Abstract
For many cell types, including pancreatic β-cells, nitric oxide is a mediator of cell death; paradoxically, nitric oxide can also activate pathways that promote the repair of cellular damage. In this report, a role for FoxO1-dependent transcriptional activation and its regulation by SIRT1 in determining the cellular response to nitric oxide is provided. In response to nitric oxide, FoxO1 translocates from the cytoplasm to the nucleus and stimulates the expression of the DNA repair gene GADD45α, resulting in FoxO1-dependent DNA repair. FoxO1-dependent gene expression appears to be regulated by the NAD+-dependent deacetylase SIRT1. In response to SIRT1 inhibitors, the FoxO1-dependent protective actions of nitric oxide (GADD45α expression and DNA repair) are attenuated, and FoxO1 activates a proapoptotic program that includes PUMA (p53-up-regulated mediator of apoptosis) mRNA accumulation and caspase-3 cleavage. These findings support primary roles for FoxO1 and SIRT1 in regulating the cellular responses of β-cells to nitric oxide.
Keywords: Apoptosis, Cell Death, Cytokine, Insulin, Nitric oxide, β-Cell
Introduction
Nitric oxide plays a central role in regulating the response(s) of pancreatic β-cells to cytokine treatment. Cytokines such as IL-1 (rat) and a combination of IL-1 + IFN-γ (mouse and human) stimulate the expression of the inducible isoform of nitric-oxide synthase (NOS) and the production of micromolar levels of nitric oxide by β-cells (1–4). Nitric oxide attenuates insulin secretion by inhibiting the oxidation of glucose to CO2 and the activity of mitochondrial iron-sulfur center containing enzymes such as aconitase and complexes of the electron transport system (5, 6). The result is a 4-fold reduction in cellular ATP concentration (2, 7) that leads to the inhibition of glucose-induced insulin secretion due to the inability to generate sufficient levels of ATP to close the ATP-sensitive K+ channels, an event required for β-cell depolarization and Ca2+-dependent exocytosis (8, 9). In addition to the inhibition of β-cell function, nitric oxide induces DNA strand breaks and oxidative DNA damage (10, 11).
The inhibitory actions of IL-1 on β-cell function and DNA damage are reversible (12, 13). The addition of a NOS inhibitor to islets pretreated for 24 h with IL-1 (without removing IL-1) results in a time-dependent recovery of insulin secretion and mitochondrial function (3) and the repair of damaged DNA (14, 15). This recovery response requires new gene expression, the activation of JNK, and can be stimulated by nitric oxide (16, 17). The ability of β-cells to recover from cytokine- and nitric oxide-induced damage is temporally limited. Following a 36-h exposure to IL-1, β-cells are no longer capable of recovering metabolic and secretory function, and the islets are committed to death (14, 18). Studies have shown that cytokines can kill β-cells by nitric oxide-dependent and -independent necrotic and apoptotic mechanisms (19–26). This dichotomy in the type of cell death that has been observed appears to reflect the temporal changes in the metabolic responses and the extent of DNA damage caused by nitric oxide. Following short exposures to cytokines, under conditions in which nitric oxide-mediated damage is reversible, biochemical assays indicate that cell death is necrotic in nature. At points in which β-cells no longer have the capacity to recover from this damage, cytokine-mediated β-cell death shifts to an apoptotic process that is associated with irreversible DNA damage and caspase activation (14). Similar to what has been observed in β-cells, nitric oxide has been implicated in the induction of both necrosis and apoptosis of multiple cell types (27–29).
Although nitric oxide is a known activator of p53 (30, 31), we recently observed that the repair of nitric oxide-damaged DNA in β-cells occurs by a p53-independent but GADD45α-dependent process (32). A number of studies have shown that GADD45α expression is regulated by p53 (33). Our findings suggest that there is an alternative pathway because p53 knockdown does not modify GADD45α expression or GADD45α-dependent DNA repair in β-cells. Members of the Forkhead family of transcription factors are known to regulate genes involved in cell cycle, stress resistance, DNA repair, and apoptosis (34, 35). FoxO1 is one member of this family whose activity is regulated by various external stimuli, including insulin, growth factors, and oxidative stress (36, 37). These stimuli can regulate FoxO1 activity by modifying its subcellular localization (cytoplasmic versus nuclear) and posttranslational modifications (37, 38). In response to insulin signaling, Akt phosphorylates FoxO1, resulting in its binding to 14-3-3 and sequestration in the cytoplasm where it is inactive (37). In contrast, stresses such as nutrient deprivation attenuate Akt-mediated phosphorylation and result in the nuclear translocation of FoxO1 (37). Once in the nucleus, the activity of FoxO1 is controlled by acetylation via the histone acetyltransferase p300/CBP and deacetylation by the NAD+-dependent deacetylase SIRT1 (39–41). Nuclear deacetylated FoxO1 promotes the transcription of genes involved in DNA repair and stress resistance, whereas acetylated FoxO1 promotes the expression of genes involved in apoptosis (39, 40). Recently, evidence has suggested that acetylation may also direct nuclear localization of FoxO1 independent of phosphorylation (38).
In this study we provide evidence that FoxO1 and its regulation by SIRT1 play primary roles in regulating the cellular responses to nitric oxide. Nitric oxide, produced in response to IL-1, or supplied exogenously, stimulates the nuclear localization of FoxO1 and the FoxO1-dependent expression of GADD45α and repair of nitric oxide-damaged DNA in β-cells. This protective response is controlled by the activity of SIRT1. Activators of SIRT1 accelerate, whereas inhibitors attenuate, the repair of nitric oxide-damaged DNA. Further, inhibition of SIRT1 is associated with the induction of apoptosis that is characterized by enhanced expression of the proapoptotic gene p53-up-regulated mediator of apoptosis (PUMA)2 and caspase activation. These findings suggest that the fate of β-cells in response to nitric oxide is controlled by the cellular localization of FoxO1 and activity of SIRT1 to either promote a protective response or induce β-cell death by apoptosis.
EXPERIMENTAL PROCEDURES
Materials
Male Sprague-Dawley rats (250–300 g) were purchased from Harlan (Indianapolis, IN). INS 832/13 cells were a gift from Chris Newgard (Duke University, NC). RPMI 1640 medium, CMRL-1066 tissue culture medium, l-glutamine, streptomycin, and penicillin were from Invitrogen. Fetal calf serum was from Sigma. Human recombinant IL-1 was purchased from PeproTech (Rocky Hill, NJ). l-NG-Monomethyl arginine (NMMA) and (Z)-1-(N,N-diethylamino)diazen-1-ium-1,2-diolate (DEANO; half-life ∼2 min) were purchased from Axxora (San Diego, CA). Splitomicin and EX-527 were obtained from Sigma. FoxO1, phospho-FoxO1, Ac-histone H3, Ac-lysine, cleaved caspase-3, cleaved caspase-9 antibodies were from Cell Signaling (Beverly, MA). GAPDH antibody was from Ambion (Austin, TX). HRP-conjugated donkey anti-rabbit and donkey anti-mouse were from Jackson Immunoresearch Laboratories, Inc (West Grove, PA). GADD45α, hsp70, PUMA, and GAPDH primers were from Integrated DNA Technologies (Coralville, IA). FoxO1 adenoviral constructs were a gift from Domenico Accili.
Islet Isolation and Cell Lines
Islets were isolated from male Sprague-Dawley rats (250–300 g) by collagenase digestion as described previously (42). Islets were cultured overnight in CMRL-1066 (containing 2 mm l-glutamine, 10% heat-inactivated fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin) at 37 °C under an atmosphere of 95% air and 5% CO2 prior to experimentation. INS 832/13 cells were removed from growth flasks by treatment with 0.05% trypsin and 0.02% EDTA for 5 min. at 37 °C, washed twice with RPMI, and plated at 200,000 cells/400 μl, unless otherwise noted. The INS 832/13 cells used in this study were glucose responsive for insulin secretion (data not shown).
Western Blot Analysis
Cells were lysed in Laemmli buffer and proteins separated by SDS-PAGE and transferred to nitrocellulose membrane under semidry transfer conditions as described previously (43). Blots were blocked with 5% milk in TBST for 1 h and then incubated overnight at 4 °C with the following dilutions for primary antibodies GADD45α (1:500), Ac-histone H3 (1:1,000), FoxO1 (1:1,000), cleaved caspase-3 (1:1,000), cleaved caspase-9 (1:1,000), or GAPDH (1:50,000) antibodies. Membranes were incubated for 1 h with HRP-conjugated donkey anti-rabbit or anti-mouse (1:7,000, 1:5,000, respectively) secondary antibodies, and antigen was detected by luminol chemiluminescence (44).
Adenoviral Transduction
INS 832/13 cells were transduced with WT-FoxO1 or Δ256-FoxO1 adenovirus constructs at 50 plaque-forming units/cell for 1 h as described previously (45). Following transduction, cells were washed three times, and experiments were started 24 h after transduction.
FoxO1 Translocation
INS 832/13 cells were seeded in chamber slides (Nunc/Thermo Fisher) and incubated for 24 h. Cells were then transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol for a surface area of 0.3 cm2. Briefly, cells were transfected with 200 ng of GFP-FoxO1 (Addgene plasmid 17551) (36) using 0.5 μl of Lipofectamine diluted in 50 μl of Opti-MEM (Invitrogen) per chamber. Cells were incubated for 24 h, the medium was then replaced with fresh medium, and cells were treated as described in the figure legends. Following treatments, cells were fixed gently to preserve GFP fluorescence using 2% paraformaldehyde for 15 min at room temperature followed by several washes with PBS. Cells were then permeabilized (0.2% Triton X-100, 150 mm NaCl, 10 mm Tris, pH 7.5) for 2 min at room temperature and washed with PBS. Nuclei were stained with DAPI (50 ng/ml in PBS) for 10 min at room temperature. Cells were imaged using Metamorph software and a Nikon eclipse 90i at 40× magnification.
Real Time PCR
RNA was isolated using the RNeasy kit (Qiagen). cDNA synthesis was performed using oligo(dT) and reverse transcriptase Superscript Preamplification System (Invitrogen) according to the manufacturer's instructions. Real time PCR was performed using the Light Cycler 480 (Roche Applied Science) and SYBR Green incorporation for product detection. The-fold-increase of GADD45α and PUMA are normalized to the housekeeping gene GAPDH as described previously (14, 32).
Comet Assay
DNA damage was assessed using the comet assay (single cell gel electrophoresis) as described previously (10, 32). Briefly, cells were harvested and embedded in 0.6% low melting agarose on slides precoated with 1.0% agar. Samples were then incubated in lysing solution (2.5 m NaCl, 100 mm EDTA, 10 mm Tris, 1% Triton X-100) overnight. Following lysis, the slides were incubated in an alkaline electrophoresis buffer (0.3 m NaOH, 1 mm EDTA, pH >13) for 40 min followed by electrophoresis at 25 volts for 20 min. Slides were washed three times in 0.4 m Tris, pH 7.5, and stained with ethidium bromide (2 μg/ml). Comet images were captured using a Nikon eclipse 90i and comets were quantified using the CASP program as the mean tail moment from 30–50 cells/condition.
Immunoprecipitation of Acetylated Lysine
To isolate crude nuclei, ∼107 cells were washed with PBS (4 °C) and collected in nuclear lysis buffer (10 mm Tris, pH 7.5, 10 mm NaCl, 3 mm MgCl2, 0.05% Nonidet P-40, 1 mm EGTA, with protease and phosphatase inhibitors). The lysate was then centrifuged at 2,700 × g for 10 min, and the nuclei-containing pellet was retained. The nuclei were washed twice in nuclear lysis buffer. The nuclei were then lysed and the proteins denatured by the addition of SDS to 1% followed by boiling for 5 min. 100 μg of the nuclear lysate was then diluted to 200 μl with PBS/0.1% BSA for immunoprecipitation. Immunoprecipitations were performed using sheep anti-mouse IgG Dynabeads (Invitrogen). The Dynabeads, 10 μl of slurry/sample, were washed twice in PBS/0.1% BSA using a magnetic particle separator. The beads were incubated with 4 μl of anti-acetylated lysine/sample in 200 μl of PBS/BSA at 4 °C with end-over-end mixing overnight. The beads were then washed twice in PBS/BSA followed by the addition of cell lysate (100 μg of protein). The beads plus lysate were incubated at 4 °C for 2 h with end-over-end mixing. The beads were then washed three times with PBS/BSA. The proteins were eluted from the beads by the addition of Laemmli sample buffer and boiling for 5 min.
siRNA Transfection
siRNA transfection was performed using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's instructions. Lipofectamine (1.5 μl/well) and siRNA were diluted in OptiMem (Invitrogen) to give a final concentration of 100 nm siRNA. The diluted Lipofectamine-siRNA complex was added to each well and overlaid with 200,000 cells in 400 μl. Experiments were started 48 h following transfection. The following Silencer® Select Pre-designed siRNA for SIRT1, 5′-GAUCAAGAGAUGGUAUUUAtt-3′ and 5′-CGAUAUUGAGUAUUUUAGAtt-3′ were obtained from Ambion.
SIRT1 Activity
The enzymatic activity of SIRT1 was evaluated using the Fluor-de-lys Sirt1 fluorometric drug discovery assay kit (Enzo Life Sciences, Plymouth Meeting, Pa).
Statistics
Statistical comparisons were made between groups using one-way ANOVA. Significant differences between groups (p < 0.05) were determined by Newman-Keuls post hoc analysis.
RESULTS
Nitric Oxide Stimulates the Nuclear Translocation of FoxO1
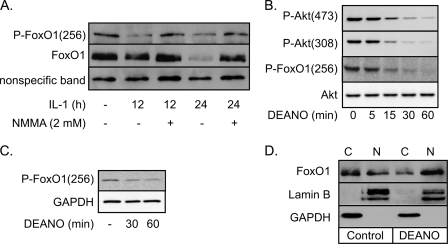
Akt-mediated phosphorylation at serine residue 256 (Ser-256) retains FoxO1 in the cytoplasm and attenuates FoxO1-dependent transcription (37). Cellular stresses, including nutrient deprivation and oxidative stress, attenuate Ser-256 phosphorylation, resulting in FoxO1 translocation to the nucleus where it is transcriptionally active (36, 37, 46). Nitric oxide, produced endogenously by the inducible isoform of NOS following 12- or 24-h IL-1 treatments, attenuates Ser-256 phosphorylation of FoxO1 (Fig. 1A). The NOS inhibitor NMMA prevents the loss of Ser-256 phosphorylation of FoxO1 in response to IL-1. Kitamura et al. (41) have shown that nuclear, transcriptionally active FoxO1 is rapidly ubiquitinated and degraded by the proteasome. Consistent with these findings, there is a significant reduction in the levels of total FoxO1 in INS 832/13 cells treated for 24 h with IL-1, and this loss of FoxO1 is prevented by NMMA (Fig. 1A). Akt-mediated Ser-256 phosphorylation of FoxO1 and Akt phosphorylation itself are reduced in INS 832/13 cells (Fig. 1B) and rat islets (Fig. 1C) treated with the nitric oxide donor DEANO. These findings indicate that nitric oxide is a mediator of the actions of IL-1 on FoxO1 activation.
FIGURE 1.
Nitric oxide stimulates the nuclear translocation of FoxO1. A and B, INS 832/13 cells were treated with IL-1 (10 units/ml) ± NMMA (2 mm) for 12 or 24 h (A) or DEANO (1 mm) for the indicated times (B). Western blot analysis was performed for phospho-FoxO1, phospho-Akt, total FoxO1, and total Akt. C, rat islets were treated with DEANO (1 mm) for the indicated times, and Western blot analysis was used to examine Ser(P)-256 FoxO1. GAPDH was used as a loading control. D, INS 832/13 cells were treated with DEANO (1 mm) for 1 h followed by cellular fractionation. Western blot analysis for total FoxO1 was performed. Lamin B (nuclear, N) and GAPDH (cytoplasmic, C) were used as subcellular fractionation markers. Results are representative of at least three independent experiments.
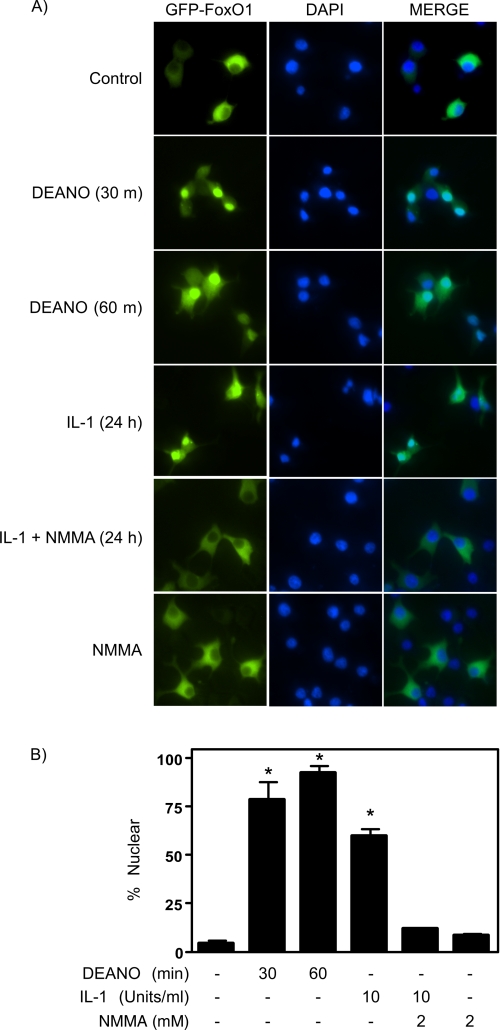
To confirm that the loss of Ser-256 phosphorylation in response to nitric oxide results in FoxO1 nuclear localization, cytoplasmic and nuclear fractions were prepared from INS 832/13 cells treated for 1 h with DEANO. In untreated control cells FoxO1 is localized predominantly in cytoplasmic fractions (Fig. 1D). Following the 1-h treatment with the nitric oxide donor there is a shift in the localization of FoxO1 to the nucleus (Fig. 1D). As a second measure of FoxO1 translocation, a GFP-FoxO1 construct was transfected into INS 832/13 cells. Both DEANO (30, 60 min) and IL-1 (24 h) stimulate GFP-FoxO1 nuclear localization (Fig. 2). The percentage of INS 832/13 cells containing nuclear FoxO1 was 79 ± 9% at 30 min and 94 ± 3% following a 60-min incubation (B, p < 0.05). Consistent with the effects of the nitric oxide donor, IL-1 stimulates the nuclear localization of FoxO1 in 60 ± 3% of INS 832/13 cells, and this nuclear translocation is attenuated to control levels by NMMA (Fig. 2). These findings show that nitric oxide stimulates the loss of Ser-256 phosphorylation and promotes the nuclear localization of FoxO1 in β-cells.
FIGURE 2.
Nuclear translocation of FoxO1 in response to IL-1. A, INS 832/13 cells expressing wild-type FoxO1 tagged with green fluorescent protein (GFP-FoxO1) were treated for 30 or 60 min with 1 mm DEANO or for 24 h with 10 units/ml IL-1 with or without NMMA. Following treatments GFP-FoxO1 cellular localization was evaluated by fluorescence microscopy (green). Nuclei were labeled using DAPI (blue). In response to DEANO or IL-1, in a nitric oxide-dependent manner, FoxO1 is localized to the nucleus whereas in untreated cells FoxO1 displays cytoplasmic localization. B, nuclear localization was quantified. Results are representative (A) or the average ± S.E. (error bars) of three independent experiments (B,*, p < 0.05 versus control).
FoxO1 Is Required for Nitric Oxide-induced GADD45α Expression and DNA Repair
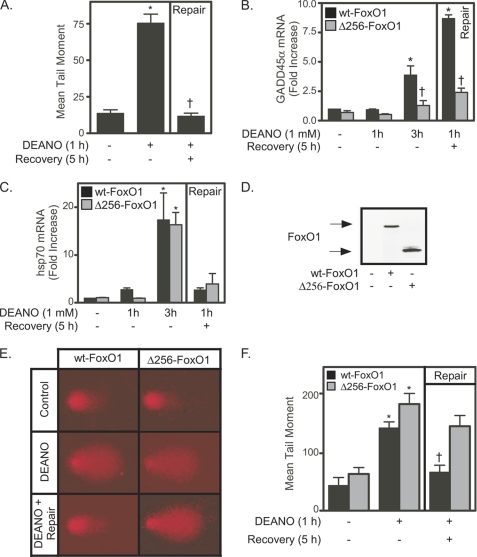
Nitric oxide is a known inducer of oxidative DNA damage (10, 11, 47). The comet assay was used to evaluate both the extent of nitric oxide-mediated DNA damage and the ability of β-cells to repair this damage (32). Treatment of INS 832/13 cells with DEANO for 1 h results in DNA damage as indicated by a 4-fold increase in mean tail moment (Fig. 3A). Removal of the donor by washing and continued culture for 5 h (in the absence of nitric oxide) results in the repair of this DNA damage (mean tail moment returns to control levels; Fig. 3A). The repair of nitric oxide-damaged DNA in β-cells does not require the presence of p53; however, GADD45α is required for DNA repair (32). Because both p53 and FoxO1 participate in the transcriptional regulation of GADD45α, yet p53 is not necessary for the repair of nitric oxide-damaged DNA in β-cells, the role of FoxO1 as a regulator of GADD45α expression by β-cells was examined. INS 832/13 cells were transduced with an adenovirus expressing either a wild-type FoxO1 (WT-FoxO1) or a dominant negative FoxO1 (Δ256-FoxO1) mutant lacking the transactivating domain (45). In cells expressing WT-FoxO1, DEANO stimulates an ∼4-fold increase in GADD45α mRNA accumulation following a 3-h incubation. There is an ∼8-fold increase in GADD45α mRNA accumulation in response to DNA repair conditions (1-h incubation with DEANO, followed by washing to remove the donor and an additional 5 h of incubation in the absence of donor; Fig. 3B). Nitric oxide-stimulated GADD45α mRNA accumulation is attenuated in INS 832/13 cells expressing the Δ256-FoxO1 mutant (Fig. 3B, gray bars). The inhibitory effects of the Δ256-FoxO1 mutant are selective because this mutant does not modify the expression of hsp70, a FoxO-independent gene whose expression is stimulated by nitric oxide (16) (Fig. 3C). In these experiments, WT-FoxO1 and Δ256-FoxO1 were expressed to similar levels (Fig. 3D). The effects of WT-FoxO1 on DEANO-induced gene expression are shown as a control for these studies because it takes into account the potential effects of the vector and the increased expression of the target protein. There were no differences in the levels of nitric oxide-induced mRNA accumulation in cells transfected with the empty vector compared with WT-FoxO1 (control data not shown).
FIGURE 3.
FoxO1 is required for nitric oxide-induced GADD45α expression. A, INS 832/13 cells were treated with DEANO (1 mm) for 1 h followed by washing and 5-h incubation in the absence of DEANO. The comet assay was used to examine DNA damage. B and C, INS 832/13 cells were transduced with Δ256-FoxO1 or WT-FoxO1 adenoviral constructs. Twenty-four hours after adenoviral transduction the cells were treated with DEANO (1 mm) for 1 h, 3 h, or 1 h followed by wash and 5-h recovery incubation. Total RNA was isolated, and real time PCR for GADD45α (B) or hsp70 (C) was performed and normalized to actin levels. D, alternatively, after adenoviral transduction, FoxO1 expression was examined by Western blot analysis. E and F, INS 832/13 cells transduced with Δ256-FoxO1 or WT-FoxO1 adenoviral constructs were treated with DEANO (1 mm) for 1 h or treated for DEANO for 1 h followed by washing and 5-h recovery incubation. The comet assay was used to assess DNA damage, and representative comet tails are shown (E) or quantified as the mean tail moment (F). Results are the average ± S.E. (error bars) of three independent experiments (A, B, C, and F; or representative of three independent experiments (D and E). Statistical significance is indicated (*, p < 0.05 compared with untreated control, †, p < 0.05 compared with WT-FoxO1 transduced cells).
Because FoxO1 regulates nitric oxide-induced GADD45α expression (Fig. 3B) and GADD45α is required for the repair of nitric oxide-damaged DNA (32), the role of FoxO1 in the repair of nitric oxide-damaged DNA in β-cells was examined. Treatment of INS 832/13 expressing a WT-FoxO1 or Δ256-FoxO1 for 1 h with DEANO results in DNA damage as evidenced by the large comet tail and ∼4-fold increase in the mean tail moment (Fig. 3, E and F). This DNA damage is repaired when nitric oxide is removed by washing and the cells are cultured for an additional 5 h in the absence of nitric oxide (mean tail moment returns to levels observed in untreated WT-FoxO1-expressing cells). INS 832/13 cells expressing the Δ256-FoxO1 mutant fail to repair nitric oxide-damage as indicated by a mean tail moment that is not significantly reduced compared with the DEANO-treated cells. In fact, nitric oxide appears to be more damaging to DNA in INS 832/13 cells expressing the Δ256-FoxO1 mutant than the levels observed in INS 832/13 cells expressing WT-FoxO1 (Fig. 3, E and F).
Inhibition of SIRT1 Attenuates Nitric Oxide-induced GADD45α Expression
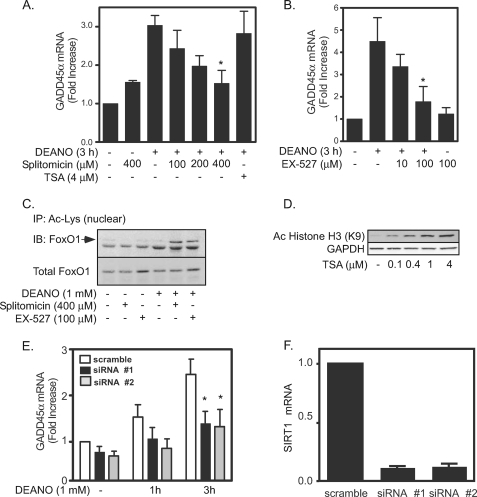
FoxO-dependent transcription is controlled, in part, by the posttranslational modification acetylation (36, 39). When deacetylated by SIRT1, FoxO transcription factors activate the expression of genes involved in the resistance to cellular oxidative stress (e.g. GADD45α) (39). In contrast, under conditions in which SIRT1 is inhibited, acetylated FoxO directs the expression of proapoptotic genes such as PUMA, NOXA, and BIM (39, 48). To evaluate the potential role of SIRT1 in the regulation of FoxO1-dependent gene expression, we evaluated the actions of two structurally different inhibitors of SIRT1 on nitric oxide-stimulated GADD45α mRNA accumulation. In a concentration-related manner, the SIRT1 inhibitors splitomicin and EX527 attenuate nitric oxide (DEANO)-induced GADD45α mRNA accumulation (Fig. 4, A and B). The inhibitory actions of SIRT1 inhibition on GADD45α mRNA accumulation correlate with an increase in the acetylation of FoxO1 (Fig. 4C), and this occurs at concentrations that do not modify histone acetylation (data not shown). As a control for potential nonspecific actions of SIRT1 inhibitors, the class I and II HDAC inhibitor trichostatin A (TSA), at concentrations that enhance histone acetylation (Fig. 4D), does not modify DEANO-induced GADD45α mRNA accumulation (Fig. 4A).
FIGURE 4.
Inhibition of SIRT1 attenuates nitric oxide-induced GADD45α expression. A and B, INS 832/13 cells were treated with DEANO (1 mm) for 3 h + splitomicin (A), TSA (A), or EX527 (B) at the indicated concentrations. Total RNA was isolated, and GADD45α mRNA accumulation was detected by real time PCR and normalized to GAPDH levels (*, p < 0.05). C, effects of splitomicin (400 μm) and EX-527 (100 μm) on FoxO1 acetylation were examined by immunoprecipitating Ac-K and immunoblotting for FoxO1. D, effects of TSA on the acetylation of histone H3 were examined by Western blot analysis. E and F, INS 832/13 cells were transfected with two different siRNAs for SIRT1 and scrambled siRNA for 48 h, DEANO (1 mm) was added, and the cells were cultured for 1 or 3 h as indicated. GADD45α and SIRT1 mRNA levels were evaluated by real time PCR (E, *, p < 0.05, versus scramble). Results are the average ± S.E. (error bars) of three or four independent experiment (A, B, E, and F) or representative of three independent experiments (C and D).
Consistent with the effects of pharmacological inhibitors of SIRT1, gene knockdown also attenuates GADD45α mRNA accumulation in INS 832/13 cells. Treatment of these cells for 1 or 3 h with DEANO results in the accumulation of GADD45α mRNA (Fig. 4E). In cells depleted of SIRT1 mRNA (Fig. 4F) there is a statistically significant attenuation in DEANO-induced GADD45α mRNA accumulation following 3-h incubation. Nitric oxide-stimulated GADD45α mRNA is also attenuated following a 1-h incubation, but at this time point the inhibitory actions of SIRT1 siRNA depletion did not achieve statistical significance. Taken together, the findings presented in Fig. 4 support a role for SIRT1 in the regulation of GADD45α gene expression in response to nitric oxide.
Regulation of DNA Damage and Repair by SIRT1
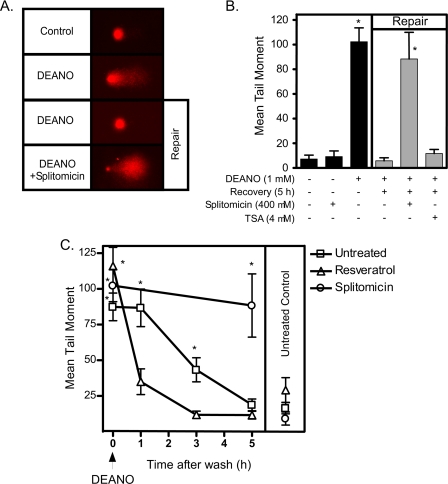
To determine whether SIRT1 activity is required for DNA repair, INS 832/13 cells were treated with DEANO for 1 h, the donor was removed by washing, and the cells were incubated for an additional 5 h in fresh medium in the presence or absence of splitomicin. DEANO induces comet formation (Fig. 5A) and a 5-fold increase in the mean tail moment (Fig. 5B). This DNA damage is repaired when nitric oxide is removing by washing and the INS 832/13 cells are cultured for an additional 5 h in the absence of nitric oxide (repair conditions). Cells treated with splitomicin during the DEANO treatment and 5-h recovery incubation fail to repair nitric oxide-damaged DNA (Fig. 5, A and B). Not only do these cells fail to repair DNA damage in the presence of a SIRT1 inhibitor, the absolute levels of DNA damage exceed the levels induced by nitric oxide alone. This is reflected in the large comet tail and small comet head in cells treated with splitomicin and DEANO (Fig. 5A). The DNA damage under these conditions was so extensive that in >20% of cells examined, the CASP program used to quantify DNA damage was not capable of distinguishing comet heads from tails. Thus, the level of DNA damage shown in Fig. 5B likely underestimates the actual level of DNA damage observed in INS 832/13 cells treated with nitric oxide and splitomicin. As a control, TSA does not affect DNA repair, indicating that the actions of splitomicin are not mediated by nonselective inhibition of deacetylation (Fig. 5B).
FIGURE 5.
Regulation of DNA damage and repair by modulation of SIRT1 activity. A and B, INS 832/13 cells were treated with DEANO (1 mm) for 1 h ± splitomicin or treated with DEANO (1 mm) for 1 h ± splitomicin, washed, and allowed to recover for an additional 5 h in the presence or absence of splitomicin. DNA damage was assessed using the comet assay. Representative comets from each condition are shown in A and quantified as the average (B). C, INS 832/13 cells were treated with DEANO for 1 h + resveratrol (10 μm) or treated with DEANO for 1 h + resveratrol, washed, and allowed to recover for 1, 3, or 5 h ± resveratrol. DNA damage was quantified using the comet assay. Results are the average ± S.E. (error bars) of three independent experiments (B and C) or representative of three independent experiments (A). Statistical significance is indicated (*, p < 0.05 compared with untreated control).
If inhibition of SIRT1 prevents the repair of nitric oxide-damaged DNA, then activation of this deacetylase should enhance the rate of DNA repair. Consistent with this hypothesis, activation of SIRT1 with resveratrol enhances the rate DNA repair in INS 832/13 cells (Fig. 5C). The repair of nitric oxide-mediated DNA damage is half-maximal following a 3-h incubation and nearly complete following a 5-h incubation (Fig. 5C). In the presence of resveratrol (at a concentration of 10 μm, which enhances SIRT1 enzymatic activity >2-fold), the time required to achieve half-maximal repair is less than 1 h, and complete repair of DNA damage is observed by 3 h (Fig. 5C). These findings provide experimental evidence that inhibition of SIRT1 attenuates DNA repair, whereas activation enhances the rate at which β-cells repair nitric oxide-damaged DNA.
Regulation of Cell Fate Decisions Is FoxO1-dependent
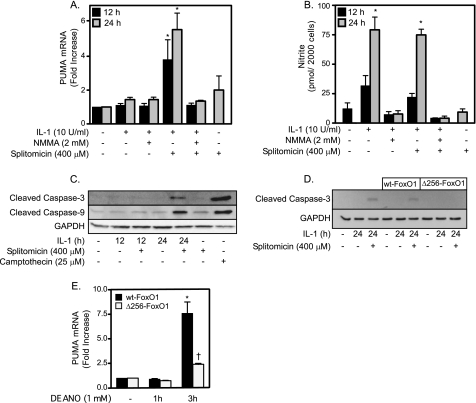
Short exposures of islets to cytokines for 12–24 h result in islet cell death that is consistent with necrosis as determined by the release of HMGB1 (19). Under these conditions there is little evidence of apoptotic cell death because caspases are not active or processed to their active form (19, 20). In contrast, prolonged exposures to cytokines (36 h or longer) result in islet cell death that is associated with an increase in caspase activity and cleavage (14). The shift to apoptosis is associated with irreversible inhibition of oxidative metabolism and DNA damage and the increased expression of PUMA (14). Consistent with these findings, IL-1 fails to stimulate PUMA expression (Fig. 6A) in INS 832/13 following a 24-h incubation, under conditions in which the INS 832/13 cells produce nitric oxide (Fig. 6B). In contrast, when SIRT-1 is inhibited using splitomicin, IL-1 induces a rapid 3-fold increase in the accumulation of PUMA mRNA following a 12-h incubation and >5-fold increase following a 24-h incubation (Fig. 6A). The accumulation of PUMA mRNA in response to IL-1 + splitomicin is sensitive to inhibition by NMMA, indicating that nitric oxide is required for induction of this proapoptotic factor (Fig. 6A). Splitomicin does not modify IL-1-induced nitrite production by β-cells (Fig. 5B) or stimulate PUMA mRNA accumulation in the absence of IL-1 (Fig. 6A). These results suggest that SIRT1 may protect β-cells from IL-1-induced apoptosis, as the inhibition of SIRT1 enhances PUMA expression following cytokine treatments.
FIGURE 6.
Inhibition of SIRT1 induces FoxO1-dependent apoptosis in response to cytokines. A and B, INS 832/13 cells were treated with IL-1 for 12 or 24 h ± NMMA or splitomicin. Total RNA was isolated, and PUMA expression was examined by real time PCR (A). Culture supernatants were examined for nitrite production (B) (*, p < 0.05). C and D, INS 832/13 cells (C) or INS 832/13 cells transduced with WT- or Δ256-FoxO1 (D) were treated with IL-1 (10 units/ml) for 12 or 24 h with or without splitomicin, and caspase-3 and caspase-9 cleavage was determined by Western blot analysis. Camptothecin was used as a positive control for caspase cleavage, and GAPDH was used as a loading control. E, INS 832/13 cells were transduced with Δ256-FoxO1 or WT-FoxO1 adenoviral constructs followed by 1- and 3-h incubation with 1 mm DEANO. Total RNA was isolated, and PUMA expression was examined by real time PCR and normalized to GAPDH levels (*, p < 0.05 compared with untreated control, †, p < 0.05 compared with WT-FoxO1 transduced cells). Results are the average ± S.E. (error bars) of three independent experiments (A, B, and E) or representative of three independent experiments (C and D).
In further support of this hypothesis, nitric oxide- and SIRT1-dependent induction of PUMA expression in response to IL-1 correlates with the cleavage of caspase-3 and caspase-9. Following a 24-h incubation, IL-1 fails to stimulate caspase-3 and caspase-9 cleavage; however, in combination with splitomicin, IL-1 induces the cleavage of both caspase enzymes to their active forms (Fig. 6C). The levels of caspase-3 and -9 cleavage are similar to the levels induced by the topoisomerase inhibitor camptothecin, a known apoptosis inducer. Alone, splitomicin does not stimulate caspase-3 or caspase-9 cleavage (Fig. 6C), and caspase-3 and caspase-9 cleavage in response to IL-1 + splitomicin is sensitive to inhibition by NMMA (data not shown).
FoxO1 is required for the activation of caspase-3 in response to a 24-h incubation with IL-1 + splitomicin. Treatment of INS 832/13 cells expressing the Δ256-FoxO1 mutant with IL-1 + splitomicin for 24 h fails to stimulate the cleavage of caspase-3; however, the combination of this cytokine and SIRT1 inhibitor stimulates caspase-3 cleavage in INS 832/13 cells expressing WT-FoxO1 (Fig. 6D). Consistent with a role for FoxO1 in regulating caspase activation, we show that nitric oxide-induced PUMA mRNA accumulation following a 3-h incubation is attenuated in INS 832/13 cells expressing Δ256-FoxO1 compared with cells expressing WT-FoxO1 (Fig. 6E). These findings support a role for FoxO1-dependent gene expression in both the protective and apoptotic responses of β-cells to nitric oxide.
DISCUSSION
The mechanisms regulating the response of cells to high output (micromolar levels) nitric oxide have yet to be fully identified. It has been reported that cytokines stimulate nitric oxide-dependent cell death by necrosis or apoptosis and in many cases in the same cell type (29). In pancreatic β-cells and insulinoma cell lines, cytokines have been shown to induce an early necrotic response that can be prevented by inhibitors of NOS (19, 20). Others have shown that prolonged incubation with cytokines, such as IL-1, stimulates an apoptotic cascade that is associated with the induction of endoplasmic reticulum stress and can be prevented using inhibitors of NOS (22–26, 49). Prolonged exposures of islets for 7–9 days with cytokines have been reported to stimulate nitric oxide-independent apoptosis (21). Complicating these divergent responses are reports that β-cells have the capacity to restore metabolic function and repair damaged DNA if the source of NO is removed and the cells are allowed a reasonable amount of time to recover (8 h for maximal recovery/repair) (3, 32).
The findings presented in this study and in two recent reports (14, 32) provide evidence in support of a molecular mechanism that may explain these divergent cellular responses to nitric oxide. The central contributors to these responses appear to be the Forkhead transcription factor FoxO1 and the regulation of FoxO1-dependent transcription by SIRT1. FoxO1 is a transcription factor that is activated in response to various stress stimuli to alert the cell to damage. The cellular response is to initiate repair/protective pathways or, when the damage is too extensive, induce cell death by apoptosis. Growth factors and hormones, such as insulin, stimulate Ser-256 phosphorylation by Akt, inactivating FoxO1 by sequestering this transcription factor in the cytoplasm (37). In response to nitric oxide (either endogenously produced following cytokine treatment or exogenously applied using a donor), FoxO1 is dephosphorylated on Ser-256 and translocates to the nucleus (Figs. 1 and 2). Once in the nucleus, deacetylated FoxO1 directs the expression of genes involved in stress resistance and repair (39). Recently, we have shown that the repair of nitric oxide-damaged DNA in β-cells requires the expression of GADD45α by a mechanism that is at least partially dependent on JNK (32). In this study, we now show that nitric oxide activates GADD45α expression in a FoxO1-dependent manner. We are currently examining the potential role of JNK in the regulation of FoxO1.
The regulation of FoxO1 transcriptional activity is controlled by the NAD+-dependent deacetylase SIRT1 (34, 39, 51). The role of SIRT1 in the regulation of stress responses has been described previously; however, studies have yet to examine its role in the response of cells to nitric oxide. When active, SIRT1 functions to deacetylate FoxO1 resulting in the expression of protective stress-resistant genes (39, 51). Consistent with a role for FoxO1 deacetylation in the repair pathway, inhibitors of SIRT1, which increase FoxO1 acetylation, attenuate the repair of nitric oxide-damaged DNA. Under these conditions, the extent of DNA damage is enhanced above the levels induced by nitric oxide alone. In contrast, the SIRT1 activator resveratrol enhances the rate of DNA repair by 3-fold (Fig. 5). The specificity of resveratrol has been questioned because it has been shown to activate targets in addition to SIRT1 (e.g. AMPK) (52), and the activation of these nonselective targets may contribute to the enhancement in DNA repair (Fig. 5C). Further, we have recently shown that nitric oxide is an activator of AMPK and that AMPK participates in the recovery of islet metabolic function (53). In an attempt to control for these potential nonselective actions of resveratrol, the effects of SIRT1 inhibition and activation on the repair of DNA damage have been evaluated. In addition, we have examined the effects of siRNA knockdown of SIRT1. Although we have taken both molecular and pharmacological approaches, it is possible that additional SIRT1-dependent or -independent factors, such as AMPK activation, also contribute to this DNA repair process.
In addition to activating this recovery response, SIRT1 protects β-cells from cytokine-mediated apoptosis. Consistent with previous studies (19, 20), IL-1 fails to stimulate apoptosis following a 24-h incubation; however, when SIRT1 is inhibited, there is an accumulation of PUMA mRNA and the activation of caspases-3 and -9 by cleavage (Fig. 6). Under these conditions, the inhibition of SIRT1 does not modify IL-1 induced nitric oxide production, indicating that changes in nitric oxide levels are not responsible for the differential gene expression and caspase activation under conditions of SIRT1 inhibition. Rather, we favor changes in the activation state of SIRT1 as the mechanism governing whether FoxO1 activates a protective or apoptotic transcriptional response to nitric oxide. Recently, we have shown the induction of caspase-3 activity, caspase-3 cleavage, and PUMA mRNA accumulation when islets are cultured for 36 h with IL-1 (14). These effects, which are not observed following a 24-h incubation with the cytokine, are prevented by inhibitors of NOS. It is likely that as damage induced by nitric oxide becomes more extensive, SIRT1 is inhibited, resulting in a shift in the FoxO1 transcriptional program from protection to the induction of apoptosis, as evidenced by PUMA expression and caspase-3 and -9 activation. Currently, the mechanisms responsible for inhibition of SIRT1 by nitric oxide are unknown. One potential target is the activation of polyADP-ribose polymerase, which uses NAD+ as a substrate (54). Thus, overactivation of polyADP-ribose polymerase resulting from DNA damage could result in the reduction in cellular NAD+ levels (15). Reductions in cellular NAD+ levels have been shown to inhibit the deacetylase activity of the NAD+-dependent enzyme, SIRT1 (55, 56). It is also possible that enhanced production of nitric oxide results in the direct modifications in SIRT1 through S-nitrosation or nitration of tyrosine residues, attenuating SIRT1 activity (57). We are currently examining these potential mechanisms responsible for the regulation of SIRT1 activity by nitric oxide.
We are also exploring the mechanisms responsible for the activation of FoxO1 by nitric oxide. The regulation of FoxO1 is likely controlled by Akt. Nitric oxide has been shown to inhibit Akt activity (58, 59), and we show that nitric oxide attenuates the phosphorylation of FoxO1 on the Akt-mediated site, Ser-256 (Fig. 1). JNK is also a known activator of FoxO-dependent gene expression and JNK participates in the GADD45α-dependent repair of nitric oxide-mediated DNA damage (32, 46). Although we have yet to observe a direct effect of JNK on FoxO1 nuclear localization,3 there is potential cross-talk of transcription factors that can be activated by JNK, such as ATF-2 (50), with FoxO1 in the regulation of expression of potential recovery factors such as GADD45α. Additional studies will be required to determine the mechanism of action of JNK in both the induction of a defense/repair response and the induction of apoptosis.
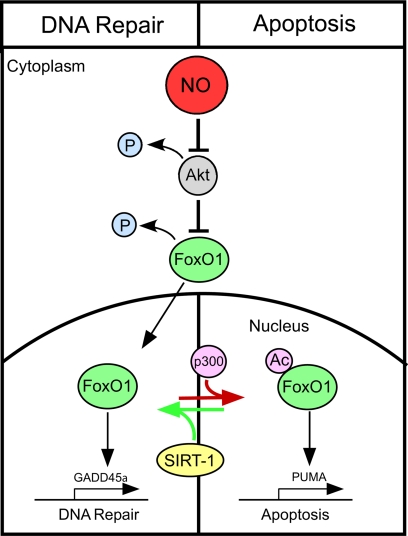
Overall, this study provides evidence that FoxO1 regulates both DNA repair and the induction of β-cell apoptosis in response to nitric oxide. The proposed mechanism that controls these pathways is outlined in Fig. 7. Central to this regulation of FoxO1 is the activity of the NAD+-dependent deacetylase SIRT1. We hypothesize that when SIRT1 is active, it deacetylates FoxO1, thereby directing a transcriptional program that attenuates apoptosis and stimulates the expression of factors that protect β-cells from nitric oxide. In contrast, when SIRT1 is less active, FoxO1 becomes more acetylated and directs a proapoptotic transcriptional program resulting in apoptosis. These findings provide a working model to explain how it is possible that nitric oxide induces a protective response as well as cell death by either necrosis or apoptosis in the same cell type and in response to the same stimuli.
FIGURE 7.
Proposed mechanism that controls β-cell fate in response to nitric oxide. In response to nitric oxide (supplied exogenously using donor molecules or endogenously following cytokine treatment) there is the loss of Akt-dependent FoxO1 phosphorylation that correlates with FoxO1 nuclear localization. In response to SIRT1 inhibitors, a proapoptotic transcriptional program appears to be activated as indicated by enhanced PUMA mRNA accumulation. If SIRT1 is activated, a FoxO1-dependent DNA repair response is activated as indicated by GADD45 mRNA accumulation.
Acknowledgments
We thank Colleen Kelly Bratcher for expert technical assistance and Dr. Michael Moxley for helpful discussion related to these studies.
This work was supported, in whole or in part, by National Institutes of Health Grants DK52194 and AI-44458 (to J. A. C.).
K. J. Hughes and J. A. Corbett, unpublished observation.
- PUMA
- p53-up-regulated mediator of apoptosis
- DEANO
- (Z)-1-(N,N-diethylamino)diazen-1-ium-1,2-diolate
- NMMA
- l-NG-monomethyl arginine
- TSA
- trichostatin A
- AMPK
- AMP-activated protein kinase.
REFERENCES
- 1. Southern C., Schulster D., Green I. C. (1990) FEBS Lett. 276, 42–44 [DOI] [PubMed] [Google Scholar]
- 2. Eizirik D. L., Flodström M., Karlsen A. E., Welsh N. (1996) Diabetologia 39, 875–890 [DOI] [PubMed] [Google Scholar]
- 3. Corbett J. A., McDaniel M. L. (1994) Biochem. J. 299, 719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Corbett J. A., Tilton R. G., Chang K., Hasan K. S., Ido Y., Wang J. L., Sweetland M. A., Lancaster J. R., Jr., Williamson J. R., McDaniel M. L. (1992) Diabetes 41, 552–556 [DOI] [PubMed] [Google Scholar]
- 5. Corbett J. A., Wang J. L., Sweetland M. A., Lancaster J. R., Jr., McDaniel M. L. (1992) J. Clin. Invest. 90, 2384–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Welsh N., Eizirik D. L., Bendtzen K., Sandler S. (1991) Endocrinology 129, 3167–3173 [DOI] [PubMed] [Google Scholar]
- 7. Corbett J. A., Wang J. L., Hughes J. H., Wolf B. A., Sweetland M. A., Lancaster J. R., Jr., McDaniel M. L. (1992) Biochem. J. 287, 229–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Koster J. C., Marshall B. A., Ensor N., Corbett J. A., Nichols C. G. (2000) Cell 100, 645–654 [DOI] [PubMed] [Google Scholar]
- 9. Heitmeier M. R., Corbett J. A. (2000) in Nitric Oxide Biology and Pathobiology (Ignarro L. J. ed) pp. 785–810, Academic Press, San Diego, CA [Google Scholar]
- 10. Delaney C. A., Green M. H., Lowe J. E., Green I. C. (1993) FEBS Lett. 333, 291–295 [DOI] [PubMed] [Google Scholar]
- 11. Eizirik D. L., Delaney C. A., Green M. H., Cunningham J. M., Thorpe J. R., Pipeleers D. G., Hellerström C., Green I. C. (1996) Mol. Cell. Endocrinol. 118, 71–83 [DOI] [PubMed] [Google Scholar]
- 12. Palmer J. P., Helqvist S., Spinas G. A., Mølvig J., Mandrup-Poulsen T., Andersen H. U., Nerup J. (1989) Diabetes 38, 1211–1216 [DOI] [PubMed] [Google Scholar]
- 13. Comens P. G., Wolf B. A., Unanue E. R., Lacy P. E., McDaniel M. L. (1987) Diabetes 36, 963–970 [DOI] [PubMed] [Google Scholar]
- 14. Hughes K. J., Chambers K. T., Meares G. P., Corbett J. A. (2009) Am. J. Physiol. Endocrinol. Metab. 297, E1187–E1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rosales A. L., Cunningham J. M., Bone A. J., Green I. C., Green M. H. (2004) Free Radic. Res. 38, 665–674 [DOI] [PubMed] [Google Scholar]
- 16. Scarim A. L., Heitmeier M. R., Corbett J. A. (1998) Endocrinology 139, 5050–5057 [DOI] [PubMed] [Google Scholar]
- 17. Scarim A. L., Nishimoto S. Y., Weber S. M., Corbett J. A. (2003) Endocrinology 144, 3415–3422 [DOI] [PubMed] [Google Scholar]
- 18. Scarim A. L., Heitmeier M. R., Corbett J. A. (1997) Endocrinology 138, 5301–5307 [DOI] [PubMed] [Google Scholar]
- 19. Steer S. A., Scarim A. L., Chambers K. T., Corbett J. A. (2006) PLoS Med. 3, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Collier J. J., Fueger P. T., Hohmeier H. E., Newgard C. B. (2006) Diabetes 55, 1398–1406 [DOI] [PubMed] [Google Scholar]
- 21. Liu D., Pavlovic D., Chen M. C., Flodström M., Sandler S., Eizirik D. L. (2000) Diabetes 49, 1116–1122 [DOI] [PubMed] [Google Scholar]
- 22. Mandrup-Poulsen T. (2003) Biochem. Pharmacol. 66, 1433–1440 [DOI] [PubMed] [Google Scholar]
- 23. Oyadomari S., Takeda K., Takiguchi M., Gotoh T., Matsumoto M., Wada I., Akira S., Araki E., Mori M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10845–10850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grunnet L. G., Aikin R., Tonnesen M. F., Paraskevas S., Blaabjerg L., Størling J., Rosenberg L., Billestrup N., Maysinger D., Mandrup-Poulsen T. (2009) Diabetes 58, 1807–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thomas H. E., McKenzie M. D., Angstetra E., Campbell P. D., Kay T. W. (2009) Apoptosis 14, 1389–1404 [DOI] [PubMed] [Google Scholar]
- 26. Gurzov E. N., Ortis F., Cunha D. A., Gosset G., Li M., Cardozo A. K., Eizirik D. L. (2009) Cell Death Differ. 16, 1539–1550 [DOI] [PubMed] [Google Scholar]
- 27. Brüne B. (2005) Antioxid. Redox Signal. 7, 497–507 [DOI] [PubMed] [Google Scholar]
- 28. Brown G. C., Borutaite V. (2002) Free Radic. Biol. Med. 33, 1440–1450 [DOI] [PubMed] [Google Scholar]
- 29. Calabrese V., Cornelius C., Rizzarelli E., Owen J. B., Dinkova-Kostova A. T., Butterfield D. A. (2009) Antioxid. Redox Signal. 11, 2717–2739 [DOI] [PubMed] [Google Scholar]
- 30. Wang X., Michael D., de Murcia G., Oren M. (2002) J. Biol. Chem. 277, 15697–15702 [DOI] [PubMed] [Google Scholar]
- 31. McLaughlin L. M., Demple B. (2005) Cancer Res. 65, 6097–6104 [DOI] [PubMed] [Google Scholar]
- 32. Hughes K. J., Meares G. P., Chambers K. T., Corbett J. A. (2009) J. Biol. Chem. 284, 27402–27408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carrier F., Smith M. L., Bae I., Kilpatrick K. E., Lansing T. J., Chen C. Y., Engelstein M., Friend S. H., Henner W. D., Gilmer T. M., et al. (1994) J. Biol. Chem. 269, 32672–32677 [PubMed] [Google Scholar]
- 34. Giannakou M. E., Partridge L. (2004) Trends Cell Biol. 14, 408–412 [DOI] [PubMed] [Google Scholar]
- 35. Huang H., Tindall D. J. (2007) J. Cell Sci. 120, 2479–2487 [DOI] [PubMed] [Google Scholar]
- 36. Frescas D., Valenti L., Accili D. (2005) J. Biol. Chem. 280, 20589–20595 [DOI] [PubMed] [Google Scholar]
- 37. Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. (1999) Cell 96, 857–868 [DOI] [PubMed] [Google Scholar]
- 38. Qiang L., Banks A. S., Accili D. (2010) J. Biol. Chem. 285, 27396–27401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., Hu L. S., Cheng H. L., Jedrychowski M. P., Gygi S. P., Sinclair D. A., Alt F. W., Greenberg M. E. (2004) Science 303, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 40. Greer E. L., Brunet A. (2005) Oncogene 24, 7410–7425 [DOI] [PubMed] [Google Scholar]
- 41. Kitamura Y. I., Kitamura T., Kruse J. P., Raum J. C., Stein R., Gu W., Accili D. (2005) Cell Metab. 2, 153–163 [DOI] [PubMed] [Google Scholar]
- 42. Kelly C. B., Blair L. A., Corbett J. A., Scarim A. L. (2003) Methods Mol. Med. 83, 3–14 [DOI] [PubMed] [Google Scholar]
- 43. Heitmeier M. R., Scarim A. L., Corbett J. A. (1997) J. Biol. Chem. 272, 13697–13704 [DOI] [PubMed] [Google Scholar]
- 44. White B. H., Kaczmarek L. K. (1997) J. Neurosci. 17, 1582–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nakae J., Kitamura T., Silver D. L., Accili D. (2001) J. Clin. Invest. 108, 1359–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Essers M. A., Weijzen S., de Vries-Smits A. M., Saarloos I., de Ruiter N. D., Bos J. L., Burgering B. M. (2004) EMBO J. 23, 4802–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Burney S., Caulfield J. L., Niles J. C., Wishnok J. S., Tannenbaum S. R. (1999) Mutat. Res. 424, 37–49 [DOI] [PubMed] [Google Scholar]
- 48. Kobayashi Y., Furukawa-Hibi Y., Chen C., Horio Y., Isobe K., Ikeda K., Motoyama N. (2005) Int. J. Mol. Med. 16, 237–243 [PubMed] [Google Scholar]
- 49. Oyadomari S., Araki E., Mori M. (2002) Apoptosis 7, 335–345 [DOI] [PubMed] [Google Scholar]
- 50. Hayakawa J., Depatie C., Ohmichi M., Mercola D. (2003) J. Biol. Chem. 278, 20582–20592 [DOI] [PubMed] [Google Scholar]
- 51. Guarente L. (2007) Cold Spring Harbor Symp. Quant. Biol. 72, 483–488 [DOI] [PubMed] [Google Scholar]
- 52. Baur J. A. (2010) Biochim. Biophys. Acta. 1804, 1626–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Meares G. P., Hughes K. J., Jaimes K. F., Salvatori A. S., Rhodes C. J., Corbett J. A. (2010) J. Biol. Chem. 285, 3191–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Szabó C. (2005) Pharmacol. Res. 52, 60–71 [DOI] [PubMed] [Google Scholar]
- 55. Pillai J. B., Isbatan A., Imai S., Gupta M. P. (2005) J. Biol. Chem. 280, 43121–43130 [DOI] [PubMed] [Google Scholar]
- 56. Zhang J. (2003) Bioessays 25, 808–814 [DOI] [PubMed] [Google Scholar]
- 57. Kornberg M. D., Sen N., Hara M. R., Juluri K. R., Nguyen J. V., Snowman A. M., Law L., Hester L. D., Snyder S. H. (2010) Nat. Cell Biol. 12, 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Størling J., Binzer J., Andersson A. K., Züllig R. A., Tonnesen M., Lehmann R., Spinas G. A., Sandler S., Billestrup N., Mandrup-Poulsen T. (2005) Diabetologia 48, 2039–2050 [DOI] [PubMed] [Google Scholar]
- 59. Yasukawa T., Tokunaga E., Ota H., Sugita H., Martyn J. A., Kaneki M. (2005) J. Biol. Chem. 280, 7511–7518 [DOI] [PubMed] [Google Scholar]