization studies, treatment of BLMs with either BAKΔC or tBID alone induced little (i.e. an increase in membrane noise but without significant fluctuations) if any electrical activity, and recordings could frequently continue for 10s of minutes without rupture of the membrane (Fig. 7, B and C, left). Also consistent with data gathered in liposome experiments, adding BAKΔC/tBID to BLMs in which CL was substituted for PS while maintaining membrane net electrical charge did not produce membrane conductance changes leading to rapid membrane rupture (Fig. 7D, left). The conductance events were further analyzed by the construction of all-point histograms, which allows for a comparison among different experiments. The consequent conductance showed a wide distribution of amplitudes ranging over several hundreds picosiemens (Fig. 7A, right).
FIGURE 7.
Electrophysiological analysis of BAKΔC and tBID-mediated effect on planar BLMs. A–D, shown are representative current traces (shown as conductance G = I/V), and all point histograms corresponding to all tracings were obtained from at least three independent recordings at the conditions illustrated (left panels). The peak at ≈0 picosiemens (pS) represents the base-line conductance of the untreated BLMs. Black lines show Gaussian multi-pick fit (Origin software, Microcal); p > 0.99 for all fits.
The electrical response induced by tBID-activated BAKΔC in BLM differs in two fundamental aspects from that typically observed with toxins forming large proteinaceous channels, exemplified by cholesterol binding pore-forming toxins. First, when BLM are treated with toxins forming large proteinaceous channels, sudden conductance changes between relatively stable levels are detected that do not increase gradually with time, resulting in a comparatively homogeneous distribution of large conductance states (51, 52). Second, unlike the situation found with tBID-activated BAKΔC, nanomolar concentrations of pore-forming toxins do not decrease BLM stability.
Next, we performed additional experiments to further examine whether BAKΔC forms pores of a discrete, large size, or alternatively, pores whose diameters are continuously increasing. On the one hand, we encapsulated fluorescent markers of different sizes within the liposomes (ANTS, ≈0.4 kDa; FD, 20 kDa; FD, 250 kDa) and examined marker release upon treatment of liposomes with low (1/2500), medium (1/500), and high (1/100) protein/lipid molar ratios. On the other hand, we used cryo-EM in an attempt to directly visualize the membrane permeability pathway under these conditions. In addition, the effect exerted by tBID-activated BAKΔC on liposomes was compared in parallel with that of the cholesterol binding pore-forming toxin tetanolysin using the same experimental approach to assess the extent of similarity in their mechanism of pore formation.
As shown in Fig. 8, A–C, LUVs treated with tetanolysin exhibited ring-shaped structures in the membrane, the number of which augmented progressively as the tetanolysin:lipid ratio increased (white arrows). At each tetanolysin/lipid ratio examined, the three markers entrapped within LUV were released practically to the same extent despite their largely different sizes (Fig. 8, A–C, bar graphs below cryo-EM images). This set of results is fully consistent with the notion that tetanolysin permeabilizes the membrane by forming large proteinaceous channels of relatively uniform diameter.
The effect exerted by tBID-activated BAKΔC in LUVs differed in several aspects from that observed with tetanolysin (Fig. 8, Ai–Ci). First, at a low BAKΔC/lipid ratio, the vast majority of LUV did not show detectable pores in the membrane, although a minor fraction of the liposomes exhibited apparent membrane discontinuity in localized areas of high curvature (Fig. 8Ai, black arrows). Under these conditions, LUV released on average a substantial fraction of ANTS, although virtually all FD-20 and FD-250 remained encapsulated within the vesicle interior. At an intermediate BAKΔC/lipid molar ratio, the fraction of liposomes showing high curvature areas in the membrane increased (Fig. 8Bi, black arrows). At the same time, visually detectable pores of sizes as large as 10s of nanometers appeared in some vesicles. Under these conditions, LUVs released on average most ANTS and FD-20 but still retained a substantial fraction of FD-250 encapsulated. Finally, at the highest BAKΔC/lipid molar ratio examined, the LUV population was dominated by vesicles exhibiting large pores of different sizes in the membrane (Fig. 8Ci and supplemental Fig. 3B). Under these conditions, liposomes released the three fluorescent markers nearly to completion. Of note, unlike the situation found with tetanolysin, ring-shaped structures were never observed in the membrane of LUVs treated with tBID-activated BAKΔC. Also, unlike tetanolysin, the pores created in the LUV membrane by tBID-activated BAKΔC did not exhibit a uniform diameter (Fig. 8, Bi and Ci and supplemental Fig. 3B). In control experiments, no pores were distinguished in LUV treated with BAKΔC or tBID alone nor in liposomes lacking CL (supplemental Fig. 3, C–F).
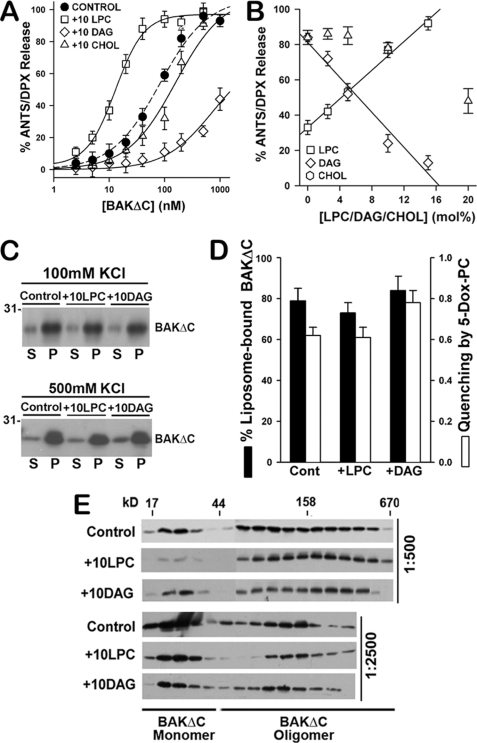
In summary, results gathered in electrophysiological, cryo-EM, and fluorescent marker release studies strongly argue against the notion that tBID-activated BAKΔC permeabilizes the membrane by forming large proteinaceous channels of fixed structure as seen with pore-forming toxins such as tetanolysin. On the other hand, the above results are consistent with the hypothesis that BAKΔC forms structurally dynamic proteolipidic pores. We and others have previously proposed the proteolipidic pore model to explain the permeabilizing mechanism of different proteins/peptides, including BAX (14, 16, 18, 20, 52–58). Theoretical studies indicate that the energy for proteolipidic pore formation/expansion depends critically in membrane monolayer intrinsic curvature, an elastic property of the bilayer that in turn depends on its lipid composition (55, 59). Thus, to further test the validity of the proteolipidic pore model for the case of BAKΔC, we examined whether three different lipids known to change membrane monolayer intrinsic curvature (LPC, DAG, and CHOL) (55, 59) affect BAKΔC-permeabilizing activity in our reconstituted system.
Incorporation of LPC (10 mol%) with positive intrinsic curvature in CL-enriched MOM-like LUVs produced an ≈6.3-fold decrease in the concentration of BAKΔC required for half-maximal release of ANTS/DPX (EC50), whereas incorporation of DAG (10 mol%) and CHOL (10 mol%) with negative intrinsic curvature increased the EC50 value ≈8.2- and ≈1.6-fold for BAKΔC-induced vesicular content release (Fig. 9A). The effect exerted by LPC and DAG on BAKΔC-induced vesicular contents release appeared specific as both lipids produced minor changes in liposome permeabilization induced by tetanolysin (supplemental Fig. 4A). Importantly, a linear correlation was found between BAKΔC-induced vesicular ANTS/DPX release and the molar amount of LPC/DAG incorporated in the liposomal membrane (Fig. 8B), in agreement with the known linear dependence of monolayer intrinsic curvature on membrane LPC/DAG molar content (55, 59). Therefore, a quantitative relationship was established between BAKΔC-induced vesicular contents release and membrane monolayer intrinsic curvature. The dependence of BAKΔC-permeabilizing activity on CHOL concentration was more complex, suggesting CHOL does not primarily inhibit BAKΔC-induced vesicular contents release via changes in membrane monolayer intrinsic curvature, at least within the range of concentrations examined here.
FIGURE 9.
LPC and DAG modulate BAKΔC-mediated LUV permeabilization. A, extents of vesicular ANTS/DPX release obtained upon incubation of increasing concentrations of BAKΔC plus tBID (100 nm) with CL-enriched MOM-like LUV of the following lipid compositions is shown: 25PC/35PE/30CL/10PI (30CL Lipos.), 20PC/30PE/30CL/10PI/10LPC (+10LPC), 20PC/30PE/30CL/10PI/10DAG (+10DAG), and 20PC/30PE/30CL/10PI/20CHOL (+10CHOL). Experimental data were fitted with a sigmoidal dose-response nonlinear regression model with Sigma Plot software to estimate the concentration of BAKΔC where 50% of the ANTS/DPX leaked out (EC50 values): Control LUV = 82.3 ± 7.4 nm; 10LPC LUV = 14.8 ± 2.4 nm; 20CHOL LUV = 174.4 ± 23.9 nm; 10DAG LUV = 739.3 ± 29.7 nm. Data represent the mean values and S.E. from at least two independent experiments. B, dose dependence of the effect elicited by LPC, DAG, and CHOL on the vesicular ANTS release induced by BAKΔC plus tBID is shown. Continuous and dashed lines represent least-square linear fits to the plots of extents of vesicular ANTS/DPX release versus molar percent of LPC (R2 = 0.97) and DAG (R2 = 0.95), respectively. Data represent the mean values and S.E. of 2–5 independent experiments. C, BAKΔC binding to liposomes of the indicated lipid composition assessed as explained in Fig. 2B is shown. S, supernatant; P, pellet. D, a comparison of the effects exerted by LPC and DAG on BAKΔC:liposome association in 500 mm KCl buffer and on the degree of quenching of BAKΔC tryptophan fluorescence intensity by 5-doxyl-PC is shown. Data represent the mean values and S.E. of at least two independent experiments. E, shown is the effect of LPC and DAG on tBID-activated BAKΔC oligomeric status assessed by gel filtration chromatography on Superdex 200 column as explained in Fig. 2C. BAKΔC was incubated with LUVs at protein:lipid ratios, indicated at the right of the immunoblots.
Additional experiments were also performed to check whether LPC and/or DAG alters membrane physical properties other than monolayer intrinsic curvature when incorporated in CL-enriched MOM-like LUVs. We found no evidence indicating that LPC and/or DAG induce lipid domains in the membrane, alter global vesicle morphology, or modify liposome size distribution (supplemental Fig. 4, B and C, and Table IV). A final set of studies was conducted to examine whether changes in BAKΔC membrane association, membrane insertion, and/or oligomerization could account for the modulation exerted by LPC/DAG on BAKΔC-induced vesicular ANTS release. Results gathered in liposome sedimentation (Fig. 9C), doxyl-5-PC-mediated quenching (Fig. 9D), and size-exclusion chromatography (Fig. 9E) assays did not support either of these alternative possibilities. A cautionary note here is that these experiments do not allow us to determine the degree of penetration into or interaction within the membrane of specific BAKΔC segments. However, a priori there is no reason to expect that LPC/DAG could modify these parameters in the quantitative manner correlating with monolayer intrinsic curvature observed in experiments of vesicular contents release, although we cannot exclude a fortuitous coincidence. Thus, the simplest explanation for the set of results obtained with LPC/DAG is that these two curvature-inducing lipids modulate BAKΔC-permeabilizing function by changing the energetic proclivity for proteolipidic pore formation.
Concluding Remarks
In this study we report a novel in vitro reconstituted system that faithfully recapitulates the core features of BAK proapoptotic function, consisting of recombinant human BAKΔC and tBID proteins in combination with MOM-like vesicles enriched in CL. The results gathered with this minimalist reconstituted system have several important mechanistic implications for our understanding of BAK-driven MOMP pathway.
First, the liposomal assays allowed us to explicitly test the capacity of different putative BAK activators for triggering BAK-permeabilizing function in the absence of other mitochondrial proteins. Among all agents examined, only tBID sufficed to trigger BAKΔC membrane insertion, oligomerization, and pore formation in MOM-like LUV. Because the number of putative BAK/BAX activators continues to expand, however, it is likely that in addition to tBID, other functional activators of BAK exist in the cell. Nevertheless, according to our results, tBID unambiguously qualifies as a direct functional activator of BAK, which still remains a major contentious issue in the field (1–4). However, our studies do not exclude the possibility that tBID also operates as an indirect activator of BAK via neutralization of antiapoptotic BCL-2 members (7), nor do we rule out that under certain conditions BAK could be autoactivated through homotypic BAK-BAK interactions.
Second, we obtained direct proof by cryo-EM that tBID-activated BAKΔC can puncture liposome membranes by forming pores of very large sizes (10s of nanometers in diameter). Analogous results have been previously obtained by others regarding tBID-activated BAX (20, 31). Although the precise structure of the apoptotic pore formed by BAK/BAX still needs to be determined, our studies provide quite compelling evidence that BAK does not operate by forming large channels with a fixed structure, as in the case of tetanolysin and related pore-forming toxins. Rather, our results suggest that BAK permeabilizes membranes by forming structurally dynamic proteolipidic pores that change in size depending on environmental conditions. Conceivably, this feature of the BAK pore could also be of importance in regulating the release of differently sized apoptogenic proteins from mitochondria. It remains to be determined, however, whether BAK forms pores at the MOM level exactly as described in our reconstituted system.
An additional important contribution of our investigations is the identification of two different potential roles played by selected mitochondrial lipids along the BAKΔC-driven MOMP pathway. On the one hand, we found that BAKΔC directly interacts with CL, leading to a rearrangement of the protein at the N terminus that resembles a conformational change observed with endogenous mitochondrial BAK early during apoptosis (4, 13, 47). The structural basis for the specific recognition of CL by BAK remains to be elucidated, as does the exact functional consequence of CL-induced BAK conformational change. This is the subject of ongoing work. Nevertheless, our results are compatible with a model wherein CL acts as an initiator of the molecular pathway for BAK activation by “priming” BAK for effective collaboration with downstream partners such as tBID. A corollary of this speculative line of reasoning is that BAK may be kept inactive at the MOM until a sufficient amount of CL accumulates in its vicinity. In the context of such a model, an important question that still remains open is how CL (and related lipids) is redistributed within MOM during apoptosis (60–62).
Outside the interaction of BAKΔC with CL, we obtained diverse lines of evidence indicating that LPCs and DAGs modulate the BAKΔC-driven membrane permeabilization process at the stage of pore formation by changing membrane monolayer intrinsic curvature. These results are consistent with (but do not prove) the notion that BAK permeabilizes membranes by forming proteolipidic pores, a model that has been previously invoked to explain the permeabilizing activity of BAX, certain pore-forming toxins, and a large variety of antimicrobial peptides (14, 16, 18, 20, 52–58). Considering that not only CL, but also LPC and DAG are present in mitochondrial membranes at degrees that can change before or concomitantly with the onset of MOMP (62–64), it is tempting to speculate that the modulatory role exerted by LPC and DAG on BAKΔC-mediated liposome permeabilization may also be relevant in the case of endogenous BAK under certain apoptotic settings. The current reconstitution system will provide an invaluable aid to further advance in our knowledge of the mechanisms by which different mitochondrial factors participate in proapoptotic BAK action and may also serve as a convenient platform with which to identify and evaluate compounds of potential therapeutic use targeting the BAK-driven MOMP pathway.
Supplementary Material
Acknowledgments
We thank Drs. B. Antonsson (Merck Serono Pharmaceuticals) and D. Huang (Walter-Eliza Institute, Australi) for the generous gifts of plasmids for expression of tBID, BCL-XLΔC, MCL-1ΔC, and BCL-2ΔC proteins as well as J. Sot and Miguel García Porras (Unidad de Biofísica, Consejo Superior de Investigaciones Cientificas-Universidad del Pais Vasco/Euskal Herriko Unibertsitatea) for technical assistance with fluorescence microscopy and protein purification studies, respectively.
This work was supported by Grants BFU2008–01637 from the Ministerio de Ciencia e Innovación (to G. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods, Tables I–IV, and Figs. 1–4.
- MOMP
- mitochondrial outer membrane (MOM) permeabilization
- LUV
- large unilamellar vesicle
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PI
- phosphatidylinositol
- CL
- heart cardiolipin
- PG
- phosphatidylglycerol
- PA
- phosphatidic acid
- PS
- phosphatidylserine
- PI
- phosphatidylinositol
- CHOL
- cholesterol
- BrPC
- 1,2-di-(9,10-dibromooctadecanoyl)-sn-glycero-3-phosphocholine
- MCL
- monolysocardiolipin
- mCL
- myristoylated CL
- ANTS
- 1,3,6-aminonaphtalene-trisulfonate
- DPX
- p-xilene-bisdipicolinic acid
- CuPhe
- copper (II) 1,10-phenanthroline
- ITC
- isothermal titration calorimetry
- LCL
- monolysocardiolipin
- SPR
- surface plasmon resonance
- LPC
- lysophosphatidylcholine
- DAG
- diacylglycerol
- BLM
- bilayer membrane.
REFERENCES
- 1. Youle R. J., Strasser A. (2008) Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]
- 2. Tait S. W., Green D. R. (2010) Nat. Rev. Mol. Cell Biol. 11, 621–632 [DOI] [PubMed] [Google Scholar]
- 3. Hardwick J. M., Youle R. J. (2009) Cell 138, 404, 404.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dewson G., Kluck R. M. (2009) J. Cell Sci. 122, 2801–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Letai A., Bassik M. C., Walensky L. D., Sorcinelli M. D., Weiler S., Korsmeyer S. J. (2002) Cancer Cell 2, 183–192 [DOI] [PubMed] [Google Scholar]
- 6. Kuwana T., Bouchier-Hayes L., Chipuk J. E., Bonzon C., Sullivan B. A., Green D. R., Newmeyer D. D. (2005) Mol. Cell 17, 525–535 [DOI] [PubMed] [Google Scholar]
- 7. Willis S. N., Fletcher J. I., Kaufmann T., van Delft M. F., Chen L., Czabotar P. E., Ierino H., Lee E. F., Fairlie W. D., Bouillet P., Strasser A., Kluck R. M., Adams J. M., Huang D. C. (2007) Science 315, 856–859 [DOI] [PubMed] [Google Scholar]
- 8. George N. M., Evans J. J., Luo X. (2007) Genes Dev. 21, 1937–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Terrones O., Etxebarria A., Landajuela A., Landeta O., Antonsson B., Basañez G. (2008) J. Biol. Chem. 283, 7790–7803 [DOI] [PubMed] [Google Scholar]
- 10. Gavathiotis E., Suzuki M., Davis M. L., Pitter K., Bird G. H., Katz S. G., Tu H. C., Kim H., Cheng E. H., Tjandra N., Walensky L. D. (2008) Nature 455, 1076–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lovell J. F., Billen L. P., Bindner S., Shamas-Din A., Fradin C., Leber B., Andrews D. W. (2008) Cell 135, 1074–1084 [DOI] [PubMed] [Google Scholar]
- 12. Kim H., Tu H. C., Ren D., Takeuchi O., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. (2009) Mol. Cell 36, 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dewson G., Kratina T., Czabotar P., Day C. L., Adams J. M., Kluck R. M. (2009) Mol. Cell 36, 696–703 [DOI] [PubMed] [Google Scholar]
- 14. Basañez G., Nechushtan A., Drozhinin O., Chanturiya A., Choe E., Tutt S., Wood K. A., Hsu Y., Zimmerberg J., Youle R. J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 5492–5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Saito M., Korsmeyer S. J., Schlesinger P. H. (2000) Nat. Cell Biol. 2, 553–555 [DOI] [PubMed] [Google Scholar]
- 16. Kuwana T., Mackey M. R., Perkins G., Ellisman M. H., Latterich M., Schneiter R., Green D. R., Newmeyer D. D. (2002) Cell 111, 331–342 [DOI] [PubMed] [Google Scholar]
- 17. Roucou X., Montessuit S., Antonsson B., Martinou J. C. (2002) Biochem. J. 368, 915–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Terrones O., Antonsson B., Yamaguchi H., Wang H. G., Liu J., Lee R. M., Herrmann A., Basañez G. (2004) J. Biol. Chem. 279, 30081–30091 [DOI] [PubMed] [Google Scholar]
- 19. Martinez-Caballero S., Dejean L. M., Kinnally M. S., Oh K. J., Mannella C. A., Kinnally K. W. (2009) J. Biol. Chem. 284, 12235–122245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schafer B., Quispe J., Choudhary V., Chipuk J. E., Ajero T. G., Du H., Schneiter R., Kuwana T. (2009) Mol. Biol. Cell 20, 2276–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Montero J., Morales A., Llacuna L., Lluis J. M., Terrones O., Basañez G., Antonsson B., Prieto J., García-Ruiz C., Colell A., Fernández-Checa J. C. (2008) Cancer Res. 68, 5246–5256 [DOI] [PubMed] [Google Scholar]
- 22. Lucken-Ardjomande S., Montessuit S., Martinou J. C. (2008) Cell Death Differ. 15, 929–937 [DOI] [PubMed] [Google Scholar]
- 23. Lucken-Ardjomande S., Montessuit S., Martinou J. C. (2008) Cell Death Differ. 15, 484–493 [DOI] [PubMed] [Google Scholar]
- 24. Christenson E., Merlin S., Saito M., Schlesinger P. (2008) J. Mol. Biol. 381, 1168–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Siskind L. J., Feinstein L., Yu T., Davis J. S., Jones D., Choi J., Zuckerman J. E., Tan W., Hill R. B., Hardwick J. M., Colombini M. (2008) J. Biol. Chem. 283, 6622–6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Etxebarria A., Terrones O., Yamaguchi H., Landajuela A., Landeta O., Antonsson B., Wang H. G., Basañez G. (2009) J. Biol. Chem. 284, 4200–4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Montessuit S., Somasekharan S. P., Terrones O., Lucken-Ardjomande S., Herzig S., Schwarzenbacher R., Manstein D. J., Bossy-Wetzel E., Basañez G., Meda P., Martinou J. C. (2010) Cell 142, 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Basañez G., Hardwick J. M. (2008) PLoS Biol. 6, e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Billen L. P., Kokoski C. L., Lovell J. F., Leber B., Andrews D. W. (2008) PLoS Biol. 6, e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oh K. J., Barbuto S., Meyer N., Kim R. S., Collier R. J., Korsmeyer S. J. (2005) J. Biol. Chem. 280, 753–767 [DOI] [PubMed] [Google Scholar]
- 31. Bleicken S., Classen M., Padmavathi P. V., Ishikawa T., Zeth K., Steinhoff H. J., Bordignon E. (2010) J. Biol. Chem. 285, 6636–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sot B., Freund S. M., Fersht A. R. (2007) J. Biol. Chem. 282, 29193–29200 [DOI] [PubMed] [Google Scholar]
- 33. Kudla G., Montessuit S., Eskes R., Berrier C., Martinou J. C., Ghazi A., Antonsson B. (2000) J. Biol. Chem. 275, 22713–22718 [DOI] [PubMed] [Google Scholar]
- 34. Leu J. I., Dumont P., Hafey M., Murphy M. E., George D. L. (2004) Nat. Cell Biol. 6, 443–450 [DOI] [PubMed] [Google Scholar]
- 35. Konishi A., Shimizu S., Hirota J., Takao T., Fan Y., Matsuoka Y., Zhang L., Yoneda Y., Fujii Y., Skoultchi A. I., Tsujimoto Y. (2003) Cell 114, 673–688 [DOI] [PubMed] [Google Scholar]
- 36. Pardo J., Urban C., Galvez E. M., Ekert P. G., Müller U., Kwon-Chung J., Lobigs M., Müllbacher A., Wallich R., Borner C., Simon M. M. (2006) J. Cell Biol. 174, 509–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cheng E. H., Sheiko T. V., Fisher J. K., Craigen W. J., Korsmeyer S. J. (2003) Science 301, 513–517 [DOI] [PubMed] [Google Scholar]
- 38. Yin X. M. (2006) Gene 369, 7–19 [DOI] [PubMed] [Google Scholar]
- 39. Gillick K., Crompton M. (2008) J. Cell Sci. 121, 618–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dewson G., Kratina T., Sim H. W., Puthalakath H., Adams J. M., Colman P. M., Kluck R. M. (2008) Mol. Cell 30, 369–380 [DOI] [PubMed] [Google Scholar]
- 41. Chen L., Willis S. N., Wei A., Smith B. J., Fletcher J. I., Hinds M. G., Colman P. M., Day C. L., Adams J. M., Huang D. C. S. (2005) Mol. Cell 17, 393–403 [DOI] [PubMed] [Google Scholar]
- 42. Willis S. N., Chen L., Dewson G., Wei A., Naik E., Fletcher J. I., Adams J. M., Huang D. C. (2005) Genes Dev. 19, 1294–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lewis R. N., McElhaney R. N. (2009) Biochim. Biophys. Acta 1788, 2069–2079 [DOI] [PubMed] [Google Scholar]
- 44. Cocklin S., Jost M., Robertson N. M., Weeks S. D., Weber H. W., Young E., Seal S., Zhang C., Mosser E., Loll P. J., Saunders A. J., Rest R. F., Chaiken I. M. (2006) J. Mol. Recognit. 19, 354–362 [DOI] [PubMed] [Google Scholar]
- 45. Bakrac B., Gutiérrez-Aguirre I., Podlesek Z., Sonnen A. F., Gilbert R. J., Macek P., Lakey J. H., Anderluh G. (2008) J. Biol. Chem. 283, 18665–18677 [DOI] [PubMed] [Google Scholar]
- 46. Rich R. L., Myszka D. G. (2008) J. Mol. Recognit. 21, 355–400 [DOI] [PubMed] [Google Scholar]
- 47. Griffiths G. J., Corfe B. M., Savory P., Leech S., Esposti M. D., Hickman J. A., Dive C. (2001) Oncogene 20, 7668–7676 [DOI] [PubMed] [Google Scholar]
- 48. Moldoveanu T., Liu Q., Tocilj A., Watson M., Shore G., Gehring K. (2006) Mol. Cell 24, 677–688 [DOI] [PubMed] [Google Scholar]
- 49. Yethon J. A., Epand R. F., Leber B., Epand R. M., Andrews D. W. (2003) J. Biol. Chem. 278, 48935–48941 [DOI] [PubMed] [Google Scholar]
- 50. Walensky L. D., Pitter K., Morash J., Oh K. J., Barbuto S., Fisher J., Smith E., Verdine G. L., Korsmeyer S. J. (2006) Mol. Cell 24, 199–210 [DOI] [PubMed] [Google Scholar]
- 51. Shepard L. A., Shatursky O., Johnson A. E., Tweten R. K. (2000) Biochemistry 39, 10284–10293 [DOI] [PubMed] [Google Scholar]
- 52. Kristan K. C., Viero G., Dalla Serra M., Macek P., Anderluh G. (2009) Toxicon 54, 1125–1134 [DOI] [PubMed] [Google Scholar]
- 53. Lee M. T., Chen F. Y., Huang H. W. (2004) Biochemistry 43, 3590–3599 [DOI] [PubMed] [Google Scholar]
- 54. García-Sáez A. J., Coraiola M., Dalla Serra M., Mingarro I., Menestrina G., Salgado J. (2005) Biophys. J. 88, 3976–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McIntosh T. J., Simon S. A. (2006) Annu. Rev. Biophys. Biomol. Struct. 35, 177–198 [DOI] [PubMed] [Google Scholar]
- 56. Sobko A. A., Kotova E. A., Antonenko Y. N., Zakharov S. D., Cramer W. A. (2006) J. Biol. Chem. 281, 14408–14416 [DOI] [PubMed] [Google Scholar]
- 57. García-Sáez A. J., Chiantia S., Salgado J., Schwille P. (2007) Biophys. J. 93, 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Qian S., Wang W., Yang L., Huang H. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17379–17383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zimmerberg J., Kozlov M. M. (2006) Nat. Rev. Mol. Cell Biol. 7, 9–19 [DOI] [PubMed] [Google Scholar]
- 60. Schug Z. T., Gottlieb E. (2009) Biochim. Biophys. Acta 1788, 2022–2231 [DOI] [PubMed] [Google Scholar]
- 61. Sorice M., Manganelli V., Matarrese P., Tinari A., Misasi R., Malorni W., Garofalo T. (2009) FEBS Lett. 583, 2447–2450 [DOI] [PubMed] [Google Scholar]
- 62. Cristea I. M., Degli Esposti M. (2004) Chem. Phys. Lipids 129, 133–160 [DOI] [PubMed] [Google Scholar]
- 63. Sandra F., Degli Esposti M., Ndebele K., Gona P., Knight D., Rossenquist M., Khosravi-Far R. (2006) Cancer Res. 65, 2668–26674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sato M., Ueda Y., Umezawa Y. (2006) Nat. Methods 3, 797–799 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.