Abstract
The ATP-binding cassette (ABC) transporter ABCB6 is a mitochondrial porphyrin transporter that activates porphyrin biosynthesis. ABCB6 lacks a canonical mitochondrial targeting sequence but reportedly traffics to other cellular compartments such as the plasma membrane. How ABCB6 reaches these destinations is unknown. In this study, we show that endogenous ABCB6 is glycosylated in multiple cell types, indicating trafficking through the endoplasmic reticulum (ER), and has only one atypical site for glycosylation (NXC) in its amino terminus. ABCB6 remained glycosylated when the highly conserved cysteine (Cys-8) was substituted with serine to make a consensus site, NXS. However, this substitution blocked ER exit and produced ABCB6 degradation, which was mostly reversed by the proteasomal inhibitor MG132. The amino terminus of ABCB6 has an additional highly conserved ER luminal cysteine (Cys-26). When Cys-26 was mutated alone or in combination with Cys-8, it also resulted in instability and ER retention. Further analysis revealed that these two cysteines form a disulfide bond. We discovered that other ABC transporters with an amino terminus in the ER had similarly configured conserved cysteines. This analysis led to the discovery of a disease-causing mutation in the sulfonylurea receptor 1 (SUR1)/ABCC8 from a patient with hyperinsulinemic hypoglycemia. The mutant allele only contains a mutation in a conserved amino-terminal cysteine, producing SUR1 that fails to reach the cell surface. These results suggest that for ABC transporters the propensity to form a disulfide bond in the ER defines a unique checkpoint that determines whether a protein is ER-retained.
Keywords: ABC Transporter, Disulfide, Membrane Proteins, Protein Degradation, Protein Folding, Protein Motifs, Protein Stability
Introduction
ATP-binding cassette (ABC)2 transporters utilize ATP to facilitate the transmembrane movement of a variety of biologically important molecules (1). ABC transporters are required for many essential biological processes such as heme biosynthesis, [Fe-S] cluster formation, antigen presentation, and insulin secretion. Some point mutations in ABC genes produce only alterations in substrate specificity (e.g. P-glycoprotein (ABCB1) and ABCG2), whereas others cause profound conformational changes producing defects in trafficking (e.g. Δ508-CFTR) (2, 3). The endoplasmic reticulum (ER) has a protein quality control system that monitors conformational changes in proteins. Membrane proteins have multiple domains (cytoplasmic, ER lumen, and membrane-spanning) that are recognized by the ER and are currently being elucidated. Some of these domains may determine the fate of a proteins within the ER (e.g. retention or degradation), and it is likely that ABC transporters contain characteristic domains determining their fate.
Recent studies suggested that the ER contains multiple protein “quality control” checkpoints, each with defined criteria for recognizing protein folding (4–6). In membrane proteins, an initial ER checkpoint appears to require interrogation of the cytoplasmic domains to scan for lesions in folding that could activate protein degradation processes. A second checkpoint monitors domains located in the ER lumen. Some post-translational modifications occurring in the ER (e.g. disulfide bond formation) may be required by a domain to ensure proper folding, avoid activation of a checkpoint, and escape ER-associated protein degradation. Typically, disulfide bonds are maintained by the oxidizing environment of the ER and protein-disulfide isomerases. If the necessary disulfide bonds are not formed, the proteins might be retained in the ER and then degraded (7). Among the ABC transporters, it was discovered that ABCG2 contains an intramolecular disulfide bond, which is critical for protein stability (8). In the absence of one of the cysteines in the disulfide bond, ABCG2 was retained in the ER and then degraded. One explanation for ER retention and degradation of ABCG2 may be the presence of the “free” cysteine thiols. Unpaired cysteine thiols have been shown to produce ER retention of adiponectin (9) and IgM (10–12), and this process is referred to as “thiol retention.”
Our previous studies demonstrated that the porphyrin transporter ABCB6 is a homodimer that localizes to the mitochondrial outer membrane where it facilitates heme biosynthesis (13). Although others have also shown mitochondrial localization of ABCB6 (14, 15), it has also been reported to localize to the plasma membrane (15). One explanation for this could be cell context. It is not known whether ABCB6 traffics the same in all cells or if its pattern of trafficking depends upon cell type. Previous studies in overexpression systems have shown that ABCB6 contains N-glycans, indicating that ABCB6 traffics to the ER (15–17). In this study, we extended this to show that endogenous ABCB6 is a glycoprotein. Notably, we demonstrate that ABCB6 uses a single atypical but evolutionarily conserved glycosylation site (NXC) site. Although these NXC glycosylation sites are rare in the glycoproteome (18), this ER luminal cysteine (Cys-8) in ABCB6 is not required for glycosylation; however, it is required for ABCB6 stability because it forms a disulfide bond with another conserved luminal cysteine (Cys-26), which is also required for ABCB6 stability. Upon close examination of ABCC subfamily members with an amino terminus predicted to localize in the ER lumen, we discovered similar conserved cysteines that were separated by non-conserved amino acids. The biological significance of these cysteines was then demonstrated for sulfonylurea receptor 1 (SUR1)/ABCC8 (19, 20) because we discovered a single mutation in one of these cysteines in SUR1/ABCC8 from a patient with a defective SUR1-regulated K+ channel. In addition, we show that MSD0 of MRP1, which has been shown to traffic to the plasma membrane (21), requires the amino-terminal cysteines for proper trafficking. In total, our studies reveal that, in ABC transporters, conserved ER-localized cysteines form an important ER checkpoint.
EXPERIMENTAL PROCEDURES
Cell Culture
NIH3T3, HEK293, Mel, and K562 cells were maintained in DMEM containing 4500 mg/liter glucose, 10% FBS (HyClone, Logan, UT), 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin in humidified 5% CO2 at 37 °C. NIH3T3 or HEK293 cells were transiently transfected with expression plasmids by using Lipofectamine Plus (Invitrogen) according to the manufacturer's protocols. Where specified, NIH3T3 cells were incubated with 50 μg/ml cycloheximide, 10 μm MG132, or 25 mm NH4Cl for indicated durations. K562 cells were transduced with MSCV-IRES-GFP containing no insert or inserts encoding ABCB6-FLAG, ABCB6-C26A-FLAG, or ABCB6-Walker A mutant-FLAG by retroviral gene transfer (13). Protoporphyrin IX (PPIX) levels were measured in K562 cells with high GFP signal by FACS flow cytometry (13).
Site-directed Mutagenesis and Sequence Analysis
Analysis of gene sequence conservation was performed by using LASERGENE 7.2 software (DNASTAR Inc., Madison, WI). Plasmids pcDNA3-hABCB6-FLAG and pcDNA3.1-hABCB6-V5-His were designed to express human ABCB6 with a carboxyl-terminal tag (FLAG or V5, respectively) as described (13). A point mutation was introduced into pcDNA3.1-hABCB6-V5-His by using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and primers listed in the supplemental table. All genes were sequenced after mutagenesis. The pcDNA3.1-hABCB6-C8S/C26A-V5-His was generated by introducing a point mutation into pcDNA3.1-hABCB6-C8S-V5-His at Cys-26. To generate truncated mutant constructs to determine disulfide bond formation, point mutations were introduced into construct pcDNA3.1-hABCB6-V5 or pcDNA3.1-hABCB6-C8S-V5 at Cys-50 and then at Cys-120 using primers listed in the supplemental table. The region encoding the amino-terminal 210 amino acids was then amplified with a FLAG tag at the carboxyl terminus from pcDNA3.1-hABCB6-C50A/C120A-V5 or pcDNA3.1-hABCB6-C8S/C50A/C120A-V5 by using primers (sense, 5′-GCCATGGTGACTGTGGGCAACTACTGCGAGGCCG-3′; antisense, 5′-CCTACTTATCGTCGTCATCCTTGTAATCACGAAGTCCAGGGGCCCAGAG-3′) under the following conditions: heating at 94 °C for 4 min, 30 cycles of denaturation at 94 °C for 30 s, annealing at 56 °C for 30 s, and reaction at 68 °C for 1 min. The PCR product was purified from an agarose gel and TA-cloned into pCR2.1-TOPO vector (Invitrogen). The inserts were subcloned into pcDNA3 using HindIII/XbaI sites to obtain pcDNA3.1-hABCB6-C50A/C120AN1–210-FLAG or pcDNA3.1-hABCB6-C8S/C50A/C120AN1–210-FLAG constructs.
The amino-terminal 203-amino acid segment of hMRP1 (hMRP1-MSD0) (21) was cloned from a pool of cDNA generated from K562 cells by using PCR with the following primers: sense, 5′-GCCACCGGCATGGCGCTCCGGGGCTTC-3′; antisense, 5′-CGTGGATGGTTTCCGAGAACAGGGGTGA-3′. The purified product was TA-cloned into pcDNA3.1/CT-GFP-TOPO (Invitrogen) to generate the pcDNA3.1-hMRP1-MSD0-GFP construct. Using this wild-type construct as a template, we introduced a point mutation at Cys-32 to generate hMRP1-MSD0-C32A-GFP by using the QuikChange mutagenesis kit and the primers listed in the supplemental table. Oligonucleotide synthesis and DNA sequencing were performed by Hartwell Center for Bioinformatics and Biotechnology (St. Jude Children's Research Hospital).
Immunoblotting
Twenty-four hours after transfection, cells were washed with PBS, scraped into 1 ml of cold PBS containing 1× protease inhibitor mixture (Complete EDTA-free, Roche Applied Science) and 10 mm N-ethylmaleimide where indicated, pelleted by centrifugation at 1,000 × g for 4 min at 4 °C, and solubilized in buffer A (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 10% glycerol, 1% Nonidet P40, and 1× commercial protease inhibitor mixture) or Nonidet P-40 lysis buffer containing 10 mm N-ethylmaleimide. Cell lysates were centrifuged at 17,000 × g for 15 min at 4 °C to remove cell debris. For immunoblotting, Laemmli sample buffer containing β-mercaptoethanol or DTT (where indicated) was added to the supernatant followed by addition of N-ethylmaleimide. The samples were fractionated by SDS-PAGE on a 7.5, 10, or 12.5% gel. Proteins were transferred to a nitrocellulose membrane (GE Healthcare) and immunoblotted as described previously (13) using anti-GFP-HRP (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), anti-FLAG (M2 or polyclonal, Sigma), anti-V5 polyclonal (Medical Biological Laboratories Co., Ltd., Nagoya, Japan), anti-ABCB6 monoclonal (developed in our laboratory), anti-ABCB6 polyclonal (Rockland Inc., Gilbertsville, PA), or anti-apoptosis inducing factor (AIF) polyclonal (Chemicon International, Billerica, MA) antibody. The intensity of the bands was quantified using ImageJ.3 A non-linear regression curve was generated using GraphPad Prism 5 (GraphPad Software, La Jolla, CA) to estimate half-life in the experiments using cycloheximide.
Glycosidase Digestion
Lysates prepared as described above from NIH3T3, K562, or Mel cells were denatured in 1× denaturing buffer (0.5% SDS and 1% β-mercaptoethanol) and incubated at 37 °C for 1 h in reaction buffer (50 mm Na2PO4 (pH 7.5) and 1% Nonidet P-40) with or without PNGase F (New England Biolabs, Beverly, MA) according to the manufacturer's recommendations. For Endo H digestion, the denatured samples were incubated with or without the enzyme in 50 mm sodium citrate buffer. Crude mitochondria prepared from liver from C57Bl/6/129 female mice (2–6 months of age) using MITOISO1 (Sigma) were treated with glycosidases as described as above. The Abcb6 knock-out mouse line was established in our laboratory and will be described in details elsewhere. Where indicated, Mel cells were treated with DMSO (vehicle) or 1 μg/ml brefeldin A for 24 h prior to mitochondrial preparations.
Pulse-Chase
K562 vector or ABCB6-FLAG cells (∼2 × 106 cells) were washed with warm 1× Hanks' buffer and incubated in labeling medium comprising DMEM without l-methionine or l-cysteine (Cellgro, Manassas, VA), 10% dialyzed FBS (HyClone), and 2 mm l-glutamine containing 0.1 mCi/ml 35S (Tran35S-label; l-[35S]methionine and l-[35S]cysteine, 1175 Ci/mmol; MP Biomedicals, Inc., Irvine, CA) for 20 or 5 min at 37 °C. Cells were then washed once with chase medium (complete DMEM supplemented with 2 mm l-methionine and l-cysteine) and chased for the times indicated in the figures. At the end of each chase time interval, cells were washed with cold PBS, pelleted, flash frozen in liquid N2, and stored at −80 °C until use. Cells were lysed in Nonidet P-40 lysis buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.5% deoxycholate, and 0.5% Nonidet P-40) for 15 min on ice, and cell debris were removed by centrifugation at 15,000 × g for 10 min at 4 °C. The lysate was incubated with 20 μl of anti-FLAG M2 antibody-conjugated agarose (50% slurry; Sigma) for 2 h at 4 °C with constant rotation. The beads were washed three times with washing buffer (50 mm Tris-HCl (pH 7.5), 400 mm NaCl, 0.5% deoxycholate, and 0.5% Nonidet P-40) and then resuspended in 2× Laemmli SDS sample buffer containing β-mercaptoethanol. Samples were separated by SDS-PAGE. The gel was fixed, stained with Coomassie G-250 (GelCode Blue, Pierce), and incubated with Amplify fluorographic reagent (Amersham Biosciences, GE Healthcare). 35S-Labeled proteins were detected by phosphorimaging (Storm 860, GE Healthcare). Where quantification was required, the immunoprecipitated samples were treated with PNGase F prior to the addition of Laemmli sample buffer to remove all the glycans.
Indirect Immunofluorescence Microscopy
HEK293 cells were seeded on poly-l-lysine-coated coverslips 24 h before transfection with plasmid DNA (50 ng) encoding hMRP1-MSD0-GFP or hMRP1-MSD0-C32A-GFP by using Lipofectamine Plus according to the manufacturer's protocols. Cells were washed with PBS 24 h post-transfection, fixed with 4% paraformaldehyde in phosphate buffer (pH 7.4), washed with phosphate buffer (pH 7.4), and permeabilized with 0.1% Triton X-100 in phosphate buffer (pH 7.4) for 10 min. Coverslips were washed again with phosphate buffer (pH 7.4), incubated with blocking buffer (2% normal goat serum in PBS), and incubated with rabbit anti-protein-disulfide isomerase antibodies (Stressgen, Ann Arbor, MI) diluted in blocking buffer. After extensive washing with PBS, coverslips were incubated with Alexa Fluor 555-conjugated goat anti-rabbit IgG antibodies (Invitrogen) in blocking buffer, washed with PBS, stained with DAPI, and mounted on slides with ProLong Gold (Invitrogen). Fluorescence signals were detected and acquired by using a Zeiss LSM 510 Meta confocal microscope system.
RESULTS AND DISCUSSION
ABCB6 Is Glycosylated at Atypical NXC Site
We have previously demonstrated that ABCB6 expression increases during erythroid differentiation and that overexpression of ABCB6 increases porphyrin synthesis (13). In one of our models (the erythroid progenitor Mel cells), we extended this to show that erythroid differentiation by hexamethylene bisacetamide produced the expected increase in endogenous ABCB6 expression, which is highly correlated with increased intracellular PPIX concentrations (supplemental Fig. 1A). An analysis of ABCB6 expression suggested glycosylation (in erythroid and non-erythroid cell types) based upon the formation of a broad band on immunoblots as well as previous results with exogenously expressed ABCB6 (15–17); therefore, we tested whether endogenous ABCB6 is also modified by N-linked glycans. To analyze endogenous ABCB6, mitochondria were purified from Mel cells and mouse liver and treated with either Endo H, which only reacts with the high mannose-containing proteins that remain in the ER, or PNGase F to remove high mannose and complex glycans from asparagine residues (22–26). Immunoblot analysis revealed faster migration of ABCB6 only after PNGase F but not Endo H treatment (Fig. 1, A and B), indicating that ABCB6 is modified by N-glycans and traffics through the ER to the Golgi. The glycanase sensitivity of a previously established functional tagged ABCB6 (13) after either stable expression in Mel or K562 cells or transient expression in NIH3T3 cells demonstrated a similar sensitivity to PNGase F (supplemental Fig. 1, B, C, and D). Moreover, increasing the amount of Endo H by 4-fold did not produce cleavage of N-glycans from ABCB6 (supplemental Fig. 1B). Notably, inherent differences in glycosylation between human and mouse cells were demonstrated when the same ABCB6 expression vector was introduced into either murine (Mel) or human (K562) cells (Fig. 1C). Using tunicamycin to disrupt the first step in N-glycosylation provided additional confirmation that ABCB6 is a glycoprotein (Fig. 1D). We further tested the requirement for ER to Golgi trafficking in the maturation of ABCB6 by using brefeldin A to disrupt ER to Golgi transit (27). Brefeldin A prevented the formation of Endo H-resistant mature ABCB6 in Mel cells, further demonstrating the requirement for Golgi-mediated processing and maturation of the glycan status of ABCB6 (Fig. 1E). Pulse-chase analysis revealed that the conversion of ABCB6 to the 94-kDa mature form occurred over 1–2 h (Fig. 1F). A PROSITE motif search (28) identified four potential consensus NX(S/T) (29) N-glycosylation sites in the ABCB6 sequence (Fig. 2A). However, the available topological predictions (17, 30, 31) (supplemental methods) place these sites in the cytosol or in the transmembrane helices but not in ER lumen where N-glycosylation occurs, suggesting that these sites are unlikely to be used (supplemental Fig. 2A). To rule out these consensus NX(S/T) sites as the potential N-glycosylation sites, we performed site-directed mutagenesis yielding glutamine substitution at the asparagines in each of the four consensus glycosylation motifs and at both residues 447 and 498 (supplemental Fig. 2) or at all four consensus residues (“Q4” mutant) (Fig. 2B). The wild-type and mutant ABCB6 expression plasmids were transiently expressed in NIH3T3 cells. The Q4 mutant had a migration pattern identical to that of wild-type ABCB6 in the absence of glycanase treatment. The sensitivity of the Q4 mutant to PNGase F (Fig. 2B) indicates that other non-consensus glycosylation site(s) exist, which is supported by the topological predictions. Moreover, the acquisition of Endo H resistance indicates that the Q4 mutant leaves the ER.
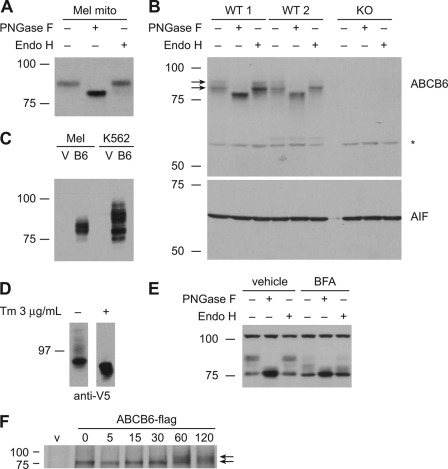
FIGURE 1.
ABCB6 is glycosylated and traffics through ER. Crude mitochondrial (mito) preparations from murine erythroleukemia (Mel) cells (A) and liver isolated from two wild-type (WT) mice and one Abcb6 knock-out (KO) mouse (B) were treated with PNGase F or Endo H, and ABCB6 was detected by immunoblotting using an anti-ABCB6 antibody. Immunoblotting was performed on one set of samples with biological duplicates. Apoptosis inducing factor (AIF) is shown as a control for equal loading of the proteins. An asterisk indicates nonspecific bands that appear both in WT and knock-out. C, analysis of hABCB6-FLAG in crude mitochondria preparations from Mel (mouse) and K562 (human) cells using an anti-FLAG antibody shows different glycosidation patterns. D, ABCB6 was no longer glycosylated when K562 cells expressing ABCB6-V5 were treated with tunicamycin. An immunoblot from a representative experiment is shown. E, crude mitochondrial preparations from Mel cells treated with vehicle (DMSO) or 1 μg/ml brefeldin A (BFA) for 24 h were analyzed for glycosidase sensitivity using an anti-ABCB6 antibody. F, pulse-chase assay using K562 cells expressing ABCB6-FLAG showed the glycosylated ABCB6 at 60 min. Tm, tunicamycin. Images are from at least two independent experiments unless otherwise noted.
FIGURE 2.
ABCB6 is modified at single conserved atypical glycosylation site. A, ABCB6 contains four consensus N-glycosylation (NX(S/T)) motifs and a single atypical N-glycosylation motif. B, ABCB6 with amino acid substitution of all four asparagines in the consensus motif (Q4) was analyzed for PNGase F and Endo H sensitivity by immunoblotting using anti-V5 antibody. The presence of glycans was detected by protein mobility shifts. The migration patterns of all mutants were similar to those of the wild-type protein. C, NIH3T3 cells were transiently transfected with single mutant N6Q, Q4, or Q5 in which glutamine was substituted for all five asparagine residues. Cell lysates were treated with the indicated glycosidases and immunoblotted with anti-V5 antibody (lower arrow, non-glycosylated ABCB6; upper arrow, glycosylated ABCB6). D, multiple sequence alignment of ABCB6 shows two conserved cysteines (red boxes) and one N-glycosylation site (green box) among the species. Each experimental result was independently performed in all iterations shown at least twice with a single representative blot from one experiment used. vec, vector; H.sapiens, Homo sapiens; P.troglodytes, Pan troglodytes; B.taurus, Bos taurus; M.musculus, Mus musculus; R.norvegicus, Rattus norvegicus; D.rerio, Danio rerio. Pro, protein (amino acid) sequence.
An atypical N-glycosylation motif containing a cysteine residue (NXC rather than the conventional NX(S/T)) has been reported in a few proteins (18, 32–36). We identified one such atypical motif (NYC) starting at position 6 in the ABCB6 amino terminus. Glutamine was substituted for Asn-6 in the wild-type and Q4 ABCB6 expression plasmids to generate N6Q and Q5 ABCB6, both of which showed faster electrophoretic mobility than wild-type ABCB6. Moreover, their mobility was identical to that of the PNGase F-treated wild-type protein and was unchanged by either PNGase F or Endo H treatment (Fig. 2C), indicating that this atypical site is the sole glycosylation site in ABCB6. This finding also supports that the ABCB6 amino terminus is in the ER lumen (17). Although the cysteine residue is highly conserved among human, chimpanzee, mouse, rat, and zebrafish ABCB6, the asparagine residue for glycosylation (NXC) is not conserved in zebrafish (Fig. 2D).
Cys-8 Is Dispensable for Glycosylation but Critical to Avoid ER Retention and Proteasomally Mediated Degradation
Other proteins reported to have an atypical NXC have additional typical glycosylation sites; thus, ABCB6 with only one such atypical site presented an opportunity to test whether the cysteine in NXC was required for glycosylation. We developed the ABCB6 substitution mutants described in Fig. 3A. The cysteine residue was substituted with a serine residue to form the consensus glycosylation site NXS (labeled C8S). The C8S mutant was transiently expressed in NIH3T3 cells, and glycosylation status was evaluated by PNGase F treatment. The ABCB6-C8S was PNGase F-sensitive, indicating that glycosylation does not require the cysteine (Fig. 3A, right panel). As a control, we showed that glycine substitution to form a non-consensus site caused a loss of glycosylation, which demonstrates that ABCB6 glycosylation requires either the consensus NX(S/T) or NXC motif. Unexpectedly, substitution of this cysteine resulted in decreased protein levels despite glycosylation (Fig. 3, A and B) as well as ER retention as shown by Endo H sensitivity (Fig. 3C). Notably, the point mutation disrupting the Walker A (13) did not result in reduced ABCB6 expression. Our multiple sequence alignment of ABCB6 homologs (Fig. 2D) revealed a conserved downstream cysteine residue at position 26, and a transmembrane (TM) helix prediction algorithm, TMHMM, predicted that Cys-26 is in the ER lumen (30). Proteins containing free cysteine thiol residues in the ER lumen are one cause of ER retention (11) and may explain the ER retention of ABCG2 after mutation of one cysteine in an intramolecular disulfide bond created a free thiol (8). Therefore, we next determined whether ABCB6 engineered to contain free thiols in the ER was ER-retained by generating the ABCB6 mutants C26A and C26S as well the mutants lacking both cysteine residues, C8S/C26S. Both the single cysteine mutants and the double mutants, which now lack a thiol in the ER lumen, were Endo H-sensitive, indicating ER retention. This finding excludes free thiols in ABCB6 as a mechanism of ER retention. Nonetheless, the expression of these cysteine mutants was reduced (Fig. 3, B and C), indicating that both Cys-8 and Cys-26 are required for maximal ABCB6 expression. We next hypothesized that we could rescue the trafficking defect and restore protein expression by adding a cysteine residue downstream from the serine now at position 8; therefore, a cysteine residue was inserted between Glu-9 and Ala-10 to create C8S/C10insertion mutant. However, this construct failed to restore maximal ABCB6 protein expression and was also ER-retained as shown by Endo H sensitivity. This finding indicates that either the spacing between the two cysteines and/or their context is important for expression (Fig. 3C).
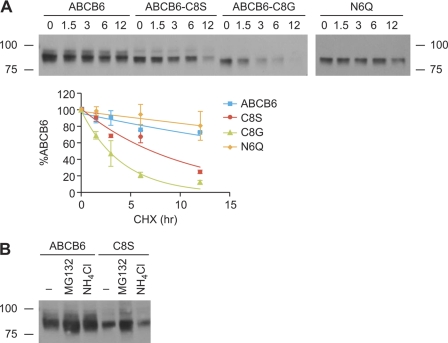
FIGURE 3.
Cys-8 is dispensable for glycosylation but is unstable. A, Cys-8 was substituted with serine or glycine, and the mutant proteins were transiently expressed in NIH3T3 cells. The glycan modification of the mutants was determined by PNGase F sensitivity and analyzed by immunoblotting using anti-V5 antibody. ABCB6 was still glycosylated when Cys-8 was substituted with Ser to make a consensus N-glycosylation motif. B, K562 cells were transduced with plasmids containing IRES-GFP and wild-type ABCB6-, Walker A mutant (mt)-, C8S-, or C26A-FLAG. Cells were sorted for GFP fluorescence by FACS and analyzed by immunoblotting using anti-FLAG antibody for the expression of ABCB6 proteins. Cysteine mutants C8S and C26A were expressed at low levels. An immunoblot from a single experiment is shown. C, serine was substituted for Cys-8 and/or Cys-26 in ABCB6, or a cysteine residue was inserted between positions 9 and 10 in the C8S mutant (C8S/C10in). Endo H sensitivity indicates that all cysteine mutants failed to exit the ER. Cys-10 insertion did not restore the protein expression level nor the impaired ER exit. D, K562 cells stably expressing wild-type ABCB6, C8S, or C26A were labeled with [35S]Met/Cys for 5 min, washed, and chased for the indicated times. Cells were harvested, and immunoprecipitated FLAG-tagged proteins were separated by SDS-PAGE and detected by phosphorimaging. The amount of ABCB6 proteins was analyzed by densitometry and plotted as percentage of ABCB6 protein at 0 h for each construct. Values shown are mean with the S.D. indicated by the error bars from three independent experiments. Unless otherwise noted, all experiments were repeated at least twice. A representative image from one complete experiment is shown.
To determine whether ABCB6 with cysteine substitutions at either position 8 or 26 was unstable, we performed pulse-chase experiments using K562 cells expressing either ABCB6, C8S, or C26A. The half-life for wild-type ABCB6 could not be estimated but appeared to be greater than 24 h. The initial rise in labeling of wild-type ABCB6 was unexpected but may be due to its long half-life. In contrast, both C8S-ABCB6 and C26A-ABCB6 had almost identical decay curves with an estimated half-life of 9.4 and 8.0 h, respectively (Fig. 3D). Collectively, these results demonstrate that loss of either Cys-8 or Cys-26 renders ABCB6 unstable; both mutants exhibit kinetically similar degradation curves, suggesting a similar mechanism.
We next determined whether N-glycosylation altered the stability of ABCB6. We compared wild-type ABCB6 and ABCB6 mutants lacking either Cys-8 (C8S), N-glycosylation (N6Q), or both (C8G). Cells were transiently transfected with either wild-type ABCB6 or ABCB6 harboring either the C8S or C8G substitution. At 17 h post-transfection, cycloheximide was added to inhibit protein synthesis, and cells were harvested at the indicated times (Fig. 4A). ABCB6 protein expression was evaluated by immunoblotting. Notably, the degradation of wild type assessed by this method confirms the pulse-chase, which showed an extremely long half-life for ABCB6 (>∼24 h), and C8S appears to be just as unstable in this assay as in the pulse-chase. Non-glycosylated C8G exhibited an even shorter half-life (2.9 h) compared with C8S (7.1 h). The absence of glycosylation of ABCB6 (N6Q) did not alter the wild-type ABCB6 degradation pattern as it had a degradation pattern similar to that of the wild-type protein (Fig. 4A). Therefore, lack of glycosylation affects the stability of ABCB6 only in the absence of the conserved cysteine. This suggests that the amino-terminal cysteines are a dominant factor stabilizing ABCB6 and that glycosylation modifies this but only in the context of the ABCB6 cysteine mutants.
FIGURE 4.
Glycosylation is not critical for ABCB6 instability, and MG132 partially rescues C8S instability. A, NIH3T3 cells were transfected with ABCB6-V5, C8S-V5, C8G-V5, or N6Q-V5, and cycloheximide (final concentration, 50 μg/ml) was added 17 h post-transfection. Cells were harvested at the indicated times and analyzed for ABCB6 proteins using anti-V5 antibody. Intensity of the bands was analyzed using densitometry and expressed as percentage of ABCB6 protein at 0 h for each construct. Curve fitting was performed by non-linear regression analysis using GraphPad Prism. Values shown are the mean from two independent experiments with the range indicated by the error bars. B, NIH3T3 cells were transiently transfected with ABCB6-V5 or C8S-V5, incubated with 10 μm MG132 or 25 mm NH4Cl for 12 h, and analyzed for ABCB6 proteins using an anti-V5 antibody. CHX, cycloheximide. A representative image from two separate experiments is shown.
To understand the pathway of degradation of cysteine mutant ABCB6, we used either MG132 or NH4Cl to inhibit the proteasomal or the lysosomal degradation pathway, respectively. Cells transiently expressing either wild-type ABCB6 or C8S mutant were incubated with MG132 for 12 h and analyzed for ABCB6 proteins. Although untreated cells express C8S protein at a lower level (51.6 ± 5.4% of wild-type protein), the protein expression of C8S mutant was comparable with that of untreated wild-type ABCB6 after MG132 incubation (112.6 ± 5.2%; Fig. 4B). However, incubation with NH4Cl failed to restore the C8S protein expression (Fig. 4B), indicating that the mutant ABCB6 is not degraded by the lysosomal pathway. These studies demonstrate that the cysteines in the amino terminus of ABCB6 are important for stability by escaping proteasomal degradation.
Cys-8 Is Not Required for Substrate Binding of ABCB6 but Forms a Disulfide Bond with Cys-26
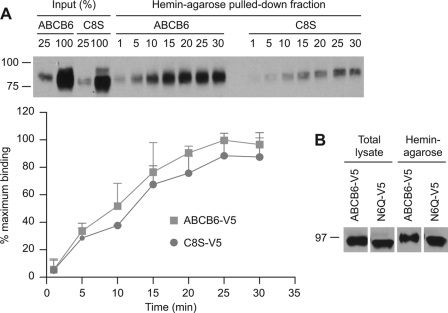
The reduced stability of ABCB6 cysteine mutants and Endo H sensitivity indicate ER retention and degradation. The signal for this degradation process is protein misfolding. However, the determinants of the stability of a misfolded protein can be location-specific (e.g. cytosol or ER lumen) (6). Because substrate binding requires a native conformation, we tested whether cysteine substitution (Cys-8) altered substrate binding. We interrogated ABCB6 substrate binding using hemin coupled to hemin-agarose in a pulldown assay. The C8S-ABCB6 protein produced in NIH3T3 cells was competent at substrate (hemin) binding. Moreover, the kinetics of hemin-agarose association between ABCB6 and ABCB6-C8S showed an identical rate of hemin association with ABCB6-C8S, which suggests that the substrate binding domain is properly folded despite being unstable (Fig. 5A). We extended this to show that the non-glycosylated form of ABCB6, N6Q, also bound hemin-agarose, suggesting that a lack of N-glycosylation does not affect substrate binding (Fig. 5B).
FIGURE 5.
Heme binding by ABCB6 does not require Cys-8 or glycosylation. A, ABCB6-V5 or ABCB6-C8S-V5 was transiently expressed in NIH3T3 cells. The cell lysates were incubated with hemin-agarose for the indicated periods (min), and the beads were washed. ABCB6 protein bound to the hemin-agarose was analyzed by immunoblot. The intensity of the immunoblot bands was analyzed by densitometry and is shown as percentage of maximum binding. Values shown are the mean from two independent experiments with the range indicated by the error bars. B, ABCB6-V5 or N6Q-V5 was transiently expressed in NIH3T3 cells, and the lysates were subjected to hemin-agarose pulldown assays. An immunoblot from a single experiment that was replicated many times (13) is shown.
The ER retention and instability of ABCB6-C8S, -C26S, and -C8S/C26S suggests that these residues have a crucial role in the folding of the ABCB6 amino terminus, a domain that appears to be important in determining the stability of ABCB6. Because both cysteine residues (Cys-8 and Cys-26) are predicted to reside in the ER lumen and the ER has a unique oxidizing potential, we postulated that they form an intramolecular disulfide bond. Depending upon the distance between the two cysteine residues such bonds cause proteins to migrate faster in SDS-PAGE under non-reducing conditions. We reasoned that the small number of amino acids separating the two conserved cysteines (17 amino acids) would not produce a readily detectable band shift in a 94-kDa ABCB6. Therefore, we generated chimeric expression plasmids that contained segments of varying length of ABCB6 protein fused with GFP. Each of these constructs was compared with the full-length ABCB6-GFP with respect to Endo H and PNGase F sensitivity, and the constructs with sensitivity comparable with full length were used to evaluate the disulfide bond for mutation (supplemental Fig. 2). Two of the constructs were poorly expressed (1–50 and 1–90), whereas a third did not achieve the required Endo H resistance. Only the construct containing the amino-terminal 210 amino acids (N1–210) mimicked the wild-type protein by showing PNGase F sensitivity and Endo H resistance (supplemental Fig. 3). To rule out the contribution of the two additional cysteines (Cys-120 and Cys-150) present in this segment, we then developed a chimeric expression plasmid encoding the amino-terminal 210 amino acids of ABCB6 fused to a carboxyl-terminal FLAG epitope where both Cys-120 and Cys-150 were substituted with Ala (ABCB6-C50A/C120AN1–210-FLAG). A schematic drawing shows the predicted location of the amino-terminal cysteines in the ER lumen (Fig. 6A).
FIGURE 6.
ABCB6 amino-terminal cysteines form a disulfide bond. A, an ABCB6-FLAG chimera including amino acids N1–210 is predicted by TMHMM to contain five TM helices. The diagram is not drawn to scale. The glycosylation site (Asn) and two cysteines are labeled. B, ABCB6-C50A/C120AN1–210-FLAG and ABCB6-C8S/C50A/C120AN1–210-FLAG were expressed in NIH3T3 cells, treated with N-ethylmaleimide to prevent spontaneous disulfide bonds, separated by SDS-PAGE with or without DTT, and analyzed by immunoblotting. The identity of each band was confirmed by glycosidase sensitivity. Band I, mature ABCB6; band II, immature ABCB6 with high mannose modification; band III, nonglycosylated ABCB6. Asterisks indicate oxidized ABCB6 (faster migration in the absence of DTT). The right panel depicts a longer film exposure; however, all samples were analyzed on a single gel (blot). Representative images from two separate experiments are shown. C, intracellular PPIX content in K562 cells transduced with GFP and with ABCB6-FLAG, the Walker A mutant (mt)-FLAG, or ABCB6-C26A-FLAG was measured by flow cytometric analysis of GFP-positive cells. The values shown are mean with the S.D. indicated by the error bars from three independent experiments. Representative histograms are shown with a representative FACS dot plot that was used to generate these data depicted in the supplemental material.
To evaluate the presence of disulfide bonds, we eliminated spontaneous disulfide bond formation by N-ethylmaleimide treatment. We next compared the electrophoretic migration of ABCB6-C50A/C120AN1–210-FLAG and the cysteine mutant ABCB6-C8S/C50A/C120AN1–210-FLAG in the presence versus absence of the reducing agent DTT (Fig. 6B). Glycosidase treatment with either PNGase F or Endo H distinguished the ABCB6 location in either the ER or Golgi. In addition, we noted that the N1–210 ABCB6 was partially Endo H-sensitive, which is probably due to the amount of plasmid transfected. ABCB6-C50A/C120AN1–210-FLAG exhibited three faster migrating bands in the absence of DTT (Fig. 6B). However, consistent with disulfide bond formation, addition of DTT reduced migration of each form of ABCB6. In contrast, the migration of ABCB6-FLAG with the C8S mutation was not affected by DTT (Fig. 6B, right panel, band II, lane 3 and 4). The ABCB6 with a C8S mutation exhibited no Endo H resistance, demonstrating ER retention like the full-length ABCB6 with amino-terminal cysteine substitution. We previously found that ABCB6 overexpression enhances de novo porphyrin biosynthesis monitored by an increase in fluorescent heme precursor PPIX (13). We hypothesized that this effect would be absent if ABCB6 was retained in the ER. As ABCB6 is expressed almost exclusively in the mitochondria of K562 cells (13, 37), these cells were transduced with retroviruses expressing ABCB6-FLAG, ABCB6-K629G-FLAG (a non-functional Walker A lysine mutant (13)), or ABCB6-C26A/S-FLAG by using plasmids containing IRES-GFP. PPIX concentration was measured in cells with comparable GFP expression by flow cytometry. The mean PPIX fluorescence in cells expressing the ABCB6-C26A mutant or nonfunctional ABCB6 was reduced in cells expressing ABCB6 (Fig. 6C and supplemental Fig. 4), indicating a loss of ability to stimulate porphyrin synthesis. Because these mutants still bind porphyrins (Fig. 5), the loss of function can be attributed to ER retention.
Only ER Luminal Cysteines Affect Stability and ER Retention
We next evaluated whether the localization of the amino-terminal cysteines is an important factor regulating either protein stability or ER exit. ABCA1 is essential for the transport of lipids across membranes and for the formation of high density lipoprotein. Our sequence analysis showed that ABCA1 contained the conserved amino-terminal cysteines (Cys-3 and Cys-23; supplemental Fig. 5); however, the predicted topology indicates that these residues reside in the cytoplasm and not the ER (38). We therefore used ABCA1 to test, regardless of predicted subcellular localization, whether amino-terminal cysteines affect protein stability. We combined confocal microscopy and differential glycosidase sensitivity to assess ER exit (supplemental Fig. 5). When the cysteines at ABCA1 residues 3 and 23 were substituted with alanine, confocal microscopic analysis showed that the mutant C3A/C23A-ABCA1 localized to both the plasma membrane and intracellular compartments (supplemental Fig. 5A). Immunoblot analysis showed resistance to Endo H digestion, confirming that both mutant and wild-type ABCA1 had exited the ER (supplemental Fig. 5B). Moreover, the expression level of the mutant protein was comparable with that of the wild-type protein. Therefore, this finding supports the concept that location of the amino-terminal cysteines is an important determinant in ABC protein trafficking.
Two Amino-terminal Cysteines Are Conserved among Multiple ABC Transporters
Because of the conservation of this cysteine motif among ABCB6 homologs (Fig. 2D), we investigated whether cysteine motifs were found in other ABC transporters. We used the predicted topology of 43 additional human ABC transporters (31) to identify candidates with amino-terminal cysteines predicted to be in the ER. To determine whether two amino-terminal cysteines with the appropriate spacing were conserved, we visually examined the amino acid sequences of ABC transporters and other transporters (see supplemental Fig. 6) aligned by using ClustalW. Because the carboxyl-terminal nucleotide binding domain in most ABC transporters resides in the cytoplasm, an odd number of TM helices suggests projection of the amino terminus into the ER lumen, and therefore, we included transporters whose amino termini do and do not project into the ER. We found the two cysteines to be conserved but separated by 17–25 non-conserved amino acids in the so-called “long multidrug-related proteins (MRPs)” that are members of the ABCC subfamily, including ABCC1, ABCC2, ABCC3, ABCC6, ABCC8 (SUR1), and ABCC9 (SUR2) (Fig. 7C). These transporters contain an additional membrane-spanning domain0 (MSD0) at the amino terminus with five TMs, causing the amino terminus to reside in the ER lumen during maturation. Disease-causing mutations have been identified in these ABC transporters, and therefore, it is of interest to determine whether these cysteines affect the folding and ultimately trafficking of the proteins.
FIGURE 7.
Two amino-terminal cysteine residues are conserved and functional in ABCC transporters. A, a point mutation resulting in Cys-6 to Ser substitution in SUR1/ABCC8 was identified in one allele of a hyperinsulinemic patient as shown in the electropherogram. B, SUR1 and mutant SUR1 constructs containing various cysteine substitutions were co-expressed with KIR6.2 in COSm6 cells. SUR1 proteins were labeled with 125I-azidoglibenclamide and detected by autoradiography after SDS-PAGE. Mature SUR1 that traffics to the cell surface was identified by its slower electrophoretic migration. C, multiple sequence alignment of SUR1 and ABCC1/MRP1 show that the amino-terminal cysteine residues (boxed in red) are conserved among the species. D, MRP1-MSD0-GFP containing either wild-type MRP1 or a C32A-MRP1 mutant was transiently expressed in HEK293 cells. Merged indirect immunofluorescence microscopy images (left) show the wild-type protein (green) on the cell surface, whereas the C32A mutant protein remains in the ER where it co-localizes with the ER protein protein-disulfide isomerase (PDI) (red, which is shown as yellow in the co-localized cell). Scale bar, 10 μm. Proteins were treated with the indicated glycosidases and analyzed by immunoblotting with anti-GFP antibody (right). The presence of glycans was detected by protein mobility shifts. Two independent experiments were performed with all iterations. Representative images of a complete experiment are shown. E, cysteine residues between TM5 and TM6 of ABCG2 that form an intramolecular disulfide bond are boxed in red. A cysteine residue that participates in intermolecular disulfide bond formation is boxed in blue. ABCG5 also contains two cysteine residues in a similar region. H.sapiens, Homo sapiens; B.taurus, Bos taurus; M.musculus, Mus musculus; R.norvegicus, Rattus norvegicus; D.rerio, Danio rerio; G.gallus; Gallus gallus; C.lupus; Canis lupus. Pro, protein (amino acid) sequence.
This analysis enabled us to identify a role for the previously unrecognized defective SUR1/ABCC8 allele in a patient with hyperinsulinemic hypoglycemia, a recessive genetic disease where a point mutation results in a Cys-26 to serine substitution (Fig. 7A). The patient was a compound heterozygote: one mutant ABCC8/SUR1 allele was a previously reported truncation (19), whereas the other had only a C26S substitution. The disease is caused by a lack of functional KATP channels composed of SUR1/K+ channels (KIR) at the cell surface. Therefore, we tested whether this mutation affects SUR1 maturation. We generated SUR1 with alanine or serine substitutions for Cys-6 and/or Cys-26, substituting alanine or serine for the remaining 27 cysteine residues. As KIR6.2 is an obligatory heterooligomeric partner required for ER exit (39), the SUR1 wild-type and mutant proteins were co-expressed with KIR6.2 in COSm6 cells and labeled with 125I-azidoglibenclamide, a substrate/ligand of SUR1 (40). Mature (fully glycosylated) SUR1, which is expressed on the surface membrane and possesses KATP channel activity, was identified by reduced mobility on SDS-PAGE (Fig. 7B) (40). ABCC8/SUR1 with Cys-6 and Cys-26 bound glibenclamide in the absence of KIR6.2 (Fig. 7B). Surprisingly, when alanine was substituted for cysteine at either of these two positions, SUR1 retained glibenclamide binding capability but did not mature even in the presence of KIR6.2. Similar to the case of ABCB6, SUR1 mutants with a double cysteine mutation and no free thiols failed to mature (Fig. 7B, lane 4), suggesting that the formation of an intramolecular bond was required to meet the quality checkpoint.
To further evaluate whether the ER luminal amino-terminal cysteines have a more general role in this ER checkpoint of the ABC transporters, we extended this analysis to investigate the role of these conserved cysteines in ABCC1/MRP1, which has important roles in resistance to chemotherapy and the inflammatory response (1, 41). We used the MSD0 (amino acids 1–203) of MRP1 because it mimics full-length MRP1 by localizing to the plasma membrane and is required for plasma membrane localization (its deletion prevents plasma membrane localization) (21), and we reasoned that the potential differences in size after glycanase treatment would be more apparent in this construct. HEK293 cells were transfected with a plasmid encoding an MRP1-MSD0-GFP chimera containing either wild-type MSD0 or MSD0 with a C32A substitution. Fluorescence microscopy showed the wild-type construct in the plasma membrane, whereas the mutant construct was retained in the ER (Fig. 7D, left). Immunoblot analysis was then performed after digestion with either PNGase F or Endo H. The wild-type MRP1-MSD0-GFP was sensitive to PNGase F, as shown by increased mobility, but not to Endo H, indicating that the wild-type MRP1-MSD0-GFP exited the ER (Fig. 7D, right). In contrast, the construct with the C32A substitution was Endo H-sensitive. Together, these findings indicate that loss of one of the two conserved amino-terminal cysteines results in ER retention of MRP1.
Conclusions
We have determined that ABCB6 undergoes glycan modification in many cell types at a conserved single atypical amino-terminal NXC site. This cysteine forms an intramolecular disulfide bond in the ER that appears to be a key event in the trafficking of ABCB6 as mutation of either cysteine led to ER retention. Quality control checkpoints in the ER are required to ensure that properly folded proteins reach their final destinations in the appropriate conformation to guarantee normal biological activity. The topological ER checkpoints for membrane proteins comprise both ER luminal and cytoplasmic signals, but the formation and type of integral membrane domains might also be a signal (6). Our studies reveal a conserved amino-terminal disulfide bond as an important conserved ER luminal checkpoint for many ABC transporters. Our analysis facilitated the identification of a new ABCC8/SUR1 mutant allele that produces ER retention as the cause of hyperinsulinemic hypoglycemia.
Intriguingly, an intramolecular disulfide bond has been previously identified in ABCG2 that when disrupted (by mutation) produces ER retention, although mechanistically it is unclear whether ER retention was due to an unpaired thiol causing thiol retention or the lack of disulfide bond formation (8). In the case of ABCB6 and ABCC8/SUR1, we showed that it was not thiol retention but instead loss of the disulfide bond. To further extend the idea that the disulfide bond might be conserved among ABC transporters, an amino acid sequence alignment of ABCG subfamily members revealed that ABCG5 also contains two cysteines in the region similar to ABCG2 (Fig. 7E). Collectively, these findings suggest that the formation of an intramolecular disulfide bond in an ER luminal loop may be a general conformational requirement for ER exit of many ABC transporters. Moreover, we showed that a subtle change in the topology of a short ER luminal segment is an important checkpoint that does not appear to appreciably alter transporter-ligand interactions. Finally, although disruption of this disulfide bond led to ER retention and, in the case of ABCB6 and ABCG2, proteasomal degradation (8), it is not clear whether this is the case for all ABC transporters with disruption of ER luminal domains. For example, SUR1 appears to be relatively stable but does not leave the ER. This finding suggests that some ABC transporters may only be retained in the ER when their ER luminal disulfide bond is disrupted, and other factors may determine whether they are degraded. If this is true, it raises the possibility that changes in ER oxidation state could impact the trafficking of many ABC transporters, and this might be the basis for some cases of loss of function.
Supplementary Material
Acknowledgments
We thank Dr. Kazumitsu Ueda for the kind gift of ABCA1-GFP plasmid, Sam Connell and Dr. Jennifer Peters for assistance with immunofluorescence microscopy, and Sharon Naron and Angela McArthur for editorial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants ES058571, P30 CA21745, and CA21865. This work was also supported by the American Lebanese Syrian Associated Charities and by a grant from the Thrasher Foundation (to L. A.-B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental methods, Figs. 1–6, and a table.
W. S. Rasband, National Institutes of Health, Bethesda, MD, 1997–2009.
- ABC
- ATP-binding cassette
- ER
- endoplasmic reticulum
- SUR1
- sulfonylurea receptor 1
- PPIX
- protoporphyrin IX
- PNGase F
- peptide-N-glycosidase F
- Endo H
- endo-β-N-acetylglucosaminidase H
- TM
- transmembrane
- IRES
- internal ribosome entry site
- MRP
- multidrug-related protein
- MSD0
- membrane-spanning domain0.
REFERENCES
- 1. Dean M. (2005) Methods Enzymol. 400, 409–429 [DOI] [PubMed] [Google Scholar]
- 2. Riordan J. R., Rommens J. M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J. L., Drumm M. L., Iannuzzi M. C., Collins F. S., Tsui L. (1989) Science 245, 1066–1073 [DOI] [PubMed] [Google Scholar]
- 3. Cheng S. H., Gregory R. J., Marshall J., Paul S., Souza D. W., White G. A., O'Riordan C. R., Smith A. E. (1990) Cell 63, 827–834 [DOI] [PubMed] [Google Scholar]
- 4. Ellgaard L., Molinari M., Helenius A. (1999) Science 286, 1882–1888 [DOI] [PubMed] [Google Scholar]
- 5. Ellgaard L., Helenius A. (2003) Nat. Rev. Mol. Cell Biol. 4, 181–191 [DOI] [PubMed] [Google Scholar]
- 6. Vashist S., Ng D. T. (2004) J. Cell Biol. 165, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Negroiu G., Dwek R. A., Petrescu S. M. (2000) J. Biol. Chem. 275, 32200–32207 [DOI] [PubMed] [Google Scholar]
- 8. Wakabayashi K., Nakagawa H., Tamura A., Koshiba S., Hoshijima K., Komada M., Ishikawa T. (2007) J. Biol. Chem. 282, 27841–27846 [DOI] [PubMed] [Google Scholar]
- 9. Wang Z. V., Schraw T. D., Kim J. Y., Khan T., Rajala M. W., Follenzi A., Scherer P. E. (2007) Mol. Cell. Biol. 27, 3716–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alberini C. M., Bet P., Milstein C., Sitia R. (1990) Nature 347, 485–487 [DOI] [PubMed] [Google Scholar]
- 11. Sitia R., Neuberger M., Alberini C., Bet P., Fra A., Valetti C., Williams G., Milstein C. (1990) Cell 60, 781–790 [DOI] [PubMed] [Google Scholar]
- 12. Isidoro C., Maggioni C., Demoz M., Pizzagalli A., Fra A. M., Sitia R. (1996) J. Biol. Chem. 271, 26138–26142 [DOI] [PubMed] [Google Scholar]
- 13. Krishnamurthy P. C., Du G., Fukuda Y., Sun D., Sampath J., Mercer K. E., Wang J., Sosa-Pineda B., Murti K. G., Schuetz J. D. (2006) Nature 443, 586–589 [DOI] [PubMed] [Google Scholar]
- 14. Mitsuhashi N., Miki T., Senbongi H., Yokoi N., Yano H., Miyazaki M., Nakajima N., Iwanaga T., Yokoyama Y., Shibata T., Seino S. (2000) J. Biol. Chem. 275, 17536–17540 [DOI] [PubMed] [Google Scholar]
- 15. Paterson J. K., Shukla S., Black C. M., Tachiwada T., Garfield S., Wincovitch S., Ernst D. N., Agadir A., Li X., Ambudkar S. V., Szakacs G., Akiyama S., Gottesman M. M. (2007) Biochemistry 46, 9443–9452 [DOI] [PubMed] [Google Scholar]
- 16. Jalil Y. A., Ritz V., Jakimenko A., Schmitz-Salue C., Siebert H., Awuah D., Kotthaus A., Kietzmann T., Ziemann C., Hirsch-Ernst K. I. (2008) Am. J. Physiol. Cell Physiol 294, C579–C590 [DOI] [PubMed] [Google Scholar]
- 17. Tsuchida M., Emi Y., Kida Y., Sakaguchi M. (2008) Biochem. Biophys. Res. Commun. 369, 369–375 [DOI] [PubMed] [Google Scholar]
- 18. Zielinska D. F., Gnad F., Wiśniewski J. R., Mann M. (2010) Cell 141, 897–907 [DOI] [PubMed] [Google Scholar]
- 19. Aguilar-Bryan L., Bryan J. (1999) Endocr. Rev. 20, 101–135 [DOI] [PubMed] [Google Scholar]
- 20. Aguilar-Bryan L., Bryan J. (2008) Endocr. Rev. 29, 265–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Westlake C. J., Cole S. P., Deeley R. G. (2005) Mol. Biol. Cell 16, 2483–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maley F., Trimble R. B., Tarentino A. L., Plummer T. H., Jr. (1989) Anal. Biochem. 180, 195–204 [DOI] [PubMed] [Google Scholar]
- 23. Kornfeld R., Kornfeld S. (1985) Annu. Rev. Biochem. 54, 631–664 [DOI] [PubMed] [Google Scholar]
- 24. Park B., Lee S., Kim E., Cho K., Riddell S. R., Cho S., Ahn K. (2006) Cell 127, 369–382 [DOI] [PubMed] [Google Scholar]
- 25. Goldenberg N. M., Grinstein S., Silverman M. (2007) Mol. Biol. Cell 18, 4762–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hernández M. V., Wehrendt D. P., Arregui C. O. (2010) Mol. Biol. Cell 21, 1387–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klausner R. D., Donaldson J. G., Lippincott-Schwartz J. (1992) J. Cell Biol. 116, 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sigrist C. J., Cerutti L., Hulo N., Gattiker A., Falquet L., Pagni M., Bairoch A., Bucher P. (2002) Brief. Bioinform. 3, 265–274 [DOI] [PubMed] [Google Scholar]
- 29. Bause E. (1983) Biochem. J. 209, 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 31. Tusnády G. E., Sarkadi B., Simon I., Váradi A. (2006) FEBS Lett. 580, 1017–1022 [DOI] [PubMed] [Google Scholar]
- 32. Titani K., Kumar S., Takio K., Ericsson L. H., Wade R. D., Ashida K., Walsh K. A., Chopek M. W., Sadler J. E., Fujikawa K. (1986) Biochemistry 25, 3171–3184 [DOI] [PubMed] [Google Scholar]
- 33. Grinnell B. W., Walls J. D., Gerlitz B. (1991) J. Biol. Chem. 266, 9778–9785 [PubMed] [Google Scholar]
- 34. Krogh T. N., Bachmann E., Teisner B., Skjødt K., Højrup P. (1997) Eur. J. Biochem. 244, 334–342 [DOI] [PubMed] [Google Scholar]
- 35. Vance B. A., Wu W., Ribaudo R. K., Segal D. M., Kearse K. P. (1997) J. Biol. Chem. 272, 23117–23122 [DOI] [PubMed] [Google Scholar]
- 36. Satomi Y., Shimonishi Y., Takao T. (2004) FEBS Lett. 576, 51–56 [DOI] [PubMed] [Google Scholar]
- 37. Lynch J., Fukuda Y., Krishnamurthy P., Du G., Schuetz J. D. (2009) Cancer Res. 69, 5560–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fitzgerald M. L., Mendez A. J., Moore K. J., Andersson L. P., Panjeton H. A., Freeman M. W. (2001) J. Biol. Chem. 276, 15137–15145 [DOI] [PubMed] [Google Scholar]
- 39. Zerangue N., Schwappach B., Jan Y. N., Jan L. Y. (1999) Neuron 22, 537–548 [DOI] [PubMed] [Google Scholar]
- 40. Sharma N., Crane A., Clement J. P., 4th, Gonzalez G., Babenko A. P., Bryan J., Aguilar-Bryan L. (1999) J. Biol. Chem. 274, 20628–20632 [DOI] [PubMed] [Google Scholar]
- 41. Wijnholds J., Evers R., van Leusden M. R., Mol C. A., Zaman G. J., Mayer U., Beijnen J. H., van der Valk M., Krimpenfort P., Borst P. (1997) Nat. Med. 3, 1275–1279 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.