Abstract
Select proteins involved in electrical and chemical neurotransmission are re-coded at the RNA level via the deamination of particular adenosines to inosine by adenosine deaminases acting on RNA (ADARs). It has been hypothesized that this process, termed RNA editing, acts to “fine-tune” neurophysiological properties in animals and potentially downstream behavioral outputs. However, the extreme phenotypes resulting from deletions of adar loci have precluded investigations into the relationship between ADAR levels, target transcripts, and complex behaviors. Here, we engineer Drosophila hypomorphic for ADAR expression using homologous recombination. A substantial reduction in ADAR activity (>80%) leads to altered circadian motor patterns and abnormal male courtship, although surprisingly, general locomotor coordination is spared. The altered phenotypic landscape in our adar hypomorph is paralleled by an unexpected dichotomous response of ADAR target transcripts, i.e. certain adenosines are minimally affected by dramatic ADAR reduction, whereas editing of others is severely curtailed. Furthermore, we use a novel reporter to map RNA editing activity across the nervous system, and we demonstrate that knockdown of editing in fruitless-expressing neurons is sufficient to modify the male courtship song. Our data demonstrate that network-wide temporal and spatial regulation of ADAR activity can tune the complex system of RNA-editing sites and modulate multiple ethologically relevant behavioral modalities.
Keywords: Drosophila Genetics, Gene Knockout, Gene Regulation, Ion Channels, Mutant, Neuroscience, RNA Editing, RNA Processing, RNA-Protein Interaction, Synapses
Introduction
Informational recoding of RNA by the catalytic deamination of adenosine to inosine proceeds through the action of ADARs3 (1). Long double strand RNA duplexes exhibiting perfect complementarity can be modified extensively by promiscuous ADAR activity. However, mRNAs may also serve as site-specific substrates for ADARs via base pairing interactions that generate short imperfect duplexes that generally include the exon destined for editing and a cis-acting complementary sequence, usually found in a neighboring intron (2, 3). Because inosine is recognized by the translation machinery as guanosine (4), A-to-I editing in mRNAs can lead to the incorporation of amino acids differing from those specified by the literal genome.
In Drosophila, the spectrum of ADAR substrates is peculiarly specific, consisting primarily of mRNAs encoding an array of voltage- and ligand-gated ion channels, as well as numerous pre-synaptic proteins involved in exo- and endocytosis of synaptic vesicles (5–8). Similarly, several mammalian ion channels and G-protein-coupled receptors are also subject to RNA editing (2, 7, 9–11). In light of the ontological class and high sequence conservation of ADAR target genes, RNA editing has been invoked as an essential function in controlling synaptic transmission and neurophysiology. Correspondingly, deletion of the single Drosophila adar locus (dAdar) results in severe adult-stage behavioral abnormalities, including extreme uncoordination, seizures and a complete lack of courtship in dAdar null (dAdar5g1) males (12), whereas mice lacking ADAR2 suffer from seizures and early mortality (13).
Due to the presence of a single X-linked adar locus and more than 100 mRNA sites of dADAR modification, Drosophila provides an ideal system to study the correlation between deaminase levels and recoding output. We have previously shown that restoration of editing in the adult nervous system partially rescues the locomotor defect of dAdar-deficient males, an effect that appears to be independent of any interactions between dAdar and the RNAi pathway (14). However, the pattern of dADAR expression and activity within the fly nervous system is currently unknown. Furthermore, although previous studies have focused on the relationship between dADAR activity and motor control, it is unclear whether complex behaviors require regulated editing and, if so, whether subpopulations of edited proteins contribute to distinct behavioral outputs.
Here, we investigate these issues using homologous recombination and a molecular reporter for RNA editing activity. Although dADAR expression can be detected in almost all neuronal nuclei, significant variation in dADAR activity exists between genetically distinct neurons. Finally, through the generation of a novel hypomorphic dAdar allele, we demonstrate an unexpectedly complex relationship between in vivo dADAR levels and deamination of specific RNA editing targets. These data, combined with neuron-specific dADAR knockdown, demonstrate that correct regulation of editing activity at both cell-autonomous and network levels is required for behavioral outputs in Drosophila and provide mechanistic insight into the complex landscape of proteomic diversity generated by RNA editing.
EXPERIMENTAL PROCEDURES
Drosophila Strains and Genetics
For a full list of Drosophila strains used, see supplemental Tables 1 and 2. Flies were raised at a constant 25 °C, on standard molasses food, and under 12-h day/night cycles. Both dAdar RNAi transgenes were obtained from the Vienna Drosophila RNAi Stock Center. Generation of the double RNAi line was described previously (14). Tissue-specific Gal4 lines were obtained from the Bloomington stock center.
Ends-out Homologous Recombination of the dAdar Locus
We performed ends out homologous recombination using a similar methodology to that reported previously (15). Briefly, we utilized the ends-out targeting vector p[w25.2] that contains the white+ selectable eye color mini-gene flanked by LoxP sites for subsequent removal by Cre-recombinase. Homology arms were cloned and sequenced in pTOPO (Invitrogen) and then shuttled into the multiple cloning sites of the vector to generate p[w25-dADAR-HA], which was then introduced into the Drosophila genome by standard transgenic methods (Genetic Services Inc.).
The cloning strategy is as follows, where all genomic coordinates are given by the Drosophila melanogaster draft, BDGP Release 5, with release 5.12 annotation provided by FlyBase at the UCSC Genome Browser. Arm 1 is the 5′ arm of p[w25-dADAR-HA] (see Fig. 1) and was generated by PCR amplification to incorporate cloning sites as follows: Arm 1, BsiWI-1,673,865–1,676,526-AscI. Arm 2 is the 3′ arm of p[w25-dADAR-HA] and was generated in two parts by PCR amplification to incorporate cloning sites and an HA epitope tag; Arm2, Acc65I-1,676,602–1677784-HA/NheI-1,677,788–1,679,400-NotI. The HA tag sequence was inserted after the terminal glutamate codon of dAdar (GAA) ending in an opal (TGA) stop codon and an NheI (gctagc) cutting site as follows: GAAtacccttacgatgttcctgattacgccagcctgTGAgctagc.
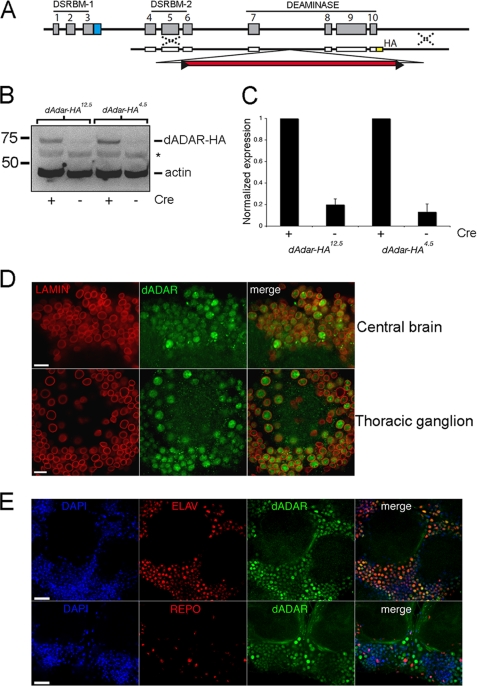
FIGURE 1.
Visualization of dADAR expression using ends-out homologous recombination. A, schematic representation of the targeting construct used to insert an HA epitope tag at the 3′ of the dAdar locus. B, representative Western blot showing HA-positive bands in two independent lines lacking the white+ mini-gene. Actin was used as a loading control. *, nonspecific labeling. This is likely to be a head/brain-specific cross-reaction because it is not observed when using whole fly tissue (see Fig. 4B). C, quantification of relative dADAR-HA levels (normalized to actin) before and after Cre expression. Values are expressed relative to the mean of each post-Cre dAdarHA line (n = 6 Western blots, three independent samples). Error bars, S.E. values. D, lamin and dADAR-HA staining in the male brain and thoracic ganglion. Scale bar, 10 μm. E, dADAR-HA co-localizes with DAPI-stained nuclei and Elav, but not Repo, in the male brain. Scale bar, 20 μm.
Targeting was performed to generate multiple independent targeting events in which the HA tag was incorporated or excluded from the recombination events. Targeted alleles were validated by amplification using primers outside the region of targeting. All targeted alleles were sequenced to verify only the presence of indicated sequences. Subsequent removal of the white+ mini-gene selectable marker was achieved by performing crosses to animals expressing Cre-recombinase and re-isolation of targeted chromosomes containing a single LoxP site. The recombinant alleles were subsequently back-crossed to Canton-S for five generations.
Behavioral Analysis
Locomotor patterns were recorded using horizontal, single fly activity monitors (TriKinetics). Flies were left to acclimatize for ∼12 h before recording was initiated. An average daily pattern was calculated for each fly by averaging data from 3 consecutive days. These values were then further averaged across the experimental population. Mating assays and song recording were performed in a custom-made chamber. For each assay, 5–7-day-old males and 3–5-day-old virgin females were used, and the time taken for male initiation of courtship (latency) and the courtship index (time spent courting/total time) was recorded over 10 min. All mating assays were performed in a narrow time window (7–10 a.m.) to minimize circadian influences on experimental outcome, blind to experimental genotype where possible. Mating songs were recorded using a MicroTrack mobile digital recorder (M-Audio) and were analyzed in Audacity. Because dAdarhyp males expressed a white+ mini-gene and dAdarWTLoxP did not, we crossed a white+ mini-gene-containing p[w25.2] vector inserted in the 3rd chromosome into the dAdarWTLoxP background to restore eye pigment expression.
RNA Editing Analysis
RNA extractions from Drosophila heads (15–20 per sample) were performed using TRIzol reagent (Invitrogen). Edited cDNAs were amplified via RT-PCR using target-specific primers (see Ref. 14 for details). Levels of editing were determined by measuring the area under A- and G-peaks in individual electropherogram traces using ImageJ. The percent editing is expressed at G/(A + G) × 100.
Western Blotting
Protein samples were prepared in buffer containing SDS and β-mercaptoethanol and electrophoresed on a 10% gel (Amresco). Anti-HA antibody (Covance) was used at 1:500; anti-actin (Millipore) was used at 1:20,000–80,000. Band intensities were quantified on a Kodak Image Station following background subtraction. For adult heads, 20 heads/100 μl of buffer were used per sample. For developmental analysis, we used 250 1st instar larvae, 10 3rd instar larvae, and 10 whole adult males per 100 μl of buffer.
Confocal Microscopy and Immunohistochemistry
A Zeiss LSM 510 meta-confocal microscope was used to obtain all images. Samples for immunohistochemistry were prepared as described previously (16). Primary antibodies were used at the following concentrations: mouse anti-Lamin, anti-Elav, anti-Repo, and anti-Dachshund (Developmental Studies Hybridoma Bank) were all used at 1:50; mouse anti-HA and rabbit anti-HA (Santa Cruz Biotechnology) were used at 1:50. Alexa-Fluor secondary antibodies (goat anti-mouse Cy3 and goat anti-mouse and anti-rabbit FITC (Invitrogen)) were used at 1:200. DAPI (Invitrogen) was used at 1:1000. Confocal images were obtained at subsaturation levels of fluorescent intensity. Images were contrast-enhanced in Adobe Photoshop. Each image shown is a representative example of n ≥ 5.
RESULTS
dADAR Is Localized to the Neuronal Nucleus in the Drosophila Brain
The endogenous dADAR protein expression pattern within the adult Drosophila nervous system has not been determined. To remedy this, we used ends-out homologous recombination (17) to generate three independent recombinant lines, two with HA epitope-tagged sequences at the 3′ end of the dAdar locus (Fig. 1, A and B) and one without. Editing levels did not significantly differ between both dAdarHA lines and w1118 controls (supplemental Fig. 1).
During homologous recombination, screening for recombinant flies is facilitated by the insertion of an ∼5-kb white+ mini-gene eye color selection cassette within an intron of the dAdar locus, subsequently removed via a Cre-recombinase step (17, 18). Western blotting using an anti-HA antibody revealed robust expression of an HA-immunoreactive protein at the predicted size of dADAR in both recombinant lines lacking the white+ mini-gene (Fig. 1C). We used these lines to detail the expression pattern of dADAR. Because dADAR-HA levels and endogenous editing were indistinguishable between the two independent lines, we use them interchangeably throughout all subsequent experiments. Confocal microscopy revealed broad expression of dADAR in the brain and thoracic ganglion (supplemental Fig. 1). Co-immunostaining for HA and the nuclear envelope protein Lamin showed that dADAR expression was prominent only within nuclei (Fig. 1D). No significant dADAR localization to the cytoplasmic, axonal, or dendritic compartments was observed. In addition, dADAR co-localized with Elav (a marker for neuronal nuclei) but not Repo (a glial nuclear marker), indicating that nuclear dADAR expression is widespread and enriched in the neuronal nucleus (Fig. 1E).
Editing Activity Varies Widely between Neuronal Subpopulations
Our initial analysis of dADAR localization revealed clear differences in dADAR protein expression even between neighboring neurons (Fig. 1, D and E), suggesting that dADAR expression is under spatial control in the Drosophila brain, as is the case in mammals (19). To investigate how dADAR activity varies in genetically defined neurons, we used a molecular reporter of editing activity based on syt-1, which contains four editing sites in exon 9, of which sites 3 and 4 are edited most robustly (Fig. 2A) (3, 7). The reporter (termed syt-T) consists of the edited exon flanked by the upstream and downstream introns and exons cloned into a pUAS vector (14), allowing targeted expression using the UAS-Gal4 binary expression system (20).
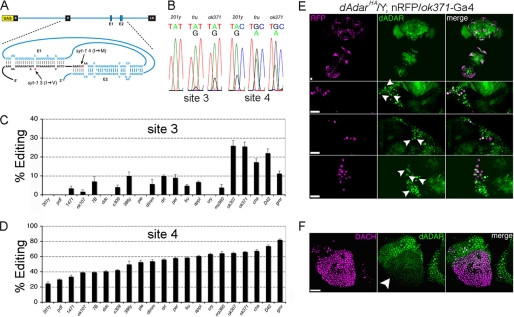
FIGURE 2.
Molecular reporter of RNA editing reveals neuron-specific patterns of dADAR activity. A, design of the reporter, termed syt-T. Exons 8–10 of syt-1, along with the intervening introns, were cloned into the pUAS expression vector. Upon transcription, the E1 and E2 elements form a pseudo-knot structure by base pairing with coding sequences in exon 9 (3), leading to formation of a dADAR substrate and editing of sites 3 and 4. B, example electropherograms showing editing of sites 3 and 4 in three genetically distinct cell types as follows: mushroom body γ neurons (201y), fruitless-positive (fru), and glutamatergic (ok371) neurons. Average editing of site 3 and 4 in 21 classes of neurons, defined by distinct Gal4 drivers, is shown in C and D. Each value is the mean of 4–6 RT-PCRs derived from males carrying each Gal4 driver and one of two independent insertions of syt-T. Error bars, S.E. values. E, dADAR expression in glutamatergic neurons. dADAR-HA and the nuclear red fluorescent protein red-stinger (21) driven by ok371-Gal4 are shown in the male brain and thoracic ganglion (upper panel), and at higher magnification in the central brain (middle) and thoracic ganglion (lower panel). F, dADAR-HA expression in Dachshund-positive Kenyon cells is clearly reduced relative to the surrounding nuclei. Scale bars, 20 μm.
We used 21 neuronal Gal4 lines to drive two independent insertions of syt-T (see supplemental Table 2 for details) and observed production of full-length transcripts with all Gal4 drivers. Sequence analysis confirmed that all full-length transcripts were the result of accurate splicing. In certain cases, a minor band corresponding to exon 9 skipping was observed. However, alterations in editing of the reporter did not correlate with alternative splicing of the edited exon (supplemental Fig. 2). Editing at site 4 was detected in all neurons defined by the library of Gal4 lines but varied widely from 27 to 82% (Fig. 2, B–D). In contrast, editing at site 3 was either undetectable or <10% in 16/21 driver lines tested, and it was only observed at robust levels (>20%) in the five subpopulations that yielded the highest editing at site 4, suggesting that although low dADAR levels are sufficient for robust editing of site 4, editing at site 3 only occurs once a certain threshold of dADAR expression has been exceeded.
To test whether dADAR expression correlated with editing of the syt-T reporter, we examined dADAR expression in two neuronal subtypes representing high and low levels of dADAR activity indicated by the syt-T reporter,: glutamatergic (ok371) and mushroom body neurons, respectively (see supplemental Table 2). Robust dADAR levels were detected in many glutamate-releasing neurons labeled with a nuclear red fluorescent protein (Fig. 2E) (21). In contrast, mushroom body cells labeled with the nuclear marker Dachshund showed strikingly lower dADAR expression levels relative to surrounding neurons (Fig. 2F). These data suggest that cell-autonomous regulation of dADAR expression contributes to neuron-to neuron variation in editing of the molecular reporter.
Stringent Reduction of dADAR Expression Reveals Differential Affinities of Edited Substrates
Initially, the two pre-Cre recombinant lines generated contain a white+ mini-gene inserted in an inverse orientation relative to dAdar transcription. Western blots revealed that prior to removal of this white+ mini-gene insertion, dADAR-HA staining was reduced by 80 ± 5% and 87 ± 7% relative to post-Cre counterparts (Fig. 1, C and D). Thus, insertion of white+ serendipitously generated independent hypomorphic alleles of dAdar (which we refer to as dAdarhyp; control flies harboring a single LoxP site in intron 7 of dAdar are referred to as dAdarWTLoxP).
The substantial reduction in dADAR expression also led to a tissue-specific decrease in auto-editing of the dAdar transcript, a developmentally regulated event that acts to down-regulate dADAR activity by re-coding a conserved serine residue to glycine near the active site (22). Amplification of the dAdar transcript revealed that auto-editing was slightly but significantly reduced in dAdarhyp heads relative to dAdarWTLoxP (WTLoxP, 53.8%; hyp, 49.1%, n = 8 RT-PCRs, p < 0.005, Mann-Whitney U test). In contrast, auto-editing in thorax tissue dropped from 38.1% in dAdarWTLoxP controls to 23.1% in dAdarhyp thoraxes (n = 5–8, p < 0.005). The decrease in ADAR protein production by the dAdarhyp allele results in comparable or only slightly reduced levels of the less active (edited) form of Drosophila ADAR protein. Thus, any system-wide changes in target deamination should be largely due to changes in ADAR levels rather than effects due to misregulated auto-editing.
Complete loss of A-to-I RNA editing in Drosophila results in multifaceted adult-stage behavioral abnormalities (12), consistent with the functional pleiotropy of dADAR. However, the extreme uncoordination exhibited by dAdar5g1 null flies has made investigations into the relation between A-to-I editing activity and complex behavior impossible. Surprisingly, despite the severe reduction in dADAR expression in dAdarhyp males, locomotor activity appeared relatively robust under casual observation (supplemental movie), and no obvious uncoordination was apparent. These observations suggested that editing might perdure in a behaviorally relevant subpopulation of adenosines in dAdar hypomorphs, despite the severe reduction in dADAR protein expression. We tested this hypothesis by comparing editing in 68 target adenosines in the hypomorph and control backgrounds. Like editing in the dAdarHA genetic background (supplemental Fig. 1), dAdarWTLoxP males did not significantly differ from wild-type Canton-S (data not shown). In dAdarhyp heads and thoraxes, however, editing was reduced by an average of 68 ± 4% and 56 ± 4%, respectively, relative to the post-Cre control (Fig. 3A). We observed a similar reduction in editing in dAdarHA males in which the white+ mini-gene was not removed (data not shown).
FIGURE 3.
Varied impacts on mRNA re-coding following reduction of dADAR expression. A, heat map representation of editing levels at 68 sites in dAdarWTLoxP heads and thoraxes, and the corresponding reduction of editing in the same tissue from dAdarhyp males. Each editing site is represented by a four-symbol code (see supplemental Table 3 for details). B and C, editing levels in female heads for 8 LE sites (B) and 10 HE sites (C). The homozygotic and heterozygotic backgrounds containing various dAdar alleles are noted below each graph. Each value is the mean of ≥3 RT-PCRs. Error bars, S.E. values.
Intriguingly, the reduction in specific editing of targets was highly nonuniform (Fig. 3A and supplemental Fig. 3, A and B). Editing at a substantial fraction of sites was reduced by 70–100% in hypomorph heads and thoraxes. In contrast, despite the extreme curtailing of wild-type dADAR levels, a subpopulation of adenosines was modified at nearly wild-type levels (Fig. 3A and supplemental Table 3). For example, in the male thorax, syt-1 site 4 and site 2 of the ard acetylcholine receptor (also known as Dβ1) are edited at similar levels (95 ± 1% and 94 ± 2%, respectively) (supplemental Table 3). However, in dAdarhyp thoraxes, editing at syt-1 site 4 was reduced by 12% relative to dAdarWTLoxP, while ard site 2 was reduced by 71%. A further striking example is site 4 of eag and the single site in Caα1T. Site 4 of eag is slightly reduced from 84 ± 0.6% to 79 ± 0.4% in dAdarhyp heads, while editing in Caα1T is completely abolished in dAdarhyp heads, despite its high level of editing in dAdarWTLoxP heads (87 ± 0.7%).
Based upon these data, we classified dADAR substrates into two groups according to their sensitivity to dADAR protein levels, which we term “high and low efficiency” (HE and LE) sites. We defined HE sites as those reduced by <30% in dAdarhyp thoraxes, while LE sites are reduced by >70%. Importantly, HE and LE sites also exhibited similar responses to dADAR reduction in male dAdarhyp head samples, although these were shifted toward slightly lower reductions in HE sites and greater reductions in LE sites (supplemental Fig. 3). Many dADAR targets are edited at higher levels in the thorax relative to head tissue (average increase, 16 ± 6% for all sites edited >10%; Fig. 3A), although notable exceptions such as shab site 5 and Caα1T were apparent. This trend may reflect increased dADAR activity in the thoracic ganglion relative to the head, perhaps due to lower levels of auto-editing in dAdarhyp thoraxes.
We took advantage of the dAdar5g1 null and hypomorphic dAdarhyp alleles to generate females with a graded range of dADAR expression, and we examined editing at 10 HE and 8 LE sites in head samples from four genetic backgrounds: hyp/hyp, 5g1/FM7 (where FM7 is a balancer chromosome with a wild-type copy of dAdar+), hyp/FM7, and WTLoxP/WTLoxP. These genotypes have predicted relative dADAR expression levels of ∼20, 50, 60, and 100%, respectively. As expected, all LE sites exhibited very low levels of editing in hyp/hyp heads (0–23%), in contrast to HE sites (30–84%) (Fig. 3, B and C). Every HE site tested showed wild-type levels of editing in 5g1/FM7 heads, and the mean reduction in hyp/hyp heads relative to wild-type controls was only 26 ± 4%. Thus, only a minimal concentration of dADAR is sufficient to yield robust editing of HE sites.
In contrast, the mean reduction in LE sites in hyp/hyp heads relative to wild-type controls was 80 ± 4%. However, within the LE sites we examined, we could delineate two subpopulations. Five of the eight sites tested (ard sites 1–3, DSC1 and Caα1T) showed wild-type editing levels in 5g1/FM7 heads, despite severely reduced editing in the dAdarhyp background. The remaining three sites (unc-13, Caα1D site 1, and site 6 of the Dα6 acetylcholine receptor) showed sequentially increased editing in proportion to higher dADAR levels and did not reach wild-type levels of editing in either 5g1/FM7 or hyp/FM7 heads. We also examined five HE and LE sites in hyp/hyp, 5g1/FM7, hyp/FM7, and WTLoxP/WTLoxP thoraxes, with similar results (supplemental Fig. 3, C and D). Finally, Western blotting indicated that there was no significant up-regulation of expression from a wild-type dAdar locus in dAdarHA/5g1 trans-heterozygote females (supplemental Fig. 3E), indicating that there does not appear to be a compensatory mechanism counteracting decreased ADAR production.
The unexpectedly complex relationship between dADAR concentration and editing levels appears to vary on a site-by-site basis. One possible explanation for the variation in the sensitivity of edited adenosines to dADAR concentration is a differential ratio of substrate mRNA to dADAR enzyme, i.e. LE sites are present in mRNAs with high steady-state expression levels, while the converse is true for HE sites. However, this explanation is not consistent with the close proximity of HE and LE sites within transcripts from a single gene, which was observed for several mRNAs, including Caα1D and Dα6 (Fig. 3, B and C), as well as shaker, shab, and eag (supplemental Table 3). For example, sites 5 and 6 of shaker are separated by just six nucleotides yet exhibit strikingly different reductions in editing in dAdarhyp heads, with site 5 reduced by 22% and site 6 by 76% (supplemental Table 3). In addition, proteins re-coded by dADAR that function in the same subcellular compartment also exhibited drastically divergent responses in dAdarhyp. Synaptotagmin-1 (syt-1) and unc-13 both act to promote vesicle release at pre-synaptic nerve terminals. Nonetheless, site 4 of syt-1 is robustly edited in dAdarhyp, while editing of unc-13 is almost abolished in both dAdarhyp heads and thoraxes (Fig. 3). Therefore, our data strongly suggest that the sensitivity of editing sites to changes in dADAR levels is a function of inherent primary sequence and/or structural properties specific to the double strand RNA structural intermediates required for ADAR-mediated deamination.
High and Low Efficiency Editing Sites Exhibit Distinct Patterns of Developmental Regulation
Editing at many, but not all, dADAR substrates is under strong temporal control, appearing predominantly at the pupal and adult stages of Drosophila development. Edited adenosines exhibiting differential developmental regulation may even be found within the same transcript (23, 24), yet how this is achieved remains unknown. Previous data has shown that dAdar transcription is low during the larval stages and rapidly peaks at the late pupal and adult stages (25). We hypothesized that adenosines showing high levels of editing throughout development represent HE sites and require low levels of dADAR expression for robust editing. Conversely, pupae/adult-specific sites would require higher concentrations of dADAR for efficient modification and populate the LE class.
We assessed whether temporal changes in dAdar transcription result in similar alterations in dADAR protein levels by comparing dADAR expression in the adult thoracic ganglion and the ventral nerve cord of 3rd instar larvae (L3) (Fig. 4A). Although strong dADAR expression was observed in neuronal nuclei in the adult thoracic ganglion, dADAR was largely undetectable by immunohistochemistry in the larval ventral nerve cord. Furthermore, Western blotting revealed robust bands corresponding to dADAR-HA isoforms in adult male tissue, which were undetectable in samples prepared from the 1st instar (L1) and L3 larval stages (Fig. 4B, inset). Thus, the increase in dAdar mRNA between the larval and adult stages is mirrored by a similar change in detectable dADAR protein expression.
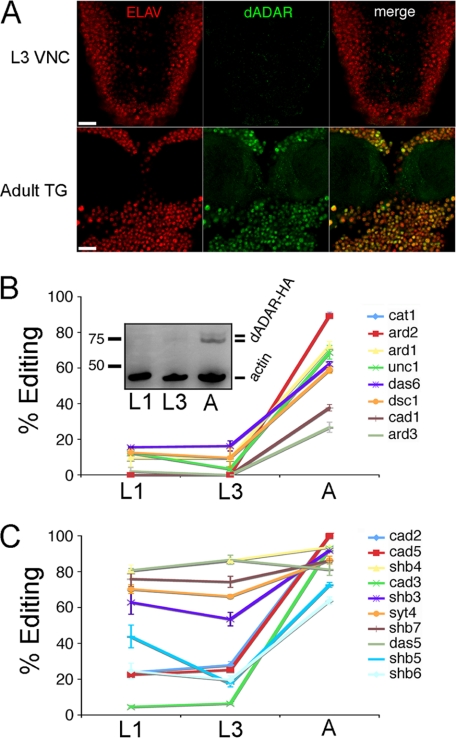
FIGURE 4.
Dynamic control of dADAR expression underlies developmental patterns of editing at low efficiency sites. A, dADAR and Elav expression in the L3 ventral nerve chord (VNC) and the adult thoracic ganglion (TG). Scale bar, 20 μm. B, LE sites are subject to strong developmental regulation. Editing at eight LE sites was examined at two larval stages (L1 and L3) and in adult males (A). Inset, representative example of n = 4 Western blots showing a strong increase in dADAR expression at the male adult-stage relative to L1 and L3. The two dADAR-HA bands likely represent dADAR proteins containing or lacking the alternatively spliced 3a exon, which is included at a higher level in the abdomen relative to the head and thorax (43). C, developmental profiles of 10 HE sites in L1, L3, and adult (A) males. Error bars, S.E. values.
We next investigated whether HE and LE sites show distinct patterns of developmental regulation, in keeping with the above hypothesis. We measured editing at the same HE and LE sites examined above (Fig. 3, B and C), amplified from Canton-S L1, L3, and adult-stage cDNAs. Editing sites representing HE and LE classes showed strikingly different developmental regulation (Fig. 4, B and C). As predicted, all LE sites tested showed clear increases in editing between the L3 and adult stages (Fig. 4B). Across the eight sites studied, editing levels at the L3 stage averaged only 8 ± 3% of the corresponding adult values. In contrast, HE sites were enriched for adenosines that were robustly edited in the early and late larval stages (Fig. 4C), with the mean values at L3 averaging 53 ± 10% of adult levels. Three of the four HE adenosines that did show developmental regulation mapped to the same transcript, encoding the Caα1D voltage-gated calcium channel. Thus, although deamination of particular transcripts may be developmentally modulated by factors distinct from dADAR itself, editing site-specific responses to dADAR protein levels explains a significant proportion of the temporal variation in editing and correlates well with our functional definition of HE and LE classes.
Reduction of dADAR Expression Affects Complex Behavior
The lack of severe uncoordination in dAdarhyp males allowed us to examine, for the first time, whether complex adult-stage behaviors are altered in a genetic background with an engineered alteration in editing levels.
Under light-dark (12:12 h) conditions, wild-type Drosophila exhibit diurnal peaks of activity centered on the lights-on (dawn) and lights-off (dusk) transitions. Importantly, spikes in activity are preceded by anticipatory increases in locomotion that are driven by an endogenous circadian clock (26). We examined rhythmic locomotor patterns using automated, single-fly activity monitors. dAdarWTLoxP males displayed peaks of activity at subjective morning and evening, as well as anticipation of both dark-light and light-dark transitions (Fig. 5A and supplemental Fig. 4). Under constant dark conditions, dAdarWTLoxP males displayed anticipation of subjective morning and night (data not shown), illustrating that the circadian clock remains intact in our control genotype. In dAdarhyp males, peaks of morning and evening activity were present but reduced in amplitude relative to dAdarWTLoxP (Fig. 5B), and anticipation of morning, but not night, was completely abolished. Importantly, this pattern of locomotor activity was distinct from dAdar5g1 males, which lack coordinated locomotor patterns (Fig. 5C and supplemental Fig. 4) (12). We quantified the degree of morning anticipation in the above three genotypes (defined as the number of beam breaks in the 3 h before lights-on normalized to the 6 h before lights-on). dAdarWTLoxP males exhibited a 60 and 45% increase respectively in the degree of morning anticipation relative to dAdarhyp and dAdar5g1 males, respectively (Fig. 5D). Thus, although limited expression of dADAR (∼20%) is sufficient to restore a degree of locomotor coordination and activity, including startle responses to changes in light stimuli, more robust dADAR expression is required for the manifestation of circadian anticipation of morning, a complex behavior. Analysis of total locomotor activity revealed that locomotion in dAdarhyp males and females was reduced by 52–58% relative to control genotypes (Fig. 5E). Interestingly, hetero-allelic females revealed that a very modest increase of dADAR expression (from ∼50% in 5g1/FM7 to ∼ 60% in hyp/FM7 females) was sufficient to completely restore wild-type locomotor levels (Fig. 5E).
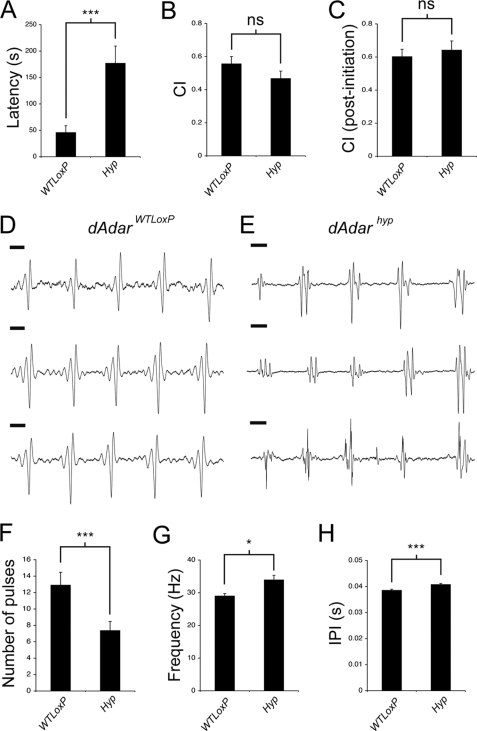
FIGURE 5.
Global reduction in dADAR activity leads to altered patterns of locomotor activity. A–C, mean activity profile of dAdarWTLoxP (A, n = 30), dAdarhyp (B, n = 30), and dAdar5g1 males (C, n = 16) under 12-h light-dark cycles (white and black bars). Each bar is an average value of time points from three consecutive days. In dAdarWTLoxP, but not dAdarhyp or dAdar5g1 males, anticipation of morning and evening can be observed under both environmental conditions (arrowheads). D, quantification of morning anticipation in the three experimental genotypes. E, mean locomotor activity in male and female dAdar allelic backgrounds. Error bars, S.E. values. **, p < 0.005; ***, p < 0.0005; not significant (ns): p > 0.05 (Mann-Whitney U test).
Temperature-sensitive paralysis and circling behavior, other hallmarks of dAdar5g1 males (12), were not observed in dAdarhyp males, although sporadic seizures were clearly apparent (see supplemental movie). Most surprisingly, hypomorphic males exhibited robust courtship toward wild-type females, a behavioral pattern completely absent in dAdar5g1 males (12). Because the relationship between RNA editing and mating behavior is unclear, we compared several courtship parameters in dAdarWTLoxP and dAdarhyp males. Although males from both genotypes court wild-type females, dAdarhyp males exhibited an ∼4-fold increase in the time taken to initiate courtship (latency) relative to dAdarWTLoxP males (p = 0.00025, Mann-Whitney U test; Fig. 6A). Despite this, the overall length of time spent courting did not significantly differ between either genotype (p = 0.33; Fig. 6, B and C).
FIGURE 6.
RNA editing is required for appropriate male courtship. A, time taken to initiate courtship (latency) is significantly higher in dAdarhyp males (n = 20) relative to dAdarWTLoxP controls (n = 15), yet the total time spent courting virgin females over a 10-min period (courtship index, CI) is not significantly different between either genotype (B and C). Courtship index was either calculated over the whole 10 min (B) or following initiation of courtship (C). Examples of three separate song trains are shown from a single dAdarWTLoxP (D) or dAdarhyp male (E). Note that although the trains from the dAdarWTLoxP male are highly stereotyped, trains from even a single dAdarhyp male show striking variability in waveform pattern. Scale bar, 10 ms. F–H, song parameters in dAdarWTLoxP (n = 26 songs, 5 males) and dAdarhyp (n = 44 songs, 9 males). Error bars, S.E. values. *, p < 0.05; ***, p < 0.0005; not significant (ns): p > 0.05 (Mann-Whitney U test).
During mating, males produce a species-specific “love song” via unilateral wing vibration, which is proposed to both facilitate female acceptance and to act as an indicator of correct species identity during courtship. Mutations in several loci that also undergo RNA editing have been shown to alter the song waveform (27). This suggested the possibility that RNA editing in neuronal mRNAs might modulate song properties. To test this, we recorded the pulse songs of dAdarWTLoxP and dAdarhyp males.
Courting dAdarWTLoxP males generated robust pulse songs with highly stereotyped waveforms similar to previously published examples from wild-type Drosophila (n = 26, Fig. 6D) (27, 28). In contrast, pulse songs from dAdarhyp males often exhibited abnormal waveforms characterized by polycyclic pulses and additional peaks (Fig. 6E). Of the 44 songs analyzed from dAdarhyp males, only 7 were similar to the dAdarWTLoxP pulse pattern. The change in waveform was accompanied by alterations in several other song parameters, including a reduced number of pulses per song train, an increased pulse frequency, and a small but highly significant increase in the inter-pulse interval (dAdarWTLoxP, 38.6 ms ± 0.4, n = 312; dAdarhyp, 40.8 ms ± 0.4, n = 281; p < 0.0001, Mann-Whitney U test) (Fig. 6, F–H). In addition, we observed striking variability in the dAdarhyp pulse waveforms, even between distinct song trains from the same male (Fig. 6E). The coefficient of variation (defined as the S.D. divided by the mean) of the pulse frequency increased from 0.121 in dAdarWTLoxP to 0.265 in dAdarhyp, but it was similar when comparing the inter-pulse intervals of the two genotypes (dAdarWTLoxP, 0.175; dAdarhyp, 0.155). Thus, in addition to influencing multiple song parameters, robust editing also appears to be required for maintaining aspects of male song pulse stereotypy.
Inhibition of RNA Editing in a Small Subset of Neurons Is Sufficient to Alter Complex Behavior
In Drosophila, the male-specific isoform of the transcription factor Fruitless (FruM) is a key mediator of male-specific behaviors, and the output of fruitless (fru) neurons is known to be essential for correct courtship behavior and generation of the mating song (29–31). Because both of these behavioral parameters were altered in dAdarhyp males, we examined the pattern and function of A-to-I editing in this behaviorally important subset of neurons.
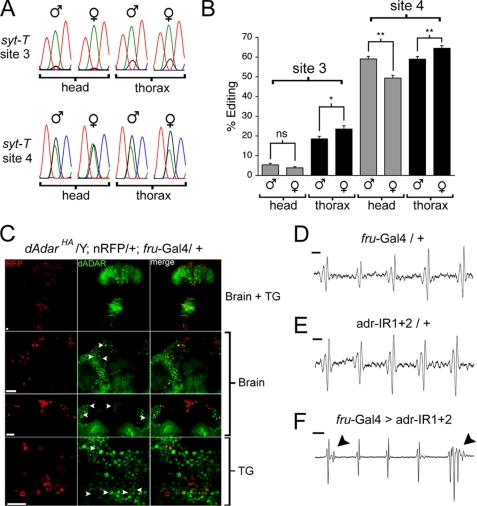
fru neurons are present in both the male and female central brain and thoracic ganglion, composing ∼2% of the total neuronal population. Although the distribution and projection patterns of fru neurons are broadly similar between male and female Drosophila (30–32), subpopulations of fru neurons have been shown to exhibit sexual dimorphism in both number and wiring (33–35). We initially tested whether editing activity in fru neurons also showed sexual dimorphism by driving the two independent insertions of the syt-T reporter (Fig. 2) using fru-Gal4 and analyzing editing at syt-T sites 3 and 4 following RT-PCR amplification from male and female head and thorax cDNA. Interestingly, editing at site 4, which is more robustly edited than site 3, indeed showed subtle but significant sexual dimorphism. Site 4 exhibited a relative increase of ∼20% in male versus female head cDNA (p = 0.004, Mann-Whitney U test). This trend was reversed in thorax cDNA, where site 4 editing in fru neurons was reduced by ∼10% in males relative to females (p = 0.03; Fig. 7, A and B,). Editing at site 3 showed a similar trend, albeit at lower levels (Fig. 7B). Furthermore, site 4 editing was statistically unchanged between fru neurons in male heads and thoraxes (p = 0.94) but increased by 30.5% between female head and thorax samples (p = 0.003; Fig. 7B). No sex-specific alternative splicing of the syt-T reporter was observed in either head or thorax tissues (supplemental Fig. 5).
FIGURE 7.
Knockdown of dADAR in fruitless-expressing neurons alters the male courtship song. A, example of electropherograms showing editing of syt-T site 3 and 4 expressed in fruitless-positive (fru) neurons within the male and female head or thorax. B, quantification of editing of two independent insertions of syt-T (n = 6–7 RT-PCRs for each value). C, dADAR expression was examined specifically in fru neurons by expressing a nuclear red fluorescent protein (23) using the fru-Gal4 driver line, in a dAdarHA background. Nuclei of fru neurons can be detected throughout the brain and thoracic ganglion (upper panel). Examples of dADAR expression in fru neurons in the dorsal anterior segment and pars intercerebralis (middle panels) and meso-thoracic ganglion (lower panel) are shown at higher magnification below. D and E, example of song trains from control males heterozygous for driver (w+; +; fru-Gal4/+, n = 26 song trains, 10 males) or RNAi transgenes (w+; adr-IR1/+; adr-IR2/+, n = 30 song trains, 10 males). Note the similarity in waveform between song trains shown in D and E compared with those from dAdarWTLoxP males (Fig. 6D). F, example of song trains from males with reduced dADAR expression in fru neurons (w+; adr-IR1/+; fru-Gal4/adr-IR2) (n = 27 song trains, 11 males). Note the extra spike in the first pulse and the polycyclic waveform in the last pulse. Scale bar, 10 ms. Error bars, S.E. values. *, p < 0.05; **, p < 0.005; not significant (ns): p > 0.05 (Mann-Whitney U test).
Because dAdar is X-linked, our results could potentially reflect sex-specific differences in dADAR expression throughout the nervous system. Thus, we examined editing of the endogenous syt-1 transcript in male and female whole head and thorax cDNA and found no significant sexual dimorphism at either site (supplemental Fig. 6). We next measured editing at a further five LE and eight HE sites (Fig. 3) in the same tissues. In this combined data set of 15 editing sites, we found a small but significant reduction in overall editing in female relative to male heads (mean reduction, 9%, p = 0.0013, paired t test). However, in contrast to editing of the syt-T reporter, there was no significant alteration in editing of endogenous mRNAs when comparing male and female thoraxes (p = 0.198) nor a significant difference in editing of the 15 sites between female head and thorax samples (p = 0.68) (supplemental Fig. 6). Thus, the female tissue-specific differences in editing of syt-T cannot be explained in terms of a global alteration in editing activity. Collectively, these data suggest that dADAR activity is differentially controlled in male and female fru neurons.
The existence of sexually dimorphic editing activity suggested a functional role in dADAR activity in fru neurons. Robust dADAR expression was detected in many fru neurons in both the male brain and the thoracic ganglion (Fig. 7C). Importantly, dADAR is expressed in fru neurons in the mesothoracic segment of the ventral nerve cord, which are thought to be a key component of the song pattern generator (Fig. 7C) (36, 37). We made use of a previously validated double-RNAi line (adr-IR1 + 2) directed against the 3′ region of the dAdar transcript and under the control of the upstream activation sequence promoter (14) to selectively reduce dADAR expression in fru neurons. Knockdown of dADAR solely in fru neurons did not significantly alter male locomotor activity, latency to court, or total time spent courting (supplemental Fig. 7). Male-male courting, a hallmark of fruitless mutants, was not observed in fru-Gal4 > adr-IR1 + 2 males (data not shown). This, as well as the robust courtship of females, indicates that the development and wiring of fru neurons are unlikely to be adversely affected by dADAR knockdown.
We next examined the mating song in the experimental and both control genotypes. Song waveforms from control males containing driver or transgenes alone were indistinguishable from dAdarWTLoxP (Fig. 7, D and E). In contrast, 12/27 song trains from males with dADAR expression inhibited in fru neurons exhibited polycyclic waveforms and/or additional peaks that were not observed in either genetic control (Fig. 7F), as was also observed in dAdarhyp males (albeit in a higher proportion of songs). This was accompanied by an increase in the average number of pulses per song train (fru-Gal4 > adr-IR1 + 2, 12.9 ± 1.7; fru-Gal4/+, 6.6 ± 1; adr-IR1 + 2/+, 8 ± 1.3; p < 0.005, Mann-Whitney U test) but no significant alteration in either pulse frequency or inter-pulse interval relative to both control genotypes. Thus, knockdown of dADAR in fru neurons can partially phenocopy a discrete subset of the multifaceted alterations in courtship behavior observed in dAdarhyp males, namely the generation of mating songs with abnormal, often polycyclic, waveforms.
DISCUSSION
Using a novel hypomorphic allele of dAdar generated through homologous recombination coupled with cell-specific dADAR knockdown, we have demonstrated that RNA editing serves a modulatory role in multiple adaptive behaviors in Drosophila. In short, we provide linkage between the loss of conserved and taxa-specific amino acid re-coding sites and alterations in wild-type ethological outputs that directly impinge on organismal fitness. Importantly, the behavioral defects observed in dAdarhyp males correlate with the severe loss of a particular subset of edited adenosines, namely those that are preferentially edited at the adult stage (Fig. 4B).
Our molecular analysis of dAdar hypomorphs revealed a striking diversity in the response of edited adenosines to changes in endogenous dADAR levels (Fig. 3). Both the local sequence surrounding edited adenosines and their predicted secondary structures vary widely between dADAR substrates, providing a potential mechanism to generate differential affinities for dADAR binding and deamination (3, 11, 37). This finding has important implications as follows. First, it provides a explanatory basis for the developmental regulation of a select population of editing sites (Fig. 4), a phenomenon common to both Drosophila and mammals (23, 24, 38, 39). Second, cell-specific variation in dADAR expression (Fig. 2) may allow spatial control of LE sites while simultaneously maintaining robust network-wide editing of HE sites, thus providing a means to fine-tune neuronal physiology through the diversification of a constrained population of proteins (see Fig. 8, for model).
FIGURE 8.
Model for neuron to neuron variation in editing levels within the Drosophila nervous system. Top panel shows a graphical representation of the change in editing of one HE site (shab site 4; shb4) and two LE sites (ard site 2; ard2, and unc-13; unc1). Shab site 4 is edited at almost wild-type levels even in genotypes with very low dADAR expression, as is the case for all HE sites (Fig. 3). Thus, editing at this, and similar sites, is unlikely to vary widely from neuron to neuron, even though dADAR activity is highly variable in different neuronal populations (Fig. 2). In contrast, editing at LE sites is likely to vary substantially in neurons with differing levels of dADAR expression. Certain LE sites only required 50% of wild-type dADAR expression for achieving wild-type editing levels, while others required more robust dADAR expression (Fig. 3). The bottom panel shows a diagrammatic representation of three distinct neuronal subtypes (derived from Fig. 2), with low, medium (med), and high relative expression of dADAR. In neurons with low dADAR activity (such as mushroom body neurons), only HE sites such as shab site 4 are likely to be strongly edited. At slightly higher levels (for example, fru neurons), both shab site 4 and ard site 2 (i.e. the “higher efficiency” LE sites) will show editing but not weak LE sites such as unc-13. Finally, in neurons with high dADAR expression (such as photo-receptors; supplemental Table 2), all subclasses may be open to robust editing.
We have previously shown that pan-neuronal expression of the two hairpin RNAi constructs used in this study reduces locomotor activity by ∼90% (14), and this effect could not be phenocopied by dADAR knockdown in any particular neuronal subset tested. Furthermore, dADAR knockdown under these conditions was robust enough to strongly reduce editing even at HE sites such as syt-1 site 4. Although knockdown is subject to the level of hairpin expression and efficiency of RNAi in particular neurons, the abrogation of ADAR expression by transgenic knockdown was clearly effective. In contrast, editing at HE sites is still maintained in our purely genetic model using dAdarhyp mutant males and females (Fig. 3), as is coordinated locomotion (albeit at lower levels; Fig. 5). Indeed, even dAdarhyp/dAdar5g1 trans-heterozygote females, predicted to express dADAR at ∼ 10% of wild-type levels, do not appear uncoordinated.4 Collectively, these data imply that network-wide editing of HE sites is sufficient to provide motor tone and prevent the extreme uncoordination observed in dAdar null flies.
Conversely, it is tempting to speculate that developmentally regulated LE sites modulate adult-specific behaviors. Indeed, we examined two ethologically relevant behaviors in dAdarhyp males, which show a severe disruption of developmentally regulated editing (Fig. 4), and we found both to be defective (Figs. 5 and 6). dAdarhyp males did not show the circadian anticipation of lights-on seen in wild-type Drosophila (Fig. 5), and multiple aspects of courtship behavior were abnormal in dAdarhyp males, including the time required to initiate courtship and the waveform of the mating song (Fig. 6). It should be stressed that defects in both of the above parameters are likely to be severely detrimental to reproductive fitness under competitive conditions in the wild.
Although our data lead us to hypothesize that loss of adult-stage LE sites may underlie the locomotor and courtship defects exhibited by dAdar hypomorphs, we cannot currently link the loss of particular editing sites to the behavioral defects seen in dAdarhyp males due to the large number of characterized dADAR substrates. Over 100 editing sites in 24 mRNAs have been identified either serendipitously or through comparative genomics approaches (7), although a recent bioinformatic screen identified a further potential 27 mRNAs subject to re-coding (40). The existence of functionally epistastic interactions between editing sites also makes it unlikely that any particular phenotype observed in dAdarhyp males can be fully mapped to the loss of a single editing site (23, 24). Rather, the relationship between re-coding and behavior can instead be viewed through the prism of the pleiotropic actions of dADAR on a wide range of RNA substrates, with many edited proteins simultaneously contributing to the total phenotype of interest.
We mapped the cellular foci for behavioral abnormalities associated with stringent loss of dADAR expression using transgenic RNAi (41). Knockdown of dADAR specifically in fruitless-expressing neurons partially recapitulated the polycyclic songs observed in dAdarhyp males (Fig. 7) but did not phenocopy alterations in other song properties or mating behavior, suggesting that these highly specific phenotypic components are influenced by editing in other fru-negative neurons and/or muscle tissue. Surprisingly, targeted expression of a molecular reporter for editing activity suggests that male and female fru neurons within both the brain and thoracic ganglion may differ in terms of dADAR activity (Fig. 7). Because only small subpopulations of fru neurons exhibit morphological sexual dimorphism, it has been hypothesized that expression of the male-specific isoform of Fruitless (FruM) may modify the physiological properties of fru neurons (29–32). Given the large number of transcripts re-coded by A-to-I editing (7), an alteration of dADAR expression or activity by FruM could hypothetically provide a means of enabling functional modulation of a wide range of ion channels and synaptic release proteins. Further experiments will be required to test whether the alterations in editing observed between male and female fru neurons represent large differences in a subset of fru neurons, subtle alterations across the fru neuron network, or are due to numerical sexual dimorphism in the fru neuron population.
That RNA editing can modulate song properties is particularly intriguing, because editing sites are not static throughout insect evolution (3, 6, 42). Indeed, even within the Drosophila lineage, we have observed species-specific changes in the magnitude of editing at orthologous adenosines in several ion channels (7, 38). Therefore, our data open the possibility that alterations in RNA editing may contribute to species-specific song waveforms, a key mechanism implicated in the reproductive isolation between Drosophilids. More broadly, our data suggest that, in principle, evolutionary divergences in RNA editing may contribute to the generation of adult-stage species-specific behavioral patterns.
Supplementary Material
Acknowledgments
We thank Cynthia Staber for expert technical assistance, members of the Reenan laboratory for comments on the manuscript, S. Baserga for providing the mouse anti-HA antibody, and M. McKeown, B. Dickson, and B. Ganetzky for Drosophila stocks.
This work was supported by an Ellison Medical Foundation Senior Scholar award.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–7, Tables 1–3, and a movie.
J. E. C. Jepson, personal observation.
- ADAR
- adenosine deaminase acting on RNA
- A-to-I
- adenosine-to-inosine
- HE
- high efficiency
- LE
- low efficiency
- L1
- 1st instar larval
- L3
- 3rd instar larval.
REFERENCES
- 1. Nishikura K. (2010) Annu. Rev. Biochem. 79, 321–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Higuchi M., Single F. N., Köhler M., Sommer B., Sprengel R., Seeburg P. H. (1993) Cell 75, 1361–1370 [DOI] [PubMed] [Google Scholar]
- 3. Reenan R. A. (2005) Nature 434, 409–413 [DOI] [PubMed] [Google Scholar]
- 4. Basillo C., Wahba A. J., Lengyel P., Speyer J. F., Ochoa S. (1962) Proc. Natl. Acad. Sci. 48, 613–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smith L. A., Peixoto A. A., Hall J. C. (1998) J. Neurogenet. 12, 227–240 [DOI] [PubMed] [Google Scholar]
- 6. Grauso M., Reenan R. A., Culetto E., Sattelle D. B. (2002) Genetics 160, 1519–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hoopengardner B., Bhalla T., Staber C., Reenan R. (2003) Science 301, 832–836 [DOI] [PubMed] [Google Scholar]
- 8. Semenov E. P., Pak W. L. (1999) J. Neurochem. 72, 66–72 [DOI] [PubMed] [Google Scholar]
- 9. Burns C. M., Chu H., Rueter S. M., Hutchinson L. K., Canton H., Sanders-Bush E., Emeson R. B. (1997) Nature 387, 303–308 [DOI] [PubMed] [Google Scholar]
- 10. Ohlson J., Pedersen J. S., Haussler D., Ohman M. (2007) RNA 13, 698–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bhalla T., Rosenthal J. J., Holmgren M., Reenan R. (2004) Nat. Struct. Mol. Biol. 11, 950–956 [DOI] [PubMed] [Google Scholar]
- 12. Palladino M. J., Keegan L. P., O'Connell M. A., Reenan R. A. (2000) Cell 102, 437–449 [DOI] [PubMed] [Google Scholar]
- 13. Higuchi M., Maas S., Single F. N., Hartner J., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., Seeburg P. H. (2000) Nature 406, 78–81 [DOI] [PubMed] [Google Scholar]
- 14. Jepson J. E., Reenan R. A. (2009) J. Biol. Chem. 284, 31391–31400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maggert K. A., Gong W. J., Golic K. G. (2008) Methods Mol. Biol. 420, 155–174 [DOI] [PubMed] [Google Scholar]
- 16. Wu J. S., Luo L. (2006) Nat. Protocols 1, 2110–2115 [DOI] [PubMed] [Google Scholar]
- 17. Rong Y. S., Titen S. W., Xie H. B., Golic M. M., Bastiani M., Bandyopadhyay P., Olivera B. M., Brodsky M., Rubin G. M., Golic K. G. (2002) Genes Dev. 16, 1568–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rong Y. S., Golic K. G. (2000) Science 288, 2013–2018 [DOI] [PubMed] [Google Scholar]
- 19. Jacobs M. M., Fogg R. L., Emeson R. B., Stanwood G. D. (2009) Dev. Neurosci. 31, 223–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brand A. H., Perrimon N. (1993) Development 118, 401–415 [DOI] [PubMed] [Google Scholar]
- 21. Barolo S., Castro B., Posakony J. W. (2004) BioTechniques 36, 436–442 [DOI] [PubMed] [Google Scholar]
- 22. Keegan L. P., Brindle J., Gallo A., Leroy A., Reenan R. A., O'Connell M. A. (2005) EMBO J. 24, 2183–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones A. K., Buckingham S. D., Papadaki M., Yokota M., Sattelle B. M., Matsuda K., Sattelle D. B. (2009) J. Neurosci. 29, 4287–4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ingleby L., Maloney R., Jepson J., Horn R., Reenan R. (2009) J. Gen. Physiol. 133, 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Palladino M. J., Keegan L. P., O'Connell M. A., Reenan R. A. (2000) RNA 6, 1004–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Allada R., Chung B. Y. (2010) Annu. Rev. Physiol. 72, 605–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peixoto A. A., Hall J. C. (1998) Genetics 148, 827–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zehring W. A., Wheeler D. A., Reddy P., Konopka R. J., Kyriacou C. P., Rosbash M., Hall J. C. (1984) Cell 39, 369–376 [DOI] [PubMed] [Google Scholar]
- 29. Clyne J. D., Miesenböck G. (2008) Cell 133, 354–363 [DOI] [PubMed] [Google Scholar]
- 30. Stockinger P., Kvitsiani D., Rotkopf S., Tirián L., Dickson B. J. (2005) Cell 121, 795–807 [DOI] [PubMed] [Google Scholar]
- 31. Manoli D. S., Foss M., Villella A., Taylor B. J., Hall J. C., Baker B. S. (2005) Nature 436, 395–400 [DOI] [PubMed] [Google Scholar]
- 32. Demir E., Dickson B. J. (2005) Cell 121, 785–794 [DOI] [PubMed] [Google Scholar]
- 33. Kimura K., Hachiya T., Koganezawa M., Tazawa T., Yamamoto D. (2008) Neuron 59, 759–769 [DOI] [PubMed] [Google Scholar]
- 34. Kimura K., Ote M., Tazawa T., Yamamoto D. (2005) Nature 438, 229–233 [DOI] [PubMed] [Google Scholar]
- 35. Datta S. R., Vasconcelos M. L., Ruta V., Luo S., Wong A., Demir E., Flores J., Balonze K., Dickson B. J., Axel R. (2008) Nature 452, 473–477 [DOI] [PubMed] [Google Scholar]
- 36. von Schilcher F., Hall J. C. (1979) J. Comp. Physiol. A 129, 85–95 [Google Scholar]
- 37. Hall J. C. (1979) Genetics 92, 437–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hanrahan C. J., Palladino M. J., Ganetzky B., Reenan R. A. (2000) Genetics 155, 1149–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rula E. Y., Lagrange A. H., Jacobs M. M., Hu N., Macdonald R. L., Emeson R. B. (2008) J. Neurosci. 28, 6196–6201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stapleton M., Carlson J. W., Celniker S. E. (2006) RNA 12, 1922–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., Couto A., Marra V., Keleman K., Dickson B. J. (2007) Nature 448, 151–156 [DOI] [PubMed] [Google Scholar]
- 42. Jin Y., Tian N., Cao J., Liang J., Yang Z., Lv J. (2007) BMC Evol. Biol. 7, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marcucci R., Romano M., Feiguin F., O'Connell M. A., Baralle F. E. (2009) Nucleic Acids Res. 37, 1663–1671 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.