Abstract
Store-operated Ca2+ entry (SOCE) represents a ubiquitous Ca2+ influx pathway activated by the filling state of intracellular Ca2+ stores. SOCE is mediated by coupling of STIM1, the endoplasmic reticulum Ca2+ sensor, to the Orai1 channel. SOCE inactivates during meiosis, partly because of the inability of STIM1 to cluster in response to store depletion. STIM1 has several functional domains, including the Orai1 interaction domain (STIM1 Orai Activating Region (SOAR) or CRAC Activation Domain (CAD)) and STIM1 homomerization domain. When Ca2+ stores are full, these domains are inactive to prevent constitutive Ca2+ entry. Here we show, using the Xenopus oocyte as an expression system, that the C-terminal 200 residues of STIM1 are important to maintain STIM1 in an inactive state when Ca2+ stores are full, through predicted intramolecular shielding of the active STIM1 domains (SOAR/CAD and STIM1 homomerization domain). Interestingly, our data argue that the C-terminal 200 residues accomplish this through a steric hindrance mechanism because they can be substituted by GFP or mCherry while maintaining all aspects of STIM1 function. We further show that STIM1 clustering inhibition during meiosis is independent of the C-terminal 200 residues.
Keywords: Calcium, Calcium Channels, Cell Cycle, Oocyte, Signal Transduction, Meiosis, Orai1, STIM1
Introduction
Store-operated Ca2+ entry (SOCE)3 is an integral Ca2+ influx pathway in non-excitable cells, activated by depletion of intracellular Ca2+ stores downstream of phospholipase C-coupled agonists (1). SOCE mediates a slow sustained entry of extracellular Ca2+ that is important for various homeostatic and signaling functions, including store refilling, T-cell activation, modulation of Ca2+ signal dynamics, and apoptosis (2). Mechanistically, SOCE activates through physical coupling between the endoplasmic reticulum (ER) Ca2+ sensor STIM1 and the SOCE channel Orai1 (3). STIM1 is a single-pass integral membrane protein that is enriched in the ER membrane and has luminal EF-hands, allowing it to monitor ER Ca2+ levels (4, 5). Current evidence argues that at low luminal Ca2+, STIM1 loses its bound Ca2+, producing a conformational change that leads to STIM1 clustering into large puncta. These puncta are stabilized in the cortical ER in very close proximity of the cell membrane (10–20 nm), allowing STIM1 to directly bind to and gate Orai1 (6–12).
STIM1 has a modular construction with a luminal EF-hand and a sterile α-motif (13). The cytosolic domain contains two predicted coiled-coil domains followed by a Ser/Pro-rich region and Lys-rich region at the end of the molecule (14). Structure-function studies have identified multiple functional domains within the STIM1 cytoplasmic domain. A minimal domain referred to as SOAR/CAD or CCb9 between residues 339 and 448 has been identified as necessary and sufficient for coupling to and activation of Orai1 (15–17). A larger domain, OASF, exhibits similar functionalities to the SOAR/CAD domain (18). Interestingly, fragments that contain the SOAR/CAD domain and extend farther C-terminally tend to decrease its activity, arguing that regions C-terminal to the SOAR/CAD domain (residues 445–485) inhibit its interaction with Orai1 (16–18). However, this modulation depends on the length of the soluble fragment used, arguing for differential folding and exposure of the SOAR/CAD domain. Two additional important domains include the STIM1 homomerization domain (SHD; residues 400–474), which is required for STIM1 oligomerization (18), and a short sequence C-terminal to the SHD that mediates the Ca2+-dependent inactivation of the STIM1-Orai1 channel complex through Ca2+ binding to a cluster of acidic amino acids (19–21). The C-terminal end of STIM1 contains a polybasic domain that has been implicated in stabilization of STIM1 puncta in the cortical ER in the absence of Orai1 (6, 15, 22). In contrast, no clear function has been assigned to the Ser/Pro-rich region in the STIM1 C terminus.
Interestingly, STIM1 loses its ability to cluster in response to store depletion during cell division (23, 24), resulting in SOCE inhibition (25–27). The mechanisms controlling this clustering inhibition are unclear and may differ between mitosis and meiosis. Smyth et al. (23) have argued that STIM1 clustering inhibition in mitosis is due to phosphorylation, whereas Yu et al. (24) have shown that phosphorylation of residues in the C-terminal end of STIM1 during meiosis does not affect STIM1 clustering. To better define the role of this C-terminal STIM1 domain in STIM1 clustering inhibition during meiosis, we generated a STIM1 deletion downstream of residue 485. Surprisingly, when expressed in Xenopus oocytes, STIM1-485 constitutively interacts with Orai1 and traps it intracellularly, hence inhibiting SOCE. STIM1-485 also disrupts the structure of the endoplasmic reticulum (ER). Replacing the C-terminal domain of STIM1(486–669) with nonspecific sequences of similar length (GFP or mCherry) produces a chimeric protein that replicates wild-type STIM1 function in terms of its ability to form puncta, translocate to the cortical ER, and bind to and gate Orai1. These results are consistent with a model in which the C-terminal end of STIM1 sterically shields the SOAR/CAD domain from constitutively interacting with Orai1 when Ca2+ stores are full. Furthermore, the STIM1-485-mCherry chimera is unable to form clusters in response to Ca2+ store depletion during meiosis, showing that inhibition of STIM1 clustering does not involve the C-terminal 200 residues of STIM1.
EXPERIMENTAL PROCEDURES
Molecular Biology
The pSGEM-mCherry-STIM1, pSGEM-mCherry-STIM1-D76A, and pSGEM-GFP-Orai1 plasmids were described previously (24). pSp64s-GFP-KDEL was a generous gift from Mark Terasaki (28). The human STIM1-YFP plasmid was obtained from Addgene (10). To construct pSGEM-mCherry-STIM1-485, an mCherry-STIM1-485 fragment was amplified by PCR using the following primers: 5′-CAATTGGCGGCCGCATGGATGTATGCGTCCGTCTTG-3′ (forward) and 5′-GAGAAGCTTCTCGAGCTACACAATCTCCTCATCCATGTCATC-3′ (reverse). The resulting PCR fragment was inserted into NotI/XhoI of pSGEM. To make pSGEM-STIM1-485-mCherry or pSGEM-STIM1-485-GFP, a STIM1-485 fragment was amplified using the following primers: 5′-CTCGAGCTCAAGCTTATGGATGTATGCGTCCGTCTTG-3′ (forward) and 5′-GAGCTCGAGAATTCGCACAATCTCCTCATCCATGTCATC-3′ (reverse). The resulting PCR fragment was inserted into XbaI/EcoRV of pSGEM in-frame with mCherry or GFP. pSGEM-STIM1-485-mCherry-polyK (amino acids 670–685 of STIM1) was generated by two mutagenesis steps with the QuikChange mutagenesis kit (Stratagene) using the following primer pairs: pair 1, 5′-ATGGACGAGCTGTACAAGGGCCGGAAGAAGTTTCCTCTCAAATAAAGCGGCCGCGACTCT-3′ (forward) and 5′-AGAGTCGCGGCCGCTTTATTTGAGAGGAAACTTCTTCCGGCCCTTGTACAGCTCGTCCAT-3′ (reverse); and pair 2, 5′-AAGAAGTTTCCTCTCAAAATCTTTAAGAAGCCTCTTAAGAAGTAAAGCGGCCGCGACTCT-3′ (forward) and 5′-AGAGTCGCGGCCGCTTTACTTCTTAAGAGGCTTCTTAAAGATTTTGAGAGGAAACTTCTT-3′ (reverse). To construct pSGEM-mCherry-KDEL, mCherry-KDEL was amplified with primers 5′-ACGCGGCCGCATGGTGAGCAAGGGCGA-3′ (forward) and 5′-GCTAGATCTTTAAAGTTCATCCTTTTTGTATAGTTCATCCATGC-3′ (reverse) and subcloned into NotI/BamHI of pSGEM. All constructs were verified by DNA sequencing or by analytical endonuclease restriction enzyme digestion. To construct pSGEM-STIM1(486–685), the STIM1(486–685) fragment was amplified using pSGEM-mCherry-STIM1 as a template and the following primers: 5′-CTCGAGAGATCTATGTCTCCCTTGTCCATGCAGTC-3′ (forward) and 5′-GAGCTCGAGAATTCGACTACTTCTTAAGAGGCTTCTTAAAG-3′ (reverse). The resulting PCR fragment was inserted into BamHI/XhoI of pSGEM.
For in vitro transcription, after linearization with NheI, capped cRNAs were transcribed using the T7 mMESSAGE mMACHINE kit (Ambion). Xenopus laevis oocytes were prepared and handled as described previously (25). Typically, for electrophysiological recording, oocytes were injected with 2 ng of RNA from GFP-Orai1 and 5 ng of RNA from Ch-STIM1 or the different mutants (STIM1-485, STIM1-485-Ch, and STIM1-485-Ch-K). For confocal imaging, cells were injected with 5 ng of GFP-KDEL RNA, 2 ng of GFP-Orai1 RNA, and/or 10 ng of STIM1 or the different mutants.
Electrophysiology
Ca2+-activated Cl− currents were used to monitor intracellular calcium elevation as described previously (25, 29, 30). Briefly, ICl-1 (activated by intracellular calcium release) is measured during a first voltage step from −30 to +40 mV. Next, a hyperpolarizing pulse to −140 mV increases the driving force for extracellular calcium entry. The third voltage step to +40 mV consequently allows the measure of the development of ICl-T that depends only on extracellular calcium influx. The value for ICl-T was corrected for the remaining ICl-1 current (end of the first pulse). To measure the SOCE current (ISOC) independently of the chloride currents, cells were injected with BAPTA (1,2-Bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid) (∼7 mm), and ISOC was measured in a solution containing 5 mm Ca2+. Current-voltage relationships were obtained using voltage ramps lasting 1.5 s from −120 to +40 mV applied every 10s. Ionomycin (10 μm) was perfused for 20 s to deplete Ca2+ stores. The amplitude of ISOC was corrected from the leak measured after inhibiting the current with 100 μm La3+. Current amplitudes are expressed as means ± S.E. Significance was tested using one-way analysis of variance, followed by Tukey's post hoc test.
Solutions
Cells were impaled with sharp microelectrodes (1–5 megohms) filled with 3 m KCl. Standard extracellular Ringer's solution contained 96 mm NaCl, 2.5 mm KCl, 1.8 mm CaCl2, 2 mm MgCl2, and 10 mm HEPES (pH 7.4). The 5 mm Ca2+ Ringer's solution to measure ISOC contained 91.2 mm NaCl, 2.5 mm KCl, 5 mm CaCl2, 2 mm MgCl2, and 10 mm HEPES (pH 7.4).
Imaging
Live cell imaging was performed on a Zeiss LSM 710 confocal using a Plan Apo 63×/1.4 oil DIC II objective. Cells were scanned in OR2 solution (82.5 mm NaCl, 2.5 mm KCl, 1 mm CaCl2, 1 mm Na2HPO4, and 5 mm HEPES (pH 7.5)) except for experiments on Ca2+ store depletion with 5 mm TPEN (Sigma) or 10 μm ionomycin (Invitrogen) in Ca2+-free OR2 solution for 5–30 min at room temperature. Images were analyzed using ZEN 2008 software (Zeiss), and figures were compiled using Adobe Photoshop.
For the time course of STIM1 punctum formation, imaging was performed on the LSM 710 confocal microscope using an EC Plan Neofluar 40×/1.30 oil DIC M27 objective with the pinhole fully open. A 744 × 744-pixel image was taken every 3 s from oocytes expressing GFP-Orai1 with STIM1-485-Ch or Ch-STIM1 or from oocytes expressing GFP-KDEL with STIM1-485-Ch-K or Ch-STIM1. Oocytes were scanned in Ca2+-free OR2 solution and store-depleted with 10 μm ionomycin. The data were collected and analyzed using ZEN2008 software. A region of interest of 300 × 300 pixels was chosen, and mCherry intensity was analyzed over time as an indicator of punctum formation.
RESULTS AND DISCUSSION
STIM1-485 Disrupts ER Structure and Interacts Constitutively with Orai1
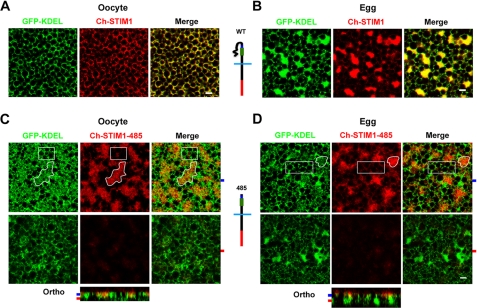
Xenopus oocytes are arrested at the G2-M transition of the cell cycle (referred to as oocytes) and progress to metaphase of meiosis II during maturation (referred to as eggs). This maturation period is essential for the egg to activate at fertilization. During meiosis, the ER restructures into large patches that are enriched in STIM1 (24, 28). This is illustrated in Fig. 1, A (oocyte) and B (egg), in cells that coexpress the ER marker GFP-KDEL and STIM1 tagged with mCherry at its N terminus (see schematic in Fig. 1, A and B). Compare the reticular ER structure observed in oocytes (Fig. 1A) with that after oocyte maturation in eggs (B), where large ER patches were formed in addition to the oocyte-like reticular ER distribution. In both oocytes and eggs, STIM1 co-localized with GFP-KDEL (Fig. 1, A and B).
FIGURE 1.
STIM1-485 disrupts ER structure. A, oocytes were injected with STIM1 (10 ng) and GFP-KDEL (5 ng) RNAs and allowed to express for 2 days. Confocal images taken deep into the cytosol show clear co-localization between the ER marker (GFP-KDEL) and STIM1. Note the reticular appearance of the ER in oocytes. B, cells were injected as described for A and then treated with progesterone to induce oocyte maturation. Eggs arrested at metaphase II of meiosis were imaged and exhibit remodeling of the ER into large patches as observed by the concurrent STIM1 and GFP-KDEL distribution. A schematic representation of the mCherry-tagged wild-type STIM1 is shown, with the SOAR/CAD domain in green, the SHD in blue, and mCherry in red. C and D, oocytes were injected with Ch-STIM1-485 (10 ng) and GFP-KDEL (5 ng) and imaged as oocytes (C) or after maturation as eggs (D). A schematic representation of Ch-STIM1-485 is also shown. Orthogonal sections (Ortho) through the z-stack of images are shown in addition to images from different focal planes, as indicated by the matching color-coded tabs to the left of the orthogonal sections. Areas of the ER that are enriched with STIM1-485 (irregular white shape, for example) show disruption of the typical reticular structure (GFP-KDEL panels), whereas areas of the ER with little STIM1-485 accumulation exhibit the normal reticular structure (white boxes). This is true in both oocytes (C) and eggs (D). Scale bars = 2 μm.
We have shown previously that activation of Cdk1 during meiosis is necessary and sufficient to down-regulate SOCE (30). Because consensus Cdk1 phosphorylation sites cluster in the C-terminal 200 residues of STIM1 and because STIM1 is hyperphosphorylated during meiosis (24), we were interested in testing the role of the C-terminal 200 residues of STIM1 in its clustering inhibition during meiosis. Alanine substitution and phosphomimetic mutations in various combinations in the Ser/Pro-rich region of STIM1 had no effect on modulating its clustering behavior during meiosis (24). Therefore, to test the role of the C-terminal 200 residues in STIM1 clustering, we generated a STIM1 deletion mutant that encompasses residues 1–485 with an mCherry tag at the luminal N terminus: Ch-STIM1-485 (see schematic in Fig. 1, C and D). Expression of STIM1-485 in oocytes resulted in disruption of ER structure (Fig. 1C), producing large amorphous structures only in areas enriched with STIM1-485. Compare the irregular white outlined shape in Fig. 1C, where STIM1-485 is enriched and ER structure is disrupted, with the white box with little STIM1-485 expression and normal ER distribution. Fig. 1C also shows an orthogonal section across the stack of z-section confocal images, with the planes displayed in the upper panel, indicated by the blue and red tabs, respectively. As illustrated in the deeper focal plane, ER regions with no significant accumulation of STIM1-485 maintain their normal structure (Fig. 1C). In eggs expressing STIM1-485, a similar disruption of ER structure was observed, with amorphous ER in areas enriched in STIM1-485 (Fig. 1D, irregular white outline) compared with areas with low STIM1-485 (white box). In addition, no ER patches were observed in areas enriched in STIM1-485 compared with the rest of the ER (Fig. 1D). As in the oocyte, deep confocal sections exhibited little STIM1-485 and normal ER structure (Fig. 1D). We interpret these data in the context of functional domains within STIM1, particularly the SHD, which has been postulated to be responsible for STIM1-STIM1 interaction following store depletion (18). STIM1-485 may have the SHD exposed, potentially leading to self-association of STIM1 molecules in trans on opposite membranes of the ER, resulting in a disrupted ER structure.
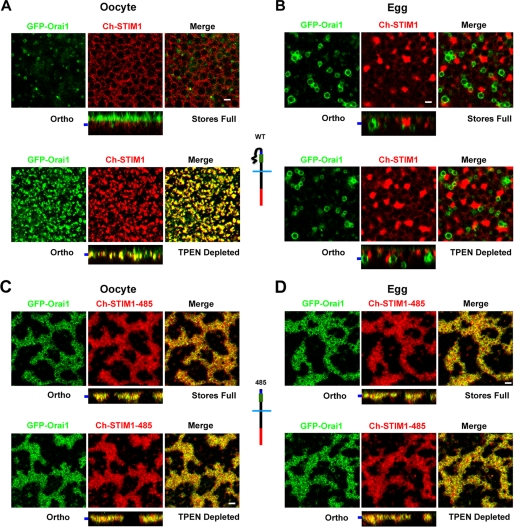
Expressing STIM1 with Orai1 shows Orai1 localization to the cell membrane (Fig. 2A, orthogonal section (Ortho)) and in recycling endosomes as illustrated in the xy focal plane view. Orai1 is removed from the cell membrane into an endosomal compartment in the egg (Fig. 2B). The recycling of Orai1 and its internalization during meiosis have been characterized previously in significant detail (24, 31). Depletion of Ca2+ stores with TPEN resulted in coincident STIM1-Orai1 puncta in oocytes (Fig. 2A) but not in eggs (Fig. 2B). Hence, store depletion in oocytes leads to STIM1 clustering into large puncta, whereas in eggs, STIM1 does not alter its subcellular distribution following store depletion (24).
FIGURE 2.
STIM1-485 constitutively associates with Orai1. A, images from oocytes expressing GFP-Orai1 and Ch-STIM1 before (upper panels) and after (lower panels) depletion with TPEN. In addition to the orthogonal sections (Ortho), focal plane images as indicated by the bar to the left of the orthogonal sections are shown. The orthogonal view of the oocyte with Ca2+ stores full (upper panels) shows Orai1 localization at cell membrane microvilli (green) and STIM1 in the ER (red). The deep focal section shows Orai1 within recycling endosomes. Depletion of Ca2+ stores with TPEN (5 mm) resulted in co-clustering of STIM1 and Orai1 in the same focal plane at the plasma membrane, as shown in the merged image and orthogonal view (lower panels). B, fully mature eggs injected and imaged as described for A. In this case, Orai1 was removed from the cell membrane and enriched in an endosomal compartment, whereas STIM1 localized to the ER, which formed patches (upper panels). Upon Ca2+ store depletion, the subcellular localization of STIM1 and Orai1 did not change, and there was no detectable interaction between STIM1 and Orai1 (lower panels). C and D, oocytes and eggs were injected with STIM1-485 and Orai1 RNAs and imaged as described for A. Confocal images and orthogonal views show that GFP-Orai1 co-localized with STIM1-485 to disrupted ER structures enriched with STIM1-485 in both oocytes and eggs. The distribution of GFP-Orai1 and Ch-STIM1-485 remained unchanged upon Ca2+ store depletion with TPEN. Schematic structures of the different constructs used are also shown. Scale bars = 2 μm.
When Orai1 was coexpressed with STIM1-485, a dramatically different picture emerged. Orai1 co-localized with STIM1-485, leading to trapping of Orai1 intracellularly and preventing its enrichment at the cell membrane (Fig. 2C). This was also true in eggs (Fig. 2D). As is the case with cells expressing STIM1-485 alone, store depletion did not significantly alter STIM1-485 distribution or Orai1 distribution (Fig. 2, C and D). The fact that STIM1-485 is capable of trapping Orai1 intracellularly shows that this mutant interacts with Orai1 in a constitutive fashion independently of store depletion, arguing that the SOAR/CAD domain is exposed and can interact constitutively with Orai1. This observation is consistent with the STIM1-485 self-association model, as it would prevent STIM1-485 from relocalizing to puncta following store depletion.
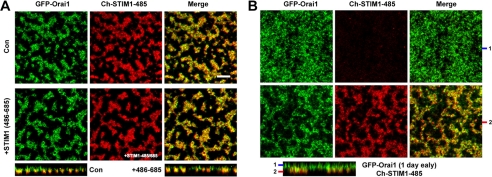
These results suggest that in the absence of the C-terminal 200 residues, STIM1-485 in oocytes can self-associate and interact with Orai1 in a constitutive fashion. To test whether the C-terminal 200 residues of STIM1 can rescue this phenotype, we overexpressed a peptide encompassing this domain of STIM1 as a separate peptide with STIM1-485 (Fig. 3A). This did not rescue the ER disruption phenotype or the constitutive interaction between STIM1-485 and Orai1 (Fig. 3A). This suggests that a soluble STIM1(486–685) fragment cannot associate with STIM1-485 to rescue its constitutive association with itself and Orai1.
FIGURE 3.
Orai1 localizes to the cell membrane when expressed prior to STIM1-485. A, overexpression of the fragment from STIM1 encompassing residues 485–685 does not rescue the ER disruption phenotype produced by STIM1-485. Oocytes were left untreated (control (Con)) or injected with RNA encoding residues 486–685 of STIM1 (20 ng) and allowed 24 h to express. Both batches were then injected with GFP-Orai1 (2 ng) and Ch-STIM1-485 (10 ng) and allowed an additional 48 h to express. Scale bar = 5 μm. B, oocytes were injected with GFP-Orai1 (2 ng) and allowed 24 h to express before injecting Ch-STIM1-485 (10 ng) and incubated for an additional 48 h. The orthogonal section and two different focal planes as indicated by the bars on the left of the orthogonal section are shown. Under those conditions, although some Orai1 was retained intracellularly, it was enriched at the cell membrane.
Our data so far argue that STIM1-485 constitutively interacts with Orai1 and traps it intracellularly. If this interpretation is correct, then expression of Orai1 before STIM1-485 should allow it to reach the cell membrane. This is indeed the case, as shown in Fig. 3B, where Orai1 expressed 1 day before STIM1-485 was enriched at the cell membrane (Fig. 3B). A proportion of the total cellular Orai1 in this case was trapped intracellularly and co-localized with STIM1-485 (Fig. 3B). This is likely due to Orai1 going through the biogenesis pathway and Orai1 recycling between the cell membrane and endosomal compartments (24, 31).
We then tested the distribution and effect on ER structure of a shorter C-terminal deletion, STIM1-535. The structure of the ER in oocytes expressing STIM1-535 was similar to that in control oocytes injected with wild-type STIM1 (supplemental Fig. 1). Furthermore, STIM1-535 responded normally to store depletion by co-clustering with Orai1 at the cell membrane focal plane. These results show that a C-terminal domain of STIM1 extending to residue 535 contains the necessary structural features necessary to maintain STIM1 in an inactive state when Ca2+ stores are full.
STIM1-485 Expression Results in an Undersized ISOC
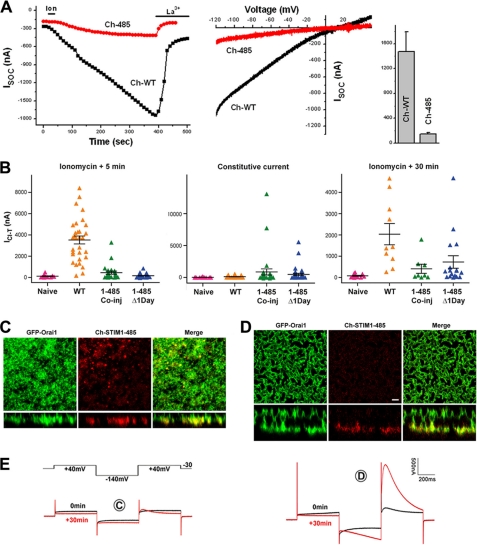
Given the subcellular localization of STIM1-485 in oocytes, it was important to characterize the effect on ISOC. Coexpression of Orai1 with STIM1-485 produced a significantly smaller ISOC than when Orai1 was expressed with full-length STIM1 (Fig. 4A). However, the STIM1-485-mediated current maintained the SOCE signature inward rectification and sensitivity to La3+ (Fig. 4A), arguing that STIM1-485 is functional but not effective. The smaller ISOC is supported by the disrupted ER phenotype following STIM1-485 expression, the ineffective formation of puncta in response to store depletion in STIM1-485-expressing cells, and the intracellular trapping of Orai1 when coexpressed with STIM1-485 (Figs. 1 and 2).
FIGURE 4.
STIM1-485 is ineffective in supporting SOCE. A, ISOC from oocytes expressing mCherry-tagged wild-type STIM1 (Ch-WT) or Ch-STIM1-485 (Ch-485). ISOC development over time, current-voltage relationships, and current amplitude (means ± S.E.) are shown (n = 5–6). B, the ICl-T was used as a readout for SOCE. See “Experimental Procedures” for additional details on ICl-T. ICl-T was measured from uninjected oocytes (Naive), oocytes injected with Orai1 plus Ch-STIM1 (WT), oocytes injected with Orai1 plus STIM1-485 (1–485 Co-inj), and oocytes that were allowed to express Orai1 for 1 day before STIM1-485 injection (1–485 Δ1Day). Shown are the currents at 5 min after store depletion with ionomycin, the constitutive ICl-T before store depletion, and 30 min after store depletion as indicated at the top of each panel. Data are presented as a scatter plot with means ± S.E. (n = 12–40). C–E, confocal images and matching current traces (E) from two representative oocytes with a small (C) or large (D) ICl-T. ICl-T was calculated as the peak transient current at the second +40-mV pulse minus the current value at the end of the first +40-mV pulse as described previously (29). Scale bar, 2 μm.
In addition, the constitutive association of STIM1-485 with Orai1 argues that with sufficient Orai1 at the cell membrane, STIM1-485 should function as a constitutively active mutant to some extent. To further explore this possibility, we used the endogenous Ca2+-activated Cl− current (ICl-T). ICl-T faithfully reproduces the dynamics and amplitude of ISOC in oocytes (29). As observed with ISOC, STIM1-485 expression produced a smaller ICl-T when coexpressed with Orai1 or when Orai1 was expressed 1 day earlier (Fig. 4B, left panel). However, in a small number of cells, a large constitutive Ca2+ influx was observed (Fig. 4B, middle panel). In addition, long-term recordings following store depletion show an increased percentage of STIM1-485 cells producing Ca2+ influx (Fig. 4B, right panel). Examples of the subcellular distribution of Orai1 and STIM1-485 from cells producing small (Fig. 4C) and large (Fig. 4D) ICl-T with the corresponding current traces (Fig. 4E) are shown. The cell with a small SOCE exhibited few STIM1-Orai1 puncta, consistent with the low current amplitude (Fig. 4C). This argues that prolonged store depletion allows a fraction of STIM1-485 to escape from the disrupted ER structures at low efficiency and form puncta to recruit Orai1 and activate ISOC. In contrast, the cell with a large ICl-T exhibited striking invaginations at the cell membrane that brought Orai1 into physical proximity to STIM1-485 in the ER, resulting in SOCE (Fig. 4, D and E). This is an interesting phenotype, which argues that, in some rare instances, invaginations of the cell membrane may be stabilized through interactions between Orai1 and STIM1-485. Together, these results suggest that STIM1-485 is still capable of sensing Ca2+ store depletion but cannot easily translocate to the cortical ER, as it appears to be trapped in amorphous ER structures (Figs. 1 and 2).
To test whether STIM1-485 interferes with full-length STIM1 function, we coexpressed both proteins in addition to Orai1 and measured ICl-T as a readout of functionality. Coexpression of full-length STIM1 with STIM1-485 did not significantly affect the levels of Ca2+ influx, with expression of full-length STIM1 alone producing an ICl-T of 5.73 ± 0.8 μA and full-length STIM1 + STIM1-485 producing a current of 4.1 ± 0.9 μA (n = 5). This argues that STIM1-485 does not interfere significantly with full-length STIM1 function.
Several groups have expressed in mammalian cells STIM1 deletions similar to STIM1-485 that are anchored in the ER membrane but have not reported any defects in ISOC levels or the subcellular distribution of these mutants (19, 21, 23). In contrast, expression of STIM1-485 in Xenopus oocytes results in a small current and disrupted ER structure. One possibility for the disparate behavior of STIM1-485 in these expression systems is the potential absence in Xenopus oocytes of accessory proteins that would interact with STIM1 deletions in mammalian cells, allowing it to function somewhat normally. This would not be unusual, as there are several precedents showing that the behavior of proteins is very much dependent on the cellular context. This is well recognized for G-protein-coupled receptors, where the same receptor links to different signaling modules depending on the cell type and expression of specific downstream interacting proteins (32). For example, expression of the pore-forming subunit of voltage-gated potassium channels in different mammalian cells leads to distinct voltage dependence and activation and inactivation kinetics because of association with an endogenous β-subunit expressed in a cell type-specific fashion (33, 34). Therefore, protein function can be modulated in a cell type-specific fashion based on the endogenous complement of interacting proteins. It is therefore not surprising that STIM1 deletions behave differently in the Xenopus oocyte versus mammalian expression systems. Nonetheless, the phenotype of STIM1-485 in Xenopus oocytes offers an opportunity to better understand STIM1 dynamics and intramolecular interactions.
Nonspecific Sequences Functionally Substitute for the STIM1 C-terminal 200 Residues
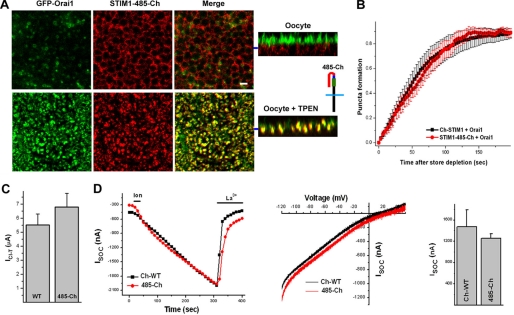
The fact that expression of the STIM1(486–685) domain does not rescue STIM1-485 suggests that this domain needs to be physically attached to the STIM1 molecule to perform its function. This raises the interesting question of whether this domain acts by physically shielding the SOAR/CAD domain and SHD and whether this effect is sequence-specific. To test this idea, we replaced this region of STIM1 with nonspecific sequences, GFP or mCherry, as both have a similar length to the STIM1 C-terminal 200 residues. Strikingly, STIM1-485-Ch with a C-terminal mCherry tag after residue 485 rescued the defects observed in the STIM1-485 deletion mutant (Fig. 5). STIM1-485-Ch did not disrupt ER structure, did not constitutively interact with Orai1, formed puncta in response to store depletion when expressed with Orai1, and was able to recruit Orai1 in a similar fashion to wild-type STIM1 (Fig. 5A). The time course of punctum formation is similar when Orai1 was expressed with either STIM1 or STIM1-485-Ch (Fig. 5B). STIM1-485-Ch produced normal SOCE when tested using either ICl-T (Fig. 5C) or ISOC (Fig. 5D) in terms of current development time course, amplitude, and current-voltage relationship. These data suggest that the C-terminal end of STIM1 maintains the SOAR/CAD domain and SHD inactive in a sequence-nonspecific fashion through a steric hindrance mechanism.
FIGURE 5.
STIM1-485-Ch coexpressed with Orai1 behaves like wild-type STIM1. A, STIM1-485-Ch was constructed by adding mCherry at the C-terminal end of STIM1-485. Coexpression of STIM1-485-Ch and Orai1 showed normal subcellular distribution of both proteins. Store depletion with TPEN led to co-clustering of STIM1-485-Ch and Orai1 in the focal plane of the cell membrane. Orthogonal sections are shown, with the focal plane of the images on the left indicated by the blue bar. Scale bar = 2 μm. B, time course of STIM1-485-Ch punctum formation as described under “Experimental Procedures” when coexpressed with Orai1 (mean ± S.E., n = 8). C, amplitude of ICl-T following the expression of full-length STIM1 (WT) or STIM1-485-Ch (485-Ch) with Orai1. D, ISOC mediated by coexpression of full-length STIM1 (Ch-WT) or STIM1-485-Ch (485-Ch) with Orai1. The SOCE development time course, current-voltage relationship, and amplitude are shown (n = 5–6).
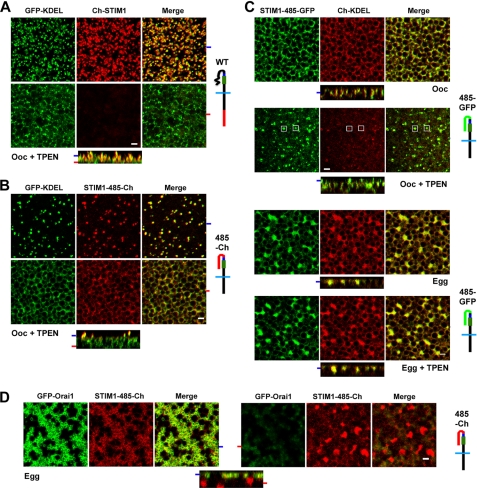
Interestingly, however, when STIM1-485-Ch was expressed without Orai1, it was not able to form stable puncta efficiently as compared with the full-length protein (Fig. 6, A and B). Following store depletion, the number of STIM1 puncta in cells expressing STIM1-485-Ch was significantly decreased (Fig. 6B) compared with cells expressing full-length STIM1 (Fig. 6A). Furthermore, a significant percentage of the STIM1-485-Ch protein remained in the deep ER and did not translocate to the cortical region (Fig. 6B). In contrast, full-length STIM1 was no longer detectable in the deep ER after store depletion (Fig. 6A). In addition, in the absence of Orai1 expression, maximal development of STIM1-485-Ch cortical puncta required ∼30 min of TPEN treatment, compared with ∼5 min when full-length STIM1 was expressed. Interestingly, however, coexpression of STIM1-485-Ch with Orai1 rescued this delay, leading to rapid punctum formation after store depletion with TPEN in ∼5 min. Similar results were observed following store depletion with ionomycin (data not shown). This phenotype is reminiscent of a STIM1 mutant missing the polybasic domain at the end of the molecule, which has been implicated in stabilizing STIM1 interactions with the cell membrane (6, 15, 35). This would argue that the delay and inefficiency in punctum formation observed with the STIM1-485-Ch mutant are due to the lack of the polybasic domain. This is indeed the case because addition of a polybasic domain at the end of STIM1-485-Ch rescued this phenotype (see below and Fig. 7).
FIGURE 6.
STIM1-485-Ch punctum formation and behavior during meiosis. A–D, oocytes (Ooc) and eggs expressing GFP-KDEL, full-length STIM1 (Ch-STIM1), STIM1-485-Ch (485-Ch), Ch-KDEL, and/or STIM1-485-GFP (485-GFP) before and after store depletion with TPEN (5 mm) as indicated. Orthogonal sections through a z-stack of images are shown in addition to images from different focal planes, as indicated by the matching tabs at the left edge of the orthogonal sections. Schematic structures of the different constructs are also shown. Scale bars = 2 μm.
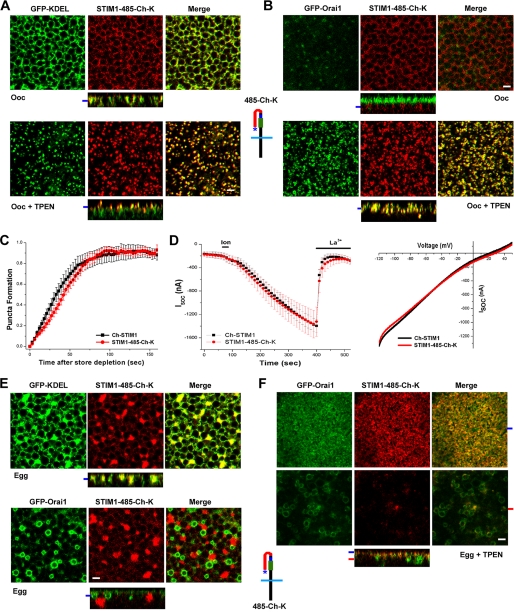
FIGURE 7.
The polylysine domain stabilizes STIM1 puncta. A, STIM1-485-Ch-K expressed with GFP-KDEL forms puncta in response to store depletion. The schematic structure of STIM1-485-Ch-K (485-Ch-K) is shown, with the polylysine domain indicated by the asterisk. B, STIM1-485-Ch-K co-clusters with Orai1 after store depletion in oocytes (Ooc). C, time course of STIM1-485-Ch-K punctum formation in oocytes expressing GFP-KDEL and STIM1-485-Ch-K (mean ± S.E., n = 8). D, ISOC from cells expressing full-length Ch-STIM1 or STIM1-485-Ch-K with Orai1. The time course of current development and current-voltage relationships are shown. E, subcellular localization of STIM1-485-Ch-K in eggs coexpressing GFP-KDEL or GFP-Orai1. F, distribution of STIM1-485-Ch-K and GFP-Orai1 following store depletion in eggs. Scale bars, 2 μm.
Substituting the STIM1 C-terminal domain (positions 486–685) with GFP also rescued the STIM1-485 defects and exhibited slow punctum formation upon expression with KDEL (Fig. 6C). Importantly, however, during meiosis, STIM1-485-GFP did not form puncta in response to store depletion (Fig. 6C, Egg+TPEN). Therefore, even in the absence of the last 200 residues of STIM1, clustering of STIM1 in response to store depletion remains inhibited, showing that clustering inhibition during meiosis is not dependent on this domain. This demonstrates that STIM1 clustering inhibition during meiosis is independent of phosphorylation within the C-terminal 200 residues of STIM1.
Interestingly, coexpression of Orai1 with STIM1-485-Ch in the egg showed trapping of Orai1 intracellularly in a manner reminiscent of that observed when STIM1-485 was expressed with Orai1 (Fig. 6D). In contrast, in the oocyte, STIM1-485-Ch behaved like full-length STIM1 (Fig. 5A). This suggests that during meiosis (egg), STIM1-485-Ch assumes a different conformation than in interphase (oocyte), leading to exposure of the SOAR/CAD domain and allowing interactions with Orai1. Consistent with these results, we have previously shown that STIM1-Orai1 interaction remains functional during meiosis (24).
To test whether the delay in punctum formation for the STIM1-485-Ch construct in the absence of Orai1 is due to the absence of the polybasic domain, we added the polylysine domain to the end of STIM1-485-Ch, thus generating a construct with residues 486–669 of STIM1 replaced with mCherry (STIM1-485-Ch-K). STIM1-485-Ch-K localized to the ER in oocytes and formed puncta with a similar time course to full-length STIM1 in the absence of Orai1 coexpression (Fig. 7, A and C). When expressed with Orai1, STIM1-485-Ch-K was able to recruit Orai1 into coincident puncta (Fig. 7B) and produced an ISOC with a similar development time course and current-voltage relationship to full-length STIM1 (Fig. 7D). During meiosis, STIM1-485-Ch-K localized to the ER and did not constitutively interact with Orai1, which was internalized and localized to endosomes (Fig. 7E) (24, 31). Therefore, addition of the polybasic domain at the end of STIM1-485-Ch is sufficient to stabilize the conformation of this construct during meiosis.
Interestingly, when Ca2+ stores were depleted in the egg, STIM1-485-Ch-K efficiently translocated to the subcell membrane focal plane (Fig. 7F). This argues that in the egg, STIM1-485-Ch-K is able to sense Ca2+ store depletion, resulting in a conformational change in the protein that exposes the polylysine domain, allowing it to interact with cell membrane lipids. However, under these conditions, STIM1-485-Ch-K did not form puncta, and a small percentage of the protein interacted with Orai1 in the endosomal compartment (Fig. 7F). This is consistent with the idea that the SOAR/CAD domain becomes available for interaction with Orai1. The lack of punctum formation demonstrates that STIM1 clustering inhibition is due to modifications outside domain 486–669, again supporting the conclusion that phosphorylation within this domain does not contribute to STIM1 clustering inhibition during meiosis.
In summary, our data support a model in which STIM1 is maintained in an inactive state in interphase with full Ca2+ stores through intramolecular shielding of the active domains (SOAR/CAD and SHD) by the C-terminal end of the molecule (positions 486–669). Surprisingly, replacing these residues (positions 486–669) with GFP or mCherry does not affect the function of STIM1, including its ability to sense Ca2+ store depletion, form puncta, translocate to the cortical ER, and bind to and gate Orai1. This would be consistent with a steric hindrance model. We have further shown that STIM1 clustering inhibition during meiosis does not require the C-terminal 200 residues of STIM1 and as such cannot be dependent on any modifications within this domain.
Supplementary Material
Acknowledgment
We thank the team of the genomics core at Weill Cornell Medical College in Qatar for sequencing various constructs.
This work was supported by Qatar National Research Fund Grants NPRP08-395-3-088 and NPRP08-138-3-050 and the Biomedical Research Program funds at Weill Cornell Medical College, a program funded by the Qatar Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- SOCE
- store-operated Ca2+ entry
- ER
- endoplasmic reticulum
- SHD
- STIM1 homomerization domain
- Ch
- mCherry
- K
- Lysine
- SOAR
- STIM1 Orai Activating Region
- CAD
- CRAC Activation Domain
- TPEN
- N,N,N′,N′-Tetrakis(2-pyridylmethyl)ethylenediamine.
REFERENCES
- 1. Parekh A. B., Penner R. (1997) Physiol. Rev. 77, 901–930 [DOI] [PubMed] [Google Scholar]
- 2. Parekh A. B., Putney J. W., Jr. (2005) Physiol. Rev. 85, 757–810 [DOI] [PubMed] [Google Scholar]
- 3. Hogan P. G., Lewis R. S., Rao A. (2010) Annu. Rev. Immunol. 28, 491–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liou J., Kim M. L., Heo W. D., Jones J. T., Myers J. W., Ferrell J. E., Jr., Meyer T. (2005) Curr. Biol. 15, 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roos J., DiGregorio P. J., Yeromin A. V., Ohlsen K., Lioudyno M., Zhang S., Safrina O., Kozak J. A., Wagner S. L., Cahalan M. D., Veliçelebi G., Stauderman K. A. (2005) J. Cell Biol. 169, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liou J., Fivaz M., Inoue T., Meyer T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9301–9306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stathopulos P. B., Li G. Y., Plevin M. J., Ames J. B., Ikura M. (2006) J. Biol. Chem. 281, 35855–35862 [DOI] [PubMed] [Google Scholar]
- 8. Vig M., Peinelt C., Beck A., Koomoa D. L., Rabah D., Koblan-Huberson M., Kraft S., Turner H., Fleig A., Penner R., Kinet J. P. (2006) Science 312, 1220–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luik R. M., Wu M. M., Buchanan J., Lewis R. S. (2006) J. Cell Biol. 174, 815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Prakriya M., Feske S., Gwack Y., Srikanth S., Rao A., Hogan P. G. (2006) Nature 443, 230–233 [DOI] [PubMed] [Google Scholar]
- 11. Wu M. M., Buchanan J., Luik R. M., Lewis R. S. (2006) J. Cell Biol. 174, 803–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yeromin A. V., Zhang S. L., Jiang W., Yu Y., Safrina O., Cahalan M. D. (2006) Nature 443, 226–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stathopulos P. B., Zheng L., Li G. Y., Plevin M. J., Ikura M. (2008) Cell 135, 110–122 [DOI] [PubMed] [Google Scholar]
- 14. Fahrner M., Muik M., Derler I., Schindl R., Fritsch R., Frischauf I., Romanin C. (2009) Immunol. Rev. 231, 99–112 [DOI] [PubMed] [Google Scholar]
- 15. Park C. Y., Hoover P. J., Mullins F. M., Bachhawat P., Covington E. D., Raunser S., Walz T., Garcia K. C., Dolmetsch R. E., Lewis R. S. (2009) Cell 136, 876–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yuan J. P., Zeng W., Dorwart M. R., Choi Y. J., Worley P. F., Muallem S. (2009) Nat. Cell Biol. 11, 337–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kawasaki T., Lange I., Feske S. (2009) Biochem. Biophys. Res. Commun. 385, 49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muik M., Fahrner M., Derler I., Schindl R., Bergsmann J., Frischauf I., Groschner K., Romanin C. (2009) J. Biol. Chem. 284, 8421–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mullins F. M., Park C. Y., Dolmetsch R. E., Lewis R. S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15495–15500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee K. P., Yuan J. P., Zeng W., So I., Worley P. F., Muallem S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14687–14692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Derler I., Fahrner M., Muik M., Lackner B., Schindl R., Groschner K., Romanin C. (2009) J. Biol. Chem. 284, 24933–24938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang G. N., Zeng W., Kim J. Y., Yuan J. P., Han L., Muallem S., Worley P. F. (2006) Nat. Cell Biol. 8, 1003–1010 [DOI] [PubMed] [Google Scholar]
- 23. Smyth J. T., Petranka J. G., Boyles R. R., DeHaven W. I., Fukushima M., Johnson K. L., Williams J. G., Putney J. W., Jr. (2009) Nat. Cell Biol. 11, 1465–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu F., Sun L., Machaca K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17401–17406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Machaca K., Haun S. (2000) J. Biol. Chem. 275, 38710–38715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Volpi M., Berlin R. D. (1988) J. Cell Biol. 107, 2533–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tani D., Monteilh-Zoller M. K., Fleig A., Penner R. (2007) Cell Calcium 41, 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Terasaki M., Runft L. L., Hand A. R. (2001) Mol. Biol. Cell 12, 1103–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Machaca K., Hartzell H. C. (1999) J. Gen. Physiol. 113, 249–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Machaca K., Haun S. (2002) J. Cell Biol. 156, 75–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu F., Sun L., Machaca K. (2010) J. Cell Biol. 191, 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wettschureck N., Offermanns S. (2005) Physiol. Rev. 85, 1159–1204 [DOI] [PubMed] [Google Scholar]
- 33. Uebele V. N., England S. K., Chaudhary A., Tamkun M. M., Snyders D. J. (1996) J. Biol. Chem. 271, 2406–2412 [DOI] [PubMed] [Google Scholar]
- 34. Petersen K. R., Nerbonne J. M. (1999) Pflugers Arch. 437, 381–392 [DOI] [PubMed] [Google Scholar]
- 35. Walsh C. M., Chvanov M., Haynes L. P., Petersen O. H., Tepikin A. V., Burgoyne R. D. (2010) Biochem. J. 425, 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.