Abstract
The activity of many proteins orchestrating different biological processes is regulated by allostery, where ligand binding at one site alters the function of another site. Allosteric changes can be brought about by either a change in the dynamics of a protein, or alteration in its mean structure. We have investigated the mechanisms of allostery induced by chemically distinct ligands in the cGMP-binding, cGMP-specific phosphodiesterase, PDE5. PDE5 is the target for catalytic site inhibitors, such as sildenafil, that are used for the treatment of erectile dysfunction and pulmonary hypertension. PDE5 is a multidomain protein and contains two N-terminal cGMP-specific phosphodiesterase, bacterial adenylyl cyclase, FhLA transcriptional regulator (GAF) domains, and a C-terminal catalytic domain. Cyclic GMP binding to the GAFa domain and sildenafil binding to the catalytic domain result in conformational changes, which to date have been studied either with individual domains or with purified enzyme. Employing intramolecular bioluminescence resonance energy transfer, which can monitor conformational changes both in vitro and in intact cells, we show that binding of cGMP and sildenafil to PDE5 results in distinct conformations of the protein. Metal ions bound to the catalytic site also allosterically modulated cGMP- and sildenafil-induced conformational changes. The sildenafil-induced conformational change was temperature-sensitive, whereas cGMP-induced conformational change was independent of temperature. This indicates that different allosteric ligands can regulate the conformation of a multidomain protein by distinct mechanisms. Importantly, this novel PDE5 sensor has general physiological and clinical relevance because it allows the identification of regulators that can modulate PDE5 conformation in vivo.
Keywords: Allosteric Regulation, Cyclic GMP (cGMP), Enzyme Inhibitors, Metals, Phosphodiesterases, GAF, PDE5, Bioluminescence Resonance Energy Transfer
Introduction
Allostery is a thermodynamic phenomenon utilized by nature to regulate the function of a large number of proteins. Thus, ligand binding at one site of a protein can alter the function of another distinct site in the protein (1–3) Due to inherent dynamics, proteins sample multiple conformations in solution (4). A “shift” in the ensemble of conformations, involving conformational selection and induced-fit mechanisms (5), is generally thought to result in allosteric regulation (6). Thermodynamically, however, allostery could manifest itself as a change in the mean conformation of the ensemble (enthalpy-driven), a change in the dynamics of the ensemble without any observable change in the backbone structure of the ensemble (entropy-driven), or a combination of the two (7, 8). In addition, the presence of multiple domains in a single polypeptide chain could allow allosteric integration of different input signals into similar outputs or allosteric dissemination of similar inputs into different output signals (9, 10).
The mammalian cyclic nucleotide phosphodiesterases (PDEs)3 serve as examples of multidomain proteins where different N-terminal regulatory domains sense a variety of ligands, and then allosterically modulate the hydrolysis of cAMP or cGMP (11). The cGMP-binding, cGMP-specific phosphodiesterase (PDE5) specifically hydrolyzes cGMP (12, 13) and is expressed in a variety of tissues (14). PDE5 has been targeted by the specific inhibitors sildenafil, vardenafil, and tadalafil (15, 16) for the treatment of erectile dysfunction and pulmonary hypertension. PDE5 is a multidomain protein containing two N-terminal tandem GAF domains (GAFa and GAFb) followed by the C-terminal catalytic domain. Cyclic GMP binding to the GAFa domain allosterically activates the catalytic domain by increasing both the Vmax and affinity for cGMP, resulting in increased cGMP hydrolysis (17–19). In addition, sildenafil binding to the catalytic domain enhances cGMP binding to the regulatory GAFa domain (20). Further, cGMP binding to the GAFa domain (21, 22) or physical interaction of the isolated GAFa domain with PDE5 (23) increases sildenafil affinity to the catalytic domain. PDE5 shows a reduced migration in native polyacrylamide gels in the presence of cGMP (24, 25), providing a qualitative indication of conformational changes in the protein. Until now, allosteric changes in PDE5 have been studied using purified preparations of the protein and therefore may not reflect the conformational states or the biochemical properties of the enzyme in the cell (26).
We asked whether we could develop a method in which conformational changes in full-length PDE5 could be measured quantitatively, not only in vitro, but also in the intact cell. Here, we report the generation of a functional reporter of PDE5 conformation that exploits intramolecular bioluminescence resonance energy transfer (BRET). The sensor reveals structural changes and associated allostery induced by cGMP and sildenafil binding to the full-length PDE5. Importantly, we show that metal ion binding to the catalytic site of PDE5 negatively modulates cGMP-induced and positively modulates sildenafil-induced conformational changes. The conformational changes induced by cGMP and sildenafil are structurally and thermodynamically distinct. This novel sensor not only monitors dynamic changes in PDE5 conformation in intact cells, but also provides new insights into allostery in a multidomain protein.
MATERIALS AND METHODS
Generation of PDE5 Sensor Constructs
The GFP gene was released from pGFP-C1 plasmid (PerkinElmer Life Sciences) with SnaBI-XhoI and cloned into similarly digested pEGFP-PDE5A2 (27) resulting in pGFP-PDE5A2-EGFP. The EGFP gene was then replaced with the Rluc gene from pRluc-N1 vector (PerkinElmer Life Sciences) with KpnI-XbaI digestion to generate pGFP-PDE5A2-Rluc. This plasmid expresses the GPF-PDE5A2-Rluc fusion protein (PDE5 sensor). The F163A mutant sensor was similarly generated from the pEGFP-PDE5A2(F163A) plasmid (27).
For mutagenesis in the catalytic domain, a HindIII-KpnI fragment was released from the pGFP-PDE5A2-Rluc plasmid and cloned into similarly digested pBlueScript II KS(+) vector (Stratagene). The single mutagenic oligonucleotide-based protocol was utilized for generation of mutations (28). Primers PDE5A2F778A (5′-CCCAAGTATGCAAGTTGGGGCCATCGATGCCATCTGCTTGC-3′), PDE5A2D612A (5′-GCTGCACTATCCCACGCTCTAGATCACCGTGGTGTG-3′), and PDE5A2G617P (5′-GCCACGATCTAGATCACCGTCCTGTGAATAACTCTTAC-3′) were used for generating F778A, D612A, and G617P mutations, respectively, and mutations were confirmed by sequencing (Macrogen). Mutant fragments in pBlueScript II KS(+) vector were then suitably subcloned to generate either single or double mutants of the PDE5 sensor.
Cell Culture and Transfection
Human embryonic kidney (HEK) 293T cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal calf serum (FCS), 120 mg/liter penicillin, and 270 mg/liter streptomycin at 37 °C in a 5% CO2 humidified incubator. Transfections were performed with polyethyleneimine lipid.
Cyclic GMP PDE Assays
Cytosol prepared from cells transfected with either the wild-type or mutant PDE5 sensors were prepared in PDE lysis buffer (50 mm HEPES (pH 7.5), 100 mm NaCl, 2 mm EDTA, 1 mm dithiothreitol (DTT), 10 mm sodium pyrophosphate, 80 μm β-glycerophosphate, 1 mm benzamidine, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 5 μg/ml soybean trypsin inhibitor, 100 μm sodium orthovanadate, and 10% glycerol) and were used for cGMP PDE assays (29). All PDE assays were performed with ∼100 nm [3H]cGMP (GE Healthcare) essentially as reported previously (30). Expression of proteins was monitored by Western blot analysis using either an antibody raised in rabbits to GFP, generated in the laboratory, or a previously described antibody to PDE5 (29).
Gel Filtration Analysis
HEK293T cells that were either untransfected or expressing pGFP-PDE5A2-Rluc were lysed in PDE lysis buffer. Cytosol from lysed cells was subjected to gel filtration (Superose 12, 10 × 30; GE Healthcare) in 50 mm Tris-HCl (pH 7.5), 100 mm NaCl, and 10% glycerol. Fractions (500 μl) were collected and analyzed for GFP fluorescence using a Victor3 microplate reader (excitation, 485 nm; emission, 510 nm; PerkinElmer Life Sciences).
Intracellular cGMP Estimation
Intracellular levels of cGMP were measured in cells transfected with either wild-type or mutant PDE5 sensors 72 h after transfection. Cells were lysed in 0.1 n HCl, and cGMP was measured by radioimmunoassay as described earlier (31). In some cases, lysates were acetylated prior to radioimmunoassay to increase the sensitivity of the radioimmunoassay.
In Vitro BRET Assays
Cells transfected with sensor plasmids were lysed in PDE lysis buffer. Following brief sonication, the lysates were centrifuged at 13,000 × g, and the cytosol was removed. Aliquots of the cytosol were incubated with the indicated amounts of ligand in a total volume of 40 μl at 37 °C for 10 min.
To determine the effect of sildenafil binding on cGMP-induced conformational change, lysates were preincubated with 100 μm sildenafil at 37 °C for 5 min followed by incubation with varying concentrations of cGMP at 37 °C for another 5 min. Similarly, to determine the effect of cGMP binding on sildenafil-induced conformational change, lysates were preincubated with 100 μm cGMP at 37 °C for 5 min followed by incubation with varying concentrations of sildenafil at 37 °C for another 5 min, and BRET was measured.
In experiments to determine the effect of EDTA and metals on the PDE5 sensor, cells were lysed in 50 mm HEPES (pH 7.5), 100 mm NaCl, 1 mm DTT, 1 mm benzamidine, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 5 μg/ml soybean trypsin inhibitor, and 10% glycerol. Lysates were incubated in the absence or presence of 2 mm EDTA and 10 mm metals in 50 mm HEPES (pH 7.5), 100 mm NaCl, 1 mm DTT, and 10% glycerol in a total volume of 40 μl at 37 °C for 10 min. To determine the concentrations of Mg2+ or Ca2+ required to inhibit the cGMP-induced decrease in BRET, lysates were preincubated with 0.1 mm EDTA and 100 μm cGMP for 5 min at room temperature followed by incubation with varying concentration of MgCl2 or CaCl2 at 37 °C for 10 min. The free metal ion concentrations were calculated with the program WinMAXC32 2.51 (C. Patton, Stanford University, Palo Alto, CA). The dependence of cGMP- or sildenafil-induced conformational changes on temperature was determined by incubating lysates with either 100 μm cGMP or sildenafil at the specified temperatures for 10 min, followed by BRET measurements.
BRET was measured by addition of 5 μm coelenterazine 400a (Molecular Imaging Products) to a final concentration of 5 μm. Readings for GFP and Rluc emission (0.8 s) were collected in the Victor3 Microplate Reader. Emission filters used for Rluc and GFP emission were 410 nm (bandpass 80 nm) and 515 nm (bandpass 30 nm), respectively. BRET was calculated as the ratio of GFP emission per second to Rluc emission per second, and the average of three such measurements is reported.
Cellular BRET Assays
BRET assays with intact cells were performed by incubating cell monolayers expressing the various sensors with Dulbecco's PBS containing 5 mm EDTA, resuspending cells in phenol red-free DMEM with 10% FCS, followed by treatment with diethylenetriamine-NO (DETA/NO;1 mm) for 5 min. To reduce the intracellular levels of metal ions, monolayers of cells were incubated in Dulbecco's PBS and 5 mm EDTA for 10 min, and then detached cells were resuspended in phenol red-free DMEM with 10% FCS. Cells were aliquoted, pelleted by centrifugation to remove the medium, and then resuspended in modified Hanks' balanced salt solution containing 0.44 mm KH2PO4, 0.34 mm Na2HPO4, 5.36 mm KCl, 137 mm NaCl, and 5.55 mm d-glucose without or with 1.26 mm CaCl2, 1 mm MgCl2, or both. The ionic strength of the medium was maintained by the addition of extra NaCl. Cells were incubated for 1 h, followed by stimulation with 1 mm DETA/NO for 5 min. BRET was determined as described in the previous section.
RESULTS
Conformational Changes in PDE5 Occur on Binding of Cyclic Nucleotides to the GAFa Domain
We generated a fusion protein with the full-length PDE5A2 isoform (all three isoforms of PDE5 have similar biochemical properties (32), but PDE5A2 is the most widely expressed (33)) sandwiched between GFP and Rluc (PDE5 sensor; Fig. 1A). Use of GFP and the modified substrate, coelenterazine 400a, provides a greater overlap of the absorption spectrum of GFP and Rluc emission spectrum and better separation of the Rluc and GFP emission spectra (34), thus providing a high signal to noise ratio. An alteration in the distance and/or relative orientation of GFP and Rluc, consequent to changes in the conformational ensemble of PDE5, is detected by a change in the ratio of GFP fluorescence to Rluc emission (BRET).
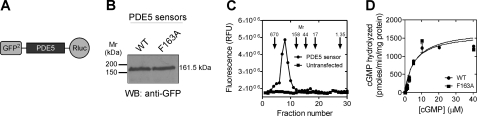
FIGURE 1.
Biochemical characterization of the PDE5 sensor. A, schematic representation of the PDE5 BRET sensor (not drawn to scale). B, Western blot analysis to detect expression of PDE5 wild-type and F163A mutant sensors in HEK293T cells using an antibody to GFP. PDE5A2 has a native molecular mass of 92 kDa, whereas the expected molecular mass of the sensor is 162 kDa. C, fluorescence in fractions eluted following gel filtration of lysates prepared from cells expressing the PDE5 sensor. Lysates from untransfected cells indicate the base-line fluorescence. Arrows indicate the positions of the molecular mass markers used for calibration of the column (thyroglobulin, 670 kDa; bovine γ-globulin, 158 kDa; chicken ovalbumin, 44 kDa; equine myoglobin, 17 kDa; and vitamin B12, 1.35 kDa). D, lysates prepared from cells expressing the PDE5 sensors assayed for PDE activity with varying concentration of cGMP in the presence of 10 mm MgCl2. The data represent mean ± S.E. of duplicate determinations of a representative experiment. Equivalent expression of the wild-type and F163A mutant proteins can be judged from the Western blot.
The PDE5 sensor expressed in HEK293T cells (Fig. 1B) was a dimer (Fig. 1C) and catalytically active (Km for cGMP 6.6 ± 0.8 μm; Fig. 1D), similar to that reported for native PDE5 (12). Addition of cGMP to lysates prepared from cells in the presence of EDTA led to a decrease (∼40%) in BRET of the wild-type sensor, whereas mutation of the Phe-163 residue (which has a stacking interaction with cGMP in the GAFa domain (27, 35)) to alanine abrogated the BRET decrease (Fig. 2A). The EC50 (0.75 ± 0.25 μm) of the cGMP-induced conformational change was similar to the binding affinity of cGMP to the GAFa domain in full-length PDE5 (36), and no apparent co-operativity in binding to the two GAFa domains in the PDE5 dimer (nH = 1.0 ± 0.1; Fig. 2B) was seen. Therefore, cGMP binding to the GAFa domain resulted in a shift in the conformational ensemble of PDE5 from a “closed” high BRET form to an “open” low BRET form (24, 25).
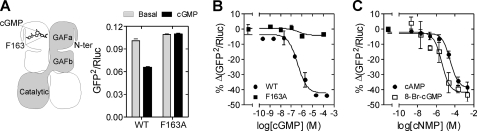
FIGURE 2.
Cyclic NMP binding to GAFa domain induces a conformational change in PDE5. A, scheme of PDE5 dimer (41) shows domain organization in the shaded monomer and relative positioning of cGMP and F163 in the GAFa domain of PDE5 (35) in the nonshaded monomer. Lysates prepared from cells expressing the wild-type and F163A mutant PDE5 sensors in PDE lysis buffer were incubated in the absence or presence of 100 μm cGMP at 37 °C for 10 min followed by BRET measurement. B, lysates prepared from cells expressing either the wild-type or F163A mutant PDE5 sensor were incubated with varying concentrations of cGMP at 37 °C for 10 min, and percentage decrease in BRET is plotted. C, lysates prepared from cells expressing the wild-type PDE5 sensor were incubated with varying concentrations of cAMP and 8-bromo-cGMP, and the percentage decrease in BRET is plotted. Data shown are the mean ± S.E. (error bars) from a representative experiment, with experiments performed at least three times with duplicate determinations.
The isolated GAFa and the tandem GAF domains of PDE5 show high specificity, with cAMP binding affinity ∼1,000-fold lower than that of cGMP (27, 37). Interestingly, cAMP reduced BRET of the wild-type sensor with an EC50 ∼40-fold higher than that of cGMP (EC50 28 ± 11 μm; Fig. 2C). 8-Bromo-cGMP was also able to decrease BRET with an EC50 of 20 ± 10 μm (Fig. 2C), similar to its binding affinity to the isolated GAFa domain (27).
Allosteric Effects Induced by Sildenafil Require H-loop Flexibility
Crystallographic studies have revealed that binding of sildenafil to the catalytic domain of PDE5 induces a major change in the positioning and secondary structure of the H-loop in the catalytic domain (38). However, no such structural changes are observed on binding of isobutylmethylxanthine (IBMX; supplemental Fig. S1). We investigated whether the conformational change induced by sildenafil could alter BRET of the PDE5 sensor. Indeed, a reduction in BRET was seen when lysates were incubated with sildenafil (∼40%) but not IBMX (Fig. 3A), even though both compounds were effective in inhibiting PDE5 catalytic activity (Fig. 3B).
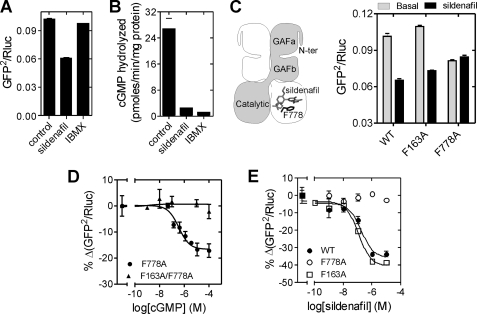
FIGURE 3.
Sildenafil induces a conformational change in PDE5. A, lysates prepared from cells expressing the PDE5 sensor were incubated with either sildenafil (100 μm) or IBMX (500 μm) at 37 °C for 10 min, and BRET was determined. B, lysates were used to determine PDE activity in the absence (control) or presence of either sildenafil (100 μm) or IBMX (500 μm). C, scheme representation of the PDE5 dimer indicates the relative positioning of sildenafil and Phe-778 in the catalytic domain (38). Lysates prepared from cells expressing wild-type, F163A, or F778A mutant PDE5 sensors in PDE lysis buffer were incubated in the absence or presence of 100 μm sildenafil at 37 °C for 10 min and BRET determined. D, lysates prepared from cells expressing either the F778A or F163A/F778A mutant PDE5 sensors were incubated with varying concentrations of cGMP followed by BRET determination. Percentage decrease in BRET in the presence of different concentrations of cGMP is plotted, and the EC50 value of the cGMP-induced conformational change was determined. Values represent mean ± S.E. (error bars) of a representative experiment with each experiment performed three times with duplicate determinations. E, lysates prepared from cells expressing the wild-type, F163A, or F778A mutant PDE5 sensor were incubated with varying concentrations of sildenafil, and percentage decrease in BRET is plotted. Data shown are mean ± S.E. from a representative experiment, with experiments performed at least three times with duplicate determinations.
To determine whether the conformational change induced by sildenafil was solely a consequence of inhibitor binding to the catalytic site, we generated a mutant sensor where Phe-778 (which shows an important stacking interaction with sildenafil (13, 39, 40)) was mutated to alanine. The F778A mutant sensor showed reduced basal BRET (Fig. 3C), indicating an alteration in PDE5 conformation due to this point mutation. Nevertheless, cGMP was able to reduce BRET by ∼20%, and the EC50 (0.81 ± 0.57 μm) of the F778A mutant sensor was similar to that of the wild-type sensor (Figs. 2B and 3D). This indicated that there was no change in the ability of the GAFa domain in the Phe-778 mutant to bind cGMP and induce a conformational change in PDE5. As expected, the F163A/F1778A double mutant PDE5 showed no response to cGMP (Fig. 3D).
In contrast, the addition of sildenafil did not result in any change in BRET of the F778A mutant sensor, whereas a change was seen with the F163A mutant sensor (Fig. 3B). The EC50 for sildenafil-induced conformational change was similar in the wild-type and F163A mutant sensors (EC50 0.68 ± 0.18 μm and 0.30 ± 0.6 μm, respectively; Fig. 3E), and binding of sildenafil showed negative co-operativity (nH = 0.7 ± 0.1). This therefore confirmed that sildenafil binding at the catalytic site and the consequent conformational change in H-loop positioning were reflected in reduced BRET, thereby further validating the use of the PDE5 sensor to monitor conformational changes in the full-length enzyme.
The movement of the H-loop is believed to be required for cGMP-mediated activation of PDE5 (25) and PDE2 (41). We mutated the conserved Gly-617 present in the beginning of the H-loop to proline, thereby restricting the flexibility of the loop without substantially affecting sildenafil binding (25). Basal BRET of the G617P mutant sensor was lower than the wild-type sensor (∼0.06 versus ∼0.09, respectively), but both the G617P mutant sensor and the F163A/G617P sensor failed to show any reduction in BRET in the presence of sildenafil (Fig. 4A). However, the EC50 of the cGMP-induced conformational change on binding to the GAFa domain remained unaltered in the G617P mutant sensor (EC50 1.45 ± 0.7 μm; Fig. 4B), and cGMP could not induce a decrease in BRET in the F163A/G617P mutant sensor. Thus, flexibility of the H-loop is required for the sildenafil-induced conformational change in PDE5, but not for that induced by cGMP, indicating distinct structural transitions induced in PDE5 by these ligands.
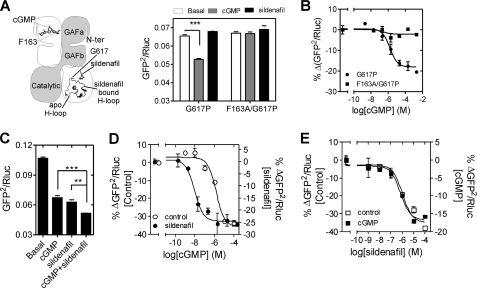
FIGURE 4.
Role of the H-loop in sildenafil-induced conformational change. A, scheme representation of the PDE5 dimer shows the relative positioning of the H-loop in the apo- and sildenafil-bound crystal structure of the catalytic domain of PDE5 (38). The positions of Gly-617 and bound sildenafil in the catalytic domain, and Phe-163 and bound cGMP in the GAFa domain are shown. Lysates prepared from cells expressing G617P or F163A/G617P mutant PDE5 sensors were incubated with 100 μm cGMP or sildenafil at 37 °C for 10 min and BRET determined. ***, p = 0.0002, Student's two-tailed t test. B, lysates prepared from cells expressing either the G617P and F163A/G617P mutant PDE5 sensors were incubated with varying concentrations of cGMP at 37 °C for 10 min in PDE lysis buffer followed by BRET determination. Percentage decrease in BRET in the presence of different concentrations of cGMP is plotted, and the EC50 value of the cGMP-induced conformational change was determined. Values represent mean ± S.E. (error bars) of a representative experiment with each experiment performed three times with duplicate determinations. C, lysates prepared from cells expressing the PDE5 sensor were incubated with either 100 μm cGMP or 100 μm sildenafil or both at 37 °C for 10 min, and BRET was determined. ** and *** denote p = 0.046 and 0.008, respectively, Student's two-tailed t test. D, lysates were preincubated with 100 μm sildenafil at 37 °C for 5 min followed by incubation with varying concentrations of cGMP at 37 °C for 5 min. The further percentage decrease in BRET was determined. E, lysates were preincubated with 100 μm cGMP at 37 °C for 5 min followed by incubation with varying concentrations of sildenafil at 37 °C for 5 min, and the percentage decrease in the BRET was determined. Lysates not preincubated with cGMP (D) or sildenafil (E) serve as controls. Values represent mean ± S.E. from a representative set of experiments performed three times with at least duplicate determinations.
In the simultaneous presence of both cGMP and sildenafil, BRET was significantly lower than that seen in the presence of the individual ligands (Fig. 4C). This indicated that the conformational changes induced in PDE5 by cGMP and sildenafil were distinct. Indeed, preincubation of lysates with sildenafil reduced the EC50 of cGMP-induced conformational change (Fig. 4D), but preincubation with cGMP did not alter the EC50 of sildenafil-induced BRET reduction (Fig. 4E). However, it is possible that cGMP competes with sildenafil for binding to the catalytic site, and cGMP binding to the catalytic site may not induce any change in BRET. If this was the case, no change in the EC50 for sildenafil would be seen.
Metal Ion Binding to the Active Site of PDE5 Regulates the cGMP-induced Conformational Change
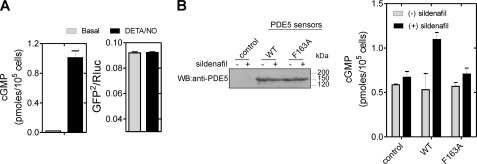
We then monitored ligand-induced conformational changes in PDE5 in intact cells. HEK293T cells were transfected with the PDE5 sensor and treated with DETA/NO. Intracellular cGMP levels increased to micromolar concentrations (assuming a cell volume of 1 pl), but no decrease in BRET was observed (Fig. 5A). Sildenafil, however, showed a decrease in BRET (supplemental Fig. S2A). The PDE5 sensor was found in the cytosol of cells (supplemental Fig. S1B), ruling out mislocalization as a cause for seeing no reduction in BRET in the presence of cGMP.
FIGURE 5.
PDE5 sensor does not respond to increases in intracellular cGMP but can act as a sink for cGMP. A, cells expressing the PDE5 sensor were treated with 1 mm DETA/NO for 5 min, and intracellular cGMP levels and BRET were measured. B, untransfected or cells transfected with the wild-type and F163A mutant PDE5 sensor plasmids were cultured in either medium alone or medium with 50 nm sildenafil for 24 h and lysed, and total intracellular cGMP was estimated by radioimmunoassay. Values represent the mean ± S.E. (error bars) from a representative experiment with triplicate determinations. Western blotting was performed using anti-PDE5 polyclonal antibody.
We checked that the GAFa domain of the PDE5 sensor could bind cGMP in intact cells by treating cells with sildenafil for 24 h. As shown in Fig. 5B, we observed an increase in intracellular levels of cGMP in cells expressing the wild-type but not the F163A mutant sensor (Fig. 5B). This is reflective of cGMP binding to the GAFa domain in this protein which acts as a sink for cGMP (27, 42). Therefore, despite the functionality of the PDE5 sensor in terms of binding cGMP in intact cells, it appeared that the conformational change (as monitored by a change in BRET) did not occur.
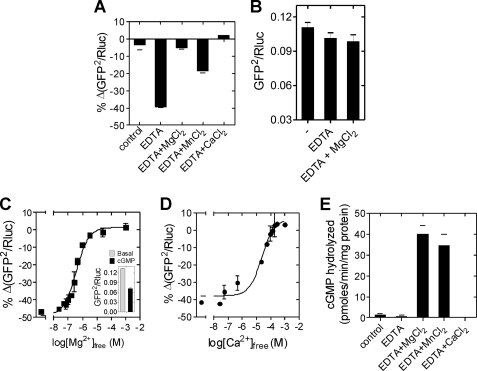
We hypothesized that factors present in intact cells constrained (or modified) the conformational changes that were seen in lysates on addition of cGMP. We inspected components of the lysis buffer that could have modulated the conformational transitions in the PDE5 sensor. Sodium ions have been shown to regulate cNMP binding to the GAF domains of CyaB2 and PDE2 (43). Analysis of the available structures of the GAFa and the catalytic domain of PDE5 (35, 38) showed potential disulfide pairing of cysteine residues, suggesting the possibility of redox regulation of PDE5 conformation in cells. Additionally, PDE5 requires two divalent cations (Mg2+ and Zn2+) bound to the catalytic domain for cGMP hydrolysis, and these metal ions could control the conformation of PDE5 in intact cells. We either removed or altered the concentrations of NaCl, DTT, or EDTA in the buffer used for cell lysis and in assays and monitored cGMP-induced BRET changes in the lysates. Changes in NaCl concentration and absence of reducing agent in the lysis buffer still permitted the cGMP-induced reduction in BRET (supplemental Fig. S3A). However, lysates prepared in a buffer without EDTA showed no cGMP-induced reduction in BRET (supplemental Fig. S3A and Fig. 6A). Addition of EDTA to the same lysates now restored the cGMP-induced conformational change, detected by a reduction in BRET (Fig. 6A). The maximum cGMP-induced BRET reduction was seen in the presence of 0.1 mm EDTA (supplemental Fig. S3B). Further, addition of MgCl2 along with EDTA abolished the cGMP-induced reduction in the BRET, and this effect was also seen in the presence of MnCl2, and CaCl2 (Fig. 6A). In the absence of cGMP, neither EDTA nor MgCl2 considerably altered BRET (Fig. 6B).
FIGURE 6.
Metal ion binding modulates the cGMP-induced conformational change in PDE5. A, lysates were prepared from cells expressing the PDE5 sensor in buffer lacking EDTA and then incubated in the absence or presence of 2 mm EDTA or with 2 mm EDTA along with the indicated salts (10 mm each). The percentage decrease in BRET in the presence of cGMP (100 μm) is plotted. B, lysates were prepared from cells expressing the PDE5 sensor in buffer lacking EDTA and then incubated with either 2 mm EDTA or 2 mm EDTA and 10 mm MgCl2 followed by BRET determination. C, lysates were prepared from cells expressing the PDE5 sensor in buffer lacking EDTA and then incubated with 0.1 mm EDTA and 100 μm cGMP for 5 min at room temperature. MgCl2 was added to generate the indicated free Mg2+ concentrations, and incubation continued at 37 °C for 10 min. The percentage decrease in BRET in the presence of indicated concentration of free metal ions was determined from that seen in the absence of EDTA and cGMP. Inset shows the reduction in BRET caused by 100 μm cGMP in the presence of 0.1 mm EDTA (mean ± S.E. (error bars)). D, lysates prepared from cells expressing the PDE5 sensor in buffer lacking EDTA were incubated with 0.1 mm EDTA and 100 μΜ cGMP for 5 min at room temperature. CaCl2 was added to generate the free Ca2+ concentrations indicated, and incubation continued at 37 °C for 10 min. The percentage decrease in BRET in the presence of the indicated concentrations of free Ca2+ is determined from that in the absence of EDTA and cGMP. E, cyclic GMP hydrolysis by the PDE5 sensor was monitored in the absence of any metal ions (EDTA) or in the presence of EDTA (2 mm) along with 10 mm total metal ions as indicated. Values represent mean ± S.E. from a representative experiment, with each experiment performed at least three times with duplicate determinations.
Mg2+ and Ca2+ inhibited the cGMP-induced conformational change with an EC50 of 1.2 ± 0.6 μm (Fig. 6C) and 19.2 ± 6 μm (Fig. 6D), respectively. Although both Mg2+ and Mn2+ hydrolyzed cGMP to similar extents (Fig. 6E), inclusion of Ca2+ in the lysate did not allow cGMP hydrolysis (44, 45). Therefore, the alleviation of the cGMP-induced BRET decrease did not result from hydrolysis of cGMP at the catalytic site, but was presumably due to a specific structural alteration brought about by the metal ions. Therefore, we suggest that intracellular Mg2+ or Ca2+ bound to PDE5 restricts the conformational change induced by cGMP binding to the GAF domain in cells, and therefore the reduction in BRET. Because the GAFa sensor was still able to respond to increases in intracellular cGMP, and in buffer without EDTA (supplemental Fig. S3, C–E), the “regulatory” metal ions are bound to PDE5 at a site distinct from the GAFa domain, perhaps in the catalytic domain.
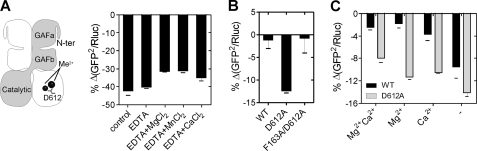
Indeed, mutation of the metal anchoring Asp-612 residue in the catalytic site (38) to alanine (D612A) permitted the cGMP-induced reduction in BRET, independent of the presence of EDTA and metal ions (Fig. 7A). Importantly, the D612A mutant PDE5 sensor was now responsive to cGMP elevation in cells (Fig. 7B), whereas responsiveness was lost in the F163A/D612A double mutant sensor. Therefore, metal ion binding at the catalytic site modified the conformational change induced on cGMP binding to the GAFa domain in PDE5.
FIGURE 7.
Catalytic metal ion binding prevents cGMP-induced conformational change in PDE5. A, scheme representation of the PDE5 dimer shows the relative positioning of the metal ions (Mg2+ and Zn2+) and Asp-612 in the crystal structure of the catalytic domain of PDE5 (38). Lysates were prepared from cells expressing the D612A mutant PDE5 sensor, and the percentage decrease in BRET from the basal levels, in the presence of 100 μm cGMP, is shown. B, cells expressing wild-type, D612A, or F163A/D612A mutant PDE5 sensors were treated with 1 mm DETA/NO for 5 min, and the percentage decrease in the BRET is plotted. C, cells expressing the wild-type or D612A mutant PDE5 sensor were incubated in Hanks' balanced salt solution and the indicated metals (as chloride salts) for 1 h. After incubation, cells were treated with 1 mm DETA/NO for 5 min, and percentage decreases in BRET are plotted. Values represent mean ± S.E. (error bars) from a representative experiment, with each experiment performed at least three times with duplicate determinations.
To demonstrate unequivocally that the presence of metal ions in cells modulates the conformation of PDE5, we reduced intracellular metal ion concentrations by incubating cells in media lacking either MgCl2, CaCl2 or both. Treatment of cells with DETA/NO caused a detectable and significant reduction in BRET in the wild-type PDE5 sensor only in cells incubated in buffer lacking both Mg2+ and Ca2+, whereas cells expressing the D612A mutant sensor showed BRET changes independent of the medium composition (Fig. 7C). The GAFa sensor was able to respond under similar conditions (supplemental Fig. S4). Therefore, a reduction in intracellular metal ion levels resulted in the generation of a fraction of metal ion-free PDE5, which was now sensitive to increased intracellular cGMP levels.
Distinct Allosteric Effects of cGMP, Sildenafil, and Catalytic Metal Ions on PDE5
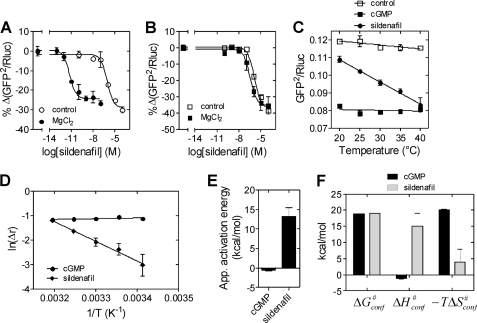
Given the allosteric effect of metal ions on the cGMP-induced structural change, we proceeded to investigate the effect of metal ions on sildenafil-induced structural change. We incubated lysates with varying concentrations of sildenafil in the presence of 10 mm MgCl2 and observed a striking decrease in the EC50 value (EC50 29.1 ± 14 pm; Fig. 8A). Incubation of the D612A mutant sensor with MgCl2 did not alter the EC50 of the sildenafil-induced conformational change (Fig. 8B), confirming that metal ion binding to the catalytic site allosterically altered ligand binding to PDE5.
FIGURE 8.
Distinct allosteric effect induced by different ligands in PDE5. A and B, lysates prepared from cells expressing wild-type (A) or D612A (B) PDE5 sensor in buffer lacking EDTA were preincubated with 10 mm MgCl2 for 5 min at room temperature, followed by incubation with varying concentrations of sildenafil at 37 °C for 10 min. The percentage decrease in BRET is plotted. Lysates not preincubated with MgCl2 serve as controls. C, lysates prepared from cells expressing the PDE5 sensor in buffer lacking EDTA were incubated in the absence or presence of either 100 μm cGMP or 100 μm sildenafil along with 2 mm EDTA for 10 min at the indicated temperatures, and BRET was determined. D, lysates prepared from cells expressing the PDE5 sensor in buffer lacking EDTA were incubated in the absence or presence of either cGMP or sildenafil (100 μm each) for 10 min at the indicated temperatures, and BRET was determined. Plot of the ln of the ratio of change in BRET in the presence of cGMP or sildenafil to basal BRET versus 1/T (K−1) is shown. E, apparent activation energies for the conformational change in PDE5 induced by cGMP and sildenafil estimated from the slope of the Arrhenius plot shown in D. Data represent mean ± S.E. (error bars) from a representative experiment performed at least twice with duplicate determinations. F, apparent free energy, enthalpy, and entropy (−TΔS) changes in the activation for the conformational change induced by cGMP and sildenafil at 37 °C. Data represent mean ± S.E. of a representative experiment, with experiments performed at least twice, with duplicate determinations.
We then extended our studies to suggest the distinct thermodynamic bases for cGMP- and sildenafil-induced conformational changes. Although the cGMP-induced conformational change occurred independently of temperature, the sildenafil-induced conformational change was temperature-dependent in the range of 20–40 °C (Fig. 8C). No considerable BRET change was observed in the absence of ligand at different temperatures, and the gross structural integrity of the PDE5 sensor in this temperature range was ascertained by similar luciferase activities at all the temperatures tested (supplemental Fig. S5).
Arrhenius plots were generated using the relative change in BRET versus 1/T, and provided an estimation of the apparent activation energies. The Eyring equation provided an estimate of apparent enthalpy and entropy changes in the activation process (supplemental Methods and Fig. 8D). The apparent activation energy for sildenafil was higher than that of cGMP (Fig. 8E), whereas the apparent free energy changes were similar. Interestingly, although sildenafil-induced conformational change involved both enthalpic and entropic changes, the cGMP-induced change was largely entropically driven (Fig. 8F). The repositioning and coil-to-310-helix transition of the H-loop, probably involving local unfolding in the catalytic domain (46), could account for the enthalpic barrier, involving an “induced-fit” mechanism in the sildenafil-induced conformational change. However, the high entropic contribution seen in the case of cGMP could arise from the “selection of conformations” with stabilized helix α4 and β2–β3 loop (including the short helix α2/3) (35) and altered the relative positioning and/or orientation of helices α2 and α5 (supplemental Fig. S1) (41, 47). Thus, although the “natural” ligand, cGMP, exploits the conformational ensemble (48) of PDE5, the “synthetic” ligand, sildenafil, requires large induced changes in the conformation, as seen in the lid-gated enzymes (49), to regulate PDE5 structure and activity.
DISCUSSION
In this study, we utilized a BRET-based sensor to monitor allosteric transitions induced in a multidomain protein and revealed novel features of allostery in PDE5. Because the BRET-based senor can report on the intracellular conformation of PDE5, it more faithfully monitors the state of the protein in its natural environment, as opposed to studying conformational alterations in highly purified preparations of the protein.
Mg2+ is known to regulate a number of cellular processes, and its intracellular free levels are tightly regulated (50, 51). Therefore, hormones and vasoactive agents (52) that regulate the free intracellular levels of Mg2+ could also alter PDE5 activity. The dramatic increase in the affinity of sildenafil in the presence of metal ions could explain the prolonged action of PDE5 inhibition (53) in vivo. Importantly, binding of Ca2+ to the catalytic domain, while not permitting catalysis, could provide an added means of PDE5 regulation, especially in the contracted state of smooth muscle cells (54) where high intracellular Ca2+ levels are seen.
Metal ion binding to the catalytic domain of PDE5 did not cause a major change in BRET (Fig. 6B), but brought about significant effects on cGMP- and sildenafil-induced conformational changes (Figs. 6, A, C, and D, and 8A). We suggest that the metal-ion induced conformational change in PDE5 could represent an entropically driven mechanism, where the conformational entropy of the protein is altered, with no marked change in the mean structure of PDE5 (7, 8). Indeed, divalent metal ion binding to an endonuclease was found to be largely entropy-driven without any gross structural change (55). This entropic effect of metal ion binding at the catalytic site of PDE5 may inhibit the conformational selection of low BRET conformers by cGMP binding at the distant GAFa domain (long range allosteric effect). However, metal ion binding facilitates the induction of conformational change by local sildenafil binding (short range allosteric effect).
Sildenafil orthosterically inhibits cGMP hydrolysis at the catalytic site, but allosterically increased the affinity of the GAFa domain for cGMP (Fig. 4B), giving rise to a “new function” for the GAFa domain to act as sink for cGMP (27, 42). Although the paradigm is to identify allosteric sites distinct from the catalytic site and target those sites for regulation of the function of the protein, in a multidomain protein such as PDE5, it appears that one can develop allosteric modulators that can bind to the catalytic site and alter the properties of a second domain in the protein.
Point mutations (F778A and G617P) altered basal BRET of the PDE5 sensor (Figs. 3 and 4). This indicates that specific mutations can shift the conformational ensemble of PDE5. Interestingly, the ability of the F163A mutant to respond to sildenafil and the ability of F778A and G617P mutants to respond to cGMP suggest that cognate ligand binding and the subsequent conformational changes brought about in otherwise allosterically coupled domains of a protein are independent of mutations in the “other” domain.
The prevention of cGMP-induced conformational change in PDE5 in the presence of metal ions (Fig. 6) and similar kinetic properties of the wild-type and F163A mutant PDE5 (Fig. 1D) raise questions as to the role of cGMP in directly regulating PDE5 catalytic activity. Indeed, cGMP binding to the GAF domains of PDE10 and PDE11 does not alter their activities (56). Cyclic GMP binding to the GAFa domain of PDE5 could, however, alter in vivo stability, allow enhanced phosphorylation by cGMP-dependent protein kinase (57), and alter the ability of PDE5 to form complexes with other proteins (58) and/or its localization in the cell (59, 60).
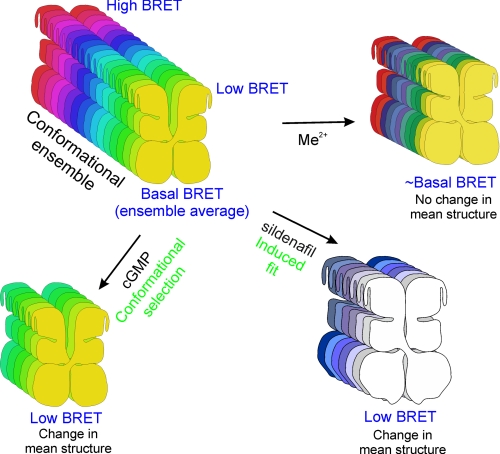
In conclusion, we propose that cGMP, cAMP, Mg2+, Ca2+, and sildenafil can regulate PDE5 conformation and hence its activity in cells. The BRET sensor that we have developed allows us to suggest the following scheme of conformational changes that may occur in PDE5. An ensemble of conformations of PDE5 can exist, with a spectrum of BRET, and basal BRET being the average (Fig. 9). Cyclic GMP may selectively bind the low BRET conformers shifting the equilibrium toward reduced average BRET. Sildenafil induces conformational changes in the protein that require high activation energy, reflected in a reduction in BRET. A change in the mean structure of the protein occurs in both the cases. Metal ions binding to the catalytic site may alter the dynamics of PDE5 without causing any considerable change in the mean structure, and therefore no reduction in BRET is observed. Sequential binding of sildenafil and cGMP, and vice versa, further changes the mean structure of the protein, reflected in a reduced BRET. Metal ion binding to the catalytic site can revert the cGMP-bound low BRET conformers to ∼basal BRET conformers. In contrast, metal ion binding to the catalytic site enhances sildenafil-induced reduction in BRET. We envisage the use of this sensor to gain further insight into the regulation of PDE5 conformation in cells that endogenously express PDE5 and for identifying allosteric modulators of PDE5 that regulate its activity, without giving rise to the sink-like property of the GAFa domain.
FIGURE 9.
Conformational transitions in PDE5. An ensemble of conformations of PDE5 can exist, with a spectrum of BRET, and the basal BRET being the average. Cyclic GMP selectively binds the low BRET conformers shifting the equilibrium toward reduced average BRET. Sildenafil induces conformational changes in the protein which require high activation energy resulting in a reduction in BRET. A change in the mean structure of the protein occurs in both the cases. Metal ions binding to the catalytic site alter the dynamics of the protein without causing any considerable change in the mean structure. Sequential binding of sildenafil and cGMP, and vice versa, further changes the mean structure of the protein, which for simplicity, is not depicted in the figure. Metal ion binding to the catalytic site reverts the cGMP-bound low BRET conformers to ∼basal BRET conformers. In contrast, metal ion binding to the catalytic site enhances sildenafil-induced reduction in BRET.
Supplementary Material
This work was supported by the Department of Biotechnology, Government of India, and the Indian Institute of Science.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5, Methods, and additional references.
- PDE
- phosphodiesterase
- BRET
- bioluminescence resonance energy transfer
- DETA/NO
- diethylenetriamine-NO
- EGFP
- enhanced green fluorescent protein
- GAF
- cGMP-specific phosphodiesterases, bacterial adenylyl cyclase, FhLA transcriptional regulator
- IBMX
- isobutylmethylxanthine
- PDE5
- cGMP-binding, cGMP-specific PDE
- Rluc
- Renilla luciferase.
REFERENCES
- 1. Changeux J. P., Edelstein S. J. (2005) Science 308, 1424–1428 [DOI] [PubMed] [Google Scholar]
- 2. Fenton A. W. (2008) Trends Biochem. Sci. 33, 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cui Q., Karplus M. (2008) Protein Sci. 17, 1295–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Henzler-Wildman K., Kern D. (2007) Nature 450, 964–972 [DOI] [PubMed] [Google Scholar]
- 5. Hammes G. G., Chang Y. C., Oas T. G. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13737–13741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hilser V. J. (2010) Science 327, 653–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cooper A., Dryden D. T. (1984) Eur. Biophys. J. 11, 103–109 [DOI] [PubMed] [Google Scholar]
- 8. Tsai C. J., del Sol A., Nussinov R. (2008) J. Mol. Biol. 378, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Del Sol A., Araúzo-Bravo M. J., Amoros D., Nussinov R. (2007) Genome Biol. 8, R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kuriyan J., Eisenberg D. (2007) Nature 450, 983–990 [DOI] [PubMed] [Google Scholar]
- 11. Conti M., Beavo J. (2007) Annu. Rev. Biochem. 76, 481–511 [DOI] [PubMed] [Google Scholar]
- 12. Turko I. V., Francis S. H., Corbin J. D. (1998) Biochemistry 37, 4200–4205 [DOI] [PubMed] [Google Scholar]
- 13. Zhang K. Y., Card G. L., Suzuki Y., Artis D. R., Fong D., Gillette S., Hsieh D., Neiman J., West B. L., Zhang C., Milburn M. V., Kim S. H., Schlessinger J., Bollag G. (2004) Mol. Cell 15, 279–286 [DOI] [PubMed] [Google Scholar]
- 14. Lin C. S. (2004) Int. J. Impot. Res. 16, S8–S10 [DOI] [PubMed] [Google Scholar]
- 15. Rotella D. P. (2002) Nat. Rev. Drug Discov. 1, 674–682 [DOI] [PubMed] [Google Scholar]
- 16. Galiè N., Ghofrani H. A., Torbicki A., Barst R. J., Rubin L. J., Badesch D., Fleming T., Parpia T., Burgess G., Branzi A., Grimminger F., Kurzyna M., Simonneau G. (2005) N. Engl. J. Med. 353, 2148–2157 [DOI] [PubMed] [Google Scholar]
- 17. Mullershausen F., Friebe A., Feil R., Thompson W. J., Hofmann F., Koesling D. (2003) J. Cell Biol. 160, 719–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rybalkin S. D., Rybalkina I. G., Shimizu-Albergine M., Tang X. B., Beavo J. A. (2003) EMBO J. 22, 469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Okada D., Asakawa S. (2002) Biochemistry 41, 9672–9679 [DOI] [PubMed] [Google Scholar]
- 20. Turko I. V., Ballard S. A., Francis S. H., Corbin J. D. (1999) Mol. Pharmacol. 56, 124–130 [DOI] [PubMed] [Google Scholar]
- 21. Blount M. A., Zoraghi R., Bessay E. P., Beasley A., Francis S. H., Corbin J. D. (2007) J. Pharmacol. Exp. Ther. 323, 730–737 [DOI] [PubMed] [Google Scholar]
- 22. Corbin J. D., Blount M. A., Weeks J. L., 2nd, Beasley A., Kuhn K. P., Ho Y. S., Saidi L. F., Hurley J. H., Kotera J., Francis S. H. (2003) Mol. Pharmacol. 63, 1364–1372 [DOI] [PubMed] [Google Scholar]
- 23. Rybalkina I. G., Tang X. B., Rybalkin S. D. (2010) Mol. Pharmacol. 77, 670–677 [DOI] [PubMed] [Google Scholar]
- 24. Francis S. H., Bessay E. P., Kotera J., Grimes K. A., Liu L., Thompson W. J., Corbin J. D. (2002) J. Biol. Chem. 277, 47581–47587 [DOI] [PubMed] [Google Scholar]
- 25. Corbin J. D., Zoraghi R., Francis S. H. (2009) Cell. Signal. 21, 1768–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gierasch L. M., Gershenson A. (2009) Nat. Chem. Biol. 5, 774–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Biswas K. H., Sopory S., Visweswariah S. S. (2008) Biochemistry 47, 3534–3543 [DOI] [PubMed] [Google Scholar]
- 28. Shenoy A. R., Visweswariah S. S. (2003) Anal. Biochem. 319, 335–336 [DOI] [PubMed] [Google Scholar]
- 29. Sopory S., Kaur T., Visweswariah S. S. (2004) Cell. Signal. 16, 681–692 [DOI] [PubMed] [Google Scholar]
- 30. Bakre M. M., Sopory S., Visweswariah S. S. (2000) J. Cell. Biochem. 77, 159–167 [PubMed] [Google Scholar]
- 31. Bakre M. M., Ghanekar Y., Visweswariah S. S. (2000) Eur. J. Biochem. 267, 179–187 [DOI] [PubMed] [Google Scholar]
- 32. Lin C. S., Lau A., Tu R., Lue T. F. (2000) Biochem. Biophys. Res. Commun. 268, 628–635 [DOI] [PubMed] [Google Scholar]
- 33. Lin C. S., Chow S., Lau A., Tu R., Lue T. F. (2002) Int. J. Impot. Res. 14, 15–24 [DOI] [PubMed] [Google Scholar]
- 34. Dacres H., Dumancic M. M., Horne I., Trowell S. C. (2009) Anal. Biochem. 385, 194–202 [DOI] [PubMed] [Google Scholar]
- 35. Heikaus C. C., Stout J. R., Sekharan M. R., Eakin C. M., Rajagopal P., Brzovic P. S., Beavo J. A., Klevit R. E. (2008) J. Biol. Chem. 283, 22749–22759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zoraghi R., Bessay E. P., Corbin J. D., Francis S. H. (2005) J. Biol. Chem. 280, 12051–12063 [DOI] [PubMed] [Google Scholar]
- 37. Sopory S., Balaji S., Srinivasan N., Visweswariah S. S. (2003) FEBS Lett. 539, 161–166 [DOI] [PubMed] [Google Scholar]
- 38. Wang H., Liu Y., Huai Q., Cai J., Zoraghi R., Francis S. H., Corbin J. D., Robinson H., Xin Z., Lin G., Ke H. (2006) J. Biol. Chem. 281, 21469–21479 [DOI] [PubMed] [Google Scholar]
- 39. Wang H., Ye M., Robinson H., Francis S. H., Ke H. (2008) Mol. Pharmacol. 73, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sung B. J., Hwang K. Y., Jeon Y. H., Lee J. I., Heo Y. S., Kim J. H., Moon J., Yoon J. M., Hyun Y. L., Kim E., Eum S. J., Park S. Y., Lee J. O., Lee T. G., Ro S., Cho J. M. (2003) Nature 425, 98–102 [DOI] [PubMed] [Google Scholar]
- 41. Pandit J., Forman M. D., Fennell K. F., Dillman K. S., Menniti F. S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18225–18230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Corbin J. D., Kotera J., Gopal V. K., Cote R. H., Francis S. H. (2003) in Handbook of Cell Signaling (Dennis E. A. ed) pp. 465–470, Academic Press, Burlington, VT [Google Scholar]
- 43. Cann M. (2007) Mol. Microbiol. 64, 461–472 [DOI] [PubMed] [Google Scholar]
- 44. Francis S. H., Colbran J. L., McAllister-Lucas L. M., Corbin J. D. (1994) J. Biol. Chem. 269, 22477–22480 [PubMed] [Google Scholar]
- 45. Francis S. H., Turko I. V., Grimes K. A., Corbin J. D. (2000) Biochemistry 39, 9591–9596 [DOI] [PubMed] [Google Scholar]
- 46. Hilser V. J., Thompson E. B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8311–8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Martinez S. E., Wu A. Y., Glavas N. A., Tang X. B., Turley S., Hol W. G., Beavo J. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13260–13265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Boehr D. D., Nussinov R., Wright P. E. (2009) Nat. Chem. Biol. 5, 789–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sullivan S. M., Holyoak T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13829–13834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wolf F. I., Trapani V. (2008) Clin. Sci. 114, 27–35 [DOI] [PubMed] [Google Scholar]
- 51. Yago M. D., Manas M., Singh J. (2000) Front. Biosci. 5, D602–D618 [DOI] [PubMed] [Google Scholar]
- 52. Touyz R. M. (2003) Mol. Aspects Med. 24, 107–136 [DOI] [PubMed] [Google Scholar]
- 53. Francis S. H., Morris G. Z., Corbin J. D. (2008) Int. J. Impot. Res. 20, 333–342 [DOI] [PubMed] [Google Scholar]
- 54. Rybalkin S. D., Yan C., Bornfeldt K. E., Beavo J. A. (2003) Circ. Res. 93, 280–291 [DOI] [PubMed] [Google Scholar]
- 55. Feng M., Patel D., Dervan J. J., Ceska T., Suck D., Haq I., Sayers J. R. (2004) Nat. Struct. Mol. Biol. 11, 450–456 [DOI] [PubMed] [Google Scholar]
- 56. Matthiesen K., Nielsen J. (2009) Biochem. J. 423, 401–409 [DOI] [PubMed] [Google Scholar]
- 57. Corbin J. D., Turko I. V., Beasley A., Francis S. H. (2000) Eur. J. Biochem. 267, 2760–2767 [DOI] [PubMed] [Google Scholar]
- 58. Wilson L. S., Elbatarny H. S., Crawley S. W., Bennett B. M., Maurice D. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13650–13655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dolci S., Belmonte A., Santone R., Giorgi M., Pellegrini M., Carosa E., Piccione E., Lenzi A., Jannini E. A. (2006) Biochem. Biophys. Res. Commun. 341, 837–846 [DOI] [PubMed] [Google Scholar]
- 60. Francis S. H., Zoraghi R., Kotera J., Ke H., Bessay E., Blount M., Corbin J. D. (2006) in Cyclic Nucleotide Phosphodiesterases in Health and Disease (Houslay M. ed) pp. 135–164, CRC Press, Boca Raton, FL [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.