Abstract
The Golgi apparatus undergoes extensive fragmentation during apoptosis due in part to caspase-mediated cleavage of its structural proteins. Significantly, the Golgi-vesicle-tethering protein p115 is cleaved at Asp757 early during apoptosis and the nuclear translocation of its 205 amino acid C-terminal fragment (CTF) precedes observable Golgi fragmentation. Nuclear localization of the p115 CTF induces apoptosis. The regulation of CTF nuclear translocation and the mechanism of its apoptotic activity however, remain unknown. Here, we demonstrate that nuclear translocation of the CTF is regulated by SUMOylation. CTF-induced apoptosis is transcription dependent and mediated by the tumor suppressor, p53. Expression of the CTF led to the phosphorylation and stabilization of p53 and results in the expression of PUMA, a pro-apoptotic target of p53. CTF-induced stabilization of p53 is sensitive to the MEK/ERK inhibitor U0126. Co-immunoprecipitation studies indicate that the p115 CTF can bind to both p53 and ERK1. The CTF is also able to form dimers and its dimerization is dependent on residues 859–884, previously determined to be required for apoptosis. Indeed, CTF expression promotes p53-ERK interaction, which is diminished upon deletion of residues 859–884. Together, our results indicate a conserved tethering function of the Golgi protein p115 CTF which promotes p53-ERK interaction for the amplification of the apoptotic signal.
Keywords: Apoptosis, Golgi, p53, Signal Transduction, Sumoylation
Introduction
The Golgi apparatus functions in the processing, sorting, and trafficking of proteins from the endoplasmic reticulum (ER)2 to various cellular destinations. In mammalian cells, the Golgi consists of a series of closely apposed stacks of membranous cisternae localized to the perinuclear region of the cell (reviewed in Ref. 1). The cis-Golgi receives newly synthesized protein and lipid cargo from the ER, which are significantly processed and modified by enzymes such as glycosyltransferases and glycosidases, as they traverse the organelle. Mature cargo proteins are then sorted in the trans-Golgi network for trafficking to the plasma membrane, lysosomal-endosomal compartment and secretory granules.
Salient to the function of the Golgi as an intracellular hub of protein and membrane trafficking is its dynamic structure, which is organized by an interplay between the microtubule and actin cytoskeletons, and resident Golgi structural proteins. A key molecule contributing to the biogenesis and maintenance of the Golgi structure is the vesicle-tethering protein p115. p115 is a 961-kDa peripheral membrane protein that contains an N-terminal globular head region and forms homodimers via its extended coiled-coil (CC) tail domain (2). It functions in Golgi-vesicle tethering and Golgi-cisternal stacking by bridging GM130 and giantin, which are found on the cis-Golgi and COPI vesicles, respectively (3). The formation of this tethering complex further facilitates subsequent membrane docking through p115 interactions with various SNAREs such as the v- and t-SNAREs GOS28 and syntaxin-5 (4). In addition, p115 also binds to the GTPase Rab1(5), COG (conserved oligomeric Golgi) complex (6) and β-COP (7), supporting its role as a tethering factor in membrane trafficking. The knockdown of p115 results in Golgi fragmentation (8), showing that its tethering properties are required for the proper maintenance of Golgi structure and morphology.
Physiological fragmentation of the Golgi apparatus occurs during mitosis and apoptosis. During mitosis, the reversible regulation of Golgi components is important to ensure proper inheritance and partitioning of Golgi fragments into daughter cells. Fragmentation of the Golgi apparatus occurs in the G2 phase and is mediated by the phosphorylation of Golgi components involving members of various kinase families such as cyclin-dependent kinases (9, 10), polo-like kinases (11, 12), and specific members of the MEK1/ERK1 pathway (13–17). While Golgi structural proteins are regulated during entry into mitosis, failure to reassemble the Golgi apparatus at the end of mitosis leads to mitotic arrest. Thus the Golgi structure can act as a signal that regulates cell cycle progression.
In contrast to mitosis, apoptotic Golgi fragmentation is irreversible, leading to the cessation of protein and membrane trafficking and facilitating the packaging of cellular components into apoptotic bodies. Apoptotic breakdown of the Golgi is mediated by the activity of caspases, which cleave its structural components including GM130 (18), GRASP65 (18), Golgin 160 (19, 20), p115 (21), syntaxin-5(22), and giantin (22). Notably, cleavage fragments of some of these Golgi proteins have been shown to act as signaling components that further regulate cell fate. For example, the N terminus caspase cleavage fragment of Golgin-160 functions as a pro-survival signal in the nucleus. Its nuclear entry is regulated by interaction with a redox-sensitive partner, GCP60 (Golgi complex-associated protein of 60kDa) (23).
In addition, our own laboratory has shown that the cleavage of p115 is required for apoptotic Golgi fragmentation. Apoptotic cleavage of p115 is an early event, occurring independently of and before the breakdown of the microtubule and actin cytoskeletons (25). p115 is cleaved at Asp757 by caspases-3 and -8, leading to the fragmentation of the Golgi apparatus and generation of a 205-residue C-terminal caspase cleavage fragment (CTF) (21). Endogenous p115 CTF can be found in the nucleus as early as 2 h after Fas receptor activation, before noticeable fragmentation of the Golgi apparatus (24). Notably, expression of the CTF (residues 758–961) is sufficient to induce apoptosis and its nuclear localization as well as a minimal domain of 26 amino acid residues (859–884) is required for the induction of apoptosis (24). Furthermore, knockdown of p115 attenuated Fas-ligand- and anisomycin-induced apoptosis, thus affecting the execution of apoptosis in response to triggers of both the extrinsic and intrinsic apoptotic pathways. The pro-apoptotic activity of the CTF within the nucleus and its mechanism of nuclear translocation however, remains elusive.
The purpose of this study was to determine the molecular basis of CTF-mediated apoptosis. We found that nuclear-CTF-induced apoptosis is transcription dependent and is mediated by the tumor suppressor p53. CTF expression leads to the phosphorylation and stabilization of p53. This stabilization however, was attenuated by the MEK1 inhibitor U0126. Co-immunoprecipitation studies showed that CTF was able to interact with p53 and with ERK-1. We observed that like full length p115, CTF was able to dimerize and the 26 amino acid minimal domain was required for CTF dimerization. Dimerization of the CTF promoted the close proximity of ERK and p53 for phosphorylation. Together, our results suggest a model where the conserved tethering function of the p115 CTF may facilitate ERK phosphorylation of p53 for apoptotic signal amplification.
EXPERIMENTAL PROCEDURES
Reagents
Rabbit polyclonal antibody to the C terminus of human p115 (amino acid 645–962) was generated as previously described (24). Polyclonal antibodies for detection of PARP(9542), phosphor-Ser15 of p53 (9284), phospho-Ser20 of p53 (9287), PUMA(4976), ERK1/2(4695), phospho-Thr180/Tyr182 of ERK1/2(4370), p38 (9212), phospho-Thr202/Tyr204 of p38 (4511), JNK(9258), phospho-Thr183/Tyr185 of JNK(4668), and Ubc-9(4918) were purchased from Cell Signaling Technology (Danvers, MA). Antibodies for p53 (sc126), p21(sc756) and murine estrogen receptor-α (sc542) were obtained from SantaCruz Biotechnology. Polyclonal GFP antibody and goat anti-rabbit IgG antibody coupled to Alexa Fluor 546 was obtained from Molecular Probes (Invitrogen). EZViewTM Red Anti-Flag® M2 Affinity Gel was purchased from Sigma. Various protein kinase inhibitors were obtained from Calbiochem, Sigma, and Fisher Scientific. Etoposide and 4-hydroxytamoxifen were obtained from Calbiochem. ICI182780, α-amanitin, and actinomycin D were obtained from Sigma.
Cell Culture
HeLa, U2OS, and Sa-OS2 cells were cultured in Dulbecco's modified Eagle's medium. HCT116 p53+/+ and p53−/− cells were gifts from Dr. Bert Vogelstein (JHUMI, Baltimore, MD) and were cultured in McCoy's 5A Medium. All media were supplemented with 10% fetal calf serum, 2 mm l-glutamine, penicillin (100 units/ml), and streptomycin (100 mg/ml) (Invitrogen). Cells were transfected by using Lipofectamine 2000 (Invitrogen).
Plasmid Constructs
The pSG5-FLAG-human p115 construct was provided by Dr. Yukio Ikehara (Fukuoka University School of Medicine, Fukuoka, Japan) and was used as a template to generate p115 constructs. pECE-HA-FOXO3a-ER was provided by Dr. Michael Greenberg (Harvard Medical School, Boston, MA) and used as a backbone to generate pECE-CTF-ER by deletion of the HA-FOXO3a sequence and insertion of a p115 CTF (758–962) fragment. pKH3-SUMO and pcDNA3.1-Ubc-9 were gifts from Dr. Moshe Sadofsky. pEGFP-c1-SUMO-CTF was constructed by sequential insertion of SUMO and p115 CTF fragments cloned from the pKH3-SUMO template and pSG5-FLAG-human p115 plasmids, respectively. pcDNA3.1 WT and mutant p53 were provided by Dr. Roger S. Y. Foo (School of Clinical Medicine, Cambridge, UK). Gal4-CTF constructs were cloned by inserting various CTF fragments into the pSG424 backbone, which expresses the Gal4-DBD under the control of the early promoter of SV40 (26).
Immunofluorescence Analysis
Cells were grown on coverslips and transfected as indicated, after which they were fixed in 3% paraformaldehyde and processed for immunofluorescence microscopy as previously described (21). Images were visualized with a Nikon Eclipse TE300 inverted fluorescence microscope using either the 60× Plan Apo/1.4NA oil DIC H or 100× Plan Apo/1.4NA oil DIC H objectives. Images were taken with a Nikon Digital Sight DS-U1 camera using the NIS-Elements Freeware software and saved as TIFF files (800 × 600 pixels, 8-bit). TIFF images were adjusted for brightness and contrast in Adobe Photoshop CS3. All images were taken at room temperature.
Luciferase Assays
A pSG424CTF expressing Gal4-CTF was co-transfected with a 5xGAL-M2-luciferase reporter that contains five Gal4 binding sites and the TATA sequence, followed by a retinoic acid receptor β2 initiator (nucleotides −5 to +6) driving the firefly luciferase gene (26) and the control plasmid pRSV β-galactosidase into HeLa cells in the presence of ZVAD to prevent apoptosis. Luciferase and β-galactosidase activities were measured at 24 h after transfection. The data were presented as the relative luciferase activity, calculated as the ratio of the activity of luciferase to the activity of β-galactosidase (values are means ± S.D.).
Annexin V Staining and FACS Analysis
After transfection cells were trypsinized and washed in PBS and resuspended in Annexin V binding buffer (10 mm HEPES, 140 mm NaCl, and 2.5 mm CaCl2, pH 7.4.). Cells were treated with Annexin V according to the manufacturer's protocol and propidium iodide added as a counterstain for necrotic cells. Cells were then analyzed by flow cytometry.
Quantitative RT-PCR
Total RNA was prepared from HCT116 p53+/+ transfected with vector alone or 3XFlagCTF in the presence of Z-VAD-fmk to prevent apoptosis. Isolated RNA was reverse transcribed with superscript II reverse transcriptase (Invitrogen). Quantitative real-time PCR was performed using a SYBR Green PCR kit (Qiagen) with primers to amplify the PUMA [F: 5′-ACGACCTCAACGCACAGTACGA-3′; R: 5′-GTAAGGGCAGGAGTCCCATGATGA-3′ (49)], NOXA [F: 5′-TGGAAGTCGAGTGTGCTACTCAACT-3′; R: 5′-AGATTCAGAAGTTTCTGCCGGAA-3′ (50)], p21 [F: 5′-GGCAGACCAGCATGACAGATT-3′; R: 5′-GCGGATTAGGGCTTCCTCTT-3′ (51)], hdm2, [F: 5′-TCCCCGTGAAGGAAACTGG-3′; R: 5′-TTTCGCGCTTGGAGTCG-3′ (52)] and actin [F: 5′-CTCTTCCAGCCTTCCTTCCT-3′; R: 5′-AGCACTGTGTTGGCGTACAG-3′ (53)] transcripts. The ΔC(t) values were normalized to those of actin by subtracting the C(t) value obtained for actin from the C(t) value obtained for a specific primer set. ΔΔC(t) was obtained from the difference between ΔC(t) values in control and 3XFlagCTF-transfected cells. The fold increase or decrease of RNA expressed were calculated by using the formula: fold change = 1−ΔΔC(t).
Protein Immunoblotting Analysis
Cells extracts were isolated in a Triton-lysis buffer containing 20 mm Tris-HCl, pH 7.4, 134 mm NaCl, 1% Triton X-100, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride and a mixture of protease inhibitors (Roche). 20 μg of each sample was loaded on a 10% polyacrylamide gel, followed by transfer to Immobilon-P membranes (Millipore Corp). Membranes were blocked in 5% nonfat milk in Tris-buffered saline containing 0.1% Tween 20 (TBST) for 1 h. Subsequently, they were probed with appropriate antibodies in TBST, washed extensively and the immunoreactive bands were visualized by Enhanced Chemiluminescence (Amersham Biosciences).
Subcellular Fractionation
Cytoplasmic and nuclear extracts were prepared according to (54). In brief, harvested cells were gently resuspended in cytoplasmic buffer (10 mm HEPES 50 mm NaCl, 0.5 m sucrose, 1 mm EDTA, 0.25 mm EGTA, 1 mm phenylmethylsulfonyl fluoride, 10 mm benzamidine, and 5 μg/ml leupeptin) containing 5% Nonidet P-40. After 10 min on ice, nuclei were pelleted at 3,000 rpm for 5 min at 4 °C and cytoplasmic extract was collected. Isolated nuclei were resuspended in nuclear extraction buffer (20 mm HEPES, pH 7.9, 0.4 m NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, 0.5 mm spermidine, 0.15 mm spermine, 25% glycerol, 1 mm phenylmethylsulfonyl fluoride, 10 mm benzamidine, and 5 μg/ml leupeptin) for 30 min, with periodic vortexing every 5 min. The sample was spun at 15,000 rpm for 15 min at 4 °C to collect nuclear extracts.
RESULTS
CTF-induced Apoptosis Is Transcription-dependent
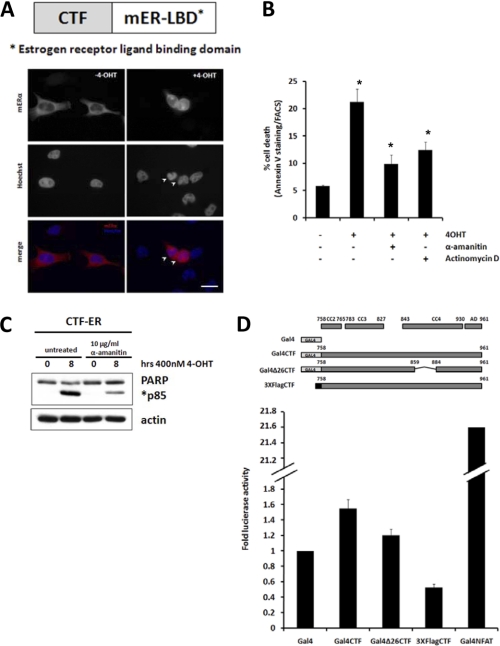
Our laboratory has previously established that nuclear localization of the p115 CTF is required for its apoptotic activity (24). This led us to hypothesize that nuclear CTF may affect gene transcription to produce this effect. To test this hypothesis, we fused the CTF with the estrogen receptor ligand-binding domain (CTF-ER) to facilitate conditional induction of CTF nuclear localization. As expected, CTF-ER was found in the cytoplasm when expressed in HeLa cells in the absence of an estrogen-receptor ligand (Fig. 1A, left panel). Upon addition of the estrogen ligand 4-hydroxytamoxifen (4-OHT), CTF-ER was found in the nucleus and nuclear resident CTF-ER induced apoptosis as measured by nuclear condensation (Fig. 1A, right panel). These data indicate that conditional induction of CTF translocation into the nucleus is pro-apoptotic. Next, we examined the effect of transcriptional inhibitors on CTF-induced apoptosis. Administration of 4-OHT to CTF-ER-expressing cells induced apoptosis as indicated by increased Annexin V staining (Fig. 1B). Pretreatment with transcriptional inhibitors α-amanitin or actinomycin D attenuated CTF-ER-mediated apoptosis. The transcriptional inhibitor α-amanitin also reduced cleavage of PARP, which was elicited by nuclear localization of CTF-ER (Fig. 1C). Together these data indicate that CTF-induced apoptosis is transcription dependent.
FIGURE 1.
CTF-induced apoptosis is transcription-dependent. A–C, HeLa cells were transiently transfected with a CTF-ER construct. A, after 12 h of transfection, cells were treated with DMSO (left panel) or 4-OHT (right panel) for 4 h, fixed, and processed for immunofluorescence microscopy. Bar, 10 μm. B, cells were pretreated for 4 h with the indicated RNA polymerase inhibitors and induced for 8 h with 4-OHT followed by staining with Annexin V and FACS analysis. C, cells were pretreated for 4 h with α-amanitin and induced with 4-OHT for 8 h, lysed, and run on SDS-PAGE. Protein levels of PARP and actin were detected by Western blot and quantified using the TINA 2.09 software program. Apoptotic index was obtained by dividing the intensity of the cleaved PARP band (p85) at the indicated time over the intensity at time 0. D, HeLa cells were transiently transfected with the indicated Gal4-tagged constructs and a luciferase reporter containing the Gal4 DNA binding element. A pRSV40-β-galactosidase construct was co-transfected as a control. After 18 h, cells were lysed and luciferase activity measured with a luminometer. Luciferase activity was normalized to β-galactosidase activity and fold-luciferase activity obtained by dividing values with the value obtained for Gal4 alone. *, p < 0.03, Student's t test.
The p115 CTF may possess intrinsic transactivation properties and directly affect apoptotic gene transcription. To test this hypothesis, we fused CTF with the Gal4 DNA binding domain (Gal4-CTF). Expression of Gal4-CTF did not significantly increase luciferase reporter activity as compared with Gal4 expression alone. In contrast, expression of Gal4-NFAT markedly increased luciferase reporter activity as reported previously (Fig. 1D) (26). These data indicate that while CTF-mediated apoptotis is transcription dependent, the CTF itself cannot directly activate gene transcription.
CTF-induced Apoptosis Is p53-dependent
Next, we tested whether the CTF induces apoptosis by indirectly modulating the activity of a known transcription factor cascade. Candidate apoptotic pathways included the tumor suppressor p53, which is involved in the transactivation of key pro-apoptotic genes in response to a variety of cellular stresses and chemotherapeutic drugs (27). Expression levels of p53 are regulated by a series of post-translational modifications including phosphorylation and ubiquitination. Under basal conditions, expression levels of p53 are low. p53 is constantly turned over due to its interaction with the ubiquitin ligase mdm2, which ubiquitinates p53 and promotes its degradation via the proteasome (reviewed in Ref. 28). Under stressed conditions, p53 levels are stabilized by phosphorylation at multiple sites, including Ser15 and Ser20, which abrogates p53 interaction with mdm2. Phosphorylation of p53, therefore, leads its accumulation and activation, which subsequently mediates gene transcription.
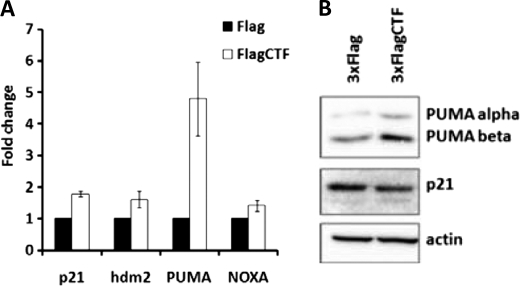
We performed quantitative-RT-PCR analysis of several p53 targeted genes in the HCT116 p53+/+ cell line which expresses wild-type p53 (Fig. 2A). Expression of the p115 CTF increased expression of the BH3-only member of the Bcl-2 family protein PUMA (p53-Up-regulated Modulator of Apoptosis). Immunoblotting analysis further confirmed increased levels of protein expression of both PUMA-α and PUMA-β in the presence of CTF (Fig. 2B). In comparison, expression of other p53 targets, such as p21, hdm2, and NOXA, however, was not affected by CTF expression (Fig. 2, A and B). These data indicate that CTF induces p53-mediated apoptotic gene transcription.
FIGURE 2.
CTF induces expression of PUMA. A, HCT116 p53+/+cells were transfected with vector alone or 3XFlagCTF in the presence of Z-VAD-fmk. RNA was collected after 18 h and subjected to quantitative RT-PCR. Fold change was calculated using −ΔΔCt normalized to actin mRNA levels. B, HCT116 p53+/+cells were transfected with a 3XFlagCTF construct or vector alone without Z-VAD-fmk treatment and lysates collected for Western blot analysis after 18 h.
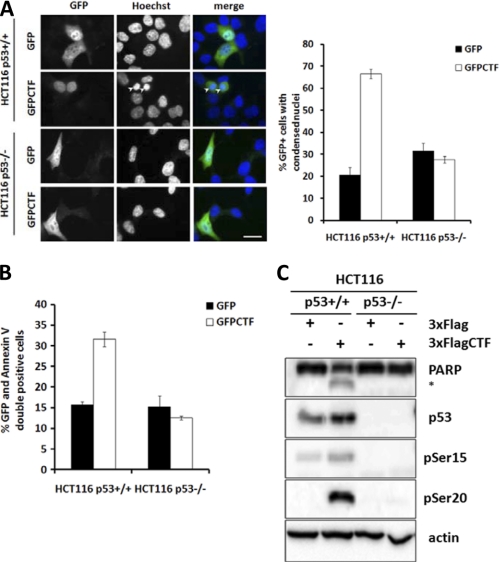
To determine whether CTF-mediated apoptosis is p53 dependent, we expressed GFP-tagged CTF (GFP-CTF) in HCT116 colon carcinoma cells that expressed wild type (p53+/+) or that was deleted for p53 (p53−/−). Immunofluorescence studies indicated that GFP-CTF was found in the nuclei of HCT116 p53+/+ and p53−/− cells (Fig. 3A). In p53+/+ cells, expression of GFP-CTF led to an increase to ∼60% rate of apoptosis compared with only ∼20% in GFP expressing cells, as evidenced by detection of condensed nuclei (Fig. 3A). In p53−/− GFP-CTF expressing cells, however, intact nuclear structure was found, suggesting that CTF-mediated apoptosis is p53 dependent. We further confirmed the lack of apoptosis in the presence of CTF in p53−/− cells by Annexin V staining (Fig. 3B) and by cleavage of PARP (Fig. 3C). Additionally, p53 levels were increased in the presence of CTF, along with levels of phospho-Ser15 and phospho-Ser20 p53. These data indicate that CTF-mediated apoptosis is p53-dependent and that expression of the CTF increases the levels of total and phosphorylated p53.
FIGURE 3.
CTF-induced apoptosis is p53-dependent. A–C, HCT116 p53+/+ and p53−/− cells were transfected with GFP or GFP-CTF, fixed for immunofluorescence and scored for number of GFP-positive cells with condensed nuclei. At least 100 cells were counted for each transfection. Experiments were repeated three times. Bar, 10 μm. B, cells were stained with Annexin V and subjected to FACS analysis. C, HCT116 p53+/+ and p53−/− cells were transfected with 3XFlag or 3XFlagCTF and lysates collected after 18 h for Western blot analysis.
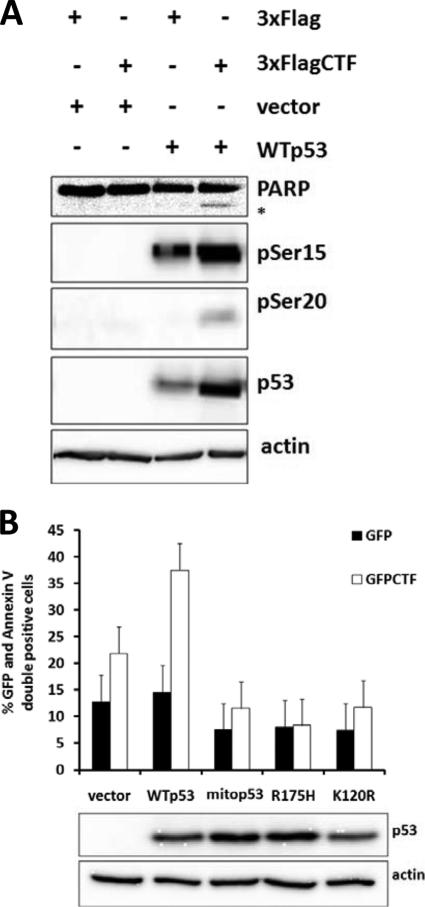
We also performed rescue experiments by expressing wild-type and mutant forms of p53 in p53−/− cells. Co-expression of wild-type p53 and GFP-CTF resulted in increased apoptosis as detected by PARP cleavage and increased levels of total and phosphorylated p53 (Fig. 4A). Expression of mutant p53 species, including mitochondria-localized p53 (mitop53), a DNA-binding defective p53 (R175H), and an acetylation mutant deficient in transactivation of pro-apoptotic, but not cell-cycle-related genes (K120R) (29), however, failed to restore the apoptotic phenotype in response to CTF expression (Fig. 4B). Together, these data demonstrate that CTF-induced apoptosis is dependent on p53, which is competent for the transcription of pro-apoptotic genes.
FIGURE 4.
Rescue of p53−/− cells with WT p53 restores sensitivity to CTF-induced apoptosis. A, HCT116 p53−/− cells were transfected with CTF or vector alone and with WTp53 or vector alone. After 18 h, lysates were collected and processed for immunoblotting. B, HCT116 p53−/− cells were transfected with GFP or GFP-CTF and either vector alone, WT p53, or various mutants of p53 (mito p53, R175H, K120R). After 18 h, cells were stained with Annexin V and scored for GFP and Annexin C double positivity.
CTF Nuclear Localization Is Regulated by SUMOylation
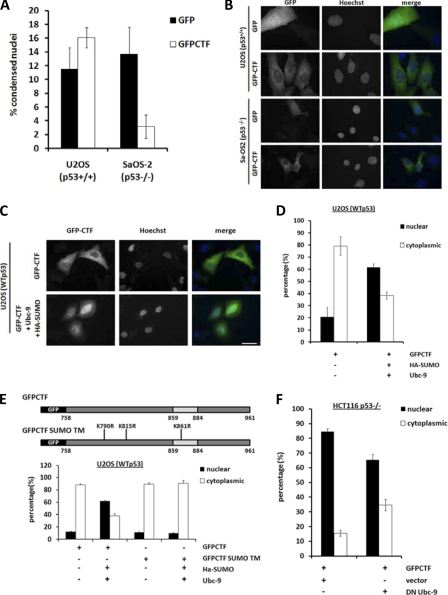
To corroborate our findings, we further examined p53 dependence of CTF-mediated apoptosis by using the U2OS (p53+/+) and Sa-OS2 (p53−/−) osteosarcoma cell lines. We observed a similar trend showing p53 dependence of CTF-induced apoptosis in U2OS (p53+/+) but not Sa-OS2 (p53−/−) cells. The rate of CTF-induced apoptosis in U2OS (p53+/+) cells expressing GFP-CTF however, was modest (∼16%) as compared with HCT116 p53+/+ (∼60%) (Fig. 3A). Immunofluorescence analyses indicated that CTF resides in the cytosol of both these osteosarcoma cell lines, instead of in the nucleus as in HCT116 cells (Fig. 5B). This observation could account for the reduced apoptosis in U2OS (p53+/+) cells and support our previous findings that nuclear localization is a pre-requisite for CTF-mediated apoptosis (24) (Fig. 1).
FIGURE 5.
CTF localization is regulated by SUMOylation. A–F, cells were transfected with the indicated constructs, fixed, and stained with Hoechst after 18 h and viewed under fluorescence microscopy. A and B, U2OS (p53+/+), Sa-OS2 (p53−/−), and HCT116 p53+/+ and p53−/− cells were transfected with GFP-CTF and quantified for GFP-positive cells with condensed nuclei. Bar, 10 μm. C and D, U2OS (p53+/+) cells were transfected with GFP-CTF alone or together with HA-SUMO and wild-type Ubc-9. Nuclear and cytoplasmically localized GFP-CTF were quantified. Bar, 20 μm. E, U2OS (p53+/+) cells were transfected with GFP-CTF or GFP-CTF SUMO TM with or without co-expression of HA-SUMO and Ubc-9. Cells were fixed and stained with Hoechst after 18 h, viewed under fluorescence microscopy and quantified for nuclear and cytoplasmic localization of the GFP-tagged constructs. F, HCT116 p53−/− cells were transfected with GFP-CTF and vector alone or together with HA-SUMO and dominant-negative Ubc-9. Cells were fixed after 18 h, stained with Hoechst and GFP-CTF localization quantified under fluorescence microscope. At least 100 cells were counted for each transfection. Data are an average of three different experiments.
We have previously shown that exogenous CTF is SUMOylated in vivo (24). Using immunoprecipitation studies, we have further observed the presence of multiple endogenously SUMOylated CTF species during apoptosis (supplemental Fig. S2). However, the role of SUMO modification in CTF function remained elusive. Given that SUMOylation affects nucleo-cytoplasmic shuttling (30), we tested the role of SUMO modification in CTF nuclear translocation. Expression of the SUMO ligase Ubc-9 to increase SUMOylation in vivo, led to nuclear accumulation of CTF in U2OS (p53+/+) (Fig. 5C). In contrast, co-expression of Ubc-9 along with a CTF SUMO mutant containing Arg replacements of putative SUMOylated Lys residues (Lys790, Lys815, and Lys861) (Fig. 5E), did not induce nuclear localization of the CTF in U2OS cells. In addition, expression of a dominant negative catalytically inactive Ubc-9 (C93S) in HCT116 cells reduced nuclear accumulation of CTF (Fig. 5F). These data indicate that SUMOylation regulates nuclear localization of CTF.
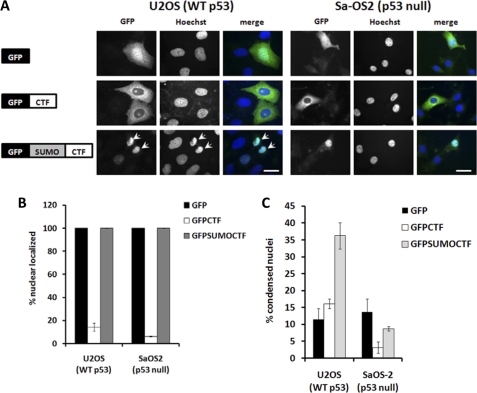
To directly examine the effect of SUMOylation in CTF nuclear localization and apoptosis, we fused the SUMO moiety in-frame with GFP-CTF to form GFP-SUMO-CTF (Fig. 6A). Immunofluorescence studies indicated that GFP-SUMO-CTF resides in the nuclei of both U2OS (p53+/+) and Sa-OS2 (p53−/−) cells (Fig. 6, A and B). GFP-SUMO-CTF expression in U2OS (p53+/+) cells led to a ∼35% rate in apoptosis compared with GFP expression alone (∼10%) (Fig. 6, A and C). In contrast, nuclear localized GFP-SUMO-CTF did not induce apoptosis in Sa-OS2 (p53−/−) cells. Together, these data demonstrate that SUMOylation is required for CTF nuclear localization and that nuclear resident CTF then promotes apoptosis via a p53-dependent mechanism.
FIGURE 6.
SUMO-CTF translocates into the nucleus and induces apoptosis in U2OS cells. A–C, U2OS (p53+/+) and Sa-OS2 (p53−/−) cells were transfected with GFP, GFP-CTF, or GFP-SUMO-CTF for 18 h, fixed, and stained with Hoechst and viewed under a fluorescence microscope. Bar, 10 μm. B, cells were quantified for nuclear or cytoplasmic localization of the GFP-tagged construct. C, cells were quantified for presence of apoptotic nuclei. At least 100 cells were counted for each transfection. Data are an average of three different experiments.
CTF Stabilizes p53 Expression
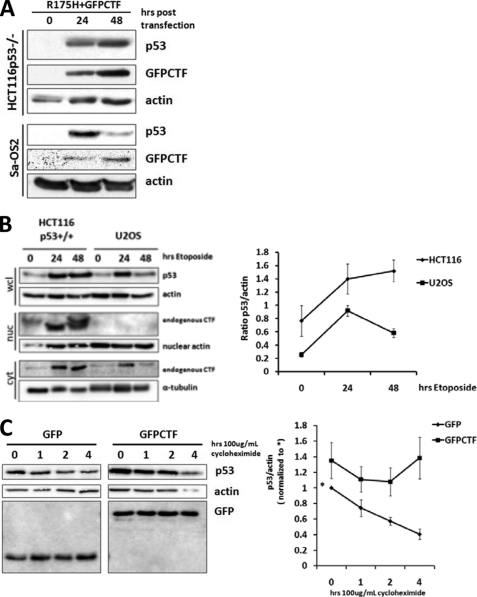
In the presence of CTF, we found that expression levels of p53 were potentiated (Figs. 3C and 4A). CTF-induced p53 stabilization could be an indirect consequence of apoptosis or directly regulated by CTF expression. We performed kinetic analysis on stabilization of DNA binding defective p53 (R175H) in HCT116 p53−/− and Sa-OS2 (p53−/−) cells. Because the R175H mutant is non-functional and cannot induce apoptosis (Fig. 4B), we reasoned that if increased levels of p53 were a consequence of apoptosis, stabilization of R175H p53 would be abrogated. On the other hand, increased levels of R175H p53 would suggest that CTF can directly regulate p53 stabilization. Kinetic analysis showed that in the presence of CTF, R175H p53 expression was sustained 48 h post-transfection in HCT116 p53−/− (Fig. 7A). In contrast, expression of p53 R175H was reduced after 24 h transfection in Sa-OS2 (p53−/−) cells in which CTF is excluded from the nucleus. These data indicate that CTF, in particular nuclear resident CTF, directly regulates p53 stabilization.
FIGURE 7.
CTF nuclear localization is required for p53 stabilization. A, HCT116 p53−/− and Sa-OS2 (p53−/−) cells were transfected with R175H p53 and GFP-CTF. B, HCT116 p53+/+ and U2OS (p53+/+) cells were treated with 50 μm etoposide for the indicated number of hours. C, HCT116 p53−/− cells were transfected with R175H mutant p53 and either GFP or GFP-CTF. After 18 h of transfection, cells were treated with cycloheximide for the indicated amount of time before being lysed. A–C, lysates were processed for SDS-PAGE and immunoblotting with the indicated antibodies. Band intensities were obtained using the TINA 2.09 software and levels of p53 protein were normalized to actin. Data are an average of three separate experiments.
Next, we exploited the differences in CTF localization between the HCT116 p53+/+ and U2OS (p53+/+) cells to investigate the endogenous effects of p115 cleavage on p53 stabilization during apoptosis. The topoisomerase II inhibitor, etoposide, was administered to induce p53-dependent apoptosis. In the presence of etoposide, DNA damage elicited apoptosis, p115 cleavage, and fragmentation of the Golgi apparatus (21). Cleavage of p115 and subsequent nuclear accumulation of the CTF was found in HCT116 p53+/+ (Fig. 7B). In contrast, while p115 was also cleaved in U2OS (p53+/+) cells, endogenous CTF was detected in the cytoplasm but not the nucleus, supporting our earlier findings shown in Fig. 5.
Etoposide treatment also led to an increase in p53 expression in both HCT116 p53+/+ and U2OS (p53+/+) cells (Fig. 7B). In HCT116 p53+/+ cells, the increase in p53 levels was sustained after 48 h of etoposide treatment. In U2OS (p53+/+) cells where the CTF was excluded from the nucleus, the initial increase in p53 levels was reduced after 24 h of etoposide treatment. These data demonstrate that sustained p53 stabilization during DNA damage requires nuclear localized CTF.
To further investigate the effect of CTF on p53 protein stability, we determined the half-life of p53 protein in CTF expressing cells using the translational inhibitor cycloheximide to inhibit de novo protein synthesis. In the presence of GFP-CTF, p53 protein levels were stabilized as compared with control cells expressing GFP alone (Fig. 7C). These data confirm that CTF mediates the stabilization of p53.
CTF Stabilizes p53 in an ERK-dependent Manner
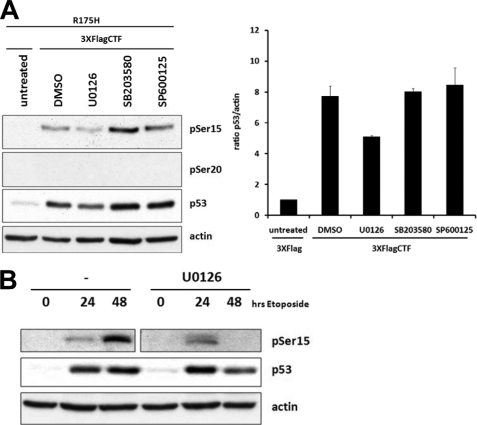
Multiple protein kinases have been shown to phosphorylate p53 and increase its stability. These include DNA-damage activated kinases such as ATM/R and Chk1/2 and the stress activated MAPKs via the ERK, p38, or JNK pathways (28). Given the role of MAPKs in mitotic Golgi fragmentation and that CTF is produced upon cleavage of p115 during stress-induced apoptosis (21), we examined the effect of MAPK inhibitors on CTF-induced p53 phosphorylation and protein stabilization. As previously observed (Fig. 7A), co-expression of CTF with R175H p53 led to an increase in total p53 and phospho-Ser15 p53 levels (Fig. 8A). However, phosphorylation of Ser20 was not detected. In the presence of MEK1 inhibitor U0126 that blocks ERK activation, phosphorylation of Ser15 and stabilization of p53 was attenuated. Inhibition of the p38 or JNK MAPKs, did not affect CTF-mediated phosphorylation of Ser15 or stabilization of p53. These data indicate that CTF-mediated stabilization of p53 specifically through Ser15 phosphorylation is ERK-dependent.
FIGURE 8.
CTF-mediated p53 stabilization is ERK-dependent. A, HCT116 p53−/− cells were transfected with R175H mutant p53 and either vector alone or 3XFlagCTF. After 24 hours of transfection, cells were treated with DMSO or the indicated kinase inhibitors for 3 hours before lysates were collected and processed for immunoblotting. B, HCT116 p53+/+ cells were treated with etoposide alone or with U0126. After the indicated time points, cells were lysed and lysates processed for immunoblotting with the indicated antibodies. Data are an average of three different experiments.
Previous studies indicated that pretreatment with U0126 inhibited etoposide-induced apoptosis (31). We asked whether U0126 specifically affected p53 stabilization elicited by etoposide. Kinetic analysis showed that etoposide treatment increased p53 and pSer15 p53 levels (Fig. 8B). In the presence of U0126, the initial stabilization of p53 was reduced at 48 h post etoposide treatment. Given the cleavage of p115 and induction of CTF nuclear accumulation by etoposide (Fig. 7), together, these data suggest that sustained p53 stabilization mediated by the CTF may be channeled through the MEK/ERK MAPK pathway.
CTF Acts as a Scaffold for ERK Activation of p53
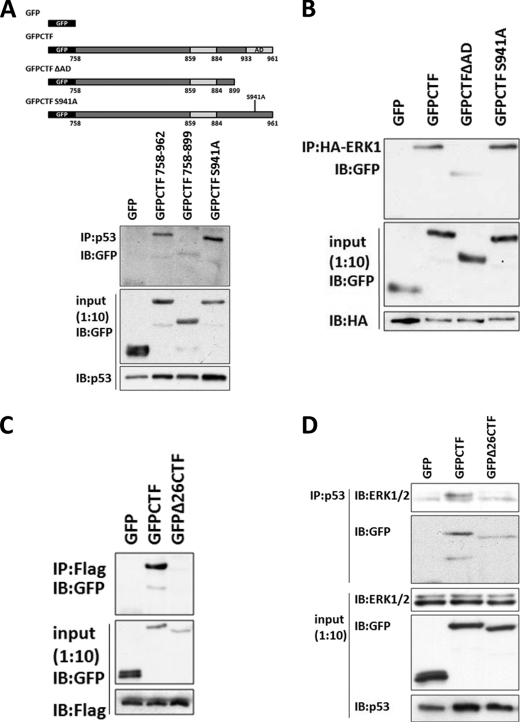
Previous studies demonstrated that ERK phosphorylates Ser15 and stabilizes p53 protein expression (32–35). To study how CTF could mediate ERK-dependent p53 phosphorylation and activation, we performed co-immunoprecipitation studies to examine protein-protein interactions between CTF, p53, and ERK1 (Fig. 9). Co-immunoprecipitation analyses showed that CTF interacted with p53 and with ERK1 (Fig. 9, A and B). The C terminus acidic domain of CTF (residues 900–961) was required for CTF-p53 and CTF-ERK1 interactions. Phosphorylation of CTF at Ser941 however, was not required for either interaction. Together, these data suggest that CTF, p53, and ERK1 may form a composite complex.
FIGURE 9.
CTF forms dimers and binds to p53 and ERK. A–D, HCT116 p53−/− cells were transiently transfected with the indicated plasmids. After 36–48 h of transfection, lysates were collected, pre-cleared and immunoprecipitated overnight with the indicated antibodies conjugated to protein G-Sepharose beads. Following extensive washing, beads were prepared for SDS-PAGE and immunoblotting with the indicated antibodies. A, cells were transfected with R175H p53 and various GFP-tagged constructs and immunoprecipitated with a monoclonal antibody against p53. B, cells were transfected with an HA-ERK1 expressing construct and the indicated GFP-tagged constructs and immunoprecipated with a monoclonal antibody against HA. C, cells were transfected with a plasmid expressing 3XFlagCTF and either GFP, GFP-CTF, or GFPΔ26CTF and immunoprecipitated with a monoclonal antibody against Flag. D, cells were transfected with R175H mutant p53 and either GFP, GFP-CTF, or GFPΔ26CTF. Lysates were immunoprecipitated with a monoclonal antibody against p53.
Full-length p115 exists as a homodimer in its native state via interaction of the coiled-coil domain (2). We speculated that the CTF, which retains part of the coiled-coil region of p115, might also form homodimers. To test this hypothesis, we conducted co-immunoprecipitation studies between differentially tagged CTF. Our results show that GFP-CTF co-immunoprecipitated with FLAG-tagged CTF (Fig. 9C). Dimerization of CTF, however, was abrogated by deletion of the 26 amino acid residue minimal domain (859–884), which forms part of coiled-coil region 4. Notably, this minimal domain is necessary to elicit apoptosis mediated by the CTF (24). Together, these data demonstrate dimerization of the CTF, which can bind to both p53 and ERK.
Binding of CTF to ERK and to p53 resembles the tethering properties of full-length p115 in bridging GM130 and giantin for Golgi-cisternal stacking and Golgi-vesicle membrane docking. We hypothesized that binding of CTF to ERK and to p53 might provide a similar tethering function and act as a scaffold to bring these entities in close proximity to facilitate efficient phosphorylation and activation of p53. We tested whether CTF may facilitate the interaction between p53 and ERK by co-immunoprecipitation analysis. In the presence of CTF, there was an increase in p53 interaction with endogenous ERK (Fig. 9D). Interaction between p53 and ERK, however, was diminished by deletion of the minimal domain encoding the coil-coiled dimerization region of CTF. These data indicate that the interaction between p53 and ERK is promoted by the dimerization of the CTF.
DISCUSSION
The Role of p115 CTF in Golgi Apoptosis Signaling
The mechanism of function of the p115 CTF in apoptosis signaling exhibits a similar pattern to a paradigm in cellular stress where a cleavage product of a cytoplasmic or organelle-associated protein is imported into the nucleus and functions to alter gene transcription. For example, in unstressed cells, activating transcription factor-6 (ATF-6) resides in the ER where it is bound by the chaperone BiP. This interaction is disrupted during ER stress. As a result, ATF-6 is allowed to traffic to the Golgi where it is cleaved by Golgi-resident proteases S1P and S2P. This releases the cytoplasmic portion of ATF-6 which then enters the nucleus and functions in the up-regulation of target genes involved in the unfolded protein response (UPR) pathway (reviewed in Ref. 36).
CTF activity follows a similar pattern although specific differences in its mechanism of action point to a unique role in apoptosis signaling. Unlike ATF-6, the CTF cannot directly activate gene transcription, as shown by a Gal4 DNA binding assay (Fig. 1D). However, use of RNA polymerase inhibitors showed that CTF-mediated apoptosis is transcription dependent (Fig. 1, A–C). This led to our speculation that the CTF may be modifying the activity of a known transcription factor such as p53. Co-expression of CTF with a K120R mutant p53 which is competent for the transactivation of cell cycle arrest but not apoptotic genes (29) did not produce apoptosis in p53−/− cells (Fig. 4). Similarly, p53 targets involved in cell cycle arrest were not up-regulated upon CTF expression (Fig. 2). Instead, our results revealed that the CTF markedly stabilized p53 protein levels (Figs. 3, 4, and 7) and led to the up-regulation of the pro-apoptotic BH3-only protein PUMA (Fig. 2). PUMA in turn promotes apoptosis by affecting the mitochondrial outer membrane permeabilization (37). In contrast, levels of NOXA, which is typically induced in response to DNA-damage and hypoxic conditions (38), remained unchanged. These data suggest that CTF modulation of p53 activity is specific for the transactivation of apoptotic genes, consistent with its role in apoptosis signaling.
Nuclear localization of the CTF is required for p53 stabilization (Fig. 7) and subsequent apoptosis (24). Compared with the cleavage fragments of ATF-6 and Golgin-160 (39, 40) which enter the nucleus via an NLS, the CTF lacks an obvious nuclear targeting sequence. Instead, we show that its nuclear import is regulated by SUMOylation (Fig. 5). Our original intention had been to use the U2OS (p53+/+) and Sa-OS2 (p53−/−) cells to validate data on the p53 dependence of CTF-induced apoptosis (Fig. 6). The unexpected finding that CTF predominantly localized in the cytoplasm of these cells allowed us to study the mechanisms involved in CTF nuclear translocation. In U2OS cells, overexpression of Ubc-9 increased CTF nuclear localization while mutation of putative SUMOylation sites prevented its nuclear entry (Fig. 5, C–E). Similarly, expression of a dominant negative, catalytically inactive Ubc-9 reduced the percentage of CTF found in the nuclei of HCT116 cells (Fig. 5F), suggesting that CTF regulation by SUMOylation may be a general mechanism occurring in a variety of cell types. Differential localization of the CTF between the osteosarcoma (U2OS and Sa-OS2) and colon carcinoma (HCT116) cells may be explained by differences in the levels or activity of SUMO ligases between these lines. Indeed, our observations indicate that endogenous Ubc-9 protein levels are relatively lower in U2OS cells compared with the HCT116 cell line (supplemental Fig. S1). The fact that CTF nuclear translocation is regulated at all lends a level of complexity to its function in apoptosis signaling and provides a means to switch its apoptotic activity on or off.
Unlike ATF-6 whose functional cytoplasmic fragment is generated by S1P and S2P proteolysis, the p115 CTF is generated by caspase-cleavage (21). This suggests that the CTF may not be initiating a stress signal, but that its main function is to potentiate and amplify existing pro-apoptotic signals. In this study, we show endogenously that this amplification signal is important to sustain a robust apoptotic response through the stabilization of p53 levels and activation of its function. In U2OS (p53+/+) cells, the initial activation of p53 after treatment with etoposide cannot be potentiated compared with HCT116 (p53+/+) cells in which CTF resides in the nucleus (Fig. 8). Similarly, inhibition of the MEK/ERK pathway by U0126 also led to down-regulation of p53 levels (Fig. 8), suggesting a role for this pathway in CTF-mediated p53 activation.
Role of MEK/ERK Signaling in Golgi-mediated Apoptosis
The involvement of the MEK/ERK pathway in CTF-mediated p53 stabilization and activation raises the question of the mechanism by which ERK is activated in response to CTF expression. In this regard, signaling via the MEK/ERK pathway has been shown to regulate Golgi morphology during mitosis (13). In particular, phosphorylation of GRASP65 and GRASP55 by ERK1 and ERK2 respectively are required for the mitotic unstacking of Golgi cisternae (14, 41). Specific isoforms of these kinases, MEK1b and ERK1c have also been found to reside in the Golgi and regulate its fragmentation during mitosis (16, 17). These studies point to the role of the MEK/ERK pathway in remodeling the Golgi structure in response to mitogen signaling (42).
Our observations on CTF-induced p53 phosphorylation support the role of ERK in CTF-mediated apoptosis. ERK has been shown to phosphorylate p53 on serine 15 in response to DNA-damaging agents (32, 33, 35, 43). In our study, we show that CTF expression induced the phosphorylation of Ser15 and Ser20 on both endogenous and overexpressed wild-type p53 (Figs. 3D and 4A). However, Ser20 phosphorylation was not detected when DNA-binding and thus apoptosis defective R175H mutant p53 was co-expressed with the CTF (Fig. 8A). This suggests that ERK and CTF activity mediates p53 stabilization specifically through the phosphorylation of Ser15 and that phosphorylation of Ser20 is an event that occurs downstream of CTF-induced apoptosis.
We speculate that through a negative feedback mechanism, Golgi-related stress and/or minor perturbations in the Golgi structure may be relayed to the MEK/ERK pathway to initiate or amplify the stress-response or apoptosis pathway. Our group previously observed that the CTF could be found in the nuclei of cells early during apoptosis, before the complete breakdown of the Golgi apparatus (24). This suggests that initially, a fraction of p115 may be cleaved early during apoptosis to generate the CTF. Simultaneous activation of the MEK/ERK pathway in response to Golgi stress may facilitate CTF amplification of the death signal in a feed-forward mechanism through its ERK/p53 tethering activity in the nucleus, leading to full blown caspase activation and apoptosis. Further work is required however, to definitively study the interaction between Golgi structure and the MEK/ERK pathway during stress and/or apoptosis.
p115 Tethering Function in Apoptosis
p115 plays a key role in the maintenance of the Golgi structure through its interaction with various peripheral and integral membrane proteins including GM130, giantin, β-COP, COG2, and various SNAREs (5–7, 44–47). During interphase, the C terminus acidic domain of p115 (931–962) mediates vesicle tethering at the Golgi through its interactions with GM130 and giantin (48). Here, we show that a region which includes these residues (900–961) is required for the binding of CTF to p53 and to ERK during apoptosis (Fig. 9, A and B). These findings suggest that cleavage of p115 by caspases may abrogate its interaction with its membrane-localized partners, thus releasing the CTF for nuclear translocation and interaction with p53 and ERK in the nuclear compartment. Interestingly, phosphorylation of serine 941 which promotes the binding of the acidic domain to GM130 and giantin (46) is not required for CTF interaction with its nuclear binding partners.
Full-length p115 exists as a homodimer and its dimerization is mediated by four coiled-coil domains (CC1–4) within its C terminus tail domain (2). In this report, we show that the p115 CTF, which encompasses part of CC2 and all of CC3 and CC4 together with the acidic domain, retains its ability to dimerize. Previously, work from our laboratory showed that a 26 amino acid stretch (residues 859–884) lying within CC4 was required for the apoptotic activity of the CTF (24). Here, we show that these same residues are required for the dimerization of the CTF (Fig. 9C), suggesting that its dimerization is required for subsequent apoptotic activity. Notably, the interaction between ERK1 and p53 is facilitated by the CTF (Fig. 9D). This interaction is diminished by the deletion of the 26-amino acid minimal domain (Fig. 9D). As such, we propose a model where the CTF acts as a tether that brings ERK and p53 together to facilitate their interaction and the subsequent phosphorylation and activation of p53 by ERK. Hence, the tethering property of the CTF is similar to the role of scaffold proteins in their function to increase the local concentration, proximity, and efficiency of phosphorylation of substrates by their kinases. However, scaffold proteins identified so far mainly function in the activation of a cascade of kinases. Here, we speculate that the scaffolding function of the p115 CTF would provide substrate specificity that directly activates an effector protein in the cell death pathway. Together, our findings suggest a conserved tethering function of the p115 CTF in facilitating ERK phosphorylation of p53 for apoptotic signal amplification.
Supplementary Material
Acknowledgments
We thank Dr. Bert Vogelstein for the gift of HCT116 cell lines, Dr. Roger S. Y. Foo for p53-encoding plasmids, Dr. Michael Greenberg for providing the pECE HA FOXO3a ER template, Dr. Moshe Sadofsky for plasmids encoding Ubc-9 and HA-SUMO, and Dr. Yukio Ikehara for the plasmid encoding p115. We also thank Dr. Chi-Wing Chow, Dr. Anne Muesch, and Dr. Maria Ruggieri for experimental suggestions and critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant DK21860 (to D. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- ER
- endoplasmic reticulum
- 4-OHT
- 4-hydroxytamoxifen
- ATF-6
- activating transcription factor-6
- CC
- coiled-coil
- COPI/II
- coat protein
- CTF
- C-terminal caspase cleavage fragment
- CTF-ER
- C-terminal caspase cleavage fragment estrogen receptor ligand binding domain
- ERK
- extracellular-signal regulated kinase
- GM130
- Golgi-matrix protein of 130kDa
- GRASP
- Golgi reassembly and stacking protein
- MEK
- mitogen-activated protein kinase kinase
- PARP
- poly(ADP-ribose) polymerase
- PUMA
- p53-up-regulated modifier of apoptosis
- ERK
- extracellular signal-regulated kinase
- SUMO
- small ubiquitin-like modifier.
REFERENCES
- 1. Warren G., Malhotra V. (1998) Curr. Opin. Cell Biol. 10, 493–498 [DOI] [PubMed] [Google Scholar]
- 2. Sapperstein S. K., Walter D. M., Grosvenor A. R., Heuser J. E., Waters M. G. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 522–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shorter J., Warren G. (1999) J. Cell Biol. 146, 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shorter J., Beard M. B., Seemann J., Dirac-Svejstrup A. B., Warren G. (2002) J. Cell Biol. 157, 45–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Allan B. B., Moyer B. D., Balch W. E. (2000) Science 289, 444–448 [DOI] [PubMed] [Google Scholar]
- 6. Sohda M., Misumi Y., Yoshimura S., Nakamura N., Fusano T., Ogata S., Sakisaka S., Ikehara Y. (2007) Traffic 8, 270–284 [DOI] [PubMed] [Google Scholar]
- 7. Guo Y., Punj V., Sengupta D., Linstedt A. D. (2008) Mol. Biol. Cell 19, 2830–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Puthenveedu M. A., Linstedt A. D. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 1253–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lowe M., Gonatas N. K., Warren G. (2000) J. Cell Biol. 149, 341–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lowe M., Rabouille C., Nakamura N., Watson R., Jackman M., Jämsä E., Rahman D., Pappin D. J., Warren G. (1998) Cell 94, 783–793 [DOI] [PubMed] [Google Scholar]
- 11. Lin C. Y., Madsen M. L., Yarm F. R., Jang Y. J., Liu X., Erikson R. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 12589–12594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Preisinger C., Körner R., Wind M., Lehmann W. D., Kopajtich R., Barr F. A. (2005) EMBO J. 24, 753–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Acharya U., Mallabiabarrena A., Acharya J. K., Malhotra V. (1998) Cell 92, 183–192 [DOI] [PubMed] [Google Scholar]
- 14. Jesch S. A., Lewis T. S., Ahn N. G., Linstedt A. D. (2001) Mol. Biol. Cell 12, 1811–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sablina A. A., Chumakov P. M., Levine A. J., Kopnin B. P. (2001) Oncogene. 20, 899–909 [DOI] [PubMed] [Google Scholar]
- 16. Shaul Y. D., Gibor G., Plotnikov A., Seger R. (2009) Genes. Dev. 23, 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shaul Y. D., Seger R. (2006) J. Cell Biol. 172, 885–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lane J. D., Lucocq J., Pryde J., Barr F. A., Woodman P. G., Allan V. J., Lowe M. (2002) J. Cell Biol. 156, 495–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maag R. S., Mancini M., Rosen A., Machamer C. E. (2005) Mol. Biol. Cell 16, 3019–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mancini M., Machamer C. E., Roy S., Nicholson D. W., Thornberry N. A., Casciola-Rosen L. A., Rosen A. (2000) J. Cell Biol. 149, 603–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chiu R., Novikov L., Mukherjee S., Shields D. (2002) J. Cell Biol. 159, 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lowe M., Lane J. D., Woodman P. G., Allan V. J. (2004) J. Cell Sci. 117, 1139–1150 [DOI] [PubMed] [Google Scholar]
- 23. Sbodio J. I., Hicks S. W., Simon D., Machamer C. E. (2006) J. Biol. Chem. 281, 27924–27931 [DOI] [PubMed] [Google Scholar]
- 24. Mukherjee S., Shields D. (2009) J. Biol. Chem. 284, 1709–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mukherjee S., Chiu R., Leung S. M., Shields D. (2007) Traffic 8, 369–378 [DOI] [PubMed] [Google Scholar]
- 26. Yang T., Davis R. J., Chow C. W. (2001) J. Biol. Chem. 276, 39569–39576 [DOI] [PubMed] [Google Scholar]
- 27. Yu J., Zhang L. (2005) Biochem. Biophys. Res. Commun. 331, 851–858 [DOI] [PubMed] [Google Scholar]
- 28. Kruse J. P., Gu W. (2009) Cell 137, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sykes S. M., Mellert H. S., Holbert M. A., Li K., Marmorstein R., Lane W. S., McMahon S. B. (2006) Mol. Cell 24, 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Salinas S., Briançon-Marjollet A., Bossis G., Lopez M. A., Piechaczyk M., Jariel-Encontre I., Debant A., Hipskind R. A. (2004) J. Cell Biol. 165, 767–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brantley-Finley C., Lyle C. S., Du L., Goodwin M. E., Hall T., Szwedo D., Kaushal G. P., Chambers T. C. (2003) Biochem. Pharmacol. 66, 459–469 [DOI] [PubMed] [Google Scholar]
- 32. She Q. B., Chen N., Dong Z. (2000) J. Biol. Chem. 275, 20444–20449 [DOI] [PubMed] [Google Scholar]
- 33. Liu J., Mao W., Ding B., Liang C. S. (2008) Am. J. Physiol. Heart Circ Physiol 295, H1956–H1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li D. W., Liu J. P., Mao Y. W., Xiang H., Wang J., Ma W. Y., Dong Z., Pike H. M., Brown R. E., Reed J. C. (2005) Mol. Biol. Cell 16, 4437–4453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brown L., Benchimol S. (2006) J. Biol. Chem. 281, 3832–3840 [DOI] [PubMed] [Google Scholar]
- 36. Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 37. Chipuk J. E., Green D. R. (2009) Cell Cycle 8, 2692–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ploner C., Kofler R., Villunger A. (2008) Oncogene. 27, Suppl. 1, S84–S92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li M., Baumeister P., Roy B., Phan T., Foti D., Luo S., Lee A. S. (2000) Mol. Cell. Biol. 20, 5096–5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hicks S. W., Machamer C. E. (2002) J. Biol. Chem. 277, 35833–35839 [DOI] [PubMed] [Google Scholar]
- 41. Bisel B., Wang Y., Wei J. H., Xiang Y., Tang D., Miron-Mendoza M., Yoshimura S., Nakamura N., Seemann J. (2008) J. Cell Biol. 182, 837–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wei J. H., Seemann J. (2009) Commun. Integr. Biol. 2, 35–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Persons D. L., Yazlovitskaya E. M., Pelling J. C. (2000) J. Biol. Chem. 275, 35778–35785 [DOI] [PubMed] [Google Scholar]
- 44. Beard M., Satoh A., Shorter J., Warren G. (2005) J. Biol. Chem. 280, 25840–25848 [DOI] [PubMed] [Google Scholar]
- 45. Diao A., Frost L., Morohashi Y., Lowe M. (2008) J. Biol. Chem. 283, 6957–6967 [DOI] [PubMed] [Google Scholar]
- 46. Dirac-Svejstrup A. B., Shorter J., Waters M. G., Warren G. (2000) J. Cell Biol. 150, 475–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nelson D. S., Alvarez C., Gao Y. S., García-Mata R., Fialkowski E., Sztul E. (1998) J. Cell Biol. 143, 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Linstedt A. D., Jesch S. A., Mehta A., Lee T. H., Garcia-Mata R., Nelson D. S., Sztul E. (2000) J. Biol. Chem. 275, 10196–10201 [DOI] [PubMed] [Google Scholar]
- 49. Garrison S. P., Jeffers J. R., Yang C., Nilsson J. A., Hall M. A., Rehg J. E., Yue W., Yu J., Zhang L., Onciu M., Sample J. T., Cleveland J. L., Zambetti G. P. (2008) Mol. Cell. Biol. 28, 5391–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Giono L. E., Manfredi J. J. (2007) Mol. Cell. Biol. 27, 4166–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Novakovic P., Stempak J. M., Sohn K. J., Kim Y. I. (2006) Carcinogenesis 27, 916–924 [DOI] [PubMed] [Google Scholar]
- 52. Wiencke J. K., Aldape K., McMillan A., Wiemels J., Moghadassi M., Miike R., Kelsey K. T., Patoka J., Long J., Wrensch M. (2005) Cancer Epidemiol. Biomarkers Prev. 14, 1774–1783 [DOI] [PubMed] [Google Scholar]
- 53. Kim J., Inoue K., Ishii J., Vanti W. B., Voronov S. V., Murchison E., Hannon G., Abeliovich A. (2007) Science 317, 1220–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Foo R. S., Nam Y. J., Ostreicher M. J., Metzl M. D., Whelan R. S., Peng C. F., Ashton A. W., Fu W., Mani K., Chin S. F., Provenzano E., Ellis I., Figg N., Pinder S., Bennett M. R., Caldas C., Kitsis R. N. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20826–20831 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.