Abstract
The Sotos syndrome gene product, NSD1, is a SET domain histone methyltransferase that primarily dimethylates nucleosomal histone H3 lysine 36 (H3K36). To date, the intrinsic properties of NSD1 that determine its nucleosomal substrate selectivity and dimethyl H3K36 product specificity remain unknown. The 1.7 Å structure of the catalytic domain of NSD1 presented here shows that a regulatory loop adopts a conformation that prevents free access of H3K36 to the bound S-adenosyl-l-methionine. Molecular dynamics simulation and computational docking revealed that this normally inhibitory loop can adopt an active conformation, allowing H3K36 access to the active site, and that the nucleosome may stabilize the active conformation of the regulatory loop. Hence, our study reveals an autoregulatory mechanism of NSD1 and provides insight into the molecular mechanism of the nucleosomal substrate selectivity of this disease-related H3K36 methyltransferase.
Keywords: Crystal Structure, Epigenetics, Gene Regulation, Gene Transcription, Histone Methylation
Introduction
Alteration of chromatin structure is an important regulatory mechanism of eukaryotic gene expression. Post-translational modifications of histones are an integral part of this epigenetic mechanism, and histone lysine methylation in particular has drawn special attention due to its complex role. Recent genome-wide studies revealed that a complex combination of histone modifications within a genomic locus is a better indicator of transcriptional state than individual modifications considered in isolation (1). Thus, a clear understanding of the entire spectrum of histone methylations and their regulation is a prerequisite for understanding the epigenetic control of gene expression.
Methylation of H3K36 is found in species ranging from yeasts to mammals. In the budding yeast Saccharomyces cerevisiae, H3K36 is trimethylated by the SET domain histone lysine methyltransferase (HKMT)2 Set2 (2, 3). Interestingly, Set2 binds to the phosphorylated C-terminal domain of RNA polymerase II and travels with the RNA polymerase during elongation (4–6). H3K36 trimethylation is found in the coding region and is most concentrated at the 3′-end of the transcribed gene, where it functions in recruiting histone deacetylase complexes to suppress intragenic transcription initiation (7–9). In addition to the yeast Set2 homolog, three mammalian SET domain proteins, NSD1, NSD2 (multiple myeloma SET domain (MMSET)/Wolf–Hirschhorn syndrome candidate 1), and NSD3 (Wolf–Hirschhorn syndrome candidate 1-like), are associated with H3K36 methylation (10–16). All three NSD proteins share a highly conserved catalytic SET domain, and they also have auxiliary domains implicated in interactions with other protein or nucleic acid components, such as the PWWP and PHD domains. Initially there was some confusion in the literature about the position of the lysine residue being methylated and the number of methyl groups attached by the NSD proteins (12, 16–19). It has become clear that NSD proteins preferentially methylate nucleosomal H3K36 and that up to two methyl groups can be attached to the target lysine (15). When a histone octamer is used as substrate, NSD proteins can also methylate histone H4K44 in vitro. Surprisingly, when a single-stranded or duplex DNA is added, H3K36 becomes the sole methylation site (15). The precise mechanism underlying the nucleosome/DNA-dependent substrate specificity of NSD proteins remains unknown.
NSD proteins have been linked to a number of human diseases. NSD1 mutations are a major cause of Sotos syndrome, which is a childhood overgrowth syndrome associated with mental retardation (20, 21); deletion of NSD2 is essential for the pathogenesis of Wolf–Hirschhorn syndrome, a disease characterized by mental retardation, epilepsy, and developmental defects (22); and NSD3 is amplified in breast cancer cell lines (23). In addition, fusion proteins NUP98-NSD1 and NUP98-NSD3 resulting from chromosomal translocations cause acute myeloid leukemia, whereas an IgH-NSD2 fusion protein is associated with multiple myeloma (14, 22, 24–26).
To better understand the molecular basis for the unusual enzymatic properties of the NSD family of H3K36 methylases and their functions in pathogenesis, we have analyzed the structure of NSD1 by x-ray crystallography and molecular dynamics simulation. Our study has uncovered a novel autoregulatory mechanism underlying H3K36 dimethylation that is closely linked to its nucleosomal substrate selectivity. The structural and biochemical information provided also gives mechanistic insights into the molecular basis of Sotos syndrome.
EXPERIMENTAL PROCEDURES
The coding sequence for the catalytic domain of human NSD1 (NSD1-CD, amino acids 1852–2082) was cloned into a pGEX6p-1 vector between the BamHI and XhoI sites. Recombinant NSD1-CD was expressed at 16 °C in the Escherichia coli strain BL21(DE3)-RIPL as a GST fusion protein. The fusion protein was first purified using glutathione-Sepharose resins followed by the removal of the GST tag and further purification on a Superdex-75 column (GE Healthcare). Purified NSD1 protein was concentrated to ∼15 mg/ml for crystallization.
NSD1-CD crystals were obtained in 0.2 m lithium sulfate, 0.1 m HEPES, pH 7.5, and 25% PEG 3350 at 20 °C. Diffraction data were collected at the Beijing and Shanghai Synchrotron Radiation Facilities (BSRF and SSRF), and data were processed using HKL2000 (27). The crystal belongs to the P212121 space group, with unit cell dimensions of a = 65.54 Å, b = 67.98 Å, and c = 68.19 Å. Multiwavelength anomalous dispersion phasing using anomalous signals from three endogenous zinc atoms was carried out using SHELX (28), and an initial model was built using ARP/wARP (29). Coot and CCP4 were used for model building and structure refinement (30, 31), and figures were prepared using PyMOL (48). Statistics from crystallographic analyses are shown in Table 1. HKMT activity assays were carried out following the procedure described by Li et al. (15).
TABLE 1.
Statistics of crystallographic analysis
Values in parentheses are those for the highest-resolution shell of reflections. Rfree was calculated using the 10% of the diffraction data set aside during refinement, and Rwork was calculated with the data used throughout the refinement. MAD, multiwavelength anomalous dispersion; r.m.s., root mean square; SAH, S-adenosyl-l-homocysteine.
| Data collection | ||||
|---|---|---|---|---|
| Space group | P212121 | |||
| Unit cell | 65.54 × 67.98 × 68.19Å | 65.86 × 67.72 × 69.09Å |
| MAD |
Native | |||
|---|---|---|---|---|
| Zinc edge | Zinc peak | Remote | ||
| Data sets | ||||
| Wave length (Å) | 1.2828 | 1.2822 | 1.0000 | 0.8103 |
| Resolution range (Å) | 50.00–2.20 (2.28–2.20) | 50.00–2.40 (2.49–2.40) | 50.00–2.40 (2.49–2.40) | 50.00–1.70 (1.76–1.70) |
| Completeness (%) | 100 (100) | 100 (100) | 100 (100) | 99.4 (99.7) |
| Rmerge | 0.063 (0.45) | 0.086 | 0.073 (0.45) | 0.086 (0.48) |
| I/σI | 33.4 (4.0) | 35.9 (7.2) | 30.9 (4.9) | 16.1 (3.3) |
| Refinement | ||||
| Rwork/Rfree (%) | 19.5/24.3 | 16.4/19.4 | ||
| r.m.s. bond length (Å) | 0.012 | 0.009 | ||
| r.m.s. bond angle (°) | 1.288 | 1.182 | ||
| B-factors (Å2) | ||||
| Main chain atoms | 33.3 | 16.0 | ||
| Side chain atoms | 35.9 | 18.7 | ||
| Zinc atoms | 46.10 | 18.3 | ||
| SAH/AdoMet atoms | 28.49 | 18.3 | ||
| Water atoms | 37.03 | 26.4 | ||
Molecular dynamics (MD) simulations were performed using the GROMACS program package, version 3.2.1 (32), and computational dockings were carried out with GOLD (33). Detailed procedures are described in supplemental material.
RESULTS
Enzymatic Properties of NSD1
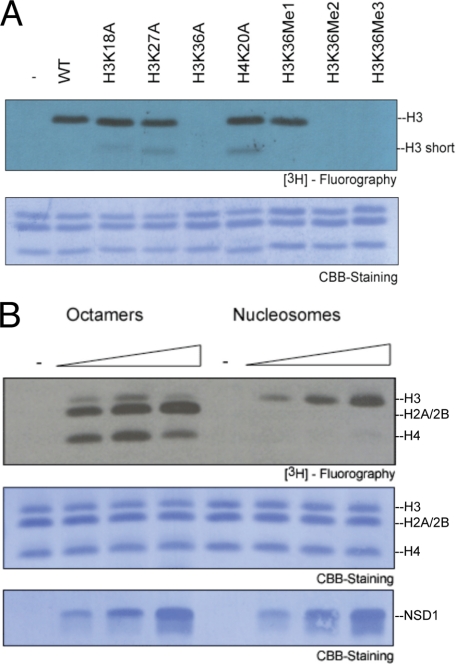
To analyze the enzymatic properties of NSD1, we expressed the catalytic domain of NSD1 (NSD1-CD, amino acids 1852–2082) encompassing the AWS (associated with SET), SET, and post-SET sequence motifs in E. coli (Fig. 1). Previously, we have shown that NSD1, NSD2, and NSD3 methylate nucleosomal H3K36 (15). As seen in Fig. 2A, the NSD1-CD did not methylate recombinant nucleosomes containing H3 K36A mutants, whereas K18A and K27A mutants of histone H3 displayed wild-type levels of methylation. Furthermore, only nucleosomes with an unmethylated H3K36 or a mimetic monomethylated H3K36, but not those with a di- or trimethylated H3K36 mimetic (34), served as substrates for NSD1 (Fig. 2A). When histone octamers were used, NSD1 also methylated histones H4 and H2A/2B, but these activities were largely eliminated when nucleosomes were used as substrates (Fig. 2B). Thus, the recombinant NSD1-CD protein fully recapitulates the enzymatic properties of NSD proteins in vitro, i.e. it dimethylates nucleosomal H3K36 and octameric histones H3, H4, and H2A/B.
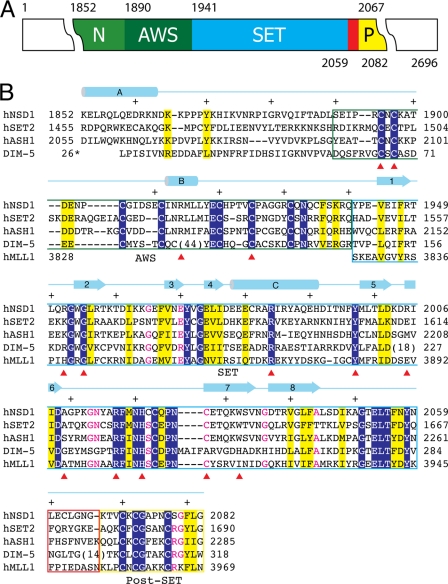
FIGURE 1.
Domain structure and sequence alignment of NSD1-CD. A, the domain structure of NSD1-CD. Each domain is represented by a colored box with the domain name indicated (N, N-terminal domain; P, post-SET domain). The numbers above and below the colored boxes indicate the numbers of the first and last residues of the domains, respectively. The red box indicates the post-SET loop connecting the SET and post-SET domains. B, structure-guided sequence alignment of the catalytic domains of human NSD1, SET2, ASH1, MLL1, and Neurospora Dim-5. Identical residues are indicated with white letters over a blue background, similar residues are highlighted in yellow, and those with all but one identical residue are shown in red. Different domains are enclosed in boxes colored as in A. A schematic diagram of the secondary structure elements of NSD1-CD is shown above the sequences, and every 10 residues is indicated with a + sign. Numbers to the left or right of the sequence indicate the numbering of the end residues. Note that the numbering for Dim-5 follows that of the PDB entry (1PEG). A red triangle below the sequences indicates the positions of missense mutations associated with Sotos syndrome.
FIGURE 2.
HKMT assay of NSD1-CD. A, nucleosomes assembled with recombinant, unmethylated wild-type or mutant histones (H3K18A, H3K27A, H3K36A, and H4K20A) or nucleosomes bearing methyl analogs mimicking different degrees of methylation on H3K36 (H3K36Me1, H3K36Me2 and H3K36Me3). Top panel, 3H fluorography using [3H]AdoMet as the methyl donor. Bottom panel, Coomassie Brilliant Blue (CBB) staining of histones. The concentration of NSD1-CD used for the assay was 0.1 μm, and that of nucleosomes in the reactions was 0.35 μm. B, HKMT activity on recombinant histone octamers and nucleosomes. Increasing concentrations (0.03, 0.1, and 0.3 μm) of NSD1-CD were titrated in the assay. Top panel, 3H fluorography shows that histones H3, H2A/2B, and H4 were methylated using histone octamers, whereas only H3K36 was methylated when nucleosomes were used as substrates. Middle panel, Coomassie Brilliant Blue staining of histones. Bottom panel, Coomassie Brilliant Blue staining of NSD1-CD.
Overall Structure
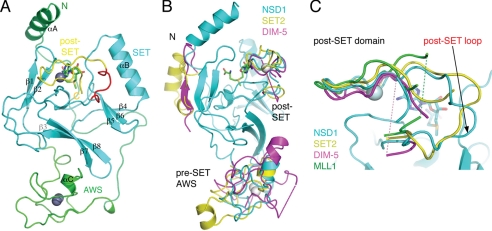
The 1.7 Å crystal structure of NSD1-CD presented here shows that the AWS, SET, post-SET motifs, and an N-terminal segment of ∼40 amino acids fold into a single compact domain (Fig. 3A). At the center of the structure lies the SET domain, which is mainly composed of three β-sheets (sheet 1, β1-β2; sheet 2, β4-β5-β6; and sheet 3, β3-β7-β8) arranged in a triangular fashion, and one mostly exposed helix (αB) is positioned adjacent to β-sheet 2. The spatial configuration is conserved among all known SET domain HKMTs (35). The SET domain is “tucked in” by an N-terminal helix (αA) at one end of the SET domain (top) next to β-sheet 1 and the AWS domain at the opposite end (bottom) next to β-sheet 3 (Fig. 3A). A 26-residue loop connecting αA and the AWS domain snakes across the SET domain on one side of the protein surface (back) opposite to the one where the post-SET loop is located (Fig. 3A). An S-adenosyl-l-methionine (AdoMet) molecule is bound in a pocket formed between the SET and the post-SET domains. Interestingly, the AdoMet molecule was found regardless of whether exogenous AdoMet was added during crystallization, indicating the E. coli origin of the AdoMet molecule. Previously, human Dot1L (hDot1L), a non-SET domain nucleosomal H3K79 HKMT, was also found to copurify with AdoMet from E. coli (36).
FIGURE 3.
Structure of NSD1-CD. A, a ribbon representation of the overall structure. Different segments of the protein are colored following the coloring scheme in Fig. 1A. The AdoMet molecule is shown as a stick model (green, carbon; blue, nitrogen; red, oxygen). Zinc ions are shown as gray spheres. Secondary structure elements and the protein termini are labeled. B, superposition of NSD1 (cyan), SET2 (yellow; PDB id: 3H6L) and Dim-5 (magenta; PDB id: 1PEG) structures. The SET domains are highly similar, and for clarity, only the NSD1 SET domain is shown. Dashed lines indicate structurally disordered regions. C, superposition of the post-SET domains of NSD1, SET2, Dim-5, and MLL1 (PDB id: 2W5Y). The magenta and green dashed lines indicate the disordered loop segments of Dim-5 and MLL1, respectively.
NSD1 belongs to a subfamily of SET domain HKMTs with cysteine-rich pre- and post-SET domains, which includes H3K9 HKMTs. A superposition of the structure of NSD1-CD on that of Dim-5, a Neurospora H3K9 HKMT (37), and human SET2 (Protein Data Bank (PDB) entry 3H6L) revealed three main differences. 1) The N-terminal helix (αA) adopts an orientation opposite to that of SET2, and in Dim-5, the helix is missing; 2) the conformation of cysteine-rich AWS/pre-SET domains of the three proteins differs significantly; and 3) a post-SET loop shows considerable conformational variation (Fig. 3B). Although the first two differences are interesting in their own right in terms of stabilizing the SET domain fold and structural diversity of SET domain HKMTs, they are not directly involved in catalysis and substrate binding, and they will not be investigated in detail here. The unusual post-SET loop conformation and its functional implication are analyzed below.
The Post-SET Domain and Its Autoinhibitory Loop
Three cysteine residues in the post-SET domain and one cysteine from the highly conserved NHS/CC motif of the SET domain coordinate the binding of a zinc ion (Figs. 1B and 3A). The zinc-coordinated post-SET domain forms one side of the AdoMet binding pocket and is critical for the catalytic competency of the enzyme. The superposition of NSD1, SET2, MLL1, and Dim-5 (38, 39) shows that the post-SET domains have homologous structures and that they are similarly positioned with respect to their SET domains (Fig. 3C). Prominently, the loops connecting the SET and post-SET domains in different enzymes have distinct conformations. This loop in MLL1, Dim5, and the mammalian H3K9 HKMT Suv39H2 (40) is disordered, whereas that in NSD1 and SET2 is ordered and occupies a position that generally binds substrate peptides in all known HKMTs (Fig. 3C). This post-SET loop conformation is not due to crystal packing, and loop conformation is quite stable, judging by the B-factors of the relevant amino acids, which are in the range of 10–28 Å2, as compared with overall B-factors of ∼20 Å2 (Table 1).
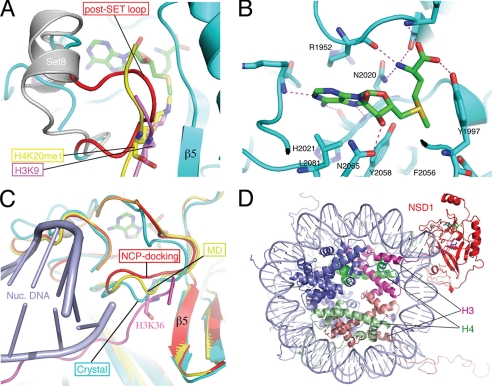
A superposition of the H3K9 peptide from the H3K9-Dim-5 complex (38), the mono-methylated H4K20 (H4K20me1) peptide bound to Pr-Set7/Set8 (38, 41, 42), and the NSD1-CD structure shows that both peptides crash into the post-SET loop of NSD1, which spans residues 2060–2066, around the substrate methyl acceptor lysine position (Fig. 4A). In the Dim-5 complex, the H3K9 peptide is present in the cleft between the SET domain and the post-SET loop, which is disordered in the structure, and makes β-sheet-like hydrogen bonds with β5. Similarly, in the Pr-Set7/Set8 complex, the H4K20me1 peptide forms three β-sheet-like hydrogen bonds with β5. In the NSD1 structure, the post-SET loop is in direct contact with β5 via two hydrogen bonds: the amide group of Leu-2063 with the carbonyl of Met-1998 and the sulfhydryl group of Cys-2062 with the carbonyl of Phe-1996. Leu-2063 also has hydrophobic interactions with residues located on β5 and αB. Thus, the post-SET loop of NSD1 is in an autoinhibitory conformation that effectively blocks the substrate binding cleft and the entrance to its methyl acceptor lysine binding channel.
FIGURE 4.
Structure of the active site and modeling of substrate binding. A, the post-SET loop of NSD1 occludes substrate binding. The H3K9 peptide (magenta) bound to Dim-5 and the H4K20me1 peptide (yellow) bound to Pr-Set7/Set8 (gray; PDB id: 2BQZ) are superimposed with the NSD1-CD structure. For clarity, the structure of Dim-5 is not shown, and only the post-SET domain of Pr-Set7/Set8 is shown. The loop colored in red represents the post-SET loop of NSD1-CD, and the rest of the protein structure is colored cyan. AdoMet is shown as a stick model (green, carbon; blue, nitrogen; red, oxygen), as are H3K9 (magenta, carbon) and H4K20me1 (yellow, carbon). B, amino acids surrounding the AdoMet molecule and their interactions with AdoMet. C, conformational dynamics of the post-SET loop. The post-SET loops from the crystal structure (cyan), from the molecular dynamics simulation at 276 ps (yellow), and from an energy-minimized model of nucleosome docking (red) are superimposed. The docked H3 tail and nucleosomal DNA are shown in magenta and light blue, respectively. H3K36 is shown as a stick model. Note that in the docked model, the post-SET domain contacts DNA. D, a global view of the modeled NSD1-CD complex with a nucleosome core particle. NSD1-CD is shown in red; two H3 molecules are shown in magenta and salmon; two H4 molecules are shown in green and light green; and H2A and H2B are shown in blue, and DNA is shown in light blue.
The Active Site and Product Specificity
NSD1 catalyzes the addition of up to two methyl groups to H3K36, whereas SET2 trimethylates H3K36. The structural basis of their ability to add different numbers of methyl groups onto substrate lysine, i.e. their product specificity, is unknown. It has previously been shown that the size of certain residues, particularly aromatic residues, in the active site influences the product specificity of HKMTs (38, 43, 44). Inspection of the NSD1 catalytic active site surrounding the AdoMet molecule reveals that residues in the close vicinity are highly conserved between NSD1 and Dim-5, an H3K9 trimethylase, in terms of both amino acid identity and spatial conformation (Fig. 4B). Of particular note is an aromatic residue, Phe-2056, near the methyl group of AdoMet, which corresponds to Phe-281 in Dim-5. In Dim-5, an F281Y mutant converts Dim-5 from a H3K9 trimethylase into a mono-/dimethylase (38). The corresponding residue in Pr-Set7/Set8 is Tyr-344, and a Y344F mutation changes it from a H4K20 monomethylase into a dimethylase (43). This phenomenon has been termed the “Phe/Tyr switch” for product specificity (35). However, all residues at positions known to affect product specificity are conserved in NSD1, SET2, and Dim-5. Thus, a new mechanism other than the Phe/Tyr switch must be involved in determining the di- versus trimethylase activities of NSD1 and SET2.
Molecular Dynamics Simulation and Nucleosome Docking
It is clear that the post-SET loop has to move aside in order for the methylation reaction to take place. To probe conformational dynamics of the post-SET loop and its role in H3K36 binding, we carried out a 2-ns MD simulation of NSD1-CD. Results show that the post-SET loop undergoes modest conformational changes, indicating that in solution, the protein molecules exist in an ensemble of post-SET loop conformations. These conformational changes, although modest, open up the lysine binding channel, allowing access of the substrate lysine to the methyl group of AdoMet. Fig. 4C shows the docking of a lysine in the canonical substrate binding channel of NSD1 in the 276-ps MD simulation. Taken together, these results show that intrinsic, spontaneous movement of the post-SET loop can expose the AdoMet molecule for methyl transfer reactions.
A more interesting question is whether certain elements in the nucleosome stabilize the active conformation of the post-SET loop as NSD proteins strongly prefer nucleosomal H3K36 as a substrate. To gain initial insights, we computationally docked NCP against the NSD1 structure (45). The following criteria guided the docking of the two structures. 1) The NCP core, i.e. NCP without the first 37 residues of the N-terminal tail, was treated as a rigid body; 2) H3K36 was bound in the canonical substrate binding channel; and 3) the rest of the H3 tail was free to adopt any conformation. Docking and energy minimization of the open conformation of NSD1 in the 276-ps MD simulation revealed that the post-SET loop contacts DNA, leading to widening of the lysine binding channel (Fig. 4, C and D). Thus, NCP contact stabilizes a post-SET loop conformation that is favorable for H3K36 methylation.
Sotos Mutations
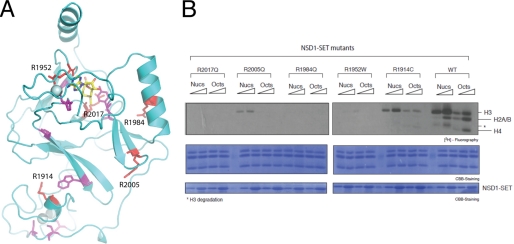
At least 14 missense mutations associated with Sotos syndrome are present in the AWS and SET domains (Fig. 1A) (46, 47). These mutations are concentrated in two areas of NSD1-CD, one surrounding the zinc ions in the AWS/pre-SET domain and another near the AdoMet binding site (Fig. 5A). Many of the mutated residues appear to play structural roles important for proper protein folding. We selected five arginine residues and changed them to those that are present in Sotos syndrome, testing their HKMT activities in vitro. Fig. 5B shows that R1914C and R2005Q have greatly reduced H3K36 HKMT activities. R1952W is barely active, and the enzymatic activities of R1984Q and R2017Q are not detectable. It is interesting to note that R1952, R1984, and R2005 are all engaged in interactions with negatively charged Asp or Glu residues in the NSD1-CD structure. R2017 occupies a central position stabilizing the conformations of three aromatic residues, Tyr-1870, Tyr-1977, and Phe-2018, via cation-π interactions, the latter two of which are highly conserved residues in SET domain proteins. Thus, the biochemical consequence of Sotos syndrome mutations appears to be a loss of or reduction in the HKMT activity of NSD1.
FIGURE 5.
Sotos syndrome missense mutations in NSD1-CD. A, amino acids at positions where the missense mutations of Sotos syndrome occur are shown as stick models (red and magenta), superimposed on a ribbon diagram of the NSD1-CD structure. AdoMet is shown as a stick model (yellow, carbon; blue, nitrogen; red, oxygen), and the spheres represent zinc ions. The residues shown in red correspond to arginines selected for in vitro mutagenesis and HKMTase assays. B, five arginine residues were individually changed to amino acids found in Sotos syndrome patients, and their HKMTase activities toward recombinant nucleosomes (Nucs) and histone octamers (Octs) were assayed using [3H]AdoMet as the methyl donor (top panel). The concentrations of NSD1-CD used in the assays were 0.04 and 0.2 μm, and those of nucleosomes and histone octamers were both 0.35 μm. Coomassie Brilliant Blue (CBB) staining of histones and the enzyme are shown in the middle and bottom panels, respectively.
DISCUSSION
In this study, we have uncovered an unusual feature of the H3K36 methylase NSD1, namely that the substrate lysine binding channel is blocked by a loop connecting the SET and post-SET domains. This is likely to be a common feature of all H3K36 methylases as an inhibitory loop conformation has been observed in human SET2, as well as in ASH1, described in an accompanying report (49). This is unusual among SET domain HKMTs as most of them are found in active conformations. The only exception known to date is the SET domain of MLL, which is known to be inactive in the absence of other subunits of the MLL complex. In general, SET domain HKMTs can be categorized into two groups based on their enzymatic activities, ones that are constitutively active without additional subunits, such as SUV39, Set7/Set8/Set9, and NSD1/SET2, and others that require the presence of auxiliary subunits, such as MLL and EZH2. In the latter group, it is clear that protein-protein interactions must play an important role in regulating their enzymatic activities. Little is known about how the HKMT activities of the first group of enzymes may be regulated. Our discovery that an autoregulatory loop gates the access of substrate lysine provides a first glimpse of potential regulatory mechanisms of the SET domain HKMTs.
It is intriguing that the post-SET loops in all known H3K36 HKMTase structures adopt an inhibitory conformation. As suggested by our molecular dynamics simulation and docking exercises, interaction with the nucleosome may stabilize an active conformation of the post-SET loop, thus promoting productive methylation of H3K36. Because H3K36 is located proximal to the nucleosomal DNA, the interaction between the enzyme and DNA appears to play a major role in stabilizing the post-SET loop. This is consistent with previous observations that the H3K36 HKMT activities of NSD1–3 and SET2 are greatly stimulated when nucleosomes are used as substrates (15). In addition, for NSD1 and NSD2, nucleosomal DNA or non-nucleosomal DNA fragments promote substrate specificity toward H3K36 and inhibit in vitro, presumably nonspecific, activity toward histone H4.
Another interesting feature about NSD1 is the structural basis for its dimethyl-lysine product specificity. Comparing it with the structures of Dim-5 and SET2, both of which are well characterized histone lysine trimethylases, revealed that there are essentially no differences between them in the area surrounding the AdoMet methyl group. On the other hand, it has been shown unambiguously that NSD1 can only add up to two methyl groups to H3K36. Our MD simulation revealed that the post-SET loop undergoes spontaneous conformational changes, opening up the substrate binding channel and allowing a snug fit of an unmethylated lysine. With the docking of the nucleosome, the post-SET loop moves further away, expanding the size of the entrance and accommodating the passage of dimethyl lysine. Thus, the determining factor for this dimethyl product specificity appears not to be the size of the active site near the methyl group of AdoMet but rather the size of the entrance of the lysine binding channel. This is in contrast to other SET domain HKMTs, where the size of the entrance is not a restriction, but the size of the active site near the AdoMet methyl group is. Thus, the structure of NSD1 provides new insights into how product specificity of SET domain HKMTases may be achieved in an alternative way.
Supplementary Material
Acknowledgments
We thank the staff at the BSRF and SSRF for help in x-ray diffraction data collection and Joy Fleming for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant 4R37G037120-24 (to D. R.). This work was also supported by the Chinese Ministry of Science and Technology (Grants 2009CB825501 and 2010CB944903 to R.-M. X.), the National Science Foundation of China (Grants 90919029 and 3098801 to R.-M. X.), and the Novo Nordisk-Chinese Academy of Sciences Foundation.
The atomic coordinates and structure factors (code 3OOI) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures.”
- HKMT
- histone lysine methyltransferase
- AdoMet
- S-adenosyl-l-methionine
- NSD
- nuclear receptor SET domain-containing protein
- MD
- molecular dynamics
- NCP
- nucleosome core particle.
REFERENCES
- 1. Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 2. Strahl B. D., Grant P. A., Briggs S. D., Sun Z. W., Bone J. R., Caldwell J. A., Mollah S., Cook R. G., Shabanowitz J., Hunt D. F., Allis C. D. (2002) Mol. Cell. Biol. 22, 1298–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Landry J., Sutton A., Hesman T., Min J., Xu R. M., Johnston M., Sternglanz R. (2003) Mol. Cell. Biol. 23, 5972–5978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Krogan N. J., Kim M., Tong A., Golshani A., Cagney G., Canadien V., Richards D. P., Beattie B. K., Emili A., Boone C., Shilatifard A., Buratowski S., Greenblatt J. (2003) Mol. Cell. Biol. 23, 4207–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li J., Moazed D., Gygi S. P. (2002) J. Biol. Chem. 277, 49383–49388 [DOI] [PubMed] [Google Scholar]
- 6. Xiao T., Hall H., Kizer K. O., Shibata Y., Hall M. C., Borchers C. H., Strahl B. D. (2003) Genes Dev. 17, 654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Keogh M. C., Kurdistani S. K., Morris S. A., Ahn S. H., Podolny V., Collins S. R., Schuldiner M., Chin K., Punna T., Thompson N. J., Boone C., Emili A., Weissman J. S., Hughes T. R., Strahl B. D., Grunstein M., Greenblatt J. F., Buratowski S., Krogan N. J. (2005) Cell 123, 593–605 [DOI] [PubMed] [Google Scholar]
- 8. Carrozza M. J., Li B., Florens L., Suganuma T., Swanson S. K., Lee K. K., Shia W. J., Anderson S., Yates J., Washburn M. P., Workman J. L. (2005) Cell 123, 581–592 [DOI] [PubMed] [Google Scholar]
- 9. Bell O., Wirbelauer C., Hild M., Scharf A. N., Schwaiger M., MacAlpine D. M., Zilbermann F., van Leeuwen F., Bell S. P., Imhof A., Garza D., Peters A. H., Schübeler D. (2007) EMBO J. 26, 4974–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun X. J., Wei J., Wu X. Y., Hu M., Wang L., Wang H. H., Zhang Q. H., Chen S. J., Huang Q. H., Chen Z. (2005) J. Biol. Chem. 280, 35261–35271 [DOI] [PubMed] [Google Scholar]
- 11. Yuan W., Xie J., Long C., Erdjument-Bromage H., Ding X., Zheng Y., Tempst P., Chen S., Zhu B., Reinberg D. (2009) J. Biol. Chem. 284, 15701–15707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nimura K., Ura K., Shiratori H., Ikawa M., Okabe M., Schwartz R. J., Kaneda Y. (2009) Nature 460, 287–291 [DOI] [PubMed] [Google Scholar]
- 13. Huang N., vom Baur E., Garnier J. M., Lerouge T., Vonesch J. L., Lutz Y., Chambon P., Losson R. (1998) EMBO J. 17, 3398–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang G. G., Cai L., Pasillas M. P., Kamps M. P. (2007) Nat. Cell Biol. 9, 804–812 [DOI] [PubMed] [Google Scholar]
- 15. Li Y., Trojer P., Xu C. F., Cheung P., Kuo A., Drury W. J., 3rd, Qiao Q., Neubert T. A., Xu R. M., Gozani O., Reinberg D. (2009) J. Biol. Chem. 284, 34283–34295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rayasam G. V., Wendling O., Angrand P. O., Mark M., Niederreither K., Song L., Lerouge T., Hager G. L., Chambon P., Losson R. (2003) EMBO J. 22, 3153–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marango J., Shimoyama M., Nishio H., Meyer J. A., Min D. J., Sirulnik A., Martinez-Martinez Y., Chesi M., Bergsagel P. L., Zhou M. M., Waxman S., Leibovitch B. A., Walsh M. J., Licht J. D. (2008) Blood 111, 3145–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kang H. B., Choi Y., Lee J. M., Choi K. C., Kim H. C., Yoo J. Y., Lee Y. H., Yoon H. G. (2009) FEBS Lett. 583, 1880–1886 [DOI] [PubMed] [Google Scholar]
- 19. Kim J. Y., Kee H. J., Choe N. W., Kim S. M., Eom G. H., Baek H. J., Kook H., Kook H., Seo S. B. (2008) Mol. Cell. Biol. 28, 2023–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Douglas J., Hanks S., Temple I. K., Davies S., Murray A., Upadhyaya M., Tomkins S., Hughes H. E., Cole T. R., Rahman N. (2003) Am. J. Hum. Genet. 72, 132–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kurotaki N., Imaizumi K., Harada N., Masuno M., Kondoh T., Nagai T., Ohashi H., Naritomi K., Tsukahara M., Makita Y., Sugimoto T., Sonoda T., Hasegawa T., Chinen Y., Tomita Ha H. A., Kinoshita A., Mizuguchi T., Yoshiura Ki K., Ohta T., Kishino T., Fukushima Y., Niikawa N., Matsumoto N. (2002) Nat. Genet. 30, 365–366 [DOI] [PubMed] [Google Scholar]
- 22. Stec I., Wright T. J., van Ommen G. J., de Boer P. A., van Haeringen A., Moorman A. F., Altherr M. R., den Dunnen J. T. (1998) Hum. Mol. Genet. 7, 1071–1082 [DOI] [PubMed] [Google Scholar]
- 23. Angrand P. O., Apiou F., Stewart A. F., Dutrillaux B., Losson R., Chambon P. (2001) Genomics 74, 79–88 [DOI] [PubMed] [Google Scholar]
- 24. Jaju R. J., Fidler C., Haas O. A., Strickson A. J., Watkins F., Clark K., Cross N. C., Cheng J. F., Aplan P. D., Kearney L., Boultwood J., Wainscoat J. S. (2001) Blood 98, 1264–1267 [DOI] [PubMed] [Google Scholar]
- 25. Rosati R., La Starza R., Veronese A., Aventin A., Schwienbacher C., Vallespi T., Negrini M., Martelli M. F., Mecucci C. (2002) Blood 99, 3857–3860 [DOI] [PubMed] [Google Scholar]
- 26. Chesi M., Nardini E., Lim R. S., Smith K. D., Kuehl W. M., Bergsagel P. L. (1998) Blood 92, 3025–3034 [PubMed] [Google Scholar]
- 27. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 28. Sheldrick G. M. (2008) Acta Crystallogr. Sect. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 29. Morris R. J., Perrakis A., Lamzin V. S. (2003) Methods Enzymol. 374, 229–244 [DOI] [PubMed] [Google Scholar]
- 30. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 31. Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 32. Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. (2005) J. Comput. Chem. 26, 1701–1718 [DOI] [PubMed] [Google Scholar]
- 33. Verdonk M. L., Cole J. C., Hartshorn M. J., Murray C. W., Taylor R. D. (2003) Proteins 52, 609–623 [DOI] [PubMed] [Google Scholar]
- 34. Simon M. D., Chu F., Racki L. R., de la Cruz C. C., Burlingame A. L., Panning B., Narlikar G. J., Shokat K. M. (2007) Cell 128, 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheng X., Zhang X. (2007) Mutat Res. 618, 102–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Min J., Feng Q., Li Z., Zhang Y., Xu R. M. (2003) Cell 112, 711–723 [DOI] [PubMed] [Google Scholar]
- 37. Zhang X., Tamaru H., Khan S. I., Horton J. R., Keefe L. J., Selker E. U., Cheng X. (2002) Cell 111, 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang X., Yang Z., Khan S. I., Horton J. R., Tamaru H., Selker E. U., Cheng X. (2003) Mol. Cell 12, 177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Southall S. M., Wong P. S., Odho Z., Roe S. M., Wilson J. R. (2009) Mol. Cell 33, 181–191 [DOI] [PubMed] [Google Scholar]
- 40. Wu H., Min J., Lunin V. V., Antoshenko T., Dombrovski L., Zeng H., Allali-Hassani A., Campagna-Slater V., Vedadi M., Arrowsmith C. H., Plotnikov A. N., Schapira M. (2010) PLoS One 5, e8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Couture J. F., Collazo E., Brunzelle J. S., Trievel R. C. (2005) Genes Dev. 19, 1455–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xiao B., Jing C., Kelly G., Walker P. A., Muskett F. W., Frenkiel T. A., Martin S. R., Sarma K., Reinberg D., Gamblin S. J., Wilson J. R. (2005) Genes Dev. 19, 1444–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Couture J. F., Dirk L. M., Brunzelle J. S., Houtz R. L., Trievel R. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20659–20664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xiao B., Jing C., Wilson J. R., Walker P. A., Vasisht N., Kelly G., Howell S., Taylor I. A., Blackburn G. M., Gamblin S. J. (2003) Nature 421, 652–656 [DOI] [PubMed] [Google Scholar]
- 45. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 46. Saugier-Veber P., Bonnet C., Afenjar A., Drouin-Garraud V., Coubes C., Fehrenbach S., Holder-Espinasse M., Roume J., Malan V., Portnoi M. F., Jeanne N., Baumann C., Héron D., David A., Gérard M., Bonneau D., Lacombe D., Cormier-Daire V., Billette de Villemeur T., Frébourg T., Bürglen L. (2007) Hum. Mutat 28, 1098–1107 [DOI] [PubMed] [Google Scholar]
- 47. Tatton-Brown K., Douglas J., Coleman K., Baujat G., Cole T. R., Das S., Horn D., Hughes H. E., Temple I. K., Faravelli F., Waggoner D., Turkmen S., Cormier-Daire V., Irrthum A., Rahman N. (2005) Am. J. Hum. Genet. 77, 193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 49. An S., Yeo K. J., Jeon Y. H., Song J. J. (2011) J. Biol. Chem. 286, 8369–8374 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.