Abstract
Proliferation of mammalian cardiomyocytes stops rapidly after birth and injured hearts do not regenerate adequately. High cyclin-dependent kinase inhibitor (CKI) levels have been observed in cardiomyocytes, but their role in maintaining cardiomyocytes in a post-mitotic state is still unknown. In this report, it was investigated whether CKI knockdown by RNA interference induced cardiomyocyte proliferation. We found that triple transfection with p21Waf1, p27Kip1, and p57Kip2 siRNAs induced both neonatal and adult cardiomyocyte to enter S phase and increased the nuclei/cardiomyocyte ratio; furthermore, a subpopulation of cardiomyocytes progressed beyond karyokynesis, as assessed by the detection of mid-body structures and by straight cardiomyocyte counting. Intriguingly, cardiomyocyte proliferation occurred in the absence of overt DNA damage and aberrant mitotic figures. Finally, CKI knockdown and DNA synthesis reactivation correlated with a dramatic change in adult cardiomyocyte morphology that may be a prerequisite for cell division. In conclusion, CKI expression plays an active role in maintaining cardiomyocyte withdrawal from the cell cycle.
Keywords: CDK (Cyclin-dependent Kinase), Cell Cycle, Cell Division, Mitosis, SiRNA, Cardiomyocytes, Cyclin-dependent Kinase Inhibitors, Terminal Differentiation
Introduction
The adult mammalian heart has very limited regenerative capacity (1). In fact, cardiomyocytes divide extensively during development and lose proliferative capacity shortly after birth in rodents and at ∼7 months of age in humans, when they withdraw from the cell cycle and remain in G0 stage of the cell cycle indefinitely (2–5). Permanently differentiated cardiac myocytes lose their ability to divide and switch from hyperplastic to hypertrophic growth in response to mitogenic stimuli (5–8). This transition is characterized by a marked increase in myofibril density, the appearance of mature intercalated discs, and the formation of binucleated cardiomyocytes (9).
Cell cycle progression is positively modulated by cyclin-cyclin-dependent kinase (CDK)2 complexes. Specifically, cyclin D-CDK4–6 and cyclin E-CDK2 complexes phosphorylate proteins of the retinoblastoma (pRb) family, releasing E2F transcription factors (3, 10, 11). Indeed, pRb phosphorylation is a prerequisite to initiate a new round of cell cycle activity allowing the cell to enter the S phase of the cell cycle (12). The expression of cyclin and CDK genes is down-modulated in neonatal hearts and further decreases in the adult. Other proliferation-promoting genes, such as E2F1 and Myc family members are similarly repressed, as demonstrated both in vitro and in vivo (13, 14). pRb and pRb family proteins also have a critical role in regulating cardiac cell cycle both in the developing and in the adult heart. pRb is barely detectable in proliferating fetal cardiomyocytes, its expression is up-regulated progressively during neonatal development, and pRb is the predominant pocket protein expressed in terminally differentiated adult cardiomyocytes. The accumulation of pRb plays a critical role in regulating cell cycle arrest associated with terminal cardiac muscle differentiation, as also observed in other in vitro culture systems, including skeletal muscle, adipocytes, and macrophages, suggesting that this may be a general phenomenon (5, 15, 16). However, other mechanisms are at play as well, as pRb phosphorylation by cyclin D/CDK4 in differentiated cardiomyocytes isolated from neonatal rat induces their hypertrophic growth but not their proliferation (17). Cyclin-CDK complexes are regulated by two structurally defined classes of inhibitors (CKIs): INK4 and CIP/KIP families. The INK4 family, which includes p15INK4B, p16INK4A, p18INK4C, and p19INK4D, specifically inhibits CDK4/6, preventing their heterodimerization with D cyclins. The CIP/KIP family comprises p21CIP1 (p21), p27KIP1 (p27), p57KIP2 (p57). These molecules display lower specificity, as they can bind and inhibit all cyclin/CDKs (5, 18). However, CIP/KIP CKI also have a positive role in the modulation of CDK activity, as they facilitate the assembly of cyclin D-CDK4 complexes (19, 20).
The expression of all CKIs is detectable during embryonic development. At later stages, the progressive withdrawal of maturing cardiac myocytes from the cell cycle coincides with increased levels of both p21 and p27, whereas p16INK4A, and p18INK4C levels are low or undetectable (9, 16, 21). Specifically, p27 seems to be critical for controlling the exit from the cell cycle, whereas p21 may maintain cell cycle arrest and prevent re-entry into cell cycle (22). In fact, p21 null mice do not show developmental defects or elevate tumor incidence. Conversely, p27 null mice are predisposed to pituitary tumors and display generalized hyperplasia with a 20% increase in heart weight (23–25). However, the lack of p21 and p27 function does not lead to gross developmental defects, suggesting the existence of compensatory mechanisms during the development. Of all the CKIs, only p57 has been shown to be essential for embryonic development. In keeping with this finding, p57 levels peak during late embryonal life and then disappear during early fetal period such that p57 protein is not detectable at all other stages (26). Ablation of p57 causes very severe developmental abnormalities. In fact, p57 null mice die in utero or soon after birth, displaying multiple developmental defect (27, 28). Deletion of both p57 and p27 accelerates the lethality, demonstrating that p27 and p57 cooperate in the control of cell cycle exit and differentiation (29).
The CKI expression pattern observed in humans is very similar to that found in rodents (30); p21 is detectable during fetal development and its level increases in post-natal life; p27 expression increases by 25 weeks of fetal life and remains constant thereafter; p57 expression decreases during development and only low levels of p57 protein are present in the adult human heart. However, in both acute and chronic heart failure, p57 increases, whereas p21 and p27 expression decreases (30, 31). Cardiomyocyte terminal differentiation may also involve other factors that can create a barrier to proliferation beyond the immediate perinatal period (31), such as telomerase reverse transcriptase (TERT) down-regulation and the resulting loss of telomerase activity (32–35). Indeed, in contrast to high activity observed at the fetal stage, adult rodent heart displays no detectable telomerase activity (36, 37). Furthermore, it has been shown that constitutive TERT expression delayed cardiomyocyte exit from the cell cycle, induced cardiac hypertrophy, and promoted the cytoprotective function of telomerase (38). Cardiac regeneration during human lifespan and upon injury largely relies on a specialized population of stem cells. However, myocardial regeneration after massive tissue loss following injuries such as myocardial infarction is very limited. Indeed, the main function of cardiac stem cell seems to be the maintenance of heart homeostasis, and their regenerative capacity may be limited to the repair of day by day wear and tear (1, 39). Thus, understanding the molecular mechanisms limiting cardiomyocyte proliferation is of paramount importance not only for cell biology but also for regenerative medicine of heart disease. In this report, we investigated the role of CKI in maintaining cardiomyocytes in a post-mitotic state and whether their down-modulation through RNAi can induce cell cycle re-entry.
EXPERIMENTAL PROCEDURES
Cardiomyocyte Isolation, Culture, and Characterization
All experimental procedures complied with the Guidelines of the Italian National Institutes of Health and with the Guide for the Care and Use of Laboratory Animals published by the U. S. National Institutes of Health (NIH publication no. 85-23, 1996) and were approved by the Institutional Animal Care and Use Committee.
Neonatal cardiomyocytes were isolated from 1–2-day-old SWISS CD1 mice as described previously (40) and specified in the supplemental “Experimental Procedures.” To minimize the number of residual proliferating cardiomyocytes and of contaminating fibroblasts, cell cultures were cultured in minimal essential medium containing 10% FBS in the presence 20 μm cytosine β-d-arabinofuranoside (AraC, C1768 Sigma) for 48 h. After a 3-day recovery time in AraC-free medium, neonatal cardiomyocytes were transfected.
Left ventricular cardiomyocytes from 8–12-week-old Wistar rats (250–350 g) were isolated from adult hearts, rapidly excised, and retrogradely perfused through the aorta as described before (41, 42) and specified in the supplemental “Experimental Procedures.” Cells were cultured for 1 day in standard serum-free medium (DMEM, 25 mm HEPES, 5 mm taurine, 5 mm creatine, 2 mm l-carnitine, 20 units/ml insulin (Sigma), 0.2% BSA, penicillin (100 units/ml), and streptomycin (100 μg/ml). Then, cardiomyocytes were transfected, and fresh serum-free medium (without BSA, containing 2 mm l-glutamine) was added every 3 days. Cardiomyocyte culture quality and purity was assayed as shown in supplemental Figs. S1 (neonatal cardiomyocytes) and S2 (adult cardiomyocytes).
RNA Interference and Cardiomyocyte Transfection
Both neonatal and adult cardiomyocytes were transfected with HiPerfect transfection reagent (Qiagen) complexed with 5 nm siRNAs (Dharmacon) as described previously (43) and specified in the supplemental “Experimental Procedures.”
Adenovirus Vectors and Cardiomyocyte Infection
Replication-deficient recombinant adenoviral vectors were prepared, stored, and administered as described before (44, 45). Both neonatal and adult cardiomyocytes were infected at 50 multiplicity of infection for 2 h, followed by transfection 12 h later.
Western Blotting and mRNA Quantification by Reverse Transcriptase Real-Time PCR (qRT-PCR)
Western blotting was performed as described previously (46) and detailed in the supplemental “Experimental Procedures.” Briefly, cells were lysed in 2× Laemmli buffer and boiled for 5 min. Equal amounts of proteins were separated by SDS-PAGE and transferred to nitrocellulose by standard procedures. Proteins of interest were detected using specific antibodies.
mRNA was quantified by reverse transcriptase real-time PCR (qRT-PCR). Total RNA was extracted using TRIzol (Invitrogen) and then used as template to synthesize cDNA using reverse transcriptase (Invitrogen) according to the manufacturer's instructions. The SYBR-GREEN qRT-PCR method was used to analyze mRNA levels, and relative expression was calculated using the comparative Ct method (2−ΔΔCt) (47). GAPDH was amplified on the same plate for each sample for normalization purposes. Primer sequences are available upon request.
Indirect Immunofluorescence Staining and BrdU Assay
Immunofluorescence stainings were performed as described previously (48). Briefly, cardiomyocytes were fixed with 4% paraformaldehyde, and immunofluorescences were performed using following antibodies: anti-Aurora B (AIM-1; BD Transduction Laboratories), anti-phospho-histone H3 (Ser10) (RR002; Upstate), anti-BrdU (ab1893; Abcam), anti-γH2AX antibody (clone JBW301; Upstate Biotechnology, Lake Placid, NY), monoclonal anti-troponin T-C (CT3) (catalog no. sc 20025; Santa Cruz Biotechnology), and anti-α-actinin (sarcomeric) (EA-53, Sigma). Cells were counter-stained with Hoechst 33342 (Sigma-Aldrich). The same number of optical fields was counted for each sample, totaling 70–200 neonatal cardiomyocytes/sample and 50–100 adult cardiomyocytes/sample. To compare the results of different experiments, data were expressed as % of control. Images were acquired by a fluorescence microscope (Axioplan2; Carl Zeiss). Images were analyzed by IAS software (Delta Sistemi), processed, and overlaid using Adobe Photoshop CS2 (Adobe). Cells were counted by two blinded readers obtaining similar results.
Apoptosis Assay
Apoptosis was assessed by measuring the amount of cytoplasmic nucleosomes generated during the apoptotic fragmentation of cellular DNA by Cell Death Detection ELISA (Roche Applied Science), according to the manufacturer's instructions.
Statistical Analysis
Variables were analyzed by Student's t test and one-way analysis of variance. A value of p ≤ 0.05 was deemed statistically significant. Results are reported as mean values ± S.E.
RESULTS
p21 and p27 Knockdown Induces Neonatal Cardiomyocyte Progression into S Phase
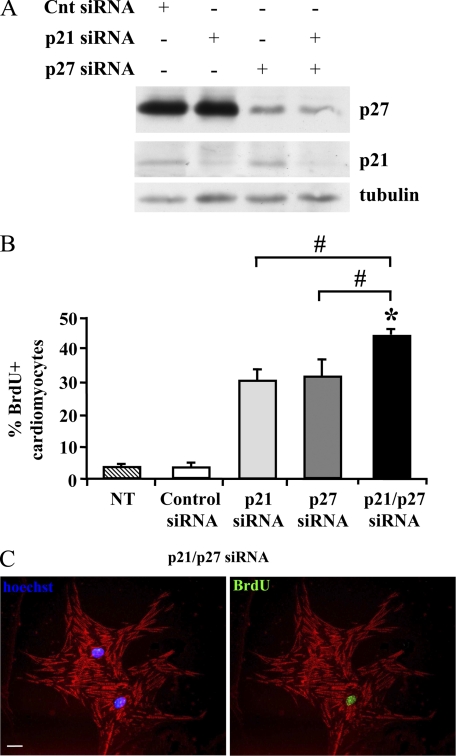
To investigate the functional role of CKIs in cardiomyocyte withdrawal from the cell cycle, neonatal mouse cardiomyocytes were transfected with siRNA targeting p21, p27, or both, as well as with a control sequence. To minimize the number of residual proliferating cardiomyocytes and of contaminating fibroblasts, cell cultures were treated with AraC, which kills proliferating cells selectively (49). AraC was added to cultures for 2 days, followed by a 3-day recovery time before transfection. Western blotting analysis of the transfected cells confirmed a p21 and p27 knockdown efficiency of >70% 2 days after transfection (Fig. 1A). To assay the cells that entered S phase upon CKI RNAi, BrdU was added at the time of transfection, and its incorporation was assayed 2 days later. Fig. 1, B and C, show that both single and double CKI knockdown significantly increased DNA replication. Interestingly, double p21 and p27 silencing was the most effective and almost half of the cardiomyocytes entered S phase. Thus, double CKI silencing was adopted for the following experiments.
FIGURE 1.
p21 and p27 knockdown induces neonatal cardiomyocyte DNA synthesis. Neonatal cardiomyocytes were transfected with p21, p27, or control (Cnt) siRNAs. A, cell lysates were prepared 2 days after transfection. Western blot analysis demonstrates efficient p21 and p27 single and double knockdown; β-tubulin shows equal loading in each lane. B and C, neonatal cardiomyocytes were transfected with p21, p27, or control siRNA. Nontransfected (NT) cells were used as additional control to exclude potential effects of the control siRNA. Then, BrdU was added. Two days later, BrdU-incorporating cells were detected by immunofluorescence using a FITC-conjugated specific antibody, whereas cardiomyocytes were stained using a Texas Red-conjugated antibody to α-sarcomeric actinin. Nuclei were detected using Hoechst 33342 (blue). B, bar graph indicating the percentage of BrdU-positive cardiomyocytes (n = 4; #, p, < 0.003; *, p < 0.00001). C, representative picture of two cardiomyocytes, one BrdU-positive and one -negative, at 2 days after transfection with p21 and p27 siRNAs. The left panel shows an α-sarcomeric actinin/Hoechst 33342 overlay; the right panel shows an α-sarcomeric actinin/BrdU overlay of the same field. Scale bar, 20 μm.
p21 and p27 Knockdown Induces Neonatal Cardiomyocyte Proliferation
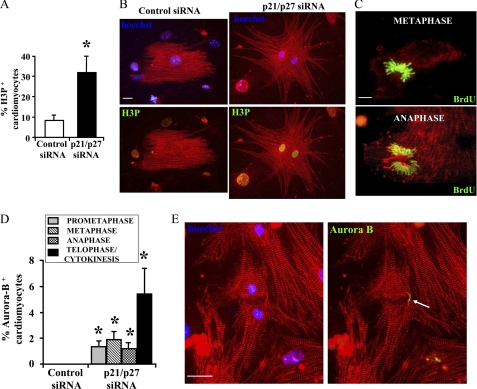
The increase in S phase induced by CKI knockdown was associated with a 2.7- ± 0.5-fold increase of bi-nucleated cardiomyocytes (CKI RNAi, 17.3 ± 3.2; control, 6.4% ± 1.7; p < 0.006, n = 4), which could account for the observed increase in DNA synthesis only in part. Hence, these data suggested that some cardiomyocytes had undergone successful cytodieresis. Thus, we assayed whether p21 and p27 knockdown stimulated not only S phase entrance but also the progression through the following phases of the cell cycle. To this aim, histone H3 phosphorylation at Ser10 (H3P), a mitotic marker, was assayed by immunofluorescence staining. Down-modulation of CKI expression more than tripled H3P-positive cardiomyocytes, which reached >30% 2 days after transfection (Fig. 2, A and B). In keeping with these findings, mitotic figures were observed in p21 and p27 knockdown cardiomyocytes (Fig. 2C) but not in controls (data not shown).
FIGURE 2.
p21 and p27 knockdown regulates cells division and induces neonatal cardiomyocyte proliferation 2 days after transfection. Neonatal cardiomyocytes were transfected with p21 and p27 or control siRNAs and labeled with BrdU for 48 h. Histone H3 phosphorylation at Ser10 (H3P), mitotic figures, BrdU incorporation, and Aurora B were detected by immunofluorescence after 2 days. Cardiomyocytes were identified using a Texas Red-conjugated antibody to α-sarcomeric actinin, and nuclei were detected using Hoechst 33342 (blue). A, a bar graph shows that p21 and p27 down-modulation increased the percentage of H3P positive cardiomyocytes (n = 3; *, p < 0.007). B, representative immunofluorescences. H3P-positive nuclei were detected using a FITC-conjugated specific antibody. Left panels show an α-sarcomeric actinin/Hoechst 33342 overlay, whereas right panels show the α-sarcomeric actinin/BrdU overlay of the same field. Scale bar, 20 μm. C, representative figures of cardiomyocytes undergoing mitosis. DNA synthesis occurred after transfection of siRNAs as assessed by anti-BrdU staining (green). Chromosome identity was confirmed by Hoechst 33342 staining. α-Sarcomeric actinin/BrdU overlays show representative metaphase (top), and anaphase (bottom) figures. Scale bar, 10 μm. D, bar graph showing Aurora B-positive cells in different mitosis phases (n = 3; *, p < 0.04). E, a representative picture displays a dividing neonatal cardiomyocyte. The white arrow points to the Aurora B-positive (green) mid-body structure. Scale bar, 20 μm.
Aurora B kinase plays a role in chromosomal condensation by phosphorylating histone H3. Indeed, Aurora B associates with centromeric heterocromatin early in mitosis (prometaphase and metaphase). Subsequently, Aurora B relocates to the central spindle during anaphase and then to the contractile ring and the mid-body during telophase representing a marker of cytokynesis (50–52). To determine whether CKI knockdown promoted cardiomyocyte cell division, we measured Aurora B by immunofluorescence staining. We found that ∼5% of the p21 and p27 knockdown cardiomyocytes displayed an Aurora B-positive mid-body (Fig. 2, D and E), whereas these structures were not present in control transfected cells.
To confirm these results, cardiomyocyte number was determined at different times after transfection. We found that the number of α-sarcomeric actinin-positive cells significantly increased 1.7- ± 0.1-fold at day 2 (n = 9; p < 0.0001).
Cell Cycle Progression Induced by CKI Knockdown Does Not Cause DNA Damage and Apoptosis
CKI knockdown in skeletal muscle myotubes is associated with induction of aberrant mitoses and apoptosis (43). However, most mitotic figures we observed in dividing cardiomyocytes were morphologically normal (Fig. 2C), and p21 and p27 double knockdown was not associated with a significant increase of cell apoptosis as assessed by TUNEL assay (supplemental Fig. S3A).
To further explore this issue, cell micronucleation was assayed by Hoechst 33342 staining in cardiomyocytes that were cultured in the presence of BrdU to monitor the cells that entered S-phase and replicated their genome over a period of 48 h. Significantly, we found that BrdU-positive cardiomyocytes displaying overt micronucleation were <2% and that the micronucleation rate did not increase following CKI knockdown (supplemental Fig. S3, B and C).
Aberrant mitosis is associated with DNA double-strand brakes (53). Thus, DNA damage in neonatal cardiomyocytes was tested measuring the formation of γH2AX foci in cells cultured in the presence of BrdU from the time of transfection. We found that, albeit virtually all γH2AX-positive cells were BrdU-positive as well, CKI interference did not increased the number of foci-positive nuclei compared with control cells (supplemental Fig. S4A). Moreover, when the percentage of γH2AX-negative cells was calculated among BrdU+ cardiomyocytes, it was found that up to 70% of the CKI knockdown cardiomyocytes underwent DNA replication in the absence of overt DNA damage (supplemental Fig. S4B). These data confirm that proliferation is compatible with cardiomyocyte differentiation program.
p57 Helps Regulating Neonatal Cardiomyocyte Proliferation
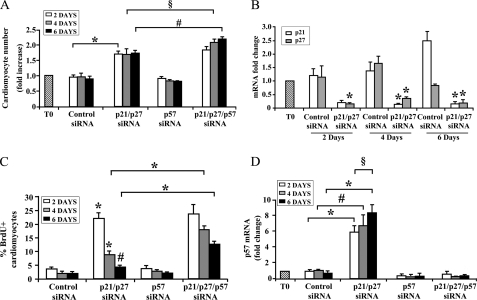
When we assayed the number of cardiomyocytes following p21 and p27 knockdown, we found that after the initial increase displayed at day 2, no further increase was observed afterward (Fig. 3A), despite sustained knockdown of both p21 and p27 (Fig. 3B). To test whether this was due to growth arrest, BrdU incorporation was assayed after an 8-h labeling pulse. It was found that, although >20% of the cardiomyocytes were BrdU-positive 2 days after p21 and p27 knockdown, DNA synthesis rate declined afterward and was very close to control at 6 days after transfection (Fig. 3C). These results suggest that p21 and p27 knockdown efficiently triggered one or few neonatal cardiomyocyte cell divisions, followed by a second arrest of the cell cycle.
FIGURE 3.
Triple p21, p27, and p57 knockdown further increases neonatal cardiomyocyte proliferation. Neonatal cardiomyocytes were transfected with p21 p27 siRNAs in the presence or absence of p57 siRNA. A, the histogram displays cardiomyocyte number at different time points expressed as fold change compared with cardiomyocyte number at the time of transfection (T0): p21 and p27 siRNA double transfection increased cardiomyocyte number at 2 days (n = 9; *, p < 0.0001), whereas p57 knockdown alone was ineffective. Differences in cardiomyocytes between double and triple CKI knockdown at 4 and 6 days are statistically significant (n = 4; #, p < 0.008; §, p < 0.05). B, RNA was extracted 2, 4, or 6 days after transfection. Bar graph shows the specific, efficient, and persistent down-modulation of p21 and p27 mRNA levels measured by qRT-PCR (n = 3; *, p < 0.001). C, cardiomyocytes were pulse labeled with BrdU for 8 h 2, 4, or 6 days after transfection. The bar graph indicates the percentage of BrdU-positive cardiomyocytes. BrdU incorporation is significantly increased between double and triple CKI siRNA at both 4 and 6 days (n = 4; *, p < 0.03). D, bar graph showing p57 mRNA levels measured by qRT-PCR. p21 and p27 double RNAi triggered a progressive increase of p57 mRNA levels and a specific siRNA to p57 prevented this increase (n = 6; #, p < 0.0008; *, p < 0.000001; §, p < 0.015).
To determine whether other CKIs may contribute to limit cardiomyocyte proliferation upon p21 and p27 knockdown, their level was measured by qRT-PCR. We found that, although the p15, p16, p18, and p19 were not affected significantly (supplemental Fig. S5), p57 was induced over time and peaked at 6 days after transfection when it increased more >7-fold (Fig. 3D).
To investigate the role played by p57, its levels were knocked down, alone or in combination with p21 and p27. Fig. 3D shows p57-siRNA targeting efficiency. p21, p27, and p57 triple knockdown more than doubled the percentage of BrdU-incorporating cardiomyocytes at 4 and 6 days after transfection compared with p21 and p27 double interference, whereas p57 knockdown alone was not effective (Fig. 3C). In keeping with these findings, p21, p27, and p57 triple siRNA further increased cardiomyocyte number at 4 and 6 days after transfection (Fig. 3A).
Cardiomyocyte terminal differentiation is associated not only with CKI induction, but also with cyclin down-modulation and consequent pRb hypophosphorylation (5, 9). Thus, we measured whether neonatal cardiomyocyte proliferation induced by CKI knockdown was associated with the reactivation of the cyclin/CDK pathway.
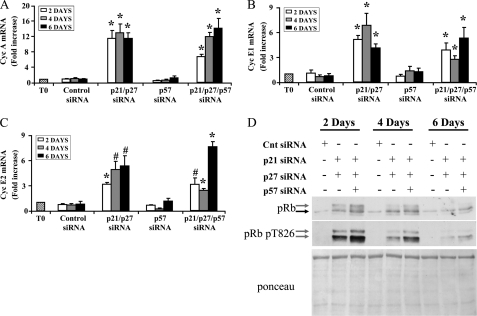
We found that, although the expression of cyclins D (D1, D2, and D3) was not affected (data not shown), CKI double or triple knockdown was associated with the induction of cyclin A, E1, and E2 (Fig. 4, A–C). In keeping with these findings, pRb hyperphosphorylation was induced, as assessed by SDS-PAGE mobility up-shift in Western blotting experiments (Fig. 4D). The phosphorylation of the crucial threonine 826 residue (54) increased as well and was more intense and permanent following p21, p27, and p57 triple knockdown compared both to p21 and p27 double knockdown and to control siRNA transfection. Furthermore, global pRb protein levels increased upon CKI knockdown, in keeping with negative pRB regulation of its own expression (55). Finally, we also found that, similarly to double CKI silencing, triple CKI knockdown also did not increase DNA damage, as assessed measuring the number of γH2AX foci (supplemental Fig. S4).
FIGURE 4.
p21, p27, and p57 knockdown induces cyclin A and E levels and pRb phosphorylation. Neonatal cardiomyocytes were transfected with p21 and p27 siRNAs in the presence or absence of p57 siRNA. Then, protein extracts and total RNA were derived and analyzed 2, 4, and 6 days after transfection. Bar graphs display relative expression of cyclin (Cyc) A (A), cyclin E1 (B), and cyclin E2 (C) measured by qRT-PCR (n = 6; *, p < 0.0001; #, p < 0,002). Primers used did not allow to distinguish between cyclin A1 and A2 isoforms. D, representative Western blotting analysis of pRb phosphorylation. Top panel, the gray and black arrows indicate the hyper- and the hypophosporylated forms of pRb, respectively. Middle panel, pRb threonine 826 phosphorylation was detected using a phospho-specific antibody. Bottom panel, Red Ponceau staining of the relevant nitrocellulose membrane, indicating similar protein loading in each lane.
p21, p27, and p57 Knockdown Promotes Proliferation of Adult Cardiomyocytes
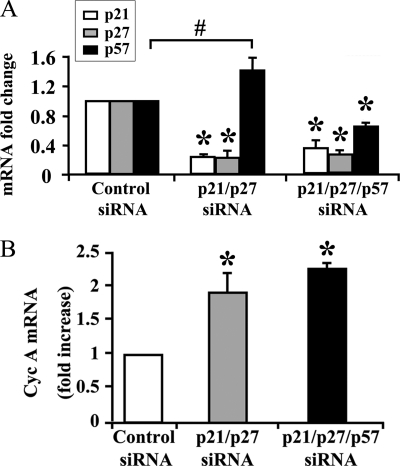
Previous studies indicate that rat cardiomyocyte proliferation arrests soon after birth (9). To evaluate the functional consequences of CKI knockdown in adult cardiomyocytes, ventricular cardiomyocytes were isolated from 8–12-week-old rats. Cardiomyocytes were transfected with siRNAs targeting p21, p27, and p57 or control. The efficiency of double and triple CKI knockdown was assayed by qRT-PCR (Fig. 5A). As observed in neonatal cardiomyocytes, p21 and p27 double knockdown up-regulated p57 expression. However, this increase was efficiently prevented when a siRNA to p57 was co-transfected (Fig. 5A). Furthermore, both double and triple CKI RNAi were accompanied by the induction of cyclin A (Fig. 5B).
FIGURE 5.
Efficient p21, p27, and p57 knockdown in adult cardiomyocytes induces cyclin A expression. Adult rat cardiomyocytes were transfected with p21 and p27 siRNAs in the presence or absence of p57 siRNA or with control siRNA. Ten days later, RNA was extracted and analyzed by qRT-PCR. A, bar graph showing the down-modulation of p21, p27, and p57 mRNAs (n = 3; *, p < 0.005). Double p21 and p27 RNAi induced p57 mRNA level increase (#, p < 0.04). B, cyclin A mRNA levels were induced by double or triple CKI knockdown (n = 3; *, p < 0.01).
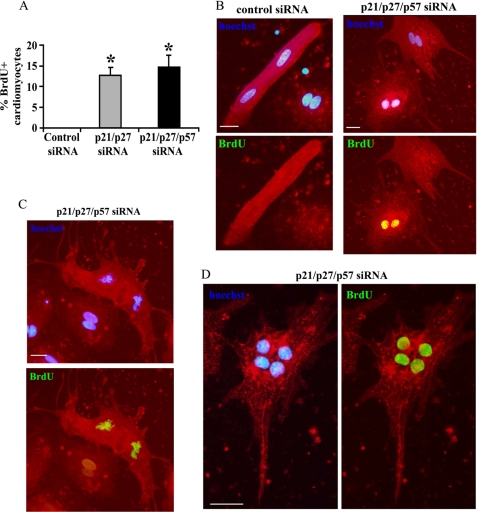
To determine whether double or triple CKI interference promoted DNA synthesis in adult cardiomyocytes even in the absence of growth factor stimulation, cells were cultured in serum-free medium (56, 57) for 10 days after transfection and then assayed for BrdU incorporation after a 24-h pulse. Double p21 and p27 CKI interference induced BrdU incorporation in >12% of the adult cardiomyocytes, whereas no BrdU-positive cells were observed among the controls (Fig. 6, A and B). However, unlike neonatal cardiomyocytes, no significant increase of BrdU incorporation was observed when p57 was silenced as well (Fig. 6A). Intriguingly, mitotic figures (Fig. 6C) and cardiomyocytes with four nuclei (Fig. 6D) were observed only in CKI knockdown cultures.
FIGURE 6.
p21, p27, and p57 triple knockdown promotes cell cycle entry of adult cardiomyocytes. Adult rat cardiomyocytes were transfected with p21 and p27 siRNAs in the presence or absence of p57 siRNA or with control siRNA. Ten days later, cells were treated with BrdU for 24 h to monitor cells undergoing DNA replication and assayed for BrdU staining by immunofluorescence (green). Cardiomyocytes were stained with α-sarcomeric actinin (red) and nuclei with Hoechst 33342 (blue) A, bar graph indicating the percentage of BrdU-positive cardiomyocytes upon double and triple CKI siRNAs (n = 4; *, p < 0.001). B, top panels show α-sarcomeric actinin/Hoechst 33342 overlays of representative control or triple CKI knockdown cardiomyocytes. Bottom panels show α-sarcomeric actinin/BrdU overlays of the same fields. Scale bar, 30 μm. C, representative adult cardiomyocyte in anaphase upon triple CKI knockdown. The top panel shows α-sarcomeric actinin/Hoechst 33342 overlay. Bottom panels show α-sarcomeric actinin/BrdU overlay of the same field. Scale bar, 20 μm. D, representative cardiomyocyte with four nuclei that underwent DNA replication and karyokinesis. Scale bar, 20 μm.
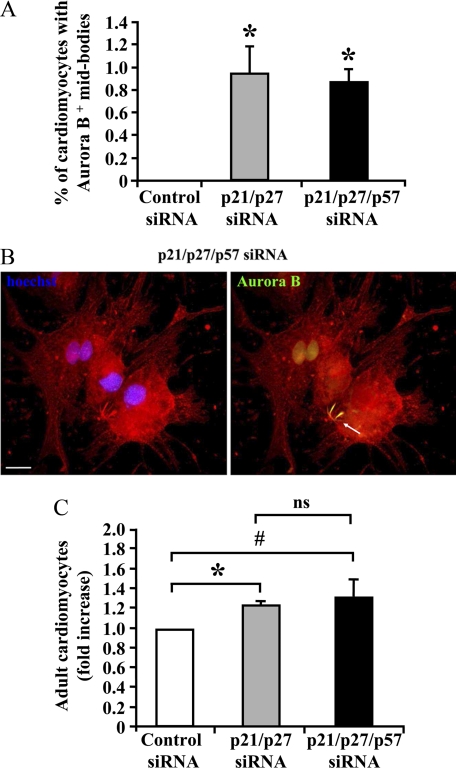
The increase in S phase induced by CKI knockdown was associated with a significant increase of cardiomyocytes displaying more than two nuclei (supplemental Table S1). However, this increase could explain the observed induction in DNA synthesis only in part. To learn whether adult mammalian cardiomyocytes can undergo cytokinesis, cardiomyocytes exhibiting an Aurora B-positive mid-body staining were evaluated (Fig. 7, A and B). Both double and triple CKI interference induced an increase in cytokinesis compared with control.
FIGURE 7.
CKI knockdown promotes adult cardiomyocytes cytokinesis and proliferation. Adult rat cardiomyocytes were transfected with p21 and p27 siRNAs in the presence or absence of p57 siRNA or with control siRNA. Ten days later, cardiomyocytes were stained with α-sarcomeric actinin (red) and assayed for Aurora B by immunofluorescence (green). Nuclei were stained with Hoechst 33342 (blue). A, bar graph showing the percentage of cardiomyocytes bearing an Aurora B-positive mid-body structure (n = 4; *, p < 0.001). B, representative picture displaying a dividing adult cardiomyocyte. The white arrow points to the Aurora B-positive mid-body structure. Scale bar, 20 μm. C, adult rat cardiomyocytes were transfected with p21 and p27 siRNAs in the presence or absence of p57 siRNA or with control siRNA. Then, cardiomyocytes were stained by α-sarcomeric actinin and Hoechst 33342 and counted 10 days later (n = 6; *, p < 0.00002; #, p < 0.005; ns, not significant).
Cardiomyocytes were also counted to measure cell proliferation. We observed that both double and triple CKI knock-down induced a small but reproducible cardiomyocyte number increase (Fig. 7C). Taken together, these results indicate that CKIs are involved in the control of adult cardiomyocyte cell cycle withdrawal.
TERT Overexpression Associated with CKI Knockdown Further Increases Proliferation in Neonatal but Not in Adult Cardiomyocytes
Down-regulation of TERT and the resulting loss of telomerase activity represents one mechanism that can cooperate together with activators and inhibitors of the CDKs to create a barrier to cardiomyocytes proliferation beyond the immediate perinatal period (32, 33). Indeed, it has been shown that TERT and telomerase activity are down-regulated in adult mouse myocardium (38).
To assess whether TERT overexpression can increase cardiomyocyte proliferation in association with CKI triple interference, neonatal and adult cardiomyocytes were infected with adenovirus expressing either human TERT (Ad-hTERT) or LacZ (Ad-LacZ, negative control). Eight hours later, the cells were transfected with siRNAs to CKIs (p21, p27, and p57) or control. Both neonatal and adult cardiomyocytes were efficiently transduced as assessed by qRT-PCR to hTERT and CKIs (data not shown).
We observed that hTERT expression further increased both neonatal cardiomyocyte S phase entry and proliferation induced by CKI knockdown (supplemental Fig. S3, A and B). However, this additional induction exerted by hTERT was very modest and was not found in adult cardiomyocytes (supplemental Fig. S6, C and D).
Moreover, it has been shown that Notch1 overexpression can induce the proliferation of neonatal cardiomyocytes (58–60). However, we found no additional increase of adult cardiomyocytes proliferation when CKI knockdown was associated with Notch pathway stimulation obtained by overexpressing Notch1 intracellular domain (data not shown).
CKI Knockdown Induces Neonatal Gene Expression
We observed that CKI knockdown induced a drastic change of adult cardiomyocyte morphology. Fig. 6B shows that, although control transfected cells retained principally the characteristic rod shape, CKI siRNA-transfected cardiomyocytes assumed a stellate morphology resembling that of neonatal cardiomyocytes. Also myofibrils organization was disarrayed, assuming a configuration reminiscent of neonatal cardiomyocytes.
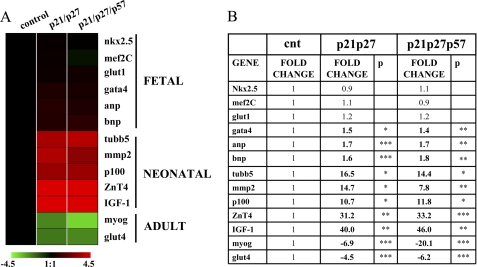
Thus, the expression of a subset of genes that are differentially expressed in fetal, neonatal, and adult cardiomyocytes was measured. We found that CKI knockdown sharply decreased the levels of adult genes mRNA Glut4 (glucose transport 4) and myoglobin (61, 62) and induced the expression of neonatal genes, tubulin β5, matrix metalloproteinase 2 (MMP2), p100 co-activator, ZnT4, and IGF-1 (Insuline-like Growth Factor-1) receptor (62). Conversely, the expression of fetal/hypertrophic markers glucose transport 1 (Glut1) (61, 63), nkx2.5, and mef2C (myocyte enhancer factor 2C) was unaffected, whereas GATA4 and atrial and B-type natriuretic factors ANP and BNP displayed only minor increases (Fig. 8).
FIGURE 8.
p21, p27, and p57 knockdown in adult cardiomyocytes induces the re-expression of neonatal genes and inhibits adult genes. Adult cardiomyocytes were transfected with p21, p27, and p57 or control siRNAs. Ten days later, total RNA was extracted, and adult, neonatal, and fetal cardiomyocyte markers were measured by qRT-PCR. A, a heat map representing gene expression average modulations in CKI knockdown cardiomyocytes compared with control. Values are expressed using a log2 scale (−ΔΔCt), and green and red indicate down- and up-regulation, respectively. B, a table showing the same values displayed in A in a linear scale. Statistical significance was calculated versus values of control transfected cells. Significantly different values are in boldface type. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
DISCUSSION
Cardiomyocyte terminal differentiation makes the heart a particularly vulnerable organ, especially toward ischemic, toxic, and inflammatory events. Thus, following massive injury, the adult heart is unable to replace necrotic or damaged tissue. Indeed, cardiac regeneration granted by endogenous cardiac stem cells and other regenerative mechanisms is limited and, in most circumstances, is insufficient to replace a large myocardial tissue loss (64, 65). Therefore, reinduction of cell cycle progression in terminally differentiated cardiomyocytes may represent an attractive approach to treat heart disease (4).
Although cardiomyocyte terminal differentiation has long been regarded as a condition of irreversible proliferation arrest or postmitotic state (66), the molecular bases of such a condition has remained unclear. High CKI expression levels have been observed in cardiomyocytes (9, 67), and although it has been speculated that the CKIs are at least partially responsible for the maintenance of the cardiomyocyte postmitotic state, data have mostly been correlative. On the other side, convincing evidence indicates that adult cardiomyocytes still hold signaling pathways capable of efficiently resuming their proliferative potential (57, 68–70). In this study, we found that cardiomyocyte postmitotic state depends on the constant expression of CKIs, demonstrating that permanent proliferation arrest cannot be maintained in their absence. These results are in keeping with a previous study of our group, demonstrating a critical requirement of CKI in sustaining skeletal muscle myofibers terminal differentiation (43). Thus, cardiomyocyte growth arrest should not be regarded simply as the consequence of the absence of proliferation-promoting regulators (cyclin/CDKs). On the contrary, it is an active state that requires continuous CKI expression to be maintained.
A clear efficiency difference in the proliferation rate was observed between neonatal and adult cardiomyocytes following CKI knockdown. Indeed, >20% of the neonatal cardiomyocytes and >10% of the adults displayed DNA synthesis upon CKI knockdown. This massive effect is unlikely due to the potential presence of small stem-like populations in the culture. Thus, CKIs seem to play an active role in maintaining the permanent withdrawal from the cell cycle in both neonatal and adult cardiomyocytes, paving the way to the investigation of further anti-proliferative mechanisms acquired during development. Indeed, we found that pRB phosphorylation decreased over time, despite sustained CKI knockdown, suggesting the presence of additional CDK regulatory mechanisms.
We found CKI down-modulation increased both cyclin E and A levels, whereas cyclin D levels were not affected. Although this result may seem surprising, it should be considered that D cyclins are not strictly required for the proliferation of most cell types. In fact, mice lacking D cyclins or CDK4/6 display relatively mild phenotypes. Triple D cyclin mice KO die in utero at embryonic day 17.5. Heart development is aberrant, but myocytes are clearly present, indicating that cyclins E and A may be sufficient to allow cardiomyocyte proliferation (71, 72).
One possible mechanism restraining cardiomyocyte proliferation was represented by TERT down-modulation and the resulting loss of telomerase activity after the immediate perinatal period (31). To test this idea, we associated CKI knockdown to TERT overexpression. Although a small increase in cell cycle induction was observed in neonatal cardiomyocytes, no difference was observed in adults, possibly owing to the absence of other telomere-associated factors in adult cardiomyocytes (73).
Cardiomyocytes are polyploid and polynucleated cells. Although ploidy was not measured, we found that most neonatal cardiomyocytes were mononucleated, and the majority of the adult cardiomyocytes were bi-nucleated, as observed previously in rodents (74). As expected for a proliferation promoting intervention, CKI knockdown increased the nuclei/cardiomyocyte ratio in both neonatal and adult cells. However, at least a sub-population of the cardiomyocytes further progressed after karyokynesis, as assessed by the detection of mid-body structures and by straight cardiomyocyte counting.
Intriguingly, neonatal cardiomyocyte proliferation induced by CKI knockdown was not associated with massive increase of DNA damage, micronucleation, and apoptosis, in sheer contrast with the skeletal muscle, where CKI knockdown induces DNA replication followed by mitotic catastrophe (43). Although the molecular mechanism underpinning this difference is not clear, it is worth noting that CKI knockdown and DNA synthesis reactivation correlated with a change in adult cardiomyocyte morphology that may be a prerequisite for division. CKI knockdown in serum-free medium induced a neonatal-like morphology and the re-expression of neonatal genes in adult cardiomyocytes, suggesting the involvement of CKIs in cardiomyocyte maturation. Intriguingly, well established hypertrophy markers such as ANP and BNP were induced only marginally, and no overt reactivation of fetal gene program was observed.
Intriguingly, it has been shown that serum stimulation of adult cardiomyocyte promotes cell spreading and loss of sarcomeric structure, resembling a spontaneous dedifferentiation to a neonatal-like state (75–77). One may speculate that, in the serum-free culture conditions we adopted, CKI knockdown may, at least in part, be a substitute for growth factor stimulation. Further studies are needed to better assess the consequence of cell cycle reactivation on genome integrity in both neonatal and adult cardiomyocytes.
Our findings are in keeping with other studies showing that cell cycle molecular machinery of the cardiomyocytes can be successfully reprogrammed, such that partial cell cycle reactivation and, less frequently, full cell cycle reactivation is observed (78). In some instances, for example upon overexpression of E2F1 or E2F3, reactivation of the cell cycle was observed, but it was followed by apoptosis (14). Overexpression of c-Myc, E2F2, or D-type cyclins promotes cell cycle reactivation in neonatal cardiomocytes and hypertrophic growth in adult cardiomyocytes (13, 14, 79, 80). Moreover, reinitiation of the cell cycle in cardiomyocytes promoted by hCDC5, jumonji (JMJ), or p38 activity, require expression of additional proteins, such as E1A/E1B, E2F, or growth factor stimulation (5, 57, 81, 82). Conversely, cyclin A2 overexpression seems to be sufficient to induce cardiomyocyte proliferation and induces cardiac regeneration after myocardial infarction (83, 84). Among several possible mechanisms of action, one may envision that CKI knockdown leads to a derepression of cyclin A2 activity. In a reciprocal perspective, it is also possible that cyclin A2 overexpression induced p21 family CKI titration, yielding an intracellular mileu similar to that induced by CKI knockdown.
The therapeutic potential of the cell cycle machinery manipulation still needs to be assessed. However, it is worth noting that RNAi therapeutics of cardiac diseases has gained considerable momentum, being burdened by lower safety concerns compared with other gene therapy approaches (85). Furthermore, one may consider that an attractive in vivo cell target is represented by cardiac stem cells or newly formed cardiomyocytes derived from the stem cells, whose amplification may be increased by CKI knock down.
Supplementary Material
Acknowledgments
We thank Dr. Germana Zaccagnini (Istituto Dermopatico dell'Immacolata-IRCCS, Rome, Italy) for critical reading of the manuscript as well as Drs. Deborah Pajalunga and Germana Falcone (Istituto Superiore di Sanità, Rome, Italy), Dr. Giuliana Di Rocco (Centro Cardiologico Monzino-IRCCS, Milan, Italy), and Dr. Chiara Collesi and Dr. Lorena Zentilin (International Centre for Genetic Engineering and Biotechnology) for sharing experimental protocols and reagents.
This work was supported in part by Ministero della Salute (RF06, RF07, and RC09-11).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Table S1, Figs. S1–S6, and additional references.
- CDK
- cyclin-cyclin-dependent kinase
- CKI
- cyclin-dependent kinase inhibitor
- TERT
- telomerase reverse transcriptase
- H3P
- histone-H3 phosphorylation
- qRT-PCR
- quantitative real-time PCR.
REFERENCES
- 1. Leri A., Kajstura J., Anversa P., Frishman W. H. (2008) Curr. Probl. Cardiol. 33, 91–153 [DOI] [PubMed] [Google Scholar]
- 2. Poolman R. A., Brooks G. (1998) J. Mol. Cell Cardiol. 30, 2121–2135 [DOI] [PubMed] [Google Scholar]
- 3. Busk P. K., Hinrichsen R. (2003) Cell Cycle 2, 91–95 [PubMed] [Google Scholar]
- 4. Engel F. B. (2005) Cell Cycle 4, 1360–1363 [DOI] [PubMed] [Google Scholar]
- 5. Bicknell K. A., Coxon C. H., Brooks G. (2007) J. Mol. Cell Cardiol. 42, 706–721 [DOI] [PubMed] [Google Scholar]
- 6. Capasso J. M., Li P., Zhang X., Anversa P. (1992) Am. J. Physiol. 262, H486–495 [DOI] [PubMed] [Google Scholar]
- 7. Li G., Li R. K., Mickle D. A., Weisel R. D., Merante F., Ball W. T., Christakis G. T., Cusimano R. J., Williams W. G. (1998) Circulation 98, II144–149 [PubMed] [Google Scholar]
- 8. Bicknell K. A., Surry E. L., Brooks G. (2003) J. Pharm. Pharmacol. 55, 571–591 [DOI] [PubMed] [Google Scholar]
- 9. Pasumarthi K. B., Field L. J. (2002) Circ. Res. 90, 1044–1054 [DOI] [PubMed] [Google Scholar]
- 10. Sears R. C., Nevins J. R. (2002) J. Biol. Chem. 277, 11617–11620 [DOI] [PubMed] [Google Scholar]
- 11. Sherr C. J., McCormick F. (2002) Cancer Cell 2, 103–112 [DOI] [PubMed] [Google Scholar]
- 12. Weinberg R. A. (1995) Cell 81, 323–330 [DOI] [PubMed] [Google Scholar]
- 13. Tamamori-Adachi M., Goto I., Yamada K., Kitajima S. (2008) Cell Cycle 7, 3768–3774 [DOI] [PubMed] [Google Scholar]
- 14. Ebelt H., Hufnagel N., Neuhaus P., Neuhaus H., Gajawada P., Simm A., Müller-Werdan U., Werdan K., Braun T. (2005) Circ. Res. 96, 509–517 [DOI] [PubMed] [Google Scholar]
- 15. MacLellan W. R., Garcia A., Oh H., Frenkel P., Jordan M. C., Roos K. P., Schneider M. D. (2005) Mol. Cell Biol. 25, 2486–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ahuja P., Sdek P., MacLellan W. R. (2007) Physiol. Rev. 87, 521–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hinrichsen R., Hansen A. H., Haunsø S., Busk P. K. (2008) Cell Prolif. 41, 813–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harper J. W. (1997) Cancer Surv. 29, 91–107 [PubMed] [Google Scholar]
- 19. LaBaer J., Garrett M. D., Stevenson L. F., Slingerland J. M., Sandhu C., Chou H. S., Fattaey A., Harlow E. (1997) Genes Dev. 11, 847–862 [DOI] [PubMed] [Google Scholar]
- 20. Bockstaele L., Coulonval K., Kooken H., Paternot S., Roger P. P. (2006) Cell Div. 1, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koh K. N., Kang M. J., Frith-Terhune A., Park S. K., Kim I., Lee C. O., Koh G. Y. (1998) J. Mol. Cell Cardiol. 30, 463–474 [DOI] [PubMed] [Google Scholar]
- 22. Poolman R. A., Gilchrist R., Brooks G. (1998) Int. J. Cardiol. 67, 133–142 [DOI] [PubMed] [Google Scholar]
- 23. Fero M. L., Rivkin M., Tasch M., Porter P., Carow C. E., Firpo E., Polyak K., Tsai L. H., Broudy V., Perlmutter R. M., Kaushansky K., Roberts J. M. (1996) Cell 85, 733–744 [DOI] [PubMed] [Google Scholar]
- 24. Nakayama K., Ishida N., Shirane M., Inomata A., Inoue T., Shishido N., Horii I., Loh D. Y. (1996) Cell 85, 707–720 [DOI] [PubMed] [Google Scholar]
- 25. Kiyokawa H., Kineman R. D., Manova-Todorova K. O., Soares V. C., Hoffman E. S., Ono M., Khanam D., Hayday A. C., Frohman L. A., Koff A. (1996) Cell 85, 721–732 [DOI] [PubMed] [Google Scholar]
- 26. Westbury J., Watkins M., Ferguson-Smith A. C., Smith J. (2001) Mech. Dev. 109, 83–89 [DOI] [PubMed] [Google Scholar]
- 27. Yan Y., Frisén J., Lee M. H., Massagué J., Barbacid M. (1997) Genes Dev. 11, 973–983 [DOI] [PubMed] [Google Scholar]
- 28. Zhang P., Liégeois N. J., Wong C., Finegold M., Hou H., Thompson J. C., Silverman A., Harper J. W., DePinho R. A., Elledge S. J. (1997) Nature 387, 151–158 [DOI] [PubMed] [Google Scholar]
- 29. Zhang P., Wong C., DePinho R. A., Harper J. W., Elledge S. J. (1998) Genes Dev. 12, 3162–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Burton P. B., Yacoub M. H., Barton P. J. (1999) Eur. Heart J. 20, 604–611 [DOI] [PubMed] [Google Scholar]
- 31. Haley S. A., Zhao T., Zou L., Klysik J. E., Padbury J. F., Kochilas L. K. (2008) BMC Physiol. 8, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bodnar A. G., Ouellette M., Frolkis M., Holt S. E., Chiu C. P., Morin G. B., Harley C. B., Shay J. W., Lichtsteiner S., Wright W. E. (1998) Science 279, 349–352 [DOI] [PubMed] [Google Scholar]
- 33. Hahn W. C., Stewart S. A., Brooks M. W., York S. G., Eaton E., Kurachi A., Beijersbergen R. L., Knoll J. H., Meyerson M., Weinberg R. A. (1999) Nat. Med. 5, 1164–1170 [DOI] [PubMed] [Google Scholar]
- 34. McEachern M. J., Krauskopf A., Blackburn E. H. (2000) Annu. Rev. Genet. 34, 331–358 [DOI] [PubMed] [Google Scholar]
- 35. Shay J. W., Wright W. E. (2001) Science 291, 839–840 [DOI] [PubMed] [Google Scholar]
- 36. Borges A., Liew C. C. (1997) J. Mol. Cell Cardiol. 29, 2717–2724 [DOI] [PubMed] [Google Scholar]
- 37. Kajstura J., Rota M., Urbanek K., Hosoda T., Bearzi C., Anversa P., Bolli R., Leri A. (2006) Antioxid. Redox Signal 8, 2125–2141 [DOI] [PubMed] [Google Scholar]
- 38. Oh H., Taffet G. E., Youker K. A., Entman M. L., Overbeek P. A., Michael L. H., Schneider M. D. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10308–10313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Urbanek K., Cesselli D., Rota M., Nascimbene A., De Angelis A., Hosoda T., Bearzi C., Boni A., Bolli R., Kajstura J., Anversa P., Leri A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9226–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Orlandi A., Pagani F., Avitabile D., Bonanno G., Scambia G., Vigna E., Grassi F., Eusebi F., Fucile S., Pesce M., Capogrossi M. C. (2008) Am. J. Physiol. Heart Circ. Physiol. 294, H1541–1549 [DOI] [PubMed] [Google Scholar]
- 41. Engel F. B., Hauck L., Boehm M., Nabel E. G., Dietz R., von Harsdorf R. (2003) Mol. Cell Biol. 23, 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Engel F. B., Hsieh P. C., Lee R. T., Keating M. T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15546–15551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pajalunga D., Mazzola A., Salzano A. M., Biferi M. G., De Luca G., Crescenzi M. (2007) J. Cell Biol. 176, 807–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gowdak L. H., Poliakova L., Wang X., Kovesdi I., Fishbein K. W., Zacheo A., Palumbo R., Straino S., Emanueli C., Marrocco-Trischitta M., Lakatta E. G., Anversa P., Spencer R. G., Talan M., Capogrossi M. C. (2000) Circulation 102, 565–571 [DOI] [PubMed] [Google Scholar]
- 45. Mühlhauser J., Merrill M. J., Pili R., Maeda H., Bacic M., Bewig B., Passaniti A., Edwards N. A., Crystal R. G., Capogrossi M. C. (1995) Circ. Res. 77, 1077–1086 [DOI] [PubMed] [Google Scholar]
- 46. Zaccagnini G., Martelli F., Fasanaro P., Magenta A., Gaetano C., Di Carlo A., Biglioli P., Giorgio M., Martin-Padura I., Pelicci P. G., Capogrossi M. C. (2004) Circulation 109, 2917–2923 [DOI] [PubMed] [Google Scholar]
- 47. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 48. Zaccagnini G., Martelli F., Magenta A., Cencioni C., Fasanaro P., Nicoletti C., Biglioli P., Pelicci P. G., Capogrossi M. C. (2007) J. Biol. Chem. 282, 31453–31459 [DOI] [PubMed] [Google Scholar]
- 49. Sacco A., Siepi F., Crescenzi M. (2003) Oncogene 22, 4027–4034 [DOI] [PubMed] [Google Scholar]
- 50. Wheatley S. P., Carvalho A., Vagnarelli P., Earnshaw W. C. (2001) Curr. Biol. 11, 886–890 [DOI] [PubMed] [Google Scholar]
- 51. Giet R., Glover D. M. (2001) J. Cell Biol. 152, 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Murata-Hori M., Tatsuka M., Wang Y. L. (2002) Mol. Biol. Cell 13, 1099–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rogakou E. P., Boon C., Redon C., Bonner W. M. (1999) J. Cell Biol. 146, 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rubin S. M., Gall A. L., Zheng N., Pavletich N. P. (2005) Cell 123, 1093–1106 [DOI] [PubMed] [Google Scholar]
- 55. Shan B., Lee W. H. (1994) Mol. Cell Biol. 14, 8166–8173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schlüter K. D., Simm A., Schäfer M., Taimor G., Piper H. M. (1999) Am. J. Physiol. 276, H1655–1663 [DOI] [PubMed] [Google Scholar]
- 57. Engel F. B., Schebesta M., Duong M. T., Lu G., Ren S., Madwed J. B., Jiang H., Wang Y., Keating M. T. (2005) Genes Dev. 19, 1175–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Campa V. M., Gutiérrez-Lanza R., Cerignoli F., Díaz-Trelles R., Nelson B., Tsuji T., Barcova M., Jiang W., Mercola M. (2008) J. Cell Biol. 183, 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Boni A., Urbanek K., Nascimbene A., Hosoda T., Zheng H., Delucchi F., Amano K., Gonzalez A., Vitale S., Ojaimi C., Rizzi R., Bolli R., Yutzey K. E., Rota M., Kajstura J., Anversa P., Leri A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15529–15534 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60. Collesi C., Zentilin L., Sinagra G., Giacca M. (2008) J. Cell Biol. 183, 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Razeghi P., Young M. E., Alcorn J. L., Moravec C. S., Frazier O. H., Taegtmeyer H. (2001) Circulation 104, 2923–2931 [DOI] [PubMed] [Google Scholar]
- 62. Chen H. W., Yu S. L., Chen W. J., Yang P. C., Chien C. T., Chou H. Y., Li H. N., Peck K., Huang C. H., Lin F. Y., Chen J. J., Lee Y. T. (2004) Heart 90, 927–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Oka T., Xu J., Kaiser R. A., Melendez J., Hambleton M., Sargent M. A., Lorts A., Brunskill E. W., Dorn G. W., 2nd, Conway S. J., Aronow B. J., Robbins J., Molkentin J. D. (2007) Circ. Res. 101, 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bergmann O., Bhardwaj R. D., Bernard S., Zdunek S., Barnabé-Heider F., Walsh S., Zupicich J., Alkass K., Buchholz B. A., Druid H., Jovinge S., Frisén J. (2009) Science 324, 98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hosoda T., Kajstura J., Leri A., Anversa P. (2010) Circ. J. 74, 13–17 [DOI] [PubMed] [Google Scholar]
- 66. Olson E. N., Schneider M. D. (2003) Genes Dev. 17, 1937–1956 [DOI] [PubMed] [Google Scholar]
- 67. von Harsdorf R., Hauck L., Mehrhof F., Wegenka U., Cardoso M. C., Dietz R. (1999) Circ. Res. 85, 128–136 [DOI] [PubMed] [Google Scholar]
- 68. Tseng A. S., Engel F. B., Keating M. T. (2006) Chem. Biol. 13, 957–963 [DOI] [PubMed] [Google Scholar]
- 69. Bersell K., Arab S., Haring B., Kühn B. (2009) Cell 138, 257–270 [DOI] [PubMed] [Google Scholar]
- 70. Kühn B., del Monte F., Hajjar R. J., Chang Y. S., Lebeche D., Arab S., Keating M. T. (2007) Nat. Med. 13, 962–969 [DOI] [PubMed] [Google Scholar]
- 71. Kozar K., Ciemerych M. A., Rebel V. I., Shigematsu H., Zagozdzon A., Sicinska E., Geng Y., Yu Q., Bhattacharya S., Bronson R. T., Akashi K., Sicinski P. (2004) Cell 118, 477–491 [DOI] [PubMed] [Google Scholar]
- 72. Kozar K., Sicinski P. (2005) Cell Cycle 4, 388–391 [DOI] [PubMed] [Google Scholar]
- 73. Serrano A. L., Andrés V. (2004) Circ. Res. 94, 575–584 [DOI] [PubMed] [Google Scholar]
- 74. Kang M. J., Koh G. Y. (1997) J. Mol. Cell Cardiol. 29, 1767–1777 [DOI] [PubMed] [Google Scholar]
- 75. Poindexter B. J., Smith J. R., Buja L. M., Bick R. J. (2001) Cell Calcium 30, 373–382 [DOI] [PubMed] [Google Scholar]
- 76. Bird S. D., Doevendans P. A., van Rooijen M. A., Brutel de la Riviere A., Hassink R. J., Passier R., Mummery C. L. (2003) Cardiovasc. Res. 58, 423–434 [DOI] [PubMed] [Google Scholar]
- 77. Stephen M. J., Poindexter B. J., Moolman J. A., Sheikh-Hamad D., Bick R. J. (2009) Open Cardiovasc Med. J. 3, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bicknell K. A., Brooks G. (2008) Curr. Opin. Pharmacol. 8, 193–201 [DOI] [PubMed] [Google Scholar]
- 79. Zhong W., Mao S., Tobis S., Angelis E., Jordan M. C., Roos K. P., Fishbein M. C., de Alborán I. M., MacLellan W. R. (2006) EMBO J. 25, 3869–3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Busk P. K., Hinrichsen R., Bartkova J., Hansen A. H., Christoffersen T. E., Bartek J., Haunsø S. (2005) Exp. Cell Res. 304, 149–161 [DOI] [PubMed] [Google Scholar]
- 81. Williams G. T., Hughes J. P., Stoneman V., Anderson C. L., McCarthy N. J., Mourtada-Maarabouni M., Pickard M., Hedge V. L., Trayner I., Farzaneh F. (2006) Gene Ther. Mol. Biol. 10, 255–262 [PMC free article] [PubMed] [Google Scholar]
- 82. Toyoda M., Shirato H., Nakajima K., Kojima M., Takahashi M., Kubota M., Suzuki-Migishima R., Motegi Y., Yokoyama M., Takeuchi T. (2003) Dev. Cell 5, 85–97 [DOI] [PubMed] [Google Scholar]
- 83. Woo Y. J., Panlilio C. M., Cheng R. K., Liao G. P., Suarez E. E., Atluri P., Chaudhry H. W. (2007) J. Thorac. Cardiovasc. Surg. 133, 927–933 [DOI] [PubMed] [Google Scholar]
- 84. Cheng R. K., Asai T., Tang H., Dashoush N. H., Kara R. J., Costa K. D., Naka Y., Wu E. X., Wolgemuth D. J., Chaudhry H. W. (2007) Circ. Res. 100, 1741–1748 [DOI] [PubMed] [Google Scholar]
- 85. Tiemann K., Rossi J. J. (2009) EMBO Mol. Med. 1, 142–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.