Abstract
Lipoic acid is a covalently attached cofactor essential for the activity of 2-oxoacid dehydrogenases and the glycine cleavage system. In the absence of lipoic acid modification, the dehydrogenases are inactive, and aerobic metabolism is blocked. In Escherichia coli, two pathways for the attachment of lipoic acid exist, a de novo biosynthetic pathway dependent on the activities of the LipB and LipA proteins and a lipoic acid scavenging pathway catalyzed by the LplA protein. LipB is responsible for octanoylation of the E2 components of 2-oxoacid dehydrogenases to provide the substrates of LipA, an S-adenosyl-l-methionine radical enzyme that inserts two sulfur atoms into the octanoyl moiety to give the active lipoylated dehydrogenase complexes. We report that the intact pyruvate and 2-oxoglutarate dehydrogenase complexes specifically copurify with both LipB and LipA. Proteomic, genetic, and dehydrogenase activity data indicate that all of the 2-oxoacid dehydrogenase components are present. In contrast, LplA, the lipoate protein ligase enzyme of lipoate salvage, shows no interaction with the 2-oxoacid dehydrogenases. The interaction is specific to the dehydrogenases in that the third lipoic acid-requiring enzyme of Escherichia coli, the glycine cleavage system H protein, does not copurify with either LipA or LipB. Studies of LipB interaction with engineered variants of the E2 subunit of 2-oxoglutarate dehydrogenase indicate that binding sites for LipB reside both in the lipoyl domain and catalytic core sequences. We also report that LipB forms a very tight, albeit noncovalent, complex with acyl carrier protein. These results indicate that lipoic acid is not only assembled on the dehydrogenase lipoyl domains but that the enzymes that catalyze the assembly are also present “on site.”
Keywords: Energy Metabolism, Enzyme Catalysis, Enzymes, Proteomics, Pyruvate Dehydrogenase Complex, Lipoic Acid
Introduction
Lipoic acid ((R)-5-(1,2-dithiolan-3-yl) pentanoic acid) is a sulfur-containing cofactor covalently attached to and essential for the function of several key enzymatic complexes of central metabolism. These include the pyruvate dehydrogenase (PDH),2 2-oxoglutarate dehydrogenase (OGDH), branched-chain 2-oxoacid dehydrogenase, acetoin dehydrogenase, and the glycine cleavage system (GCV) (1). Escherichia coli contains only the PDH, OGDH, and GCV enzyme complexes, the first two of which are essential for aerobic metabolism. In each of these enzymes, a specific component (E2 in the 2-oxoacid dehydrogenases, H protein in the glycine cleavage system) is modified by the covalent attachment of lipoic acid to the ϵ-amino group of a conserved lysine residue via an amide bond. The lysine residues protrude from the surfaces of the highly conserved domains, which are generally called lipoyl domains (LD). The LD-bound coenzyme shuttles reaction intermediates among the multiple active sites of these multiprotein complexes in a type of covalent substrate channeling (1).
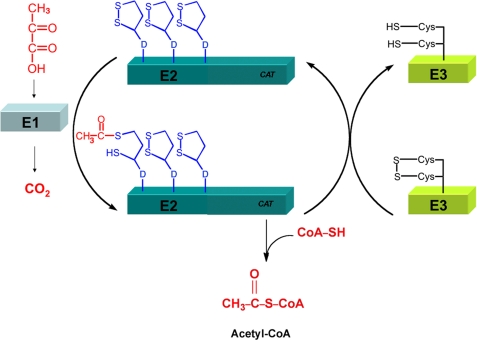
PDH catalyzes the oxidative decarboxylation of pyruvate to form acetyl-CoA (Fig. 1) (1). This very large enzyme complex (larger than the ribosome) consists of multiple copies of three components encoded by the aceEF lpd operon. The first component, E1p, encoded by aceE is a thiamine pyrophosphate-dependent decarboxylase that catalyzes the decarboxylation of pyruvate and the reductive acetylation of the lipoyl moiety of the second component E2p. The E2p component (encoded by aceF) is the dihydrolipoyl acetyltransferase responsible for the transfer of the acetyl group from the lipoyl moiety to CoA to produce the essential metabolic intermediate acetyl-CoA. The third component, E3, encoded by lpd, is the common component of all three lipoic acid-dependent enzymes and is the dihydrolipoyl dehydrogenase responsible for oxidizing the disulfide bond of lipoate to prepare the enzyme complex for another round of catalysis. The OGDH complex, which is closely related to the PDH complex, catalyzes the decarboxylation of 2-oxoglutarate to succinyl-CoA in the citric acid cycle. It also contains three components as follows: Lpd, a succinate decarboxylase component; E1o, encoded by sucA and a succinyltransferase component; and E2o, encoded by sucB. The third E. coli lipoylated complex is the glycine cleavage system (GCV), which catalyzes the reversible oxidation of glycine, yielding carbon dioxide, ammonia, and a C1 moiety in the form of methylene tetrahydrofolate. It consists of Lpd plus three proteins T, H, and P encoded by the gcvT gcvH gcvP operon where gcvH encodes the lipoic acid-modified H protein.
FIGURE 1.
PDH reaction. Pyruvate (upper left corner) is decarboxylated by the E1 component (AceE) in a thiamine pyrophosphate-dependent reaction that results in the reductive acetylation of the lipoyl group bound to an LD (designated D) of the second component, E2p (AceF). The acetylated lipoyl moiety of the LD then interacts with the E2 catalytic site (CAT) located at the C terminus of the component where the acetyl group is transferred to CoA. Finally, the reduced LD lipoyl moiety is oxidized by the E3 component (LpdA) to reset the dehydrogenase for another cycle of acetyl-CoA synthesis. The OGDH reaction proceeds by the same mechanism with 2-oxoglutarate in place of pyruvate and produces succinyl-CoA. However, the OGDH E2 component (SucB) contains only a single LD.
In all 2-oxoacid dehydrogenase (2-OADH) complexes, the E2 components constitute the inner core of the complexes and play vital roles in both assembly of the complexes and in substrate channeling (1). The E2s are highly segmented proteins composed of modular, independently folded domains linked together by Ala-Pro-rich hinge segments. The N-terminal lipoyl domains (1–3 domains depending on the enzyme and organism) are followed by a domain that binds the E1 and E3 components. The C-terminal half of the protein contains the acyltransferase catalytic domain with the active site residues located close to the C terminus. The E2 components are organized into either octahedral or icosahedral symmetries depending on the organism (24-mer octahedral symmetry in Gram-negative bacteria; 60-mer icosahedral symmetry in Gram-positive bacteria and mitochondria). Multiple copies of the E1 and E3 components bind tightly to the E2 core to form the outer shell of the complexes. The gap between the inner and outer shells (75–90 Å) is spanned by the flexible, but extended, conformations of the linker polypeptides that radiate outward from the inner E2 core (1, 2). The lipoyl moiety of the lipoyl domains, together with the linker regions connecting the lipoyl domains, make up long swinging arms that are able to carry the reaction intermediates to distant E1 and E3 active sites (1). In contrast to the 2-OADH complexes, the lipoylated H protein is a relatively small protein of 14 kDa found free in cell extracts (the well characterized GCV complexes are known to readily dissociate) (3).
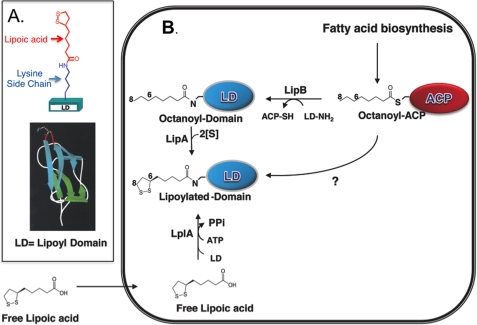
In the current model of lipoic acid biosynthesis and attachment (Fig. 2), an octanoyl moiety is first transferred from the octanoyl-acyl carrier protein (ACP) intermediate of fatty acid biosynthesis to the apoproteins by LipB (an octanoyl-ACP:protein-N-octanoyltransferase) (4). LipA then catalyzes sulfur insertion into positions 6 and 8 of the octanoyl moiety (4–8). Therefore, lipoic acid is assembled on its cognate proteins in an unusual type of biosynthetic pathway in which an inactive cofactor precursor is attached to the apoprotein and subsequently converted to the active species in a separate enzymatic step. E. coli also has the ability to scavenge free lipoic acid from the medium and attach it to lipoyl domains. This process is due to a single ATP-dependent enzyme, LplA, the paradigm lipoate protein ligase which is also active with octanoate (9–11).
FIGURE 2.
Lipoic acid biosynthetic and salvage pathways. A, mode of attachment of lipoic acid and the structure of the innermost LD of the E. coli PDH E2 protein (AceF). In this depiction, the lipoylated lysine is at the top of the structure (the protruding β-turn) and the N and C termini of the domain are at the bottom of the structure. B, biosynthetic pathway is at the top of the figure, and the LplA-dependent salvage pathway is at the bottom. Lipoic acid is synthesized as an offshoot of fatty acid biosynthesis by the consecutive activities of LipB and LipA. LipB transfers an octanoyl moiety from octanoyl-ACP to the ϵ-amino group of a specific lysine residue of the highly conserved lipoyl domains of the PDH, OGDH, and Gcv complexes. LipA then inserts two sulfur atoms at positions 6 and 8 of the octanoyl moiety of the LipB product to form the functional enzyme complexes. The curved arrow is the putative pathway that would proceed by sulfur insertion into octanoyl-ACP (see Introduction). Carbons 6 and 8 of the octanoyl moiety are labeled.
In this study, we report the unexpected finding that E. coli LipB and LipA form tight noncovalent interactions with the PDH and OGDH complexes, but not with GcvH, the GCV lipoylated protein. In contrast, the LplA lipoate ligase showed no evidence of association with the 2-OADH complexes. We provide genetic and biochemical evidence that LipB and LipA interact with the complexes through binding to the E2 components. These data indicate that the highly dynamic nature of the 2-OADH complexes serves not only to greatly increase the catalytic rates of the dehydrogenase complexes but also facilitates the assembly of the essential cofactor required by the complexes to allow aerobic metabolism.
These interactions were discovered in the process of testing a possible alternative lipoic acid assembly pathway (Fig. 2) in which LipA would act on octanoyl-ACP to produce lipoyl-ACP followed by LipB-catalyzed transfer of the lipoyl moiety to unmodified lipoyl domains. Two in vitro results argued for the alternative pathway. These were the observations that LipB utilizes lipoyl-ACP in vitro (7, 12) and that traces of S-adenosyl-l-methionine consumption in the presence of LipA, octanoyl-ACP, and LD (although no lipoylated LD was observed) (4). To test the alternative pathway in vivo, we took advantage of the finding that octanoyl transfer proceeds though an acyl enzyme intermediate in which the LipB active site residue, Cys-169, is transiently acylated (13). LipB was overexpressed and purified, and the enzyme-bound acyl intermediate was found to be octanoic acid. We also report the isolation and properties of a very stable complex between LipB and ACP.
EXPERIMENTAL PROCEDURES
Materials
Anti-FLAG M2 affinity gel and monoclonal anti-HA-agarose conjugate clone HA-7 were purchased from Sigma. [1-14C]Octanoate was purchased from American Radiolabeled Chemicals. The anti-lipoic acid rabbit antibody was from Calbiochem. Vivapure Maxi spin columns type QH were purchased from Sartorius. Tris(2-carboxyethyl phosphine) hydrochloride was from Pierce. The oligonucleotides used for cloning and gene deletions were synthesized by Integrated DNA Technologies. The FLAG (DYKDDDDK) and HA (YPYDVPDYA) peptides were synthesized by the Carver Metabolomics Center at the University of Illinois, Urbana-Champaign.
Bacterial Strains, Growth Conditions, and Media
The bacterial strains used in this study were all derivatives of E. coli K-12 (Table 1). Strains KER184 and KER62 were described previously (14). Strain Top10 was from Invitrogen. Strain MC1061 was from our laboratory collection and was used to construct gene deletions using the λred recombinase method (15). Cassettes encoding resistance to either kanamycin or chloramphenicol were amplified by PCR using plasmids pKD3 and pKD4 and electroporated into induced electrocompetent MC1061 cells transformed with pKD46. Using this method, ΔaceE, ΔaceF, ΔaceEF, and Δlpd mutant strains carrying the chloramphenicol resistance cassette in place of the coding sequences were obtained. A strain with ΔsucAB deletion with a kanamycin resistance cassette was also obtained. All constructed strains were verified by PCR and by requirements for acetate and/or succinate for growth. Double mutant strains were obtained by P1 transduction of ΔsucAB::Kan allele into strain YYC186 (ΔaceEF). Strain BH284 was constructed by transduction of the lipA::chloramphenicol allele from strain ZX219 into strain QC144. Plasmid pCP20 was used to remove the antibiotic resistance cassettes where needed. LB and minimal medium E were the growth media routinely used for growth of bacterial strains. Supplements were added as necessary at the following concentrations: sodium acetate, 5 mm; sodium succinate, 5 mm; thiamine, 1 μg/ml; glucose (0.2%). Antibiotics were added at the following concentrations as necessary (in mg/liter): sodium ampicillin, 100; chloramphenicol, 20; kanamycin sulfate, 50; spectinomycin sulfate, 100; and tetracycline hydrochloride, 15.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant genotype or characteristic | Source or Ref. |
|---|---|---|
| Strains | ||
| MC1061 | araD139 Δ(araA-leu)7697 ΔlacX74 galK16 galE15(GalS) λ- e14- mcrA0 relA1 rpsL150(strR) spoT1 mcrB1 hsdR2 | Lab collection |
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu) 7697 galU galK rpsL (StrR) endA1 nupG | Invitrogen |
| KER184 | rpsL lipB::Tn1000dkan | 27 |
| KER62 | PA360 zbd-601::Tn10 | 7 |
| YYC186 | zac::Tnl0 AaceEF | 4 |
| SI181 | ΔfadE | 45 |
| QC134 | fabA::CmR(lacZ) | 47 |
| ZX293 | KER184/pYFJ31 | 13 |
| ZX297 | XL1-Blue/pZX146 | 13 |
| ZX313 | XL1-Blue/pZX152 | 13 |
| BH48 | SI181/pYFJ31 | This study |
| BH55 | KER184/pBH101 | This study |
| BH77 | SI181/pBH101 | This study |
| BH109 | MC1061 ΔaceE::CmR | This study |
| BH110 | MC1061 ΔaceF::CmR | This study |
| BH111 | MC1061 ΔaceEF::CmR | This study |
| BH112 | MC1061 ΔsucAB::KanR | This study |
| BH114 | MC1061 Δlpd::CmR | This study |
| BH153 | MC1061 ΔsucAB (KanR cassette removed) | This study |
| BH154 | MC1061 ΔaceF (CmR cassette removed) | This study |
| BH155 | MC1061 ΔaceEF (CmR cassette removed) | This study |
| BH156 | MC1061 Δlpd (CmR cassette removed) | This study |
| BH195 | MC1061 ΔaceE (CmR cassette removed) | This study |
| BH229 | QC134(fabA)/pBH101 | This study |
| BH232 | KER62(serA1)/pBH101 | This study |
| BH235 | BH153/pBH101 | This study |
| BH236 | BH155/pBH101 | This study |
| BH237 | BH195/pBH101 | This study |
| BH238 | BH154/pBH101 | This study |
| BH239 | BH156/pBH101 | This study |
| BH245 | BH153/ pBH123+ pCY560 | This study |
| BH246 | BH155/ pBH123+ pCY560 | This study |
| BH247 | BH195/ pBH123+ pCY560 | This study |
| BH248 | BH154/ pBH123+ pCY560 | This study |
| BH249 | BH156/ pBH123+ pCY560 | This study |
| BH260 | YYC186; ΔsucAB (KanR cassette removed) | This study |
| BH278 | ΔsucA775::KanR (Keio collection) | 46 |
| BH279 | ΔsucB776::KanR (Keio collection) | 46 |
| BH284 | MG1655 ΔlplA::KanR ΔLipA::CmR | This study |
| BH285 | BH284 (KanR and CmR casettes removed) | This study |
| BH300 | YYC186; ΔsucB::KanR | This study |
| BH319 | YYC186; ΔsucA (KanR cassette removed) | This study |
| BH321 | BH155/ pBH122 and pBH146 | This study |
| BH323 | BH155/ pBH12 and pBH148 | This study |
| BH324 | BH155/ pBH122 and pBH149 | This study |
| BH325 | BH112/ pBH141 | This study |
| BH326 | BH155/ pBH141 | This study |
| BH327 | BH260/ pBH141 | This study |
| BH328 | BH319/ pBH141 | This study |
| BH329 | BH300/ pBH141 | This study |
| Plasmids | ||
| pQE-2 | N-terminal His-tag cloning vector | Qiagen |
| pET28b | N-terminal His-tag cloning vector with T7 promoter | Novagen |
| pFLAG-MAC | N-terminal Met-FLAG fusion expression vector under tac promoter | Sigma |
| pYFJ31 | LipB(wild type) cloned in pQE-2 vector | 3 |
| pZX146 | LipB(C169S) cloned in pQE-2 vector | 3 |
| pZX152 | LipB(C137A, C147A, C169A triple mutant) cloned in pQE-2 vector | 3 |
| pBH101 | C-terminal flagged LipB cloned in pQE-2 vector | This study |
| pBH104 | N-terminal His-tagged LipA under T7 promoter in pET28b | This study |
| pBH122 | LipB-FLAG under T5 promoter in pBAD33 | This study |
| pBH123 | LipA-FLAG in pET28b | This study |
| pBH141 | N-terminal FLAG-LipB in pFLAG-MAC vector | This study |
| pBH142 | LplA-FLAG in vector pQE-2 | This study |
| pBH144 | LipB-HA tagged in vector pQE-2 | This study |
| pBH145 | LplA-HA tagged vector pQE-2 | This study |
| pBH146 | E2o full length vector pQE-2 | This study |
| pBH148 | E2o-II construct vector pQE-2 | This study |
| pBH149 | E2o-I construct vector pQE-2 | This study |
| pBH158 | E2o-d construct vector pQE-2 | This study |
| pNMN108 | His-tagged E. coli GcvH protein in pET28-a | 6 |
| pCY560 | pTara-T7 RNA polymerase expression under arabinose promoter | This study |
Plasmid Constructions
Plasmids pYFJ31, pZX146, and pZX152 were described previously (13). Plasmids for the expression of the C-terminal FLAG-tagged LipB were constructed by PCR amplification of the lipB gene from pYFJ31 using primers lipB-FLAG-fwd and lipB-FLAG-rev (primer sequences are given in Table 2), where the FLAG sequence was introduced by the reverse primer. The insert carrying lipB with the C-terminal FLAG octapeptide sequence was inserted into the NdeI/HindIII sites of vector pQE-2, and the resulting plasmid was designated as pBH101. For coexpression experiments, plasmid pBH122 was constructed by moving LipB-FLAG along with the phage T5 promoter from pBH101 into the PstI plus HindIII-digested pBAD33 using primers B-pst1-xho1-fwd and B-Hind3-Sal1-rev. An N-terminal FLAG-tagged version of LipB was also inserted into the HindIII/BglII sites of pFLAG-MAC vector using FL-B-fwd and FL-B-rev primers (pBH141). LipA and LipA-FLAG were inserted into the NdeI/EcoRI sites of pET28b under the phage T7 promoter using either LipA-Fwd and LipA-Rev primers for the untagged version or LipA-Fwd and LipA-FLAG-rev primers for the FLAG-tagged version to give the plasmids pBH104 and pBH123, respectively. Plasmid pBH142 for the expression of FLAG-tagged LplA was constructed by PCR amplification using primers lplA-fwd and lplA-Flag-rev followed by insertion into the NdeI/HindIII sites of vector pQE-2. The E2o C-terminal variants (E2o-f, E2o-II, and E2o-I) were inserted into the NdeI/PstI sites of pQE-2 vector using a combination of primer SucB-fwd and the following primers SucB-rev, SucBII-rev, and SucBI-rev, which resulted in plasmids pBH146, pBH148, and pBH149, respectively. Similarly, the E2o N-terminal variant (E2o-d) was constructed using primers SucB-a-fwd and SucB-rev, and the resulting plasmid was designated pBH158. The RSF1030 origin plasmid, pCY560 was constructed in two stages. First, pTARA (16) was digested with NsiI plus NgoMIV, and the fragment containing the gene encoding phage T7 RNA polymerase plus the AraC regulatory protein was ligated to plasmid pDLK29 (17) digested with PstI and NgoMIV to give the kanamycin-resistant plasmid pCY558. Plasmid pCY558 was digested with NsiI (which cuts twice in the kanamycin-resistant cassette) and ligated to the spectinomycin resistance cassette of p34S-Sm2 (18) liberated by PstI digestion to give the spectinomycin-resistant, kanamycin-sensitive plasmid pCY560. This plasmid expresses T7 RNA polymerase under arabinose control, and the replication origin is compatible with those of the pBR322 and pACYC plasmid families. All constructs were verified by sequencing at the Carver Core DNA Sequencing Center at the University of Illinois.
TABLE 2.
Oligonucleotide primers
| Primer | Oligonucleotide sequence |
|---|---|
| lipB-FLAG-fwd | CATATGTTGTATCAGGATAAAATTCTTGTCCGCCAGCTCGGTCTTCAGCCTTACG |
| lipB-FLAG-Rev | AAGCTTTTACTTGTCGTCATCGTCCTTGTAGTCAGCGGTAATATATTCGAAGTCCGGATTG |
| LipA-Fwd | GCGGCGTCCATATGAGTAAACCCATTGTGATGGAACGC |
| LipA-Rev | GCCGGAATTCTTACTTAACTTCCATCCCTTTCG |
| LipA-FLAG-rev | CGCGAATTCTTACTTGTCGTCATCGTCCTTGTAGTCCTTAACTTCCATCCCTTTCGCCTGCAAATCGGCGTG |
| B-pst1-Xho1-fwd | GCGGCGTCCCTGCAGCTCGAGAAATCATAAAAAATTTATTTGC |
| B-Hind3-Sal1-rev | GGACGCCGCGTCGACAAGCTTTTACTTGTCGTCATCGTCCTTG |
| Fl-B-fwd | GCGGCGTCAAGCTTTTGTATCAGGATAAAATTCTTGTCCGCCAGCTCGG |
| Fl-B-rev | GACGCCGCAGATCTTTAAGCGGTAATATATTCGAAGTCCGGATTGTT TAG |
| lplA-fwd | GCGGCGTCCATATGTCCACATTACGCCTGCTCATCTCTGACTCTTACGACCCGTGG |
| lplA-FL-rev | GACGCCGCAAGCTTTTACTTGTCGTCATCGTCCTTGTAGTCCCTTACAGCCCCCGCCATCCATGCCGATAACTCCCGTAGCTC |
| LipA-HA-fwd | GCGGCGTCCATATGAGTAAACCCATTGTGATGGAACGCGGTGTTAAA TACCG |
| LipA-HA-rev | CGCTACGCCTGCAGTTATGCATAGTCAGGAACGTCATAAGGATACTT AACTTCCATCCCTTTCGCCTGCAAATCGGCGTGGTAAGAAGAGC |
| LipB-HA-fwd | GCGGCGTCCATATGTTGTATCAGGATAAAATTCTTGTCCGCCAGCTC GGTCTTC |
| LipB-HA-rev | CGCTACGCCTGCAGTTATGCATAGTCAGGAACGTCATAAGGATAAGC GGTAATATATTCGAAGTCCGGATTGTTTAGTAGCGCTAAAATAT |
| LplA-HA-fwd | GCGGCGTCCATATGTCCACATTACGCCTGCTCATCTCTGACTCTTAC GACCC |
| LplA-HA-rev | CGCTACGCCTGCAGTTATGCATAGTCAGGAACGTCATAAGGATA CCTTACAGCCCCCGCCATCCATGCCGATAACTCCCGTAGCTCTTTTTC |
| SucB-fwd | GCGGCGTCCATATGAGTAGCGTAGATATTCTGGTCCCTGAC |
| SucB-rev | CGCTACGCCTGCAGCTACACGTCCAGCAGCAGACGCGTCGGATC |
| SucBII-rev | CGCTACGCCTGCAGTTAGGCGGTGGAGTTTTTCGCTTCCAGCAGACG |
| SucBI-rev | CGCTACGCCTGCAGTTATTGCGCCGGAGTGGACGCTTTCTCTTCAGA |
| SucB-a-fwd | GCGGCGTCCATATGTCTGAAGAGAAAGCGTCCACTCCGGCGCAAC |
Acid Hydrolysis and GC-MS of LipB Extracts
Preparations of LipB (5–10 mg) purified as described previously (13) were dissolved in 6 m HCl and autoclaved for 2 h to completely hydrolyze the proteins. The hydrolysates were then extracted with methylene chloride, and these extracts were treated with trimethylsilyl diazomethane and methanol to convert fatty acids to their methyl esters (5). The derivatized samples were injected into an Agilent1 0091S GC-MS instrument by splitless injection (250 °C) into an HP-1MS column (30 m × 250 × 0.25 μm). Helium carrier gas was maintained at a constant flow of 1.5 ml/min. The initial oven temperature was maintained at 40 °C for 1 min and then increased to 180 °C at a rate of 20 degrees/min. Temperature was later increased to 270 °C at a rate of 10 degrees/min. The gas chromatograms were searched for the presence of methyl octanoate from the library or extracted for methyl lipoate ions obtained from standards (19).
Coelution Assays
Liquid cultures (typically 50–100 ml) were inoculated into LB or glucose minimal E media supplemented with acetate or succinate (or both as necessary) plus the appropriate antibiotics. Cultures were shaken at 37 °C until the A600 reached 0.6 and then isopropylthiogalactoside was added to 1 mm final concentration. Cultures were shaken for an additional 4 h and centrifuged, and the cell pellet was washed and frozen at −80 °C. The cell pellets were suspended in 50 mm Tris-HCl (pH 7.4) containing 150 mm NaCl (called TBS buffer) and lysed by addition of lysozyme to 1 mg/ml on ice and then sonicated three times for 10 s each. Cell lysates were centrifuged for 30 min, and the cleared lysates were incubated with 250 μl of agarose affinity gel and placed on a rotary mixer for 1 h of gentle mixing at 4 °C. The agarose affinity gel was poured into a column and washed with 20 column volumes of TBS and then eluted with three consecutive 250-μl washes with TBS containing 200 μg/ml of either the FLAG or HA peptides. The second and third eluates (which contained the bulk of the proteins) were usually analyzed on SDS-PAGE.
In Gel Trypsin Digestion and Protein Identification
The gel slices were destained and crushed in 25 mm ammonium bicarbonate buffer containing 50% acetonitrile and dried under vacuum. The sample was suspended in 25 μl of a 0.0125 ng/μl trypsin solution (G-Biosciences, St. Louis) and subjected to digestion using a CEM Liberty digester (Matthew, NC) for 15 min at settings of 50 watts and 55 °C. The digested peptides were extracted using 50% acetonitrile containing 5% formic acid, concentrated by evaporation under vacuum, and suspended in 13 μl of 5% acetonitrile containing 0.1% formic acid. Mass spectral analyses were done on 10 ml of the samples. Mass spectrometry was performed on a Waters Q-ToF with a Waters NanoAcquity UPLC using a linear gradient of acetonitrile with 0.1% formic acid from 0 to 50% in 50 min on a Waters Atlantis dC18 3 μm particle size, 75 μm diameter × 150 mm at a flow rate of 250 nl/min. The raw mass spectrometry results were filtered using Waters ProteinLynx Global server 2.2.5 and further analyzed using MASCOT (Matrixsciences, Cambridge, UK), and the data base searches were conducted by Blast against the NCBI nonredundant data base.
Ion Exchange and Gel Exclusion Chromatography
Ion exchange chromatography of LipB complexes was performed using a HiTrapTM Q FastFlow column on an AKTA purifier FPLC system in Tris-HCl buffer (pH 7.4) and a NaCl gradient to 1 m during 40 min at a 2-ml/min flow rate. Fractions corresponding to different peaks obtained were concentrated by ultrafiltration and analyzed by SDS-PAGE. Gel filtration chromatography of LipB complexes was performed in 50 mm sodium phosphate buffer (pH 7.0) containing 150 mm NaCl using a Superdex 75 10/300 GL column on an AKTA purifier FPLC system run at a 0.5-ml/min flow rate. Fractions corresponding to each peak were collected, concentrated, and analyzed by SDS-PAGE.
MALDI Mass Spectrometry
MALDI mass spectrometry analysis of the LipB-ACP complex was accomplished by pulling down LipB from an ΔaceEF sucAB strain grown in rich medium supplemented with glucose, acetate, and succinate. The column eluates were dialyzed against 2 mm ammonium acetate, and the proteins were concentrated by ultrafiltration, mixed with sinapinic acid matrix, and analyzed on a Voyager DE-STR MALDI mass spectrometer calibrated with enolase standards at 2 mass points (doubly charged and singly charged species) and run in the reflection mode.
RESULTS
High Molecular Weight Proteins Coelute with FLAG-tagged Versions of LipB and LipA
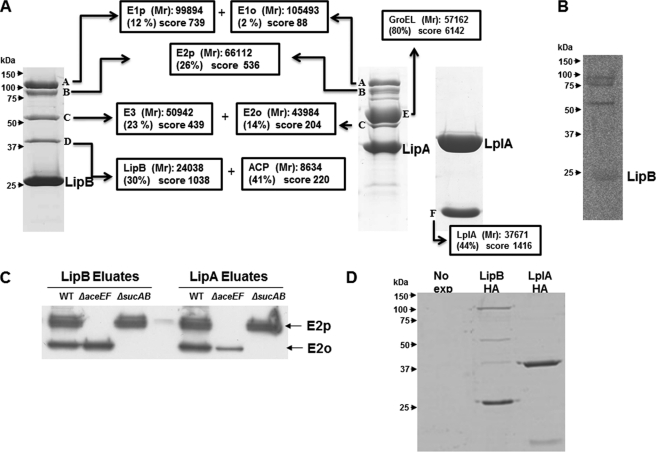
In a series of experiments to test the specificity of LipB under physiological conditions (see below), we attempted to isolate the LipB acyl-enzyme intermediate formed in vivo. To preserve the thioester linkage of the intermediate, we used LipB species tagged with the FLAG epitope rather than hexahistidine-tagged LipB because the imidazole used to elute His-tagged proteins attacks thioesters (20). Unexpectedly, upon elution from the antibody column with the FLAG peptide, the column fractions were found to contain not only FLAG-tagged LipB, but also a consistent set of higher molecular weight proteins (Fig. 3A).
FIGURE 3.
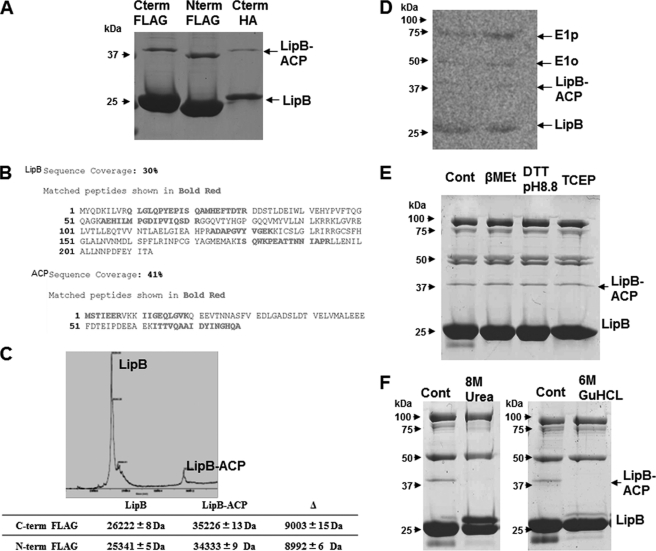
Affinity chromatography of tagged LipB, LipA, and LplA and identification of affinity-purified proteins. A, SDS-PAGE analysis of fractions from FLAG affinity purifications of LipB, LipA, and LplA and identification of the protein bands. Data obtained from Mascot search are displayed in terms of the mass of the identified protein, percentage of sequence covered, and Mascot score. B, autoradiogram of an SDS-polyacrylamide gel of affinity-purified FLAG-LipB eluates expressed in a fadE strain grown in a medium containing [1-14C]octanoate. C, Western blot analysis of LipB and LipA affinity purified proteins from aceEF and sucAB mutant strains using an anti-lipoic acid antibody. D, affinity-purified proteins of HA-tagged LipB and LplA. The 1st lane is a negative control in which the tagged protein was not expressed.
Both N- and C-terminal FLAG-tagged LipB species gave this result. One of the high molecular weight protein bands (band B of Fig. 3A) had the characteristic diffuse and complex SDS-PAGE band pattern given by the PDH E2p protein which migrates (often in multiple bands) at an apparent molecular mass of >75 kDa (E2p is a 66-kDa protein, and its unusual migration is due to atypical SDS binding by the three lipoyl domains) (21). To test the generality of copurification of lipoic acid metabolism enzymes with high molecular weight proteins, overexpression plasmids encoding FLAG-tagged versions of LipA and LplA were constructed, and the FLAG tag protein purification was repeated (Fig. 3A). Elution of FLAG-LipA from the column gave a set of accompanying proteins very similar to that seen with LipB, whereas the LplA eluates lacked higher molecular weight proteins, only LplA plus a smaller protein (later shown to be a LplA proteolysis product) were present.
Identification of the High Molecular Weight Proteins
To test the hypothesis that the high molecular weight proteins were 2-OADH components, we overexpressed LipB in a β-oxidation-defective fadE strain (used to avoid scrambling of the label) in a medium containing [1-14C]octanoate and found that bands B and C became specifically labeled (Fig. 3B) because of LplA-catalyzed octanoylation of the E2 component LDs (10). Modification of the proteins of bands B and C was confirmed by Western blotting with an anti-lipoic acid antibody (Fig. 3C). Moreover, when the FLAG-tagged LipB and LipA proteins were overexpressed in a ΔaceEF host strain (the bacterial strains are given in Table 1), which lacks the PDH E1p and E2p components, the eluates lacked band B (Fig. 3C). A similar experiment with a ΔsucAB strain showed loss of band C (Fig. 3C). To identify the other proteins that coeluted with LipB, gel slices were excised from the SDS-PAGE separations for in-gel trypsin digestion. The resulting peptides were analyzed by LC-MS as described under “Experimental Procedures” and identified by the Mascot search engine (Fig. 3A). Band A contained two proteins, the PDH E1 (pyruvate decarboxylase) component (E1p) encoded by the aceE gene plus the OGDH E1 component (E1o) encoded by the sucA gene, whereas band B contained the PDH dihydrolipoamide acetyltransferase (E2p) component, the product of the aceF gene. The two proteins found in band C were identified as the dihydrolipoamide dehydrogenase E3 component encoded by the lpd gene (the common component of the PDH, OGDH, and the GCV complexes) and the OGDH dihydrolipoamide succinyltransferase component (E2o) encoded by the sucB gene (Fig. 3A). Peptides from both LipB and ACP were found in band D indicating a LipB-ACP complex, the properties of which will be discussed below. Some of the proteins identified above comigrated on the gels (Fig. 3A), and we were unable to resolve E1o and E1p, although manipulation of the electrophoresis conditions allowed resolution of E2o and E3 (Fig. 5). Given that all components of both the PDH and OGDH complexes were present in LipB eluates, it seemed likely that the eluates would be active in the overall reactions of the PDH and OGDH complexes. Indeed, this was the case. The FLAG-LipB eluates had PDH- and OGDH-specific activities, respectively, of 50 and 21 μmol h−1 mg−1 protein in reduction of 2-acetylpyridine adenine dinucleotide (an acetylated NAD analog) in the standard 2-OADH assay (22, 23).
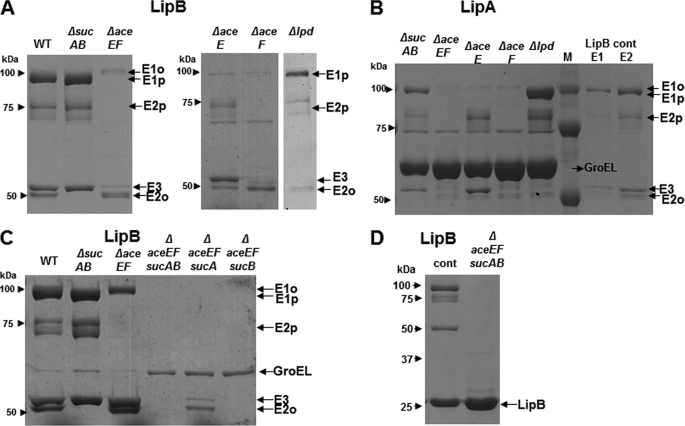
FIGURE 5.
Genetic identification of the 2-OADH component(s) responsible for LipB and LipA binding. Proteins coeluted with FLAG-tagged versions of LipB and LipA from ace and suc deletion strains are shown. A, proteins coeluted with FLAG-LipB after overexpression in different host strains as indicated. B, proteins coeluted with FLAG-LipA after overexpression in Δace or Δsuc strains. The last two lanes show proteins from a wild type that coeluted with FLAG-LipB, and two column fraction are shown. C, proteins coeluted with FLAG-tagged LipB after overexpression in different ace suc double mutant strains. D, proteins coeluted with FLAG-LipB from an ΔaceEF ΔsucAB double mutant strain, the 1st lane is a wild type control.
The FLAG-LipA eluates gave a very similar picture (Fig. 3A) except that no band D was present, and LipA was accompanied by a very abundant protein identified as GroEL, the large subunit of the GroESL chaperonin complex. Chaperonin binding can be attributed to the marked instability of LipA particularly under aerobic conditions where the Fe-S centers of the protein become oxidized (7, 24). We expect that LipA becomes oxidized and partially unfolded upon cell lysis and thereby becomes a substrate of GroESL present in the extracts (the 10-kDa GroES subunit would migrate through the gels and be lost).
In contrast to LipB and LipA, no high molecular weight proteins coeluted with FLAG-tagged LplA. Only a low molecular weight protein migrating at an apparent molecular mass of 20 kDa was present (Fig. 3A). LC-MS identified the 20-kDa band as LplA. The identified peptides spanned the length of the protein indicating that LplA had been cleaved in half. Proteolysis of LplA in crude cell extracts has been observed previously (9), and the protein is known to have a central region that is particularly susceptible to proteolytic cleavage (9, 25).
A possible cross-reactivity of the FLAG antibody with a FLAG-like sequence in the E. coli PDH E1p protein has been reported (26). However, under our experimental conditions PDH and OGDH binding required either LipB or LipA. This was shown by the finding that FLAG-LplA showed no binding of the 2-OADH proteins (Fig. 3A) and by the further chromatographic analyses described below. Moreover, LipB and LplA constructs in which a human influenza hemagglutinin antigen (HA) tag was substituted for the FLAG tag gave results essentially identical to those obtained with the FLAG-tagged proteins (7) (Fig. 3D).
Further Characterization of the Complexes
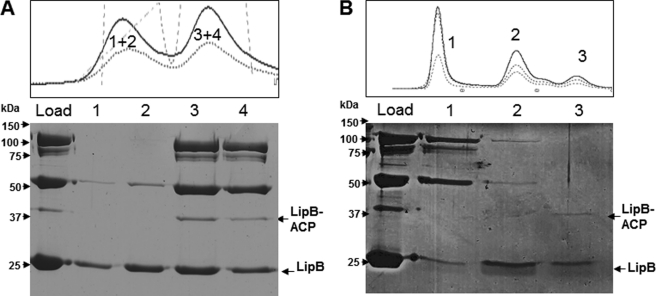
Chromatography on anion exchange and gel exclusion columns were used to further characterize the proteins that coeluted with FLAG-LipB (Fig. 4). All of the fractions containing LipB were combined and subjected to anion exchange chromatography. Only two major peaks were obtained (Fig. 4A). Analysis by SDS-PAGE showed that the first peak was composed of LipB largely unassociated with 2-OADH complexes (Fig. 4A, lanes 1 and 2), whereas the second peak contained LipB associated with the PDH and OGDH complexes (lanes 3 and 4). Because the LipB-ACP complex also eluted in the second peak, this raised the question of whether or not the LipB-ACP complex was bound to the 2-OADH complexes. To answer this question, LipB FLAG column eluates were analyzed by size exclusion chromatography, which gave three major peaks (Fig. 4B). The first peak contained PDH complexes associated with LipB, whereas the second peak contained LipB-OGDH complexes. The third peak contained free LipB plus the LipB-ACP complex showing that the LipB-ACP complex was not bound to the 2-OADH complexes. It is noteworthy that no subunits of the glycine cleavage system were found among the proteins that coeluted with LipB or LipA. This was first thought to be due to the growth conditions that could have repressed gcv gene expression. Hence, the experiments were repeated with FLAG-LipB expressed in a serA strain grown in a minimal medium with glycine as the sole serine source to ensure that the gcv genes were induced and functional (27). However, no proteins other than those characterized above were observed (data not shown). Note that GcvH is reported to be as abundant as the E2 subunits (28).
FIGURE 4.
Anion exchange and gel filtration chromatographic separations of FLAG-LipB affinity-purified proteins. A, anion exchange chromatography of proteins coeluted with FLAG-LipB on a Hitrap Q FF column as described under “Experimental Procedures.” Fractions that covered the first peak were run on SDS-PAGE in lanes 1 and 2, and fractions covering the second peak are in lanes 3 and 4. The position of the LipB-ACP complex is indicated. The lane marked load is the mixture of proteins applied to the column. B, gel exclusion chromatography of proteins coeluted with FLAG-LipB. A Superdex 75 10/300 GL column was used as described under “Experimental Procedures.” The lane marked load is the mixture of proteins applied to the column. Fractions corresponding to peaks 1–3 were concentrated and analyzed by SDS-PAGE in lanes 1–3, respectively. The position of the LipB-ACP complex is also indicated. The chromatogram traces in the upper panels are absorbances of the column effluents monitored at 280, 254, and 215 nm.
Identification of the 2-OADH Components Responsible for Binding LipA and LipB
To identify the component(s) responsible for LipB and LipA binding, we constructed E. coli mutant strains carrying in-frame deletions of the genes encoding the PDH (aceEF) and/or OGDH sucAB components as well as the E3 (lpd) component of both complexes (Table 1). Analysis of these strains confirmed the mass spectral identifications of the 2-OADH components, some of which were based on only a few peptides. Both kanamycin and chloramphenicol resistance cassettes were used to allow facile construction of double mutant strains. When necessary, the resistance cassettes were removed to allow unhindered expression of E2p and E2o from their native promoters. Plasmids expressing FLAG-tagged versions of LipB or LipA were transformed into the mutant strains (the LipA plasmid was accompanied by a phage T7 RNA polymerase expression plasmid). FLAG column eluates were analyzed by 8% SDS-PAGE under conditions where LipB and LipA migrated through the gel (Fig. 5). In the case of the LipB eluates, deletion of sucAB resulted in clear loss of the E2o component plus loss of an indistinct band at the upper edge of the E1p band (Fig. 5A). The indistinct band was the E1o component as was more clearly seen in the ΔaceEF strain where the genes encoding E1p and E2p were deleted (Fig. 5A). The level of E3 protein was markedly decreased in the ΔaceEF strain indicating that most of the E3 (Lpd) resided in the PDH complexes with the reminder being in OGDH complexes (Fig. 5A). Deletion of aceE and lpd had no effect on the levels of the other proteins indicating that E1p and E3 played no roles in LipB binding (Fig. 5A). This left only the E2 components as LipB-binding partners. This was tested more directly by individually deleting either E1p (ΔaceE) or E2p (ΔaceF). Deletion of E1p resulted in loss of only that protein, whereas deletion of E2p resulted in loss of E1p and a large decrease in E3. The FLAG-LipA eluates of the deletion strains gave a very similar picture (Fig. 5B) except for the presence of GroEL as noted above. Therefore, LipB and LipA were bound to the PDH complexes through the E2p component.
We also investigated the interaction of LipB with the OGDH complex using doubly deleted strains (Fig. 5C). The FLAG-LipB eluates from a ΔaceEF ΔsucA strain contained E2o and E3 components, whereas the eluates from a ΔaceEF ΔsucB strain showed no interaction of LipB with the remaining OGDH components indicating that the LipB contacted the OGDH complex through E2o. FLAG-LipB eluates of a ΔaceEF ΔsucAB strain showed loss of all of the 2-OADH components providing direct evidence that LipB does not interact with the E3 component (Lpd) (Fig. 5D). Hence, the LipB and LipA interactions mapped to the E2 components of PDH and OGDH, and the other components were carried through the chromatographic purifications by their association with E2p and E2o in the native complexes.
Interaction of LipB with E2o Protein Fragments in Vivo and in Vitro
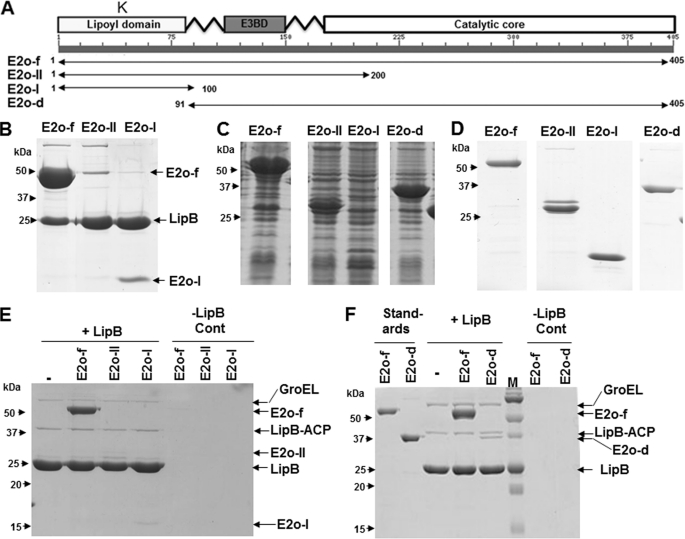
To delineate the E2-binding site(s), we studied E2o rather than E2p because it has a single lipoyl domain rather than the three present in E2p (Fig. 6A) and LipB rather than LipA to avoid the instability of LipA. We constructed plasmids encoding several truncated versions of an N-terminally hexahistidine-tagged E2o construct called E2o-f (Fig. 6A). The E2o-II protein lacked the catalytic core but retained the first 200 E2o residues, whereas the E2o-I protein lacked both the E3 binding domain and the catalytic core and thus essentially consisted of the lipoyl domain. Each of these proteins could be expressed and purified (Fig. 6, C and D). Several other constructs proved unstable and were discarded. In in vivo interaction experiments, derivatives of an ΔaceEF strain carrying compatible plasmids encoding FLAG-LipB plus E2o-f or a truncated E2o protein were induced, and the LipB eluates were examined by SDS-PAGE (Fig. 6B). The FLAG-LipB eluates contained a high level of E2o-f, the full-length tagged E2o protein. Loss of the catalytic core affected the binding properties because FLAG-LipB failed to bind the E2o-II truncation protein and seemed to bind a lower level of the E2o-I truncation protein. The coexpression experiments indicated that LipB bound the lipoyl domain. Similar results were obtained in vitro by passing purified E2o derivatives over a column of immobilized FLAG-LipB. In this assay, FLAG-LipB purified from a ΔaceEF ΔsucAB strain was bound to a FLAG affinity column followed by washing with 20 column volumes of buffer. E2o-f or its truncated derivatives were then passed through the columns followed by further washing of the columns with buffer to remove unbound proteins. The bound proteins were eluted with the FLAG peptide and analyzed by SDS-PAGE (Fig. 6E). These data suggested that another binding site was present on E2o. This was tested in vitro by construction of the E2o-d protein that lacked the lipoyl domain (the first 90 residues). The His-tagged E2o-d variant was purified from a ΔaceEF ΔsucAB strain (Fig. 6D) and used in binding assays (Fig. 6F). As expected from the in vivo data (Fig. 6B), E2o-f was strongly bound and eluted with LipB, whereas E2o-d showed an apparently weaker binding to FLAG-LipB. Taken together, we propose that LipB binding to E2o may involve two synergistic binding interactions as follows: a strong interaction with the lipoyl domain and a second with the catalytic core. It should be noted that isolated lipoyl domains of 70–80 residues are excellent substrates for octanoylation and subsequent sulfur insertion, both in vivo (4, 29, 30) and in vitro (4, 5, 13, 31), and a small octanoylated peptide has been shown to be a substrate for sulfur insertion in vitro (8, 32). Moreover, the apo form of the intact PDH complex is a substrate for lipoate assembly both in vivo and in vitro (4, 7, 13, 33).
FIGURE 6.
In vivo and in vitro mapping of LipB interactions with the E2o protein. A, schematic of the E2o component and its C- and N-terminal truncated derivatives. B, coexpression in vivo of FLAG-LipB with E2o-f and two truncated derivatives. The proteins that coeluted with FLAG-LipB were analyzed by SDS-PAGE. C, expression of E2o protein and its truncation derivatives. D, His tag purifications of E2o protein and its truncated derivatives. E and F, in vitro binding assays of FLAG-LipB to the E2o-f, E2o-II, or E2o-I proteins and E2o-f or E2o-d proteins. The proteins that coeluted with FLAG-LipB were analyzed by SDS-PAGE. In the experiments of E and F, FLAG-LipB from an aceEF sucAB background strain was applied to the column, and the column was thoroughly washed with buffer to remove any unbound proteins and then either E2o-f or one of the truncated derivatives was applied to the column followed by another round of washing. The bound proteins were then eluted with FLAG peptide and analyzed by SDS-PAGE. Control (Cont) reactions for E2o-f and E2o-d run in the absence of FLAG-LipB are in two right-hand lanes.
LipA and LipB Independently Bind the 2-OADHs
An attractive scenario is that LipA or LipB first binds a 2-OADH E2 component and thereby creates a binding site for the other enzyme. However, this was not the case. When FLAG-tagged LipB was expressed in a ΔlipA host strain, the pattern and amounts of dehydrogenase proteins that coeluted with LipB were indistinguishable from that seen with wild type strains (e.g. Fig. 3A) (data not shown). The converse experiment, expression of FLAG-LipA in a ΔlipB host strain, also gave a wild type result (data not shown).
Properties of the LipB-ACP Complex
As mentioned above, among the high molecular weight proteins an SDS-PAGE band that migrated just behind the LipB band with an apparent mass of ∼35 kDa (Figs. 3A and 7A) was present. This band was present irrespective of how LipB was tagged, and the band always migrated slower than the tagged LipB (Fig. 7A). Mascot search of the band D peptides resulted in two hits, LipB and ACP (Fig. 7B). Given these results and the fact that the SDS-PAGE mobility of band D differed from the migration rates of both LipB and ACP, the band clearly was a complex of LipB and ACP. Because the molecular weight of the LipB-ACP complex was close to those of LipA and LplA proteins, we repeated the experiment in ΔlipA, ΔlplA, and ΔlipA lplA strains and found that the LipB-ACP complex was unaffected by these mutations (data not shown). For the C-terminally FLAG-tagged LipB species, MALDI mass spectrometry gave mass values of 26,222 ± 8 and 35,226 ± 13 for LipB and the LipB-ACP complex, respectively. For the N-terminally FLAG-tagged species, the respective values were 25,341 ± 5 and 34,333 ± 9. Hence, the changes in LipB mass upon addition of ACP were 9003 ± 15 and 8,992 ± 6, mass values close to that of decanoyl-ACP. However, given the error in the mass values plus those inherent in subtraction of one large value from another, we could only conclude from these data that the ACP species is fully modified with its 4′-phosphopantetheinyl prosthetic group, and the prosthetic group is probably acylated with a fatty acid of medium chain length.
FIGURE 7.
Properties of the LipB-ACP complex. A, LipB variants with differing affinity tags (C-terminal FLAG, N-terminal FLAG, and C-terminal HA) form the complex as assayed by SDS-PAGE. B, Mascot search results for the band migrating behind LipB resulted in two hits, LipB and ACP. The peptide sequences that matched LipB and ACP are given in boldface type. Note that the central tryptic peptide of ACP is insoluble (42, 43), and thus only terminal peptides were recovered. C, MALDI-MS data for the LipB and LipB-ACP complex. The mass difference between LipB and LipB-ACP is also given. D, FLAG purification of LipB from a fadE strain grown with [1-14C]octanoate added to the medium. The approximate position of the LipB-ACP complex is indicated. E, resistance of the LipB-ACP complex to reduction by different disulfide-reducing agents. F, treatment of the LipB-ACP complex with 8 m urea or 6 m guanidine HCl (GuHCl).
To test whether the LipB-ACP complex is acylated, we purified LipB from a fadE strain grown in a medium containing [1-14C]octanoate and exposed the SDS-polyacrylamide gel to a phosphorimager storage screen. A faint ∼35-kDa band was observed between the LipB band and the E2o band where the LipB-ACP complex migrates (Fig. 7D). Its presence on these gels indicated that the LipB-ACP complex was resistant to treatment with SDS and heat in the presence of 2-mercaptoethanol. Substitution of other disulfide-reducing agents, dithiothreitol (at pH 8.8) and tris(2-carboxyethyl)phosphine, had no effect on the complex (Fig. 7E). Addition of an excess of a PDH LD to the LipB eluates had no effect on the complex indicating that it was not a reaction intermediate (data not shown). However, the LipB-ACP complex was sensitive to treatment with 8 m urea or 6 m guanidine hydrochloride (in the absence of a reducing agent) prior to SDS-PAGE indicating that the linkage of LipB to ACP is noncovalent (Fig. 7F).
LipB Functions Solely as an Octanoyltransferase in Vivo
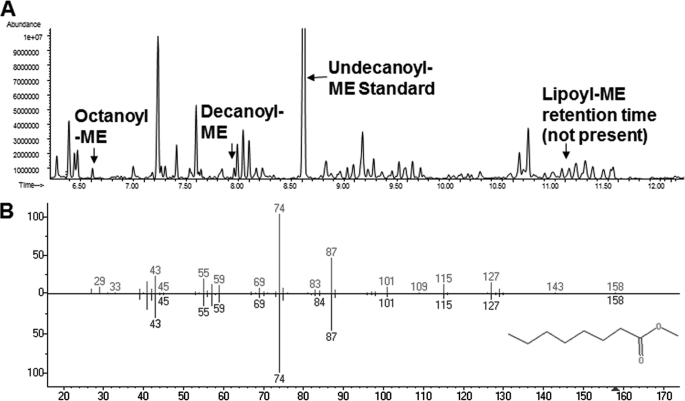
As outlined in the Introduction, octanoyl-ACP is thought to be the in vivo substrate of LipB, and lipoyl-ACP would act as an octanoyl-ACP mimic. To test this belief in vivo, we took advantage of the finding that octanoyl transfer proceeds though an acyl enzyme intermediate in which the LipB-active site residue, Cys-169, is transiently acylated (13). LipB was overexpressed such that it was in great excess over the available acceptor domains and purified. Numerous attempts were made to isolate the acyl-enzyme intermediate on a tryptic peptide. However, this was unsuccessful due to the low level of modification (presumably because of transfer back to ACP, the most abundant soluble protein of E. coli) plus the instability of the thioester bond during trypsin digestion. The ester bond of the dead-end product formed when serine was substituted for LipB Cys-169 (13) and was also labile under the proteolysis conditions. Consequently, we resorted to analysis of purified LipB by GC-MS following acid hydrolysis of the protein, extraction of fatty acids, and their conversion to methyl esters. Extracts of the hydrolyzed wild type LipB preparations were found to contain low levels (<2%) of methyl octanoate, whereas no trace of methyl lipoate was observed. Gas chromatography of extracts of the hydrolyzed C169S mutant protein showed higher methyl octanoate levels than those of the wild type protein preparations (Fig. 8), and the mass spectrum obtained was identical to the methyl octanoate spectrum of the NIST library. In agreement with the results obtained with the wild type enzyme, we found no detectable methyl lipoate, although a low level of methyl decanoate (comparable with that of methyl octanoate) and significantly higher levels of 16 and 18 carbon methyl esters. The significance of the decanoyl species was unclear, although the Mycobacterium tuberculosis LipB is known to bind C10 fatty acids (34). The long chain acids can be attributed to phospholipids from traces of membrane fragments that were carried through the LipB purification procedure.
FIGURE 8.
GC-MS chromatogram and mass spectrum of methyl octanoate extracted from purified LipB. Purified LipB proteins were acidified by addition of concentrated HCl to 6 m final concentration and then autoclaved for 2 h at 120 °C, extracted with methylene chloride, derivatized by addition of trimethylsilyldiazomethane plus methanol, and analyzed by GC-MS. A, gas chromatogram obtained from the material extracted from the hydrolyzed LipB C169S protein with the elution positions of methyl octanoate (octanoyl-ME) and methyl lipoate (lipoyl-ME) are indicated. B, mass spectrum of methyl octanoate obtained from LipB C169S (lower spectrum) compared with the methyl octanoate spectrum of the NIST library (upper spectrum).
DISCUSSION
An unexpected by-product of experiments designed to study LipB function in vivo (discussed below) was the indication that protein-protein interactions are involved in the assembly of lipoic acid. LipB and LipA were found to form noncovalent interactions with the E2 components of PDH and OGDH that are sufficiently strong to survive chromatographic purifications. These are not enzyme-substrate interactions because both the E2o and E2p components are known to be fully modified with lipoic acid in wild type cells (35), a finding consistent with our Western blotting and dehydrogenase activity results. The interactions were specific in that another enzyme of lipoic acid metabolism, LplA, showed no interactions with either PDH or OGDH. Moreover, neither LipA nor LipB bound another LD protein, the H protein of the GCV system.
The advantage of interactions of LipB and LipA with their respective substrates, the apo and octanoyl forms of the 2-OADH E2 components is straightforward. Upon assembly of complexes from the nascent 2-OADH components, LipA and LipB would become bound and therefore poised to produce the protein-bound lipoate molecules required for the activity of these key metabolic enzymes. One objection to this scenario is that the 2-OADH components are highly abundant proteins (∼5,000 molecules per cell), whereas the levels of LipA and LipB are below the limits of proteomic analyses (<300 molecules/cell) (28), a scarcity that precluded their inclusion in the E. coli global interaction network (36). However, a LipA content of 30 molecules per cell was recently reported based on a chromosomal fusion to yellow fluorescent protein (YFP) (37). (No YFP fusion data were reported for LipB, but only a fourth of the E. coli proteins were analyzed.) Hence, the only data set that includes both the 2-OADHs and a lipoic acid synthetic protein is that obtained from YFP fusions. A possible caveat is that the YFP fusion protein values for 2-OADH protein molecules per cell are 5–6- and 2–3-fold lower than those obtained by proteomic analysis (28) and two-dimensional gel electrophoresis of extracts of cells labeled with radioactive amino acids (38), respectively. However, given that the same analytical method was used for both, the ratios of the YFP fusion protein numbers should be reliable. The YFP fusion data gave values of 843 and 850 molecules per cell for the PDH E1 and E2 components and a value of 158 molecules per cell for the OGDH E1 component (37). Assuming that the OGDH E2 component is produced at the same level as the cognate E1 component, then each cell roughly contains about 1,000 E2 components to be modified by 30 molecules of LipA. These seemingly incompatible numbers are, however, offset by two properties of the 2-OADH complexes. First, each complex has 24 E2 components in an octahedral array, and therefore the 1,000 E2 components are structured into about 40 complexes, a number similar to the reported number of LipA molecules. The second property of the 2-OADH complexes is that a given E2 lipoyl domain can reach far across the protein complex. This latter property was demonstrated by the finding that PDH complexes can retain full enzymatic activity despite containing major fractions of E2 proteins that either lack lipoyl domains or contain lipoyl domains that cannot be modified (39). Because the E1 component catalyzes the rate-limiting step in the overall PDH reaction (39–41), it follows that the lipoyl domains must be able to reach distant E1 components (24 of which are in the complex). Moreover, the lipoyl domains of different E2 molecules are known to interact (39). Therefore, it seems possible that a single molecule of a lipoic acid synthetic enzyme bound to a given E2 component of the complex could catalyze assembly of lipoyl groups on the other E2 components of the complex (or that LipA and LipB dissociate from a lipoylated LD and efficiently bind a neighboring unmodified LD because of the high localized LD concentration). The highly localized concentrations of LipA and LipB may be required because the proteins are not only in short supply but are also poor catalysts (6, 31) (the turnover numbers reported for LipB and LipA are 0.2 s−1 and 0.175 min−1, respectively, although catalytic formation of lipoate by LipA has yet to be demonstrated in vitro). It should be noted that our analyses required overproduction of the lipoic acid metabolic proteins due to their scarcity in wild type cells. Overproduction undoubtedly raised the effective association constants for complex formation and thereby facilitated formation of the complexes. However, once formed, the complexes were stable in that they survived affinity chromatography as well as chromatography on ion exchange and size exclusion columns.
We attempted to delineate the site(s) of interaction of LipB with the E2o component of OGDH (Fig. 6). The expected interaction with the isolated E2o lipoyl domain was observed both in vivo and in vitro, but the levels of LipB-bound domain appeared low. However, this discrepancy is probably more apparent than real given the characteristically poor staining of lipoyl domains (because of their small sizes and markedly acidic amino acid compositions). In contrast, the E2o-d fragment would be expected to stain comparably to the full-length E2o protein, and thus the puzzling interaction of LipB with the core domain seems likely to be considerably weaker than the LipB-lipoyl domain interaction. We have attempted further study of E2o-LipB interactions by isothermal titration calorimetry, but we were unable to attain the concentrations of these proteins required for heat change measurements.
A very stable (but noncovalent) LipB-ACP complex was also detected, the physiological relevance of which is unclear. The complex seems to carry an acyl group that is probably octanoate but is unable to transfer the acyl moiety to either an LD domain or to ACP and thus is a dead-end complex. In previous work from this laboratory, purified preparations of the [1-14C]octanoyl-LipB intermediate were found to transfer the octanoyl moiety either to an E2p LD or to ACP (13). However, transfer was incomplete, and the label that failed to transfer was present in a band that migrated slightly behind LipB in SDS-PAGE, a behavior similar to that seen for the in vivo LipB-ACP complex. Given that the in vivo complex was inactive and was not associated with the 2-OADHs, the LipB-ACP complex seems unlikely to have any physiological significance. However, its presence does provide further evidence of the link between the fatty acid and lipoic acid synthetic pathways. It seems likely that the LipB-ACP complex is formed in vitro by a fraction of LipB molecules that adopt an aberrant off-pathway conformation that traps octanoyl-ACP.
Our observation that LipB accumulates an octanoyl-enzyme intermediate with no sign of a lipoyl-enzyme intermediate (Fig. 8) indicates that lipoyl-ACP is not an intermediate in lipoic acid biosynthesis. Moreover, these data are in accord with the mass spectral data that show that LipB forms a complex with an acyl-ACP (probably octanoyl-ACP) that is not lipoyl-ACP plus the lack of a LipA-ACP complex.
Acknowledgments
We thank Dr. Peter Yau of the Carver Biotechnology Center and Dr. Alexander Ulanov of the Carver Metabolomics Center at the University of Illinois, Urbana-Champaign, for their help in protein identification and the use of GC-MS equipment.
This work was supported, in whole or in part, by National Institutes of Health Grant AI15650 from the NIAID.
- PDH
- pyruvate dehydrogenase
- OGDH
- 2-oxoglutarate dehydrogenase
- GCV
- glycine cleavage system
- 2-OADH
- 2-oxoacid dehydrogenase
- LD
- lipoyl domain
- ACP
- acyl carrier protein.
REFERENCES
- 1. Perham R. N. (2000) Annu. Rev. Biochem. 69, 961–1004 [DOI] [PubMed] [Google Scholar]
- 2. Lengyel J. S., Stott K. M., Wu X., Brooks B. R., Balbo A., Schuck P., Perham R. N., Subramaniam S., Milne J. L. (2008) Structure 16, 93–103 [DOI] [PubMed] [Google Scholar]
- 3. Douce R., Bourguignon J., Neuburger M., Rébeillé F. (2001) Trends Plant Sci. 6, 167–176 [DOI] [PubMed] [Google Scholar]
- 4. Zhao X., Miller J. R., Jiang Y., Marletta M. A., Cronan J. E. (2003) Chem. Biol. 10, 1293–1302 [DOI] [PubMed] [Google Scholar]
- 5. Cicchillo R. M., Booker S. J. (2005) J Am. Chem. Soc. 127, 2860–2861 [DOI] [PubMed] [Google Scholar]
- 6. Cicchillo R. M., Iwig D. F., Jones A. D., Nesbitt N. M., Baleanu-Gogonea C., Souder M. G., Tu L., Booker S. J. (2004) Biochemistry 43, 6378–6386 [DOI] [PubMed] [Google Scholar]
- 7. Miller J. R., Busby R. W., Jordan S. W., Cheek J., Henshaw T. F., Ashley G. W., Broderick J. B., Cronan J. E., Jr., Marletta M. A. (2000) Biochemistry 39, 15166–15178 [DOI] [PubMed] [Google Scholar]
- 8. Douglas P., Kriek M., Bryant P., Roach P. L. (2006) Angew. Chem. Int. Ed. Engl. 45, 5197–5199 [DOI] [PubMed] [Google Scholar]
- 9. Green D. E., Morris T. W., Green J., Cronan J. E., Jr., Guest J. R. (1995) Biochem. J. 309, 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morris T. W., Reed K. E., Cronan J. E., Jr. (1995) J. Bacteriol. 177, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morris T. W., Reed K. E., Cronan J. E., Jr. (1994) J. Biol. Chem. 269, 16091–16100 [PubMed] [Google Scholar]
- 12. Jordan S. W., Cronan J. E., Jr. (1997) J. Biol. Chem. 272, 17903–17906 [DOI] [PubMed] [Google Scholar]
- 13. Zhao X., Miller J. R., Cronan J. E. (2005) Biochemistry 44, 16737–16746 [DOI] [PubMed] [Google Scholar]
- 14. Reed K. E., Cronan J. E., Jr. (1993) J. Bacteriol. 175, 1325–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Datsenko K. A., Wanner B. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wycuff D. R., Matthews K. S. (2000) Anal. Biochem. 277, 67–73 [DOI] [PubMed] [Google Scholar]
- 17. Phillips G. J., Park S. K., Huber D. (2000) BioTechniques 28, 400–402 [DOI] [PubMed] [Google Scholar]
- 18. Dennis J. J., Zylstra G. J. (1998) BioTechniques 25, 772–774, 776 [DOI] [PubMed] [Google Scholar]
- 19. Pratt K. J., Carles C., Carne T. J., Danson M. J., Stevenson K. J. (1989) Biochem. J. 258, 749–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stadtman E. (1954) in The Mechanism of Enzyme Action (McElroy W., Glass B. eds) pp. 581–598, Johns Hopkins Press, Baltimore, MD [Google Scholar]
- 21. Guest J. R., Lewis H. M., Graham L. D., Packman L. C., Perham R. N. (1985) J. Mol. Biol. 185, 743–754 [DOI] [PubMed] [Google Scholar]
- 22. Christensen Q. H., Cronan J. E. (2009) J. Biol. Chem. 284, 21317–21326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guest J. R., Creaghan I. T. (1973) J. Gen. Microbiol. 75, 197–210 [DOI] [PubMed] [Google Scholar]
- 24. Busby R., Schelvis J., Yu D., Babcock G., Marletta M. (1999) J. Am. Chem. Soc. 121, 4706–4707 [Google Scholar]
- 25. McManus E., Luisi B. F., Perham R. N. (2006) J. Mol. Biol. 356, 625–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Worrall J. A., Górna M., Crump N. T., Phillips L. G., Tuck A. C., Price A. J., Bavro V. N., Luisi B. F. (2008) J. Mol. Biol. 382, 870–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vanden Boom T. J., Reed K. E., Cronan J. E., Jr. (1991) J. Bacteriol. 173, 6411–6420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu P., Vogel C., Wang R., Yao X., Marcotte E. M. (2007) Nat. Biotechnol. 25, 117–124 [DOI] [PubMed] [Google Scholar]
- 29. Ali S. T., Guest J. R. (1990) Biochem. J. 271, 139–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ali S. T., Moir A. J., Ashton P. R., Engel P. C., Guest J. R. (1990) Mol. Microbiol 4, 943–950 [DOI] [PubMed] [Google Scholar]
- 31. Nesbitt N. M., Baleanu-Gogonea C., Cicchillo R. M., Goodson K., Iwig D. F., Broadwater J. A., Haas J. A., Fox B. G., Booker S. J. (2005) Protein Expr. Purif. 39, 269–282 [DOI] [PubMed] [Google Scholar]
- 32. Bryant P., Kriek M., Wood R. J., Roach P. L. (2006) Anal. Biochem. 351, 44–49 [DOI] [PubMed] [Google Scholar]
- 33. Jordan S. W., Cronan J. E., Jr. (1997) Methods Enzymol. 279, 176–183 [DOI] [PubMed] [Google Scholar]
- 34. Ma Q., Zhao X., Nasser Eddine A., Geerlof A., Li X., Cronan J. E., Kaufmann S. H., Wilmanns M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8662–8667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Packman L. C., Green B., Perham R. N. (1991) Biochem. J. 277, 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Butland G., Peregrín-Alvarez J. M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Davey M., Parkinson J., Greenblatt J., Emili A. (2005) Nature 433, 531–537 [DOI] [PubMed] [Google Scholar]
- 37. Taniguchi Y., Choi P. J., Li G. W., Chen H., Babu M., Hearn J., Emili A., Xie X. S. (2010) Science 329, 533–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smith M. W., Neidhardt F. C. (1983) J. Bacteriol. 156, 81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perham R. N. (1991) Biochemistry 30, 8501–8512 [DOI] [PubMed] [Google Scholar]
- 40. Akiyama S. K., Hammes G. G. (1980) Biochemistry 19, 4208–4213 [DOI] [PubMed] [Google Scholar]
- 41. Akiyama S. K., Hammes G. G. (1981) Biochemistry 20, 1491–1497 [DOI] [PubMed] [Google Scholar]
- 42. Vanaman T. C., Wakil S. J., Hill R. L. (1968) J. Biol. Chem. 243, 6409–6419 [PubMed] [Google Scholar]
- 43. Vanaman T. C., Wakil S. J., Hill R. L. (1968) J. Biol. Chem. 243, 6420–6431 [PubMed] [Google Scholar]
- 44. Chang Y. Y., Cronan J. E., Jr. (1988) J. Bacteriol. 170, 3937–3945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Iram S. H., Cronan J. E. (2006) J. Bacteriol 188, 599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. (2006) Mol. Syst. Biol. 2, 2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Christensen Q. C., Crona J. E. (2010) Biochemistry 49, 10024–10036 [DOI] [PMC free article] [PubMed] [Google Scholar]