Abstract
Type I collagen is a heterotrimeric extracellular matrix protein consisting of two α1(I) chains and one α2(I) chain. During liver fibrosis, activated hepatic stellate cells (HSCs) are the major source of the type I collagen that accumulates in the damaged tissue. Expression of α1(I) and α2(I) collagen mRNA is increased 60-fold compared with quiescent stellate cells and is due predominantly to post-transcriptional message regulation. Specifically, a stem-loop structure in the 5′-untranslated region of α1(I) collagen mRNA may regulate mRNA expression in activated HSCs through its interaction with stem-loop binding proteins. The stem-loop may also be necessary for efficient production and folding of the type I collagen heterotrimer. To assess the role of the stem-loop in type I collagen expression in vivo, we generated a knock-in mouse harboring a mutation that abolished the stem-loop structure. Heterozygous and homozygous knock-in mice exhibited a normal phenotype. However, steady-state levels of α1(I) collagen mRNA decreased significantly in homozygous mutant MEFs as well as HSCs; intracellular and secreted type I collagen protein levels also decreased. Homozygous mutant mice developed less liver fibrosis. These results confirm an important role of the 5′ stem-loop in regulating type I collagen mRNA and protein expression and provide a mouse model for further study of collagen-associated diseases.
Keywords: Collagen, Extracellular Matrix Proteins, Gene Knockout, Liver, mRNA, Hepatic Stellate Cells, Liver Fibrosis
Introduction
The collagen family is a group of extracellular matrix proteins involved in a variety of biological processes, including providing structural support to connective tissue and forming the basement membrane of organs (1, 2). Type I collagen is a heterotrimeric protein comprised of two α1(I) collagen chains and one α2(I) collagen chain, and although located on different chromosomes, these genes are coordinately regulated in a developmental, tissue-specific, and inducible manner (3, 4). The three peptide chains form a triple-helical structure, with several post-translational modifications resulting in multiple inter- and intrachain interactions, providing the protein with increased stability (5, 6). Increased expression and deposition of type I collagen are the most dramatic aspects of liver fibrosis, although expression of other collagens, including type III, is also increased (4). Activated hepatic stellate cells (HSCs)2 are the major source of type I collagen in the fibrotic liver (7). Expression of type I collagen is very low in quiescent HSCs, which act as a storage site of vitamin A and regulate blood flow through the hepatic sinusoid. However, following a fibrogenic stimulus, HSCs undergo an activation process, characterized by a loss of the vitamin A stores; increased cellular proliferation, migration, and contractility; and increased synthesis of both α1(I) and α2(I) collagen mRNA and type I collagen protein (8).
Type I collagen biosynthesis is a complex process that is regulated at both the transcriptional and post-transcriptional levels. Many of these regulatory processes differ in the quiescent versus activated HSC phenotype. Activated HSCs show a 50–60-fold increase in α1(I) collagen mRNA expression, with a 3-fold increase in gene transcription and a 16-fold increase in mRNA stability (9). Like many mRNAs (10) (11), the α1(I) collagen gene contains regulatory elements located within both the 3′- and 5′-UTRs, which are involved in message stability and processing (12). In particular, an evolutionarily conserved sequence located ∼75 nucleotides downstream of the 5′ 7-methylguanine cap (involved in translation initiation (13)) encompasses the translational start codon and is able to form a stem-loop (5′ SL) structure (this stem-loop is also found in α2(I) and α1(III) mRNA (14)). Reporter gene assays revealed that the 5′ SL inhibited α1(I) collagen mRNA steady-state levels in quiescent HSCs and that mutation of the sequence resulted in increased mRNA levels; this inhibition was reduced in activated HSCs through interactions of the SL with RNA-binding proteins, indicating the SL is important in regulating α1(I) collagen mRNA expression during fibrogenesis (15–17).
The type I collagen protein is also regulated at the translational and post-translational levels. Modifications such as cleavage of pro-domains, hydroxylation of proline and lysine residues in the GXY repeats of the triple-helical region (where G is glycine, and X and Y are often proline and hydroxyproline, respectively (18)), and the formation of disulfide bonds are necessary to ensure proper folding of the constituent chains and to provide intra- and interchain stabilization interactions (6). Transfection studies indicated that although the 5′ SL appeared to be inhibitory to translation in vitro, binding of proteins to the SL in vivo resulted in proper translation (19). However, mutating the 5′ SL resulted in a type I collagen species that was susceptible to protease degradation; as such, it has been suggested that the 5′ SL is also important in regulating translational efficiency of the α1(I) collagen gene (19).
Analysis of the 5′ SL of α1(I) collagen has for the most part been performed in cell culture studies using exogenous mutated mRNAs or molecular decoys. Therefore, we looked to extend the understanding of this element to a more physiologically relevant in vivo system. In this study we have generated a knock-in mouse harboring a sequence mutation in the 5′-UTR stem-loop that prevents formation of its secondary structure. Characterization of embryonic fibroblasts demonstrated that mutation of the SL resulted in decreased steady-state levels of α1(I) collagen mRNA expression and, in turn, type I collagen protein in mouse embryonic fibroblasts (MEFs) and in activated HSCs. Furthermore, mice homozygous for the SL mutation had decreased liver fibrosis and collagen accumulation in response to liver injury. Thus, the 5′ SL is critical in regulating expression of α1(I) collagen mRNA in MEFs and HSCs.
EXPERIMENTAL PROCEDURES
Generation of Targeting Construct
Briefly, a 2.8-kb HindIII/HpaI restriction fragment of mouse genomic DNA clone p1a containing 2.3 kb of upstream promoter sequence, the first exon, and 300 bp of the first intron of α1(I) collagen was cloned into HindIII/HpaI sites in the pGL3 vector (Promega, Madison, WI) (supplemental Fig. S1A). A mutation in the 5′ SL was introduced by cloning a double-stranded oligonucleotide into the BstXI and XbaI sites of the pGL3 construct. This mutation changed 21 nucleotides of the 5′ end of the stem-loop, introduced an EcoRI restriction site, and eliminated binding of LARP6 (17). From this clone, a NheI/StuI fragment was subcloned into the OSDupDel vector (a gift of Dr. Oliver Smithies, University of North Carolina) at NheI/ClaI (blunt) sites 5′ of the neomycin cassette. The second fragment, a 7.7-kb genomic StuI fragment of p1a containing the remaining 1.2 kb of the first intron and an additional 6.5 kb of downstream exons and introns, was cloned into SalI/NotI of the above OSDupDel vector 3′ of the neomycin cassette. The plasmid also contained an ampicillin resistance cassette for positive selection of clones and a herpesvirus thymidine kinase cassette for negative selection.
Generation and Selection of 5′ SL Mutant Mouse
TC1 ES cells (provided by Dr. Beverly Koller, University of North Carolina) derived from the 129 mouse strain were maintained on irradiated MEFs in ES medium (DMEM-high glucose containing 15% ES-qualified FBS (Invitrogen, Carlsbad, CA), 0.1 mm β-mercaptoethanol, and 2 mm l-glutamine). PmeI-linearized OSDupDel-SLMut plasmid was introduced to the ES cells by electroporation. The cells were suspended in 0.3 ml of PBS with 13 μg of plasmid DNA and pulsed in a 2-mm gap electroporation cuvette with 270 V, 50 microfarads, and 350 Ω for 0.4 s using an Electro Cell Manipulator 630 (BTX, Holliston, MA). The cells were immediately plated on neomycin-resistant, sublethally irradiated MEFs in ES medium. After 24 h, the medium was replaced, and the cells were grown in ES medium containing 200 μg/ml G418 (Invitrogen, Carlsbad, CA); after an additional 24 h, 1 μg/ml ganciclovir was added to the medium.
Individual G418/ganciclovir-resistant ES colonies were expanded and screened for proper recombination by Southern blotting. Genomic DNA was isolated by standard techniques (20), digested with EcoRI, separated on a 0.8% agarose Tris acetate-EDTA gel, and transferred to a nylon membrane (Millipore, Bedford, MA), followed by UV-cross-linking of the blot. The blot was hybridized with two probes to ensure homologous recombination (supplemental Fig. S3). The first probe (Recomb) binds a region of endogenous genomic DNA located outside the construct DNA and was generated by RT-PCR of mouse NIH3T3 RNA using Moloney murine leukemia virus-reverse transcriptase (Invitrogen, Carlsbad, CA) and Taq DNA polymerase (Invitrogen, Carlsbad, CA). The primer sequences are: sense, 5′-TGCCCACCCCCACATACAGC-3′, and antisense, 5′-CACGCCACGCCACGCCACCTCATCTT-3′. The second probe (Neo) binds within the neomycin resistance cassette of the construct and was generated by PCR of OSDupDel-SLMut plasmid DNA. The primer sequences are: sense, 5′-TGACGCGTGTGGCCTCGAACAC-3′, and antisense, 5′-TGAGCCTGGC GAACAGTTCGGC-3′.
The probes were labeled with [α-32P]dCTP using the random primed DNA labeling kit (Roche Applied Science, Indianapolis, IN); each hybridization was performed by incubating 12.5 × 1012 counts of probe with the membrane for 1 h at 68 °C in Rapid-Hyb buffer (GE Biosciences, Buckinghamshire, UK). The blot was washed once with 2× SSC, 0.1% SDS; washed once with 0.2× SSC, 0.1% SDS (each for 20 min at 55 °C); and analyzed the following day using a Typhoon 8600 PhosphorImager (GE Biosciences, Buckinghamshire, U.K.). Two positive heterozygous clones were weaned off feeder cells and expanded on 0.1% gelatin-coated culture plates in ES medium containing 1000 units/ml recombinant leukemia inhibitory factor (Chemicon, Temecula, CA) to maintain the cells in an undifferentiated state. Expanded clones were karyotyped to ensure the absence of chromosomal abnormalities.
Generation and Genotyping of Mouse Line
One positive undifferentiated ES clone was expanded, microinjected into blastocysts, and implanted in C57BL/6 surrogate females (Animal Models Core, University of North Carolina). Successful introduction of the ES cell DNA was determined by the appearance of chimeric offspring. Males of 70–80% chimerism were bred with wild-type C57BL/6 females. Successful integration of the mutation into the germ line was determined by the appearance of agouti (F1) offspring; genotyping by Southern blot (as described above) confirmed the mutation. F1 heterozygote males and females were intercrossed, and the resulting offspring were bred to perform MEF isolations and initial phenotyping. As before, genotyping was performed by Southern blot. All of the animal work was approved by the University's Institutional Animal Care and Use Committee.
Mouse HSC Isolation
HSCs were isolated through collagenase-Pronase perfusion of livers, followed by 8.2% Nycodenz (Accurate Chemical and Scientific Corporation, Westbury, NY) two-layer discontinuous density gradient centrifugation resulting in 99% purity of HSCs as confirmed by retinoid autofluorescence (21). After the isolation, HSCs were plated and cultured on plastic dishes with 10% FBS supplemented DMEM for 7 days, and then the cells were analyzed.
Model of Hepatic Fibrosis
Specific pathogen-free WT (Balb/c/J) and 5′ SL mutant mice backcrossed with Balb/c background eight times were used for this study. For the bile duct ligation (BDL) model, 8–12-week-old mice were anesthetized. After laparotomy, the common bile duct was ligated twice, and the abdomen was closed. The sham operation was performed similarly without BDL. The mice received humane care according to National Institutes of Health recommendations outlined in the Guide for the Care and Use of Laboratory Animals. All of the animal experiments were approved by the Columbia University and University of California, San Diego Institutional Animal Care and Use Committee.
Measurement of Hepatic Collagen Content
Hepatic hydroxyproline content was measured as described previously (21). Hepatic collagen content was also quantitated by Sirius red staining of paraffin-embedded sections. The Sirius red-positive area was analyzed in six random fields (100×) on each slide and quantified using National Institutes of Health imaging software.
Ribonuclease Protection Assays
MEFs were isolated by standard techniques (20). Total RNA was isolated from MEFs using TRIzol reagent (Invitrogen, Carlsbad, CA), and ribonuclease protection assays (RPAs) were performed as described previously (22). Antisense probes for mouse α1(I) collagen were designed by RT-PCR of NIH3T3 RNA (wild-type probe) or PCR from the OSDupDel-SLMut plasmid (mutant probe). The primer sequences are: wild type: sense, 5′-CCGCAAAGAGTCTACATGTC-3′, and antisense, 5′-ACACAGCCGTGCCATTGTGG-3′; mutant: sense, 5′-CTCGGGACGGAGCAGGAG-3′, and antisense, 5′-GAATTCTGTGTACGAGCAATTCTA-3′. Antisense probe for mouse α2(I) collagen was designed by RT-PCR of NIH3T3 RNA. The primer sequences are: sense, 5′-GGTTTCCAAGGACCTGCT-3′, and antisense, 5′-CTTTGAAGCCAGGAAGTCC-3′. The products were cloned into BlueScript SK+ (Stratagene, La Jolla, CA) under the control of the T7 promoter; an antisense probe against mouse glyceraldehyde-3-phosphate dehydrogenase (p-TRI-GAPDH; Ambion, Austin, TX) was used as a loading control. [α-32P]UTP-labeled probes were incubated with 5 μg of RNA, and the protected fragments were subjected to electrophoresis in 5% acrylamide urea sequencing gels; the gels were subsequently fixed in 10% methanol:10% acetic acid for 15 min, followed by autoradiography and quantitation on a Typhoon 8600 PhosphorImager, using 5 μg of NIH3T3 RNA as a normalization control.
Protein Extraction and Western Blotting
Whole cell protein lysate was extracted from MEFs and HSCs by a 20-min incubation at 4 °C in Dignam C buffer (10 mm HEPES, pH 7.9, 0.42 m NaCl, 1.5 mm MgCl2, 0.5 mm dithiothreitol, 25% glycerol, and 0.5% Nonidet P-40) containing 10 mm 4-nitrophenyl phosphate, 20 mm β-glycerophosphate, 500 μm Pefabloc, 2 μg/ml aprotinin, 50 μm sodium orthovanadate, and 0.5 μg/ml leupeptin. The samples were centrifuged for 10 min at 14,000 × g at 4 °C. The supernatant was collected, and the protein concentration was determined using the Bradford assay (Bio-Rad, Hercules, CA). Protein lysate was separated by 7.5% SDS-PAGE under native or denaturing conditions and transferred to nitrocellulose membrane (Schleicher & Schuell, Keene, NH). Equal protein loading was determined by Ponceau S staining (Sigma-Aldrich, St. Louis, MO) of the transfer blot and by Western blot using 1:1000 goat anti-actin (Santa Cruz Biotechnology, Santa Cruz, CA) or 1:1000 mouse anti-α-tubulin (Zymed Labs, San Francisco, CA) primary antibodies and 1:1000 rabbit anti-goat or goat anti-mouse HRP-conjugated secondary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA). Blotting was performed as described (23), using 1:1000 rabbit anti-collagen type I (Rockland, Gilbertsville, PA), and 1:1000 goat anti-rabbit HRP-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Protein expression was detected by enhanced chemiluminescence (Amersham Biosciences, Arlington Heights, IL), and expression levels were quantified by densitometry using AlphaEase software (Alpha-Innotech Corporation, San Leandro, CA).
Collagen Secretion Western Blotting
MEFs or HSCs were washed twice with PBS, fresh DMEM-high glucose medium containing 10% FBS was added to the cells, and aliquots were removed at the indicated time points. The aliquots were separated by 7.5% SDS-PAGE; blotting was performed with rabbit anti-type I collagen as described above and with 1:1000 mouse anti-fibronectin (BD Transduction Laboratories, Lexington, KY) with 1:1000 goat anti-mouse HRP-conjugated secondary antibody.
Cre-mediated Removal of Neomycin Cassette
Wild-type and mutant MEFs were plated on 60-mm cell culture dishes at a density of 20 × 104 cells/dish. The cells were transduced overnight with either AdCre or Ad-Track CMV (which expresses GFP under the cytomegalovirus promoter) adenovirus in DMEM containing 2% FBS at a multiplicity of infection of 6000, which resulted in >90% transduction efficiency (data not shown). The following day, the viral medium was removed and replaced with DMEM containing 10% FBS; 36 h post-transduction, the medium was collected, and the cells were harvested for RNA (as described under “Ribonuclease Protection Assays”). Genomic DNA was isolated from additional dishes using the Puregene Cell & Tissue DNA Purification System (Gentra, Minneapolis, MN).
Polymerase Chain Reaction Assays
To determine splicing efficiency of the mutant transcript, human 293A cells were transfected with 5 μg of OSDupDel-SLMut plasmid or empty vector using Lipofectamine reagent (Invitrogen, Carlsbad, CA); 48 h post-transfection, cellular RNA was harvested using TRIzol, and RNA samples were reverse-transcribed, followed by PCR amplification. PCR primers were designed such that the sense primer bound to a region within the first exon of the mouse α1(I) collagen gene, whereas the antisense primer bound to a region within the second exon (supplemental Fig. S2A). The primer sequences were: sense, 5′-CTCGGGACGGAGCAGGAG-3′, and antisense, 5′-ACACAGCCGTGCCATTGTGG-3′. For confirmation of neomycin cassette removal using AdCre, 100 ng of MEF genomic DNA was amplified using primers that flanked the cassette. The primer sequences were: sense, 5′-GCAGGGTTCCTCCCAGCTCT-3′, and antisense, 5′-AGCCGGGTCCTTCAAAGCCAG-3′.
mRNA Stability Assay
Wild-type or SL mutant MEFs were treated with actinomycin D (5 ng/ml) or DMSO in DMEM containing 10% FBS. The cells were lysed at 0, 4, 8, 12, 18, and 24 h after treatment. RNA was isolated by RNeasy kit (Qiagen) and reverse-transcribed using random hexamer primer. Real time quantitative PCR was performed with an ABI 7000 sequence detection system (Applied Biosystems, Foster City, CA) using an α1(I) collagen primer-probe set (Mm00801666_g1; ABI, Foster City, CA) for 40 cycles of 15 s at 95 °C and 60 s at 60 °C. Quantification was performed by comparing the Ct values of each sample to a standard curve and normalization to 18 S (primer probe set (Hs99999901_s1; ABI, Foster City, CA).
Statistical Analysis
Student's t test was used to determine statistical significance between groups. A p value < 0.05 was considered significant.
RESULTS
α1(I) Collagen 5′-UTR SL Construct
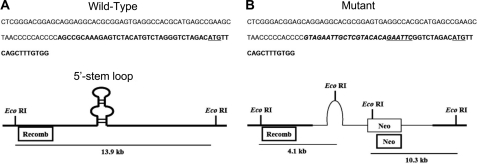
Approximately 10.5 kb of mouse genomic DNA containing the α1(I) collagen sequence, harboring a mutation in the 5′-UTR stem-loop sequence that abolished SL formation, was cloned into the OSDupDel targeting vector (supplemental Fig. S1A). Along with the 21-nucleotide mutation, an EcoRI restriction site was included, which was used for genotyping purposes (Fig. 1) but did not disrupt the translational start codon located within the loop. The short arm of the construct contained 2.8 kb of genomic DNA, incorporating 2.3 kb of upstream sequence, the first exon (including the SL mutation), and a 300-bp portion of the first intron. The long arm included the remaining 1.2 kb of the first intron and 6.5 kb of additional downstream sequence. A neomycin cassette was located between the arms and within the first intron, making the entire insert ∼12.8 kb in length. The product of a successful homologous recombination event would result in introduction of the SL mutation along with the neomycin cassette into the genome (supplemental Fig. S1B).
FIGURE 1.
DNA sequence depicting SL mutation. The entire 5′-UTRs for both the wild-type (A) and mutant (B) sequences are shown, with the stem-loop region in bold; the actual SL mutation is in italics. The translation initiation codons and the EcoRI site inserted into the mutant sequence are underlined.
The 5′-UTR SL Mutation Did Not Affect Splicing Efficiency
RT-PCR was performed on 293 cells transfected with SL mutant plasmid to determine whether insertion of the mutant construct affected the capability to splice α1(I) collagen mRNA. A PCR primer set was generated such that the sense primer bound within the first exon of the message, whereas the antisense primer bound within the second exon (supplemental Fig. S2A). If proper splicing occurred, the expected PCR product would be 290 bp; however, because of the neomycin cassette within the first intron, if the message was not efficiently or properly spliced, a product of 3020 bp would be observed. As shown in supplemental Fig. S2B, only the smaller PCR product was detected in cells transfected with the SL mutant plasmid; cells transfected with the empty vector generated no product. The efficiency of production of the large PCR product was confirmed using the SL mutant plasmid as a template.
Generation of the 5′-UTR SL Mutation ES Cells
The linearized targeting construct was electroporated into 129 strain embryonic stem cells, and clones were positively selected for homologous recombination and negatively selected against random integration. Individual clones were isolated and expanded, and homologous recombination was assessed using two Southern blot probes. Following EcoRI digestion of the ES DNA, the first probe (Recomb) bound to a region outside the recombined construct DNA and generated a 4.1-kb fragment if the SL mutation was properly inserted (supplemental Fig. S3B). In addition, because the neomycin cassette contained an EcoRI site, proper homologous recombination was further confirmed using a second probe (Neo), which bound the cassette downstream of the restriction site, generating a 10.3-kb fragment. Digested wild-type DNA would generate a 13.9-kb band with the Recomb probe and no band with the Neo probe. As shown in supplemental Fig. S3, several clones were positive for proper homologous recombination (see lanes 7 and 8). Positive heterozygous clones had normal chromosomal karyotypes (data not shown).
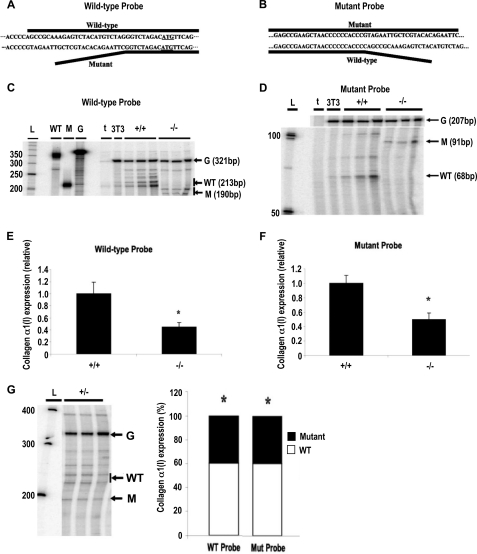
Mutation of the 5′-UTR SL Decreased Steady-state Levels of α1(I) Collagen mRNA in Mouse Embryonic Fibroblasts
Following successful generation of mice harboring the 5′ SL mutation, the mutant mice had normal development, grossly normal phenotype, and normal fertility. This was somewhat surprising, given the role of type I collagen in growth and development (see “Discussion”). RNA was harvested from MEFs isolated from wild-type and mutant mice and subjected to RPAs using α1(I) collagen-specific probes to analyze effects of the SL mutation on message expression. Two probes were designed, and both bound either genotype; however, a portion of each probe bound specifically to either wild-type or mutant sequence (Fig. 2, A and B). Digestion of the RNA-probe hybrid would thus generate protected bands of differing size depending on the genotype of the cell. The wild-type probe protected a 213-bp fragment of wild-type RNA, whereas the protected fragment of the mutant-containing RNA was 190 bp; likewise, the mutant probe protected a 91-bp fragment of mutant RNA and a 68-bp fragment of wild type. As seen in Fig. 2 (C and D), the wild-type probe protected the proper fragment for each respective genotype; similar results were obtained using the mutant probe. Quantitation of the bands revealed an ∼55% reduction in α1(I) collagen mRNA expression in mutant cells, regardless of the probe used (Fig. 2, E and F). To further investigate these findings, we isolated MEFs from heterozygous embryos and analyzed the relative contribution of the wild-type and mutant alleles to the total α1(I) collagen mRNA population (Fig. 2G, wild-type probe RPA; mutant probe RPA not shown). RPA results using either riboprobe indicated that expression of the wild-type allele was approximately twice that of the mutant, in close agreement with our findings comparing the alleles separately in homozygous MEFs. No significant difference in expression of α2(I) collagen mRNA was detected between WT and mutant MEFs (supplemental Fig. S4, A and B).
FIGURE 2.
Mutation of the 5′ SL decreased α1(I) collagen mRNA steady-state levels. A and B, RNase protection assay schematic. An antisense wild-type probe (A) was designed such that the 3′ end binds only wild-type sequence. RNase digestion of the probe-wild-type transcript duplex results in a 213-bp protected fragment; a 190-bp fragment is protected with mutant RNA (A). An antisense mutant probe (B) was designed such that the 5′ end binds only mutant sequence. Digestion of the probe-mutant transcript duplex results in a 91-bp protected fragment; a 68-bp fragment is protected in wild-type RNA (B). A nonbinding sequence is indicated by a solid bent line. For the wild-type schematic, the translation initiation codon is underlined. C and D, representative RPA using wild-type probe. RNA from wild-type (+/+) and mutant (−/−) MEFs was incubated with wild-type (C) or mutant (D) riboprobe; each lane represents a separate cell isolation. L, 50-bp DNA ladder (fragment sizes indicated to the left of the gel); WT, undigested mouse α1(I) collagen wild-type riboprobe; M, undigested mouse α1(I) collagen mutant riboprobe; G, undigested mouse GAPDH riboprobe; t, yeast tRNA; 3T3, NIH3T3 RNA. Arrow G, protected GAPDH; arrow WT, protected wild type; arrow M, protected mutant. E and F, result of four independent RPAs using wild-type (E) and mutant (F) probes. α1(I) collagen-specific bands were quantitated and normalized to GAPDH values. The data are expressed as relative amounts of message versus wild-type MEFs (n = 12). *, p < 0.05. G, analysis of heterozygote MEFs. Left panel, representative RPA using wild-type riboprobe; each lane represents a separate cell isolation. L, 100-bp DNA ladder (fragment sizes indicated to the left of the gel); other abbreviations are as described previously. Right panel, graph depicting contribution (as percentages) of each allele to total α1(I) collagen mRNA levels; expression was quantitated using both wild-type and mutant riboprobes. *, p < 0.05, wild-type transcript versus mutant transcript. For all graphs, the error bars represent S.E.
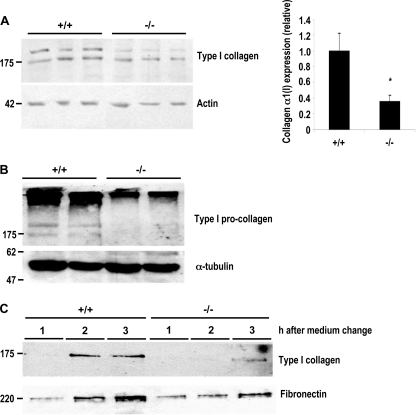
Mutation of the 5′-UTR SL Decreased Intracellular Type I Collagen Protein
Because the production and accumulation of the type I collagen heterotrimer is the end product of the pathway, we next determined what effect mutation of the stem-loop had on type I collagen protein expression. As shown in Fig. 3A, intracellular type I collagen expression was decreased ∼65% in homozygous mutant compared with wild-type MEFs. This decrease is similar to that observed in the steady-state levels of α1(I) collagen mRNA (Fig. 2), indicating that there does not appear to be increased translational activity in the mutant cells to compensate for reduced RNA levels. A similar decrease in intracellular type I pro-collagen protein was also observed under nonreducing conditions (Fig. 3B), which allows for the detection of the high molecular weight, disulfide-bonded species of collagen.
FIGURE 3.
Mutation of the 5′ SL decreased type I collagen protein levels. A and B, reduced intracellular expression of type I collagen in mutant MEFs. Intracellular protein (30 μg) was harvested from wild-type (+/+) and mutant (−/−) MEFs and separated in denaturing (A, left panel) or nonreducing denaturing (B) 7.5% acrylamide gels, followed by Western blotting for type I collagen and actin or α-tubulin. Representative Western blots are shown; each lane represents separate cell isolations. A, right panel, result of six independent Western blots. Type I collagen bands were quantitated and normalized to actin values. The data are expressed as relative amounts of protein versus wild-type MEFs. *, p < 0.05; the error bars represent S.E. C, secretion of type I collagen is reduced in mutant MEFs. Aliquots (50 μl) of media harvested 1, 2, and 3 h after a change of fresh media from wild-type (+/+) and mutant (−/−) MEFs were separated by 7.5% SDS-PAGE gels, and the blot was probed with anti-type I collagen and anti-fibronectin. Representative Western blots are shown.
We next analyzed the effect of mutating the 5′ SL on collagen secretion. Collagen levels were decreased in medium harvested from mutant MEFs over a period of 3 h compared with wild-type cells (Fig. 3C). Although collagen secretion was clearly observed after 2 h in wild-type cells, expression was detected only faintly after 3 h in mutant cells and at levels much lower than wild type at either 2 or 3 h. This decrease correlated with the decreased intracellular expression of type I collagen in mutant MEFs observed in Fig. 3 (A and B).
Removal of the Neo Cassette Had No Effect on α1(I) Collagen Expression
It is well established that introns control a variety of mRNA activities, including splicing, nuclear export, cellular localization, and translational efficiency (24). Previous studies have reported that interfering with the first intron of α1(I) collagen mRNA can have deleterious effects on collagen expression (25–27). Although these studies dealt with deletions in the first intron, evidence suggests that increased intron size can also influence transcript expression (28, 29).
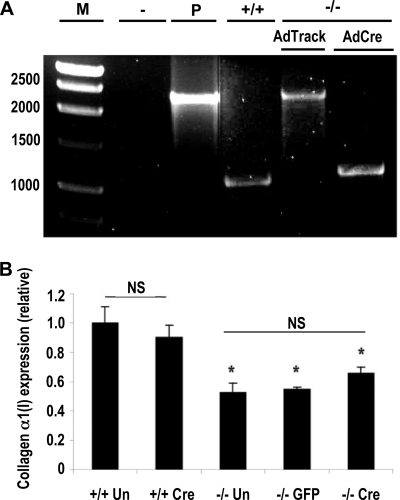
Because the neomycin cassette is located between loxP sites, mutant MEFs were transduced with a Cre recombinase-expressing adenovirus to remove the Neo gene. Transduction of mutant MEFs with AdCre resulted in complete removal of the cassette, as determined by PCR genotyping of genomic DNA (Fig. 4A). The transduced cells contained an additional 34-bp loxP sequence (compared with wild-type DNA) that remained after recombination (29). RPA analysis showed that removal of the cassette resulted in a slight and not significant increase in α1(I) collagen mRNA steady-state levels compared with mutant MEFs containing the Neo cassette (Fig. 4B). More importantly, α1(I) collagen mRNA expression in the AdCre-transduced mutant MEFs was still decreased in a statistically significant manner compared with wild-type cells. Adenoviral conditions had no effect on α1(I) collagen expression, because mutant MEFs transduced with AdTrack-GFP and wild-type cells transduced with AdCre exhibited no changes compared with controls.
FIGURE 4.
Removal of the neomycin cassette had no effect on α1(I) collagen expression. A, confirmation of cassette removal. Mutant (−/−) MEFs were transduced with adenovirus expressing GFP (AdTrack) or Cre recombinase (AdCre). Genomic DNA was isolated, and then PCR was performed using primers flanking the cassette. The presence of the Cre cassette resulted in a 2373-bp product, whereas the absence of the cassette generated a product of 1109 bp. The products were separated in a 1% agarose gel. M, DNA marker (fragment sizes indicated to the left of the gel); −, water template; P, OSDupDel targeting construct; +/+, wild-type MEFs. B, result of three independent RPAs using wild-type probe. α1(I) collagen-specific bands were quantitated and normalized to GAPDH values. The graph compares α1(I) collagen mRNA levels in untreated (+/+ Un) or AdCre-transduced (+/+ Cre) wild-type MEFs to untreated (−/− Un), AdTrack-transduced (−/− GFP), or AdCre-transduced (−/− Cre) mutant MEFs. The data are expressed as relative amounts of message versus wild-type untreated cells. *, p < 0.05; NS, not significant; the error bars represent S.E.
Mutation of the SL Did Not Affect Stability of the α1(I) Collagen Message
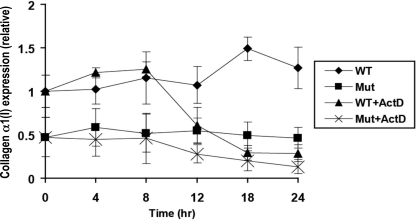
To gain an understanding of how the 5′ SL positively regulates expression of α1(I) collagen mRNA, we treated wild-type and SL mutant MEFs with actinomycin D to determine the effect of SL on message stability (Fig. 5). Somewhat unexpectedly, both messages exhibited similar half-lives; although mutation of the SL appeared to decrease message stability, this affect was minimal (t½ of 13 h versus 15 h in wild-type message).
FIGURE 5.
Mutation of the SL did not affect α1(I) collagen mRNA stability. Wild-type (WT) and mutant (Mut) MEFs were treated with 5 ng/ml actinomycin D (ActD) or DMSO. RNA was harvested 4, 8, 12, 18, and 24 h after treatment and reverse-transcribed; the resulting cDNA was analyzed by real time PCR using an α1(I) collagen-specific primer. The samples were normalized to 18 S expression levels.
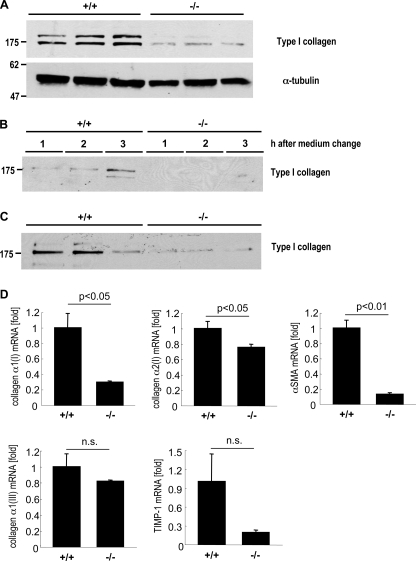
Mutation of the 5′-UTR SL Reduced Protein Expression of Type I Collagen in HSCs
Fig. 3 demonstrates that intracellular and secreted type I collagen levels were decreased in MEFs. To investigate whether type I collagen production is also reduced in mutant HSCs, HSCs were isolated from wild-type and 5′ SL mutant mice. After 7 days culturing, HSCs are fully activated. Intracellular type I collagen expression was significantly decreased to 23% in mutant compared with wild-type HSCs (Fig. 6A). Next, we analyzed the secretion of collagen in mutant HSCs. Collagen levels were significantly decreased to 33% in medium harvested from mutant HSCs for 3 h in comparison with wild-type cells (Fig. 6, B and C). These results demonstrated that the 5′ SL is crucial for collagen production in both MEFs and HSCs.
FIGURE 6.
Mutation of the 5′ SL in HSCs suppressed collagen α1(I) mRNA and protein levels. Primary HSCs were isolated from wild-type and mutant mice. A, intracellular expression of type I collagen was reduced in mutant HSCs. Intracellular protein (10 μg) was harvested from wild-type (+/+) and mutant (−/−) HSCs 7 days after culturing on plastic dishes. Western blotting for type I collagen and α-tubulin was performed. Representative Western blots are shown; each lane represents a separate cell isolation. B and C, secretion of type I collagen is reduced in mutant HSCs. Aliquots (50 μl) of media harvested 1, 2, and 3 h after change of fresh media from wild-type (+/+) and mutant (−/−) HSCs were separated by 7.5% SDS-PAGE gels, and the blot was probed with anti-type I collagen (B). Secreted type I collagen in the media 3 h after change of fresh media is shown (C). Three independent media collections. Representative Western blots are shown. D, mRNA levels of collagen α1(I), collagen α2(I), collagen α1(III), α smooth muscle actin (αSMA), and TIMP-1 in wild-type (+/+) and mutant (−/−) HSCs were measured by quantitative real time PCR. The results are shown as fold change compared with wild-type HSCs. NS, not significant. The error bars represent S.E.
We investigated mRNA levels of collagen α1(I), collagen α2(I), collagen α1(III), α-SMA, and tissue inhibitor of metalloproteinases (TIMP)-1 in wild-type and mutant HSCs (Fig. 6D). Not only collagen α1(I) but also collagen α2(I) and α-SMA mRNA levels were significantly reduced in mutant HSCs compared with wild-type HSCs (Fig. 6D). These results suggest that collagen production regulated by 5′ SL is associated with HSC activation.
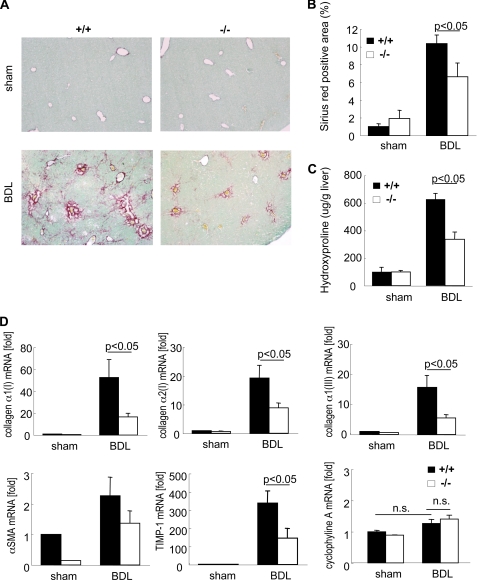
Mice Mutated in 5′ SL Showed Reduced Hepatic Fibrosis after BDL
21 days after BDL, wild-type mice had significant hepatic fibrosis, as demonstrated by Sirius red staining and hydroxyproline content. In contrast, 5′ SL mutant mice had less fibrosis, as demonstrated by reduced collagen deposition and hydroxyproline level (Fig. 7, A–C). Fibrogenic gene expression was determined at 21 days after BDL, including mRNAs for collagen α1(I), collagen α2(I), collagen α1(III), α-SMA, and TIMP-1. Wild-type mice showed an increased expression of these genes, whereas 5′ SL mutant mice had suppression in fibrogenic gene expression but not the expression of cyclophyline A (Fig. 7D). These results indicated that 5′ SL is crucial for collagen production in liver fibrosis induced by BDL.
FIGURE 7.
Reduced liver fibrosis in 5′ SL mutated mice after bile duct ligation. Wild-type (+/+) (n = 6) and 5′ SL mutant (−/−) (n = 6) mice underwent BDL. A and B, fibrillar collagen deposition was evaluated by Sirius red staining (A) and its quantification (B) 21 days after BDL. C, hepatic hydroxyproline content was measured. D, hepatic mRNA levels of collagen α1(I), collagen α2(I), collagen α1(III), α-SMA, TIMP-1, and cyclophyline A were measured in wild-type (+/+) and 5′ SL mutant (−/−) (n = 5) mice 21 days after BDL by quantitative real time PCR. These genes were normalized to 18 S and are expressed as fold change compared with sham operated wild-type mice. The original magnification is ×100 (A).
DISCUSSION
Liver fibrosis is characterized by increased synthesis and deposition of extracellular matrix proteins that disrupt the tissue structure, leading to organ dysfunction. Of the extracellular matrix proteins overproduced during liver fibrosis, type I collagen predominates. Many recent studies have examined activated HSCs, the main source of type I collagen in the fibrotic liver, as potential targets for anti-fibrotic therapies (30).
One aspect of HSC-mediated production of type I collagen occurs at the mRNA level of the constituent α1(I) and α2(I) chains. Transcription of both genes is up-regulated during fibrogenesis, and their expression is enhanced by a various post-transcriptional regulatory elements. Of particular note is the presence of a stem-loop structure located in the 5′-UTR of the two genes (31, 32), as well as in the 5′-UTR of the α1(III) gene (33). This SL encompasses the translation initiation codon (34) and is evolutionarily conserved from sea urchins (35) to zebrafish (36) to chicks (14) to frogs and humans (15), thus implicating a vital role for this structure in collagen gene expression and providing a possible therapeutic target.
To investigate the role of 5′ SL on type I collagen in vivo, we generated a knock-in mouse line harboring a mutation in the SL region that inhibited proper secondary structure formation. Although many previous studies of the 5′ SL were performed in HSCs, the concepts involved in the regulation of α1(I) collagen mRNA apply to all collagen-expressing cells, including fibroblasts and HSCs; therefore, we isolated MEFs to initially characterize effects of the SL mutation. Steady-state levels of α1(I) collagen mRNA in SL mutant MEFs were ∼50% of that found in wild-type cells, indicating the SL regulates message expression. This observation was further strengthened by an approximate 2:1 ratio of wild-type to mutant transcript expression in the RNA of heterozygote MEFs. Mutation of the SL had no effect on expression of α2(I) collagen mRNA, even though transcription of both genes is generally coordinately regulated, and the stem-loop sequence of α2(I) collagen is similar to that of α1(I) collagen. This finding was not unexpected, because the mutation does not affect the promoter region or any transcription factor-binding sequence, and the two genes do not share the same regulatory sequences (37).
Our results demonstrate that the SL is necessary for efficient and specific expression of α1(I) collagen mRNA. A previous study suggested that the SL inhibits α1(I) collagen mRNA steady-state levels in quiescent HSCs (15). However, it should be noted that the prior study utilized transient adenoviral transduction of HSCs with artificial reporter gene constructs, whereas our study utilized endogenous cells from mice with a germ line-transmitted mutation, thus better reflecting the in vivo environment. More importantly activated HSCs, more so than quiescent HSCs, phenotypically resemble collagen-producing fibroblasts and myofibroblasts (38, 39).
Another study found that expression of α1(I) collagen mRNA and α1(I) pro-collagen protein levels were reduced in activating HSCs treated with “molecular decoys” that mimic the 5′ SL and compete for SL-binding proteins, titrating out factors necessary for α1(I) collagen mRNA processing (34). This pointed to a potential correlation between α1(I) collagen mRNA abundance and message translation and indicated that the SL may be involved in a coordinated transcription/translation process, because the decoys may also compete for binding of protein factors necessary for translation. Additionally, although the SL inhibited translation of α1(I) collagen in vitro (similar to previous findings (40)), mutation of the SL resulted in inefficient formation of triple-helical type I collagen in Mov-13 fibroblasts.
We observed similar results in SL mutant MEFs and HSCs. Expression of intracellular type I collagen (both denatured and disulfide-bonded) was reduced in the mutant cells, furthering the correlation between α1(I) collagen mRNA abundance and protein expression. Additionally, secretion studies showed that mutant MEFs and HSCs exhibited a diminished and/or delayed secretion of type I collagen in comparison with wild-type cells. These data indicate that SL mutant-expressing cells do not increase their rate of protein secretion to compensate for reduced protein expression.
Stem-loop structures in the UTRs play a critical role in regulating mRNA processing (41). A canonical example of this is the interaction of the human immunodeficiency virus type-1 transactivation-responsive region with the Tat transactivator protein. Binding of Tat to a SL within the transactivation-responsive region at the 5′ end of the RNA ensures proper transcriptional elongation (42); additionally, the binding and subsequent modification of Tat is necessary for proper splicing of the viral mRNA (43). Given the importance of 5′ SLs in enhancing message stability in other systems (44, 45) and the proposed model that an uncharacterized 120-kDa SL-binding protein may interact with proteins in the 3′-UTR of α1(I) collagen mRNA to protect against exonuclease activity (15, 38), we examined whether mutation of the SL resulted in decreased mRNA stability. Unexpectedly, the mutant mRNA exhibited only a minimal decrease in stability versus wild type, indicating that the SL regulates α1(I) collagen mRNA by another mechanism.
Recently, we cloned a protein that binds the 5′ stem-loop of collagen mRNAs (17). This protein, LARP6, binds 5′ SL of collagen α1(I) and α2(I) mRNAs with similar affinity. Knockdown of LARP6 resulted in greatly reduced collagen protein synthesis, which was a combination of less efficient translation of collagen mRNAs and inefficient folding of the collagen heterotrimer. When overexpressed, LARP6 bound 5′ SL with affinity high enough to block ribosomal scanning, resulting in exclusion of collagen mRNAs from the polysomes (17). Based on these findings, we postulated that LARP6 has a role in coordination of translation of collagen α1(I) and α2(I) mRNAs (16). Its binding to 5′ SL may serve to prevent their random translation, which is a prerequisite for their subsequent coordinated translation on the membrane of the ER. Coordinated synthesis of α1(I) and α2(I) polypeptides results in higher efficiency of folding of the heterotrimer of type I collagen. These results are consistent with a lower degree of fibrosis in mice lacking the 5′ SL.
RNA elements are not greatly involved in regulating transcriptional initiation in eukaryotes, although there is some evidence of this in viruses (46); therefore, we do not expect the SL mutation to affect transcriptional activation. However, in addition to regulating message stability, the 5′- and 3′-UTRs regulate other eukaryotic mRNA metabolic processes including transcriptional elongation, post-transcriptional modifications (capping of the 5′ end and 3′ polyadenylation), splicing, nuclear export, subcellular message localization, and translation (41, 47–50). These functions are mediated by the binding of protein factors to mRNA in the nucleus, the cytoplasm, or both, resulting in the formation of a messenger ribonucleoprotein particle (41). Disruption of the messenger ribonucleoprotein particle through deletion of the 5′ SL could result in improper nuclear processing and increased degradation of the α1(I) collagen mRNA; because the 3′-UTR sequence is unaffected, polyadenylation can still occur, leaving the stability of the mutant mRNA virtually unchanged compared with wild type. Although most localization elements are located in the 3′-UTR (48), 5′-UTR elements can also regulate message localization, as is the case with Drosophila gurken mRNA (51). Indeed, one model suggests that the 5′ SL targets new α1(I) collagen mRNAs to distinct regions of the endoplasmic reticulum to ensure proper translation (19).
Additionally, the current model of RNA processing suggests that many cytoplasmic messenger ribonucleoprotein particle proteins, including several heterogeneous nuclear ribonucleoproteins and the exon junction complex, associate with the mRNA in the nucleus and accompany the molecule into the cytoplasm (41). Mutation of the 5′ SL may interfere with interaction of these proteins (or associated co-factors) with the α1(I) collagen mRNA inside the nucleus and lead to increased message degradation.
Any number of mutations in either the α1(I) or α2(I) collagen chain can impair type I collagen production in humans, resulting in the disease osteogenesis imperfecta (“brittle bone disease”) (18). Based upon the structural support role of type I collagen and the phenotypic effects of collagen mutations in osteogenesis imperfecta model mice (52–54), we were somewhat surprised that SL mutant mice developed normally with a grossly normal phenotype when compared with wild-type littermates. However, this finding is not unprecedented. Several studies using transgenic mouse models of osteogenesis imperfecta and other collagen-related diseases have reported that mutant mice can lack any readily apparent abnormal phenotypes (52, 55–57).
One potential explanation for a lack of phenotype is that baseline levels of type I collagen produced in the SL mutant mice are sufficient for normal development, breeding, and survival; additional levels of collagen in wild-type mice may simply serve to provide additional structural support required during stress. Indeed, some patients with mild forms of osteogenesis imperfecta do not exhibit any of the characteristic phenotypes of the disease, even when 75% of the α1(I) collagen chains contain mutations (18, 58). Furthermore, it has been reported that mutations that result in synthesis of longer or shorter α1(I) or α2(I) chains generate more deleterious effects than those mutations that affect expression of the constituent chains (59), such as in the case of SL mutant mice. The SL mutation is not incorporated into the type I collagen protein; therefore, the effect of the mutation on any physical phenotype may be subtle and may not present itself until the mice are physically challenged, as has been shown in other collagen-based mouse models (55, 60).
Our findings strengthen the importance of the 5′ SL of α1(I) collagen mRNA in regulating mRNA expression in vivo and its involvement in regulating type I collagen production. The fact the SL structure has been conserved throughout evolution underscores its importance. Our findings also enhance the therapeutic potential of SL decoys, which can decrease collagen synthesis from endogenous RNA, potentially attenuating liver fibrosis (34). Finally, we have developed an in vivo mouse model for studying the 5′ SL and its effects on collagen expression and production in MEFs and HSCs. Indeed, 5′ SL mutant mice showed a significant reduction of liver fibrosis after BDL, confirming our concept that 5′ SL is crucial for collagen production in vivo during liver injury.
Supplementary Material
Acknowledgments
We thank Elizabeth Livanos, Kimberly Kluckman, Randy Thresher, Beverly Koller, and Kelly Krock Parsons for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants AA010459 and DK065972 (to R. A. R.) and GM041804 (to D. A. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- HSC
- hepatic stellate cell
- 5′ SL
- 5′ stem-loop
- MEF
- mouse embryonic fibroblast
- RPA
- ribonuclease protection assay
- TIMP-1
- tissue inhibitor of metalloproteinases-1
- BDL
- bile duct ligation.
REFERENCES
- 1. Aumailley M., Gayraud B. (1998) J. Mol. Med. 76, 253–265 [DOI] [PubMed] [Google Scholar]
- 2. Badylak S. F., Freytes D. O., Gilbert T. W. (2009) Acta Biomater. 5, 1–13 [DOI] [PubMed] [Google Scholar]
- 3. Niederreither K., D'Souza R., Metsäranta M., Eberspaecher H., Toman P. D., Vuorio E., De Crombrugghe B. (1995) Matrix Biol. 14, 705–713 [DOI] [PubMed] [Google Scholar]
- 4. Milani S., Herbst H., Schuppan D., Surrenti C., Riecken E. O., Stein H. (1990) Am. J. Pathol. 137, 59–70 [PMC free article] [PubMed] [Google Scholar]
- 5. Eleftheriades E. G., Ferguson A. G., Spragia M. L., Samarel A. M. (1995) J. Mol. Cell Cardiol. 27, 1459–1473 [DOI] [PubMed] [Google Scholar]
- 6. Doyle S. A., Smith B. D. (1998) J. Cell. Biochem. 71, 233–242 [PubMed] [Google Scholar]
- 7. Friedman S. L., Roll F. J., Boyles J., Bissell D. M. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 8681–8685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brenner D. A., Waterboer T., Choi S. K., Lindquist J. N., Stefanovic B., Burchardt E., Yamauchi M., Gillan A., Rippe R. A. (2000) J. Hepatol. 32, 32–38 [DOI] [PubMed] [Google Scholar]
- 9. Stefanovic B., Hellerbrand C., Holcik M., Briendl M., Aliebhaber S., Brenner D. A. (1997) Mol. Cell. Biol. 17, 5201–5209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ross J. (1995) Microbiol. Rev. 59, 423–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shyu A. B., Wilkinson M. F., van Hoof A. (2008) EMBO J. 27, 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lindquist J. N., Marzluff W. F., Stefanovic B. (2000) Am. J. Physiol. Gastrointest Liver Physiol. 279, G471–G476 [DOI] [PubMed] [Google Scholar]
- 13. Sachs A. B., Sarnow P., Hentze M. W. (1997) Cell 89, 831–838 [DOI] [PubMed] [Google Scholar]
- 14. Yamada Y., Mudryj M., de Crombrugghe B. (1983) J. Biol. Chem. 258, 14914–14919 [PubMed] [Google Scholar]
- 15. Stefanovic B., Hellerbrand C., Brenner D. A. (1999) Mol. Cell. Biol. 19, 4334–4342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cai L., Fritz D., Stefanovic L., Stefanovic B. (2009) Expert Rev. Gastroenterol. Hepatol. 3, 1–3 [DOI] [PubMed] [Google Scholar]
- 17. Cai L., Fritz D., Stefanovic L., Stefanovic B. (2009) J. Mol. Biol. (2010) 395, 309–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Forlino A., Marini J. C. (2000) Mol. Genet Metab. 71, 225–232 [DOI] [PubMed] [Google Scholar]
- 19. Stefanovic B., Brenner D. A. (2003) J. Biol. Chem. 278, 927–933 [DOI] [PubMed] [Google Scholar]
- 20. Hogan B. (1994) Manipulating the Mouse Embryo: A Laboratory Manual, 2nd Ed., pp. 1–487, Cold Spring Harbor Laboratory, Plainview, NY [Google Scholar]
- 21. Seki E., De Minicis S., Osterreicher C. H., Kluwe J., Osawa Y., Brenner D. A., Schwabe R. F. (2007) Nat. Med. 13, 1324–1332 [DOI] [PubMed] [Google Scholar]
- 22. Rippe R. A., Almounajed G., Brenner D. A. (1995) Hepatology 22, 241–251 [PubMed] [Google Scholar]
- 23. Schnabl B., Kweon Y. O., Frederick J. P., Wang X. F., Rippe R. A., Brenner D. A. (2001) Hepatology 34, 89–100 [DOI] [PubMed] [Google Scholar]
- 24. Le Hir H., Nott A., Moore M. J. (2003) Trends Biochem. Sci. 28, 215–220 [DOI] [PubMed] [Google Scholar]
- 25. Rippe R. A., Lorenzen S. I., Brenner D. A., Breindl M. (1989) Mol. Cell. Biol. 9, 2224–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Breault D. T., Lichtler A. C., Rowe D. W. (1997) J. Biol. Chem. 272, 31241–31250 [DOI] [PubMed] [Google Scholar]
- 27. Rahkonen O., Su M., Hakovirta H., Koskivirta I., Hormuzdi S. G., Vuorio E., Bornstein P., Penttinen R. (2004) Circ. Res. 94, 83–90 [DOI] [PubMed] [Google Scholar]
- 28. Fu X. Y., Manley J. L. (1987) Mol. Cell. Biol. 7, 738–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ledermann B. (2000) Exp. Physiol. 85, 603–613 [PubMed] [Google Scholar]
- 30. Tsukada S., Parsons C. J., Rippe R. A. (2006) Clin. Chim. Acta 364, 33–60 [DOI] [PubMed] [Google Scholar]
- 31. Bernard M. P., Myers J. C., Chu M. L., Ramirez F., Eikenberry E. F., Prockop D. J. (1983) Biochemistry 22, 1139–1145 [DOI] [PubMed] [Google Scholar]
- 32. D'Alessio M., Bernard M., Pretorius P. J., de Wet W., Ramirez F. (1988) Gene 67, 105–115 [DOI] [PubMed] [Google Scholar]
- 33. Benson-Chanda V., Su M. W., Weil D., Chu M. L., Ramirez F. (1989) Gene 78, 255–265 [DOI] [PubMed] [Google Scholar]
- 34. Stefanovic B., Schnabl B., Brenner D. A. (2002) J. Biol. Chem. 277, 18229–18237 [DOI] [PubMed] [Google Scholar]
- 35. Exposito J. Y., Boute N., Deleage G., Garrone R. (1995) Eur. J. Biochem. 234, 59–65 [DOI] [PubMed] [Google Scholar]
- 36. Dubois G. M., Haftek Z., Crozet C., Garrone R., Le Guellec D. (2002) Gene 294, 55–65 [DOI] [PubMed] [Google Scholar]
- 37. Sengupta P., Xu Y., Wang L., Widom R., Smith B. D. (2005) J. Biol. Chem. 280, 21004–21014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lindquist J. N., Parsons C. J., Stefanovic B., Brenner D. A. (2004) J. Biol. Chem. 279, 23822–23829 [DOI] [PubMed] [Google Scholar]
- 39. Lang A., Brenner D. A. (1999) Ital J. Gastroenterol. Hepatol. 31, 173–179 [PubMed] [Google Scholar]
- 40. Hensold J. O., Stratton C. A., Barth D. (1997) Nucleic Acids Res. 25, 2869–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moore M. J. (2005) Science 309, 1514–1518 [DOI] [PubMed] [Google Scholar]
- 42. Karn J. (1999) J. Mol. Biol. 293, 235–254 [DOI] [PubMed] [Google Scholar]
- 43. Berro R., Kehn K., de la Fuente C., Pumfery A., Adair R., Wade J., Colberg-Poley A. M., Hiscott J., Kashanchi F. (2006) J. Virol. 80, 3189–3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fournier B., Truong-Bolduc Q. C., Zhang X., Hooper D. C. (2001) J. Bacteriol. 183, 2367–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bricker A. L., Belasco J. G. (1999) J. Bacteriol. 181, 3587–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang Y., Zhang X. (2000) J. Virol. 74, 10571–10580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mignone F., Gissi C., Liuni S., Pesole G. (2002) Genome Biol. 3, REVIEWS0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kloc M., Zearfoss N. R., Etkin L. D. (2002) Cell 108, 533–544 [DOI] [PubMed] [Google Scholar]
- 49. Kornblihtt A. R., de la Mata M., Fededa J. P., Munoz M. J., Nogues G. (2004) Rna. 10, 1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lipshitz H. D., Smibert C. A. (2000) Curr. Opin. Genet. Dev. 10, 476–488 [DOI] [PubMed] [Google Scholar]
- 51. Saunders C., Cohen R. S. (1999) Mol. Cell 3, 43–54 [DOI] [PubMed] [Google Scholar]
- 52. Pereira R., Khillan J. S., Helminen H. J., Hume E. L., Prockop D. J. (1993) J. Clin. Invest. 91, 709–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stacey A., Bateman J., Choi T., Mascara T., Cole W., Jaenisch R. (1988) Nature 332, 131–136 [DOI] [PubMed] [Google Scholar]
- 54. Forlino A., Porter F. D., Lee E. J., Westphal H., Marini J. C. (1999) J. Biol. Chem. 274, 37923–37931 [DOI] [PubMed] [Google Scholar]
- 55. Kvist A. P., Latvanlehto A., Sund M., Eklund L., Väisänen T., Hägg P., Sormunen R., Komulainen J., Fässler R., Pihlajaniemi T. (2001) Am. J. Pathol. 159, 1581–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pereira R., Halford K., Sokolov B. P., Khillan J. S., Prockop D. J. (1994) J. Clin. Invest. 93, 1765–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Helminen H. J., Kiraly K., Pelttari A., Tammi M. I., Vandenberg P., Pereira R., Dhulipala R., Khillan J. S., Ala-Kokko L., Hume E. L. (1993) J. Clin. Invest. 92, 582–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Roughley P. J., Rauch F., Glorieux F. H. (2003) Eur. Cell Mater. 5, 41–47 [DOI] [PubMed] [Google Scholar]
- 59. Kuivaniemi H., Tromp G., Prockop D. J. (1991) FASEB J. 5, 2052–2060 [DOI] [PubMed] [Google Scholar]
- 60. Hormuzdi S. G., Strandjord T. P., Madtes D. K., Bornstein P. (1999) Matrix Biol. 18, 287–294 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.