Abstract
Ribosomal protein S6 (rpS6) is a critical component of the 40 S ribosomal subunit that mediates translation initiation at the 5′-m7GpppG cap of mRNA. In response to mitogenic stimuli, rpS6 undergoes ordered C-terminal phosphorylation by p70 S6 kinases and p90 ribosomal S6 kinases on four conserved Ser residues (Ser-235, Ser-236, Ser-240, and Ser-244) whose modification potentiates rpS6 cap binding activity. A fifth site, Ser-247, is also known to be phosphorylated, but its function and regulation are not well characterized. In this study, we employed phospho-specific antibodies to show that Ser-247 is a target of the casein kinase 1 (CK1) family of protein kinases. CK1-dependent phosphorylation of Ser-247 was induced by mitogenic stimuli and required prior phosphorylation of upstream S6 kinase/ribosomal S6 kinase residues. CK1-mediated phosphorylation of Ser-247 also enhanced the phosphorylation of upstream sites, which implies that bidirectional synergy between C-terminal phospho-residues is required to sustain rpS6 phosphorylation. Consistent with this idea, CK1-dependent phosphorylation of rpS6 promotes its association with the mRNA cap-binding complex in vitro. Additionally, we show that protein phosphatase 1 (PP1) antagonizes rpS6 C terminus phosphorylation and cap binding in intact cells. These findings further our understanding of rpS6 phospho-regulation and define a direct link between CK1 and translation initiation.
Keywords: PP1, Protein Phosphorylation, Protein Synthesis, Ribosome Function, Translation Regulation, Casein Kinase 1 (CK1)
Introduction
The casein kinase 1 (CK1)2 and casein kinase 2 (CK2) families represent a group of ubiquitously expressed, constitutively active protein kinases that are involved in many biological processes including DNA repair, cell cycle control, and circadian rhythm entrainment (1, 2). Hundreds of in vitro substrates of CK1 and CK2 have been identified, and this number continues to rise (1–3). CK1 and CK2 preferentially phosphorylate Ser or Thr residues that are flanked by acidic amino acids or phosphorylated residues in the +3 or −3 position, respectively (1, 2). Thus, in many instances, CK1 and CK2 do not initiate phosphorylation of a particular substrate but instead fulfill a supportive function by phosphorylating adjacent sites. This property of CK1 and CK2 may be particularly important in those cases where a phosphorylation threshold must be surpassed to elicit a biological response.
Ribosomal protein S6 is one of 33 proteins that, together with one molecule of 18 S rRNA, comprise the small 40 S ribosomal subunit (4). rpS6 directly interacts with the m7GpppG 5′-cap-binding complex required for translation initiation and represents a point of regulatory convergence for signal transduction pathways controlling translation initiation in response to cell growth and cell proliferation cues. rpS6 undergoes inducible phosphorylation in response to mitogenic and cell growth stimuli, and this phosphorylation is conserved in vertebrates, invertebrates, plants, and fungi (5). In higher eukaryotes, phosphorylation occurs on a cluster of five serine residues at the carboxyl terminus of rpS6: Ser-235, Ser-236, Ser-240, Ser-244, and Ser-247 (6). Drosophila rpS6 contains a similar organization of five phosphorylation sites, whereas the homolog found in Saccharomyces cerevisiae contains two Ser residues corresponding to mammalian Ser-235 and Ser-236 (4). Phosphorylation of rpS6 occurs in an ordered manner, beginning with Ser-236 and followed sequentially by phosphorylation of Ser-235, Ser-240, Ser-244, and Ser-247 (7, 8). The phosphorylation of rpS6 on C-terminal residues enhances its affinity for the m7GpppG cap, which strongly implies that rpS6 phosphorylation enhances mRNA translation initiation.
The physiologic roles of rpS6 phosphorylation have been investigated through the generation of a knock-in mouse encoding a mutant rpS6 harboring Ala substitutions at all five C-terminal phosphorylation sites (9). rpS6 knock-in animals exhibit several physiologic abnormalities, including reduced overall size, glucose intolerance, and muscle weakness (9, 10). Cells derived from rpS6 knock-in mice also show reduced size, a trait that is shared by S6K-deficient flies or mice (11, 12). These findings are consistent with a model in which rpS6 phosphorylation enhances translation and cell growth. Surprisingly, however, overall protein translation was not grossly reduced in rpS6 knock-in cells, suggesting that deregulation of select mRNAs may be responsible for observed phenotypes (9).
Carboxyl-terminal phosphorylation of rpS6 is regulated by at least two signal transduction pathways. The p70 ribosomal S6 kinases, S6K1 and S6K2, play a major role in rpS6 C terminus phosphorylation in response to insulin, serum, and amino acid stimulation (4). S6K1 and S6K2 phosphorylate Ser-240 and Ser-244 but are dispensable for Ser-235 and Ser-236 phosphorylation in intact cells (13). The activities of S6K1 and S6K2 are in turn directly regulated by the mammalian target of rapamycin, mTOR, which responds to growth and mitogenic cues. Inhibition of mTOR with rapamycin causes a drastic reduction in rpS6 phosphorylation in mammalian cells (14). mTOR also phosphorylates the translational repressor 4E-BP1, causing its dissociation from the m7GpppG 5′-cap-binding complex and, through combined phosphorylation of S6Ks and 4E-BP1, mTOR positively regulates protein translation in response to favorable growth conditions. The RAS/ERK pathway also regulates rpS6 phosphorylation independent of mTOR through the activation of p90 ribosomal S6K kinases, RSK1 and RSK2 (12). RSK1 and RSK2 phosphorylate rpS6 on Ser-235 and Ser-236 in response to phorbol ester, serum, and oncogenic RAS, and the phosphorylation of both residues is required for cap binding (13).
Despite recent progress, there remain several outstanding questions regarding rpS6 function and regulation. Although the research into rpS6 phosphorylation has been focused primarily on the kinases, the phosphatases involved in dephosphorylating rpS6 have not been established. The transitory nature of rpS6 phosphorylation in serum-stimulated cells indicates that the C-terminal sites are highly susceptible to dephosphorylation. Indirect evidence has implicated protein phosphatase 1 (PP1) (15, 16), but definitive data are lacking. In addition, it is unclear whether additional kinases phosphorylate rpS6 C-terminal sites in vivo, and the function and regulation of the most C-terminal rpS6 phosphorylation site, Ser-247, have not been defined. In this report, we utilized an antibody that detects Ser-247 phosphorylation in vivo to show that CK1 regulates rpS6 phosphorylation and m7GpppG cap binding in intact cells. We further demonstrate that rpS6 C terminus phosphorylation occurs in a bidirectional fashion and that PP1 is a negative regulator of rpS6 phosphorylation and m7GpppG cap binding. These findings provide new insights into the biochemical mechanisms of rpS6 phosphorylation and implicate CK1 and PP1 in the regulation of protein synthesis.
EXPERIMENTAL PROCEDURES
DNA Constructs
The HA-rpS3 plasmid was a kind gift from Dr. Joon Kim, Korea University. The HA-rpS6 plasmid was provided by Dr. Phillipe Roux, University of Montreal. The dominant-negative PP1 plasmid harboring the D95N mutation was obtained from Dr. David Virshup at the Duke-National University of Singapore Graduate Medical School. Site-directed mutagenesis was carried out using the QuikChange method (Stratagene) and the following oligonucleotide primer pairs: rpS6S235A (5′-AAGAGACGTAGGCTGGCCTCACTGAGAGCTTC-3′ (forward) and 5′-GAAGCTCTCAGTGAGGCCAGCCTACGTCTCTT-3′ (reverse)); rpS6S236A (5′-AGACGTAGGCTGTCCGCACTGAGAGCTTCTAC-3′ (forward) and 5′-GTAGAAGCTCTCAGTGCGGACAGCCTACGTCT-3′ (reverse)); rpS6S240A (5′-TCCTCACTGAGAGCTGCTACTTCTAAGTCTGAGTC-3′ (forward) and 5′-GACTCAGACTTAGAAGTAGCAGCTCTCAGTGAGGA-3′ (reverse)); rpS6S244A (5′-GAGCTTCTACTTCTAAGGCTGAGTCCAGTCAAAAATG-3′ (forward) and 5′-CATTTTTGACTGGACTCAGCCTTAGAAGTAGAAGCTC-3′ (reverse)); rpS6S247A (5′-GAGCTTCTACTTCTAAGTCTGAGTCCGCTCAAAAATGAGTC-3′ (forward) and 5′-GACTCATTTTTGAGCGGACTCAGACTTAGAAGTAGAAGCTC-3′ (reverse)); rpS6S235A/S236A (5′-CTGGCCGCACTGAGAGCTTCTACTTCTAAGTCTGAGTCCAGTCAA-3′ (forward) and 5′-TTGACTGGACTCAGACTTAGAAGTAGAAGCTCTCAGTGCGGCCAG-3′ (reverse)); rpS6S240A/S244A (5′-CTGTCCTCACTGAGAGCTGCTACTTCTAAGGCTGAGTCCAGTCAA-3′ (forward) and 5′-TTGACTGGACTCAGCCTTAGAAGTAGCAGCTCTCAGTGAGGACAG-3′ (reverse)); rpS6S235D/S236D (5′-CTGGACGACCTGAGAGCTTCTACTTCTAAGTCTGAGTCC-3′ (forward) and 5′-GGACTCAGACTTAGAAGTAGAAGCTCTCAGGTCGTCCAG-3′ (reverse)); rpS6S240D/S244D (5′-CTGTCCTCACTGAGAGCTGATACTTCTAAGGATGAGTCCAGTCAA-3′ (forward) and 5′-TTGACTGGACTCATCCTTAGAAGTATCAGCTCTCAGTGAGGACAG-3′ (reverse)); rpS6S247D (5′-CTGTCCTCACTGAGAGCTTCTACTTCTAAGTCTGAGTCCGATCAA-3′ (forward) and 5′-TTGATCGGACTCAGACTTAGAAGTAGAAGCTCTCAGTGAGGACAG-3′ (reverse)). All oligonucleotide primers were purchased from Integrated DNA Technologies (Coralville, IA).
Cell Culture, Antisera, and Inhibitors
HEK 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 5% fetal bovine serum. The α-pPER2-662/665/668 antibody was generated by immunizing rabbits with a triply phosphorylated hPER2 peptide (KAE[pS]VA[pS]LT[pS]QC, where pS represents phosphorylated Ser) and will be described elsewhere.3 Other antibodies used in this study include α-rpS6 (Cell Signaling), α-rpS6 Ser(P)-235/236 (Cell Signaling), α-rpS6 Ser(P)-240/244 (Cell Signaling), α-eIF4E (Cell Signaling), α-S6K1 (Cell Signaling), α-HA (Millipore), α-tubulin (Millipore), α-CK1α (Santa Cruz Biotechnology), α-CK1δ (Bethyl Laboratories), α-CK1ϵ (Santa Cruz Biotechnology), α-CK2α (Santa Cruz Biotechnology), α-PP1 (Santa Cruz Biotechnology), and α-Myc (Santa Cruz Biotechnology). Chemicals and suppliers include rapamycin (30 nm), Cell Signaling; LY294002 (2-(4-morpholinyl)-8-phenylchromone (20 μm), Cell Signaling); PD98059 (2′-amino-3′-methoxyflavone (50 μm), Cell Signaling); D4476 (4-(4-(2,3-dihydrobenzo[1,4] dioxin-6-yl)-5-pyridin-2-yl-1H-imidazol-2-yl)benzamide (75 μm), Sigma); 4,5,6,7-tetrabromobenzotriazole (50 μm), EMD Biosciences; okadaic acid (100 nm), EMD Biosciences; insulin (100 nm), Sigma; and phorbol-12-myristate-13-acetate (PMA (200 nm), Sigma).
Transfections and Immunoblotting
Transfections were carried out using the calcium phosphate DNA precipitation procedure. siRNA was obtained from Dharmacon Inc. Target sequences are as follows: hCK1α 1 (5′-GCUAAAGGCUGCAACAAAG-3′); hCK1α 2 (5′-CAAUAUGACUACACAUUUG-3′); hCK1α 3 (5′-AGAGUAACAUGAAAGGUUU-3′); hPP1α 1 (5′-CAAGAGACGCUACAACAUC-3′); and hPP1α 2 (5′-GAACGACCGUGGCGUCUCU-3′). For DNA constructs and siRNA, cells were harvested 24 and 48 h after transfection, respectively. Seventy-five μg of total protein was separated on 10% SDS-polyacrylamide gels and transferred to Immobilon polyvinylidene difluoride membranes (Millipore). Membranes were blocked in Tris-buffered saline (TBS) containing 0.2% Tween 20 (TBS-T) and 5% dried milk and incubated overnight at 4 °C with the indicated primary antibodies diluted in blocking solution. After washing, the blots were incubated with horseradish peroxidase-conjugated sheep anti-mouse, goat anti-rabbit, or donkey anti-goat secondary antibodies (Jackson ImmunoResearch) and developed using the SuperSignal chemiluminescent substrate (Pierce). Where indicated, band pixel intensities were determined using the ImageJ software (National Institutes of Health).
Immunoprecipitations and m7GpppG Cap Binding Assays
Immunoprecipitations were carried out with the indicated antibody for 4 h followed by incubation with protein A-Sepharose CL-4B beads (GE Healthcare) or protein G beads (Sigma) for 1 h. For cap binding assays, 500 μg of protein was incubated with 50 μl of 7-methyl-GTP-Sepharose (GE Healthcare) for 2 h at 4 °C. Immunoprecipitates were washed in lysis buffer and subjected to SDS-PAGE and immunoblotting along with whole cell lysates.
Kinase Assays
CK1 kinase assays were performed with purified, recombinant CK1 enzyme (New England Biolabs) according to the manufacturer's conditions. Immunoprecipitated rpS6 was incubated with 500 units of CK1 and 200 μm ATP in a total reaction volume of 125 μl. Reactions were performed for 1 h at 30 °C and were terminated by the addition of 4× SDS sample loading buffer. Phosphorylation of rpS6 was monitored by immunoblotting with α-P-rpS6-235/236, α-P-rpS6-240/244, and α-P-rpS6-247 antibodies.
RESULTS
Generation and Characterization of Antibodies That Detect Ser-247-phosphorylated rpS6
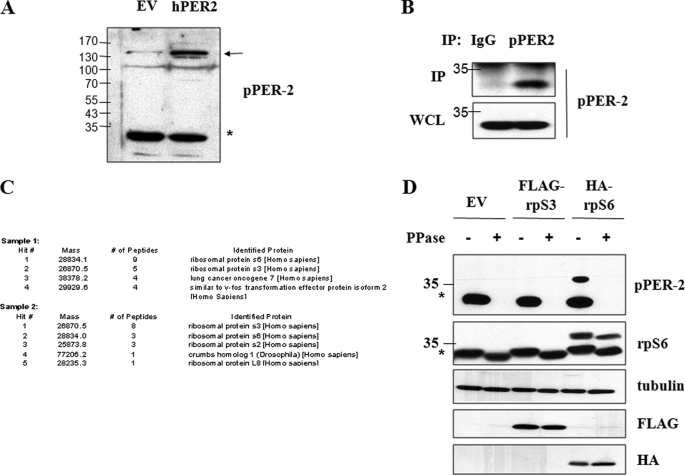
During the course of studies investigating phospho-regulation of the PERIOD2 (PER2) protein, we generated phospho-PER2 antibodies (α-pPER2) that recognize PER2 phosphorylated on Ser-662, Ser-665, and Ser-668. We found that the α-pPER2 antibody cross-reacted with an endogenous protein of ∼34 kDa that could also be efficiently immunoprecipitated from HEK 293T cell extracts (Fig. 1, A and B). We excised two replicate immunoprecipitated bands from a Coomassie Blue-stained SDS-PAGE gel and had them analyzed via LC-MS/MS. The proteomic analysis suggested that the cross-reacting protein was either ribosomal protein S3 (rpS3) or rpS6 (Fig. 1C). Transient transfection assays were then used to demonstrate that the α-pPER2 antibody directly reacted with overexpressed rpS6, but not rpS3, in a phosphatase-sensitive manner (Fig. 1D). These results indicated that the α-pPER2 antibody recognized a phosphorylated form of rpS6.
FIGURE 1.
Identification of α-pPER2 cross-reacting species as rpS6. A, α-pPER2-662/665/668 reacts with overexpressed hPER2 and an endogenous 34-kDa protein. HEK 293T cells were transfected with either empty vector (EV) or hPER2. Cell extracts were analyzed by immunoblotting with α-pPER2-662/665/668 antibody. hPER2 is indicated by an arrow, and the endogenous 34-kDa protein is indicated by an asterisk. B, immunoprecipitation of 34-kDa protein. HEK 293T cell extracts were subjected to immunoprecipitation with mouse IgG or α-pPER2-662/665/668 antibody. Immunoprecipitates (IP) and whole cell lysates (WCL) were analyzed by immunoblotting with α-pPER2-662/665/668 antibody. C, summary of proteomic analysis of cross-reacting 34-kDa protein. Two replicate immunoprecipitated 34-kDa bands were excised from Coomassie Blue-stained SDS-PAGE gels and submitted to ProtTech Inc. for analysis by LC-MS/MS. A summary of the sequencing report is shown. D, α-pPER2-662/665/668 antibody reacts with rpS6. HEK 293T cells were transfected with empty vector, HA-rpS3, or HA-rpS6. Where indicated, cell extracts were treated with λ-phosphatase (PPase) prior to analysis by SDS-PAGE and immunoblotting with α-pPER2-662/665/668, α-rpS6, α-tubulin, α-FLAG, and α-HA antibodies. These data are representative of three independent experiments.
Phospho-specific Antibodies Detect Phosphorylated Ser-247
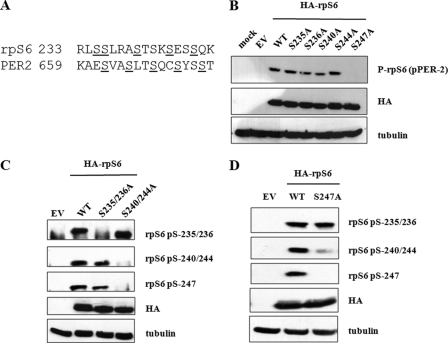
We next sought to identify which phosphorylation sites in rpS6 were detected by α-pPER2 antibodies and whether they were different from the previously established sites (Fig. 2A). We introduced individual Ser → Ala mutations at codons 235, 236, 240, 244, and 247 of rpS6 and expressed the mutant proteins in HEK 293T cells. The Ser-247 → Ala mutation ablated immunoreactivity, suggesting that the α-pPER2 antibody recognizes phosphorylated Ser-247 (Fig. 2B). This result was interesting to us because although Ser-247 has been suggested to be an in vivo rpS6 phosphorylation site (6), this assertion had not been validated using phospho-specific antibodies. Thus, these findings provide strong evidence that Ser-247 is phosphorylated in intact cells. The α-pPER2 antibody, which will be further described in a separate study, is heretofore referred to as α-P-rpS6-247.3
FIGURE 2.
Mapping of rpS6 phosphorylation sites. A, sequence overlay between mammalian rpS6 and PER2 proteins. Putative phosphorylation sites are shown in boldface and underlined. B, pPER2 antibody selectivity. HEK 293T cells were transfected with empty vector (EV), plasmids encoding HA-tagged rpS6WT, or the indicated phosphorylation site mutants. Cell extracts were then prepared and analyzed by immunoblotting using α-P-rpS6-247 (α-pPER2-662/665/668), α-HA, and α-tubulin antibodies. mock, mock-treated. C, rpS6 phosphorylation site requirements. HEK 293T cells were transfected with empty vector, plasmids encoding HA-tagged rpS6WT, or the indicated phosphorylation site mutants. Cells were harvested, and cell lysates were analyzed by immunoblotting using α-P-rpS6-235/236, α-P-rpS6-240/244, α-P-rpS6-247, α-HA, and α-tubulin antibodies. S235/236A, S235A/S236A; S240/244A, S240A/S244A. D, Ser-247 promotes phosphorylation of Ser-240/Ser-244. HEK 293T cells were transfected with empty vector or plasmids encoding HA-tagged rpS6WT or rpS6S247A. Cell extracts were analyzed by immunoblotting with α-P-rpS6-235/236, α-P-rpS6-240/244, α-P-rpS6-247, α-HA, and α-tubulin antibodies. These data are representative of three independent experiments.
It is thought that the rpS6 C terminus is phosphorylated processively, with Ser-235/236 phosphorylation occurring prior to the phosphorylation of Ser-240, Ser-244, and Ser-247. Although mutation of Ser-240 or Ser-244 alone had no effect on Ser-247 phosphorylation, mutation of both residues abolished phosphorylation of Ser-247, suggesting that prior phosphorylation of Ser-240 and Ser-244 is required for Ser-247 phosphorylation (Fig. 2C). Interestingly, we also observed that the mutation of Ser-247 inhibited phosphorylation of Ser-240 and Ser-244 but had no effect on phosphorylation of Ser-235/236 (Fig. 2D). Taken together, these results suggest that rpS6 phosphorylation may proceed in a bidirectional fashion, with phospho-Ser-240 and phospho-Ser-244 promoting phosphorylation of Ser-247 and phospho-Ser-247 in turn promoting phosphorylation of Ser-240 and Ser-244.
Regulation of rpS6 Ser-247 Phosphorylation
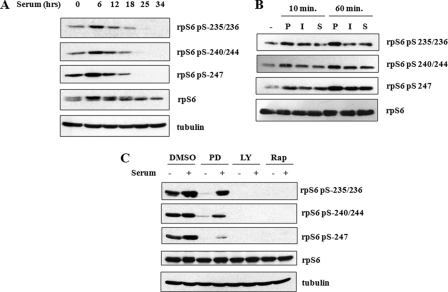
Although Ser-235/236 and Ser-240/244 are well characterized phosphorylation sites, the extent of knowledge pertaining to the regulation of Ser-247 phosphorylation is limited because of the lack of a phospho-specific antibody. Using the α-P-rpS6-247 antibody, we sought to determine whether Ser-247 is regulated differently from the four upstream phosphorylation sites. To this end, we stimulated HEK 293T cells with serum and observed rpS6 phosphorylation over time. Like Ser-235/236 and Ser-240/244, Ser-247 was phosphorylated within 6 h of serum stimulation (Fig. 3A). Ser-247 phosphorylation was more labile than either Ser-235/236 or Ser-240/244, exhibiting complete dephosphorylation 25 h after serum stimulation (Fig. 3A). Ser-247 was also phosphorylated in response to PMA and insulin with kinetics that closely paralleled phosphorylation of Ser-235/236 and Ser-240/244 (Fig. 3B). Because PMA and insulin trigger RAS/ERK/RSK1/2- and mTOR/s6K1-dependent phosphorylation of rpS6, respectively, we tested whether inhibition of these pathways affected the phosphorylation of Ser-247. We utilized the small molecule inhibitors, LY294002, PD98059 and rapamycin, which inhibit the upstream PI3K/mTOR (LY294002 and rapamycin) and ERK kinase (MEK) pathways, respectively. Phosphorylation of Ser-247 was completely abolished in response to LY294022 and rapamycin, consistent with S6K1 and S6K2 being the predominant rpS6 kinases. Inhibition of MEK by PD98059 caused a more modest reduction in rpS6 Ser-247 phosphorylation (Fig. 3C), which implies that the RAS/ERK pathway also contributes to maintenance of Ser-247 phosphorylation.
FIGURE 3.
Phospho-specific antibodies detect phosphorylated Ser-247. A, rpS6 phosphorylation time course in response to serum. HEK 293T cells were serum-starved for 24 h prior to treatment with 5% fetal bovine serum (FBS). Cell extracts were prepared and analyzed by immunoblotting using α-P-rpS6-235/236, α-P-rpS6-240/244, α-P-rpS6-247, α-rpS6, and α-tubulin antibodies. B, induction of rpS6 phosphorylation by various stimuli. HEK 293T cells were serum-starved for 24 h prior to treatment with PMA (P), insulin (I), or 5% fetal bovine serum (S). Cells were harvested at the indicated times, and extracts were analyzed by immunoblotting using α-P-rpS6-235/236, α-P-rpS6-240/244, α-P-rpS6-247, and α-rpS6 antibodies. C, inhibition of rpS6 phosphorylation. HEK 293T cells were serum-starved for 24 h and then treated with dimethyl sulfoxide (DMSO), PD98059 (PD), LY294002 (LY), or rapamycin (Rap) for 1 h. Then cells were either mock-treated or treated with 5% fetal bovine serum for 2 h, after which cells were harvested and analyzed by immunoblotting with α-P-rpS6-235/236, α-P-rpS6-240/244, α-P-rpS6-247, α-rpS6, and α-tubulin antibodies. These data are representative of three independent experiments.
PP1 Is an rpS6 Phosphatase
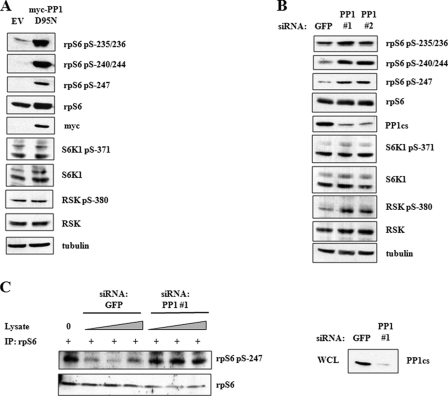
Investigation of rpS6 phosphorylation research has been heavily centered on rpS6 kinases, whereas the rpS6 phosphatase(s) have yet to be identified. Indirect evidence has suggested a role for PP1 in rpS6 dephosphorylation (15, 16), but definitive evidence is lacking. To directly test the involvement of PP1 in rpS6 phosphorylation, we transfected HEK 293T cells with a dominant-negative PP1 catalytic subunit, PP1D95N, and measured its impact on the phosphorylation of Ser-235/236, Ser-240/244, and Ser-247 by immunoblotting with phospho-specific antibodies. The expression of PP1D95N caused a strong induction in rpS6 phosphorylation at all sites tested (Fig. 4A), suggesting that PP1 is involved in rpS6 dephosphorylation. This finding was validated by siRNA-mediated knockdown of PP1 catalytic subunit α (PP1csα) (Fig. 4B). To establish whether PP1 could directly dephosphorylate the Ser-247 residue of rpS6, we carried out an in vitro phosphatase assay (Fig. 4C). In this experiment, immunoprecipitated rpS6 was mixed with whole cell lysates in which GFP or PP1 were knocked down. Immunoprecipitates (IP) mixed with lysates containing PP1 displayed decreased Ser-247 phosphorylation, whereas Ser-247 phosphorylation persisted in immunoprecipitates mixed with PP1-depleted lysates. These findings strongly suggest that PP1 actively dephosphorylates rpS6.
FIGURE 4.
PP1 regulates rpS6 dephosphorylation. A, dominant-negative PP1 induces rpS6 phosphorylation. HEK 293T cells were transfected with empty vector (EV) or plasmids encoding Myc-tagged PP1D95N for 24 h. Cell extracts were analyzed by immunoblotting using α-P-rpS6-235/236, α-P-rpS6-240/244, α-P-rpS6-247, α-rpS6, α-Myc, α-P-S6K1-371, α-S6K1, α-P-RSK-380, α-RSK, and α-tubulin antibodies. B, siRNA knockdown of PP1 induces rpS6 phosphorylation. HEK 293T cells were transfected with siRNA directed against GFP or PP1cs for 48 h. Cells were serum-starved 24 h prior to harvesting. Cell lysates were analyzed by immunoblotting with α-P-rpS6-235/236, α-P-rpS6-240/244, α-P-rpS6-247, α-rpS6, α-PP1, α-P-S6K1-371, α-S6K1, α-P-RSK-380, α-RSK, and α-tubulin antibodies. C, PP1 dephosphorylates rpS6 in vitro. HEK 293T extracts were subjected to immunoprecipitation with α-P-rpS6-235/236 antibody. Immunoprecipitates (IP) were mixed for 30 min at 37 °C with HEK 293T whole cell lysates that were transfected with siRNA directed against either GFP or PP1. Precipitates were analyzed by SDS-PAGE and immunoblotting with α-P-rpS6-247 and α-rpS6 antibodies. Whole cell lysates (WCL) from siRNA-transfected cells were analyzed by SDS-PAGE and immunoblotting with α-PP1 antibody. These data are representative of three independent experiments.
Identification of CK1 Isoforms as Endogenous rpS6 Kinases
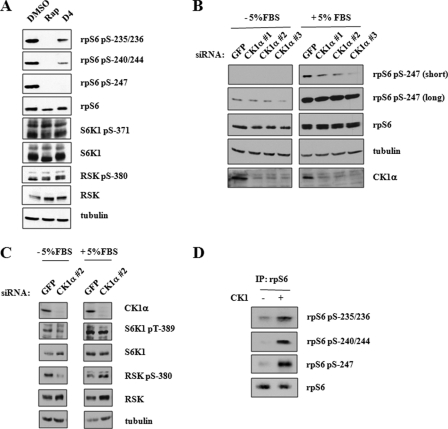
The Ser-247 phosphorylation site of rpS6 fits the (pS/pT)XX(S/T) consensus for CK1-mediated phosphorylation, where pS represents phosphorylated Ser and phosphorylated Thr represents phosphorylated Thr. To test the involvement of CK1 family members as mediators of rpS6 Ser-247 phosphorylation, we treated HEK 293T cells with the pan-CK1 inhibitor D4476 (17). D4476 treatment abolished rpS6 phosphorylation on Ser-247 and strongly suppressed the phosphorylation of upstream Ser-235/236 and Ser-240/244 sites (Fig. 5A). This latter finding implies that CK1-mediated phosphorylation of rpS6 on Ser-247 is important for maintaining phosphorylation of upstream residues. To further test CK1 involvement in rpS6 phosphorylation, we used siRNA to knockdown CK1α in HEK 293T cells. Three independent CK1α knockdowns reduced rpS6 phosphorylation on Ser-247 (Fig. 5B). siRNA-mediated knockdown of CK2α, however, did not affect the phosphorylation of rpS6 on Ser-247 or upstream sites (data not shown). Finally, to test whether CK1 could phosphorylate rpS6 directly, we performed in vitro kinase assays using recombinant CK1 and immunoprecipitated rpS6. CK1 increased the level of phosphorylation of all five sites as analyzed by immunoblotting with the indicated phospho-specific antibodies (Fig. 5D). This latter finding suggests that the activity of CK1α may be sufficient for maintenance of rpS6 phosphorylation on all five sites in asynchronously growing cells.
FIGURE 5.
rpS6 phosphorylation by CK1. A, CK1 inhibition abrogates rpS6 phosphorylation. HEK 293T cells were pretreated with rapamycin (Rap) or D4476 for 3 h and then stimulated with 5% FBS for 1 h. Cells were then harvested, and cell extracts were analyzed by immunoblotting using α-P-rpS6-235/236, α-P-rpS6-240/244, α-P-rpS6-247, α-rpS6, α-P-S6K1-371, α-S6K1, α-P-RSK-380, α-RSK, and α-tubulin antibodies. DMSO, dimethyl sulfoxide. B, CK1 isoforms phosphorylate rpS6 Ser-247. HEK 293T cells were transfected with siRNA directed against GFP and CK1α for 48 h. Where indicated, cells were then treated with 5% FBS for 1 h prior to harvesting. Cell extracts were prepared and analyzed by immunoblotting with α-P-rpS6-247, α-rpS6, α-tubulin, and α-CK1α antibodies. C, p70/p90 rpS6 kinases are unaffected by CK1 knockdown. HEK 293T cells were transfected with siRNA directed against GFP and CK1α for 48 h. Where indicated, cells were treated with 5% FBS for 1 h prior to harvesting. Cell extracts were prepared and analyzed by immunoblotting with α-CK1α, α-P-S6K1-389, α-S6K1, α-P-RSK-380, α-RSK, and α-tubulin antibodies. D, CK1 phosphorylates rpS6 C terminus in vitro. Immunoprecipitated rpS6 (IP: rpS6) was incubated with purified recombinant CK1 for 1 h at 30 °C in the presence of 200 μm ATP. rpS6 phosphorylation was monitored by immunoblotting with α-P-rpS6-235/236, α-P-rpS6-240/244, and α-P-rpS6-247 antibodies. These data are representative of three independent experiments.
CK1 and PP1 Regulate rpS6 Recruitment to 43 S Preinitiation Complex
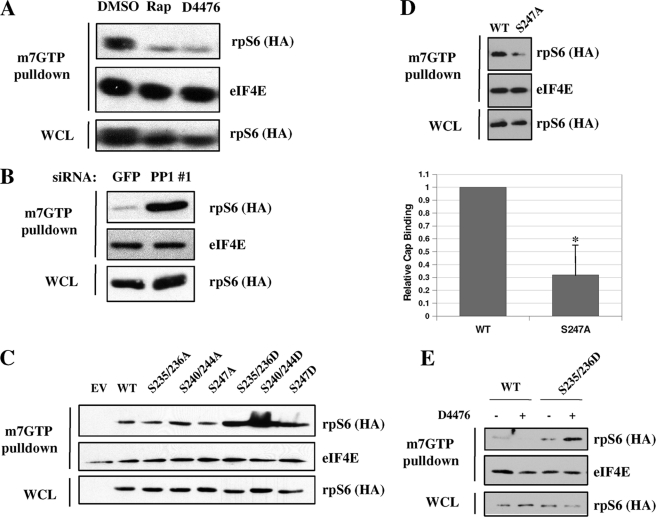
To test the functional importance of CK1-mediated rpS6 phosphorylation, we performed m7GpppG cap binding assays using extracts from cells treated with vehicle, D4476, or rapamycin. As expected, rapamycin drastically reduced rpS6 cap binding affinity in triplicate experiments (Fig. 6A). Interestingly, the inhibitory effect of D4476 on cap binding were comparable with that of rapamycin. On the other hand, knockdown of PP1cs caused a strong increase in rpS6 cap binding activity (Fig. 6B). These combined findings suggest that the CK1 and PP1 exert opposing effects on rpS6 phosphorylation and cap binding activity.
FIGURE 6.
CK1 and PP1 regulate rpS6 recruitment to 7-methylguanosine cap complex. A, inhibition of rpS6 kinases reduces rpS6 binding to 7-methylguanosine beads. HEK 293T cells were treated with rapamycin (Rap) or D4476 for 3 h prior to stimulation with 5% fetal bovine serum for 1 h. Cells were then harvested, and 500 μg of protein was incubated with 50 μl of 7-methylguanosine beads for 2 h at 4 °C. Precipitates were washed in lysis buffer and analyzed along with whole cell lysates (WCL) by immunoblotting with α-rpS6 and α-eIF4E antibodies. DMSO, dimethyl sulfoxide. B, knockdown of PP1cs promotes rpS6 recruitment to 7-methylguanosine beads. HEK 293T cells were transfected with siRNA directed against GFP or PP1cs for 48 h. Cells were serum-starved 24 h prior to harvesting. Cell extracts were prepared, and cap binding assays were performed as described. C, binding of rpS6 mutants to 7-methylguanosine cap complex. HEK 293T cells were transfected with plasmids encoding HA-tagged rpS6WT or the indicated phosphorylation site mutants. Cells were harvested 24 h later, cell extracts were prepared, and cap binding assays were performed as described. EV, empty vector. S235/236A, S235A/S236A; S240/244A, S240A/S244A; S235/236D, S235D/S236D; S240/244D, S240D/S244D. D, rpS6S247A displays decreased cap binding. HEK 293T cells were transfected with plasmids encoding HA-tagged rpS6WT or rpS6S247A. Cells were harvested 24 h later, cell extracts were prepared, and cap binding assays were performed as described. (Bars depict mean ± S.E. * indicates p < 0.05; n = 4.) E, rpS6S235D/S236D (S235/236D) cap binding is resistant to D4476. HEK 293T cells were transfected with plasmids encoding HA-tagged rpS6WT or rpS6S235D/S236D. Cells were harvested 24 h later, extracts were prepared, and cap binding assays were performed as described. Where indicated, cells were treated with D4476 for 3 h. These data are representative of at least three independent experiments.
We then tested the cap binding ability of various rpS6 phosphorylation site mutants. Interestingly, rpS6WT, rpS6S235A/S236A, and rpS6S240A/S244A were all recruited to cap beads with similar affinities (Fig. 6C), whereas rpS6S247A had significantly decreased cap binding ability when compared with rpS6WT (Fig. 6D). Cap recruitment of the phosphomimetic mutants, rpS6S235D/S236D, rpS6S240D/S244D, and rpS6S247D, was markedly increased when compared with rpS6WT (Fig. 6C). These data suggest that multisite phosphorylation of rpS6 promotes assembly of the 43 S preinitiation complex.
Finally, to show that CK1 affects cap binding by directly phosphorylating rpS6 rather than by indirect mechanisms, we tested whether or not the increased cap binding of rpS6 Ser → Asp mutants could be reduced by the CK1 inhibitor D4476, the rationale being that if CK1 was affecting cap binding by mechanisms other than rpS6 phosphorylation, D4476 would still inhibit the cap binding ability of a phospho-mimetic rpS6 mutant. D4476 abrogated the cap binding ability of rpS6WT but not rpS6S235D/S236D (Fig. 6E), which strongly suggests that D4476 inhibits cap binding by influencing the level of rpS6 C terminus phosphorylation by CK1.
DISCUSSION
In this study, we have utilized a novel α-P-rpS6-247 antibody to study the complex process of regulated rpS6 phosphorylation. Salient findings of the study include: (i) Ser-247 of rpS6 is coregulated with upstream S6K and RSK phosphorylation sites in response to mitogenic stimuli; (ii) CK1 is required for Ser-247 phosphorylation and may contribute to maintenance phosphorylation of upstream sites; (iii) CK1-dependent phosphorylation of rpS6 stimulates its cap binding activity; and (iv) PP1 antagonizes CK1-dependent phosphorylation of rpS6 in intact cells. These findings provide new insights into rpS6 phospho-regulation and provide the first evidence that CK1 regulates translation initiation.
CK1 has previously been shown to phosphorylate a number of yeast ribosomal proteins and translation factors in vitro (18, 19); however, to our knowledge, this is the first demonstration that CK1 regulates a specific step in protein translation. CK1 inhibition with D4476 or siRNA knockdown strongly suppressed serum-dependent phosphorylation of rpS6 on Ser-247, which is a consensus CK1 site. CK1 phosphorylated Ser-247 in vitro, consistent with a direct role for CK1 in rpS6 phospho-regulation. Interestingly disruption of the CK1 priming site at Ser-244 did not abolish serum-dependent phosphorylation of rpS6 on Ser-247; however, CK1 is known to phosphorylate acidic and/or heavily phosphorylated motifs in the absence of priming phosphorylation at the −3 position, and we propose a similar mechanism for CK1-dependent phosphorylation of rpS6 Ser-247 (20).
Peptide mapping and kinetic studies have suggested that phosphorylation of individual rpS6 residues takes place in an ordered fashion with phosphorylation initiated at Ser-236 followed sequentially by Ser-235, Ser-240, Ser-244, and finally Ser-247. The finding that mutation of Ser-240/244 is required for CK1-dependent phosphorylation of Ser-247 (Fig. 2C) is compatible with this model. Interestingly, mutation of Ser-247 or treatment with D4476 also partially inhibited the phosphorylation of Ser-240/244, suggesting bidirectional phosphorylation site interdependence. It is not clear how Ser-247 promotes Ser-240/244 phosphorylation; however, Ser-244 is a consensus CK2 site in the context of Ser-247-phosphorylated rpS6, and it is conceivable that CK2 contributes to Ser-244 phosphorylation in vivo. We propose that phospho-Ser-247 triggers feedback phosphorylation of Ser-240/244, which helps sustain rpS6 phosphorylation in response to mitogenic stimulation. It is also possible that CK1 directly phosphorylates the upstream Ser-235/236 and Ser-240/244 sites in intact cells given that CK1 or a closely associated kinase activity phosphorylated these sites in vitro (Fig. 5D).
The phosphatases involved in the dephosphorylation of rpS6 have yet to be identified, although reports have suggested a possible role for PP1. We found that dominant-negative PP1 as well as siRNA targeting the PP1 catalytic subunit enhanced rpS6 Ser-247 phosphorylation, providing the first direct evidence that PP1 negatively regulates rpS6 phosphorylation in intact cells (Fig. 4, A and B). Inhibition of PP1 induced the phosphorylation of all rpS6 C-terminal residues, suggesting that it is a general regulator of rpS6 C terminus phosphorylation.
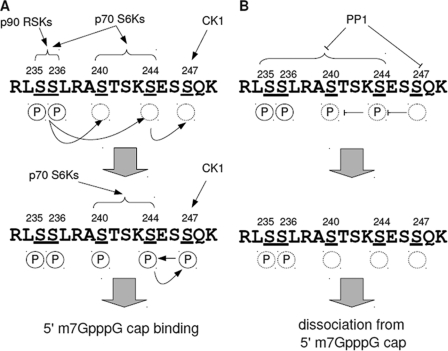
Phospho-regulation by CK1 and PP1 appears to play a modulatory role in the regulation of rpS6 cap binding activity. D4476 inhibited rpS6 association with m7GpppG-coupled beads to nearly the same extent as rapamycin without grossly impacting the activation status of RSKs or p70 S6K (Figs. 5A and 6A). A constitutively active rpS6 mutant containing phospho-mimetic Asp substitutions at Ser-235/236 was resistant to D4476, suggesting that the effects of CK1 in this assay are mediated through C-terminal rpS6 phosphorylation (Fig. 6E). A role for CK1-mediated phosphorylation of Ser-247 is also strongly suggested by the finding that mutation of this residue to Ala partially inhibited rpS6 association with m7GpppG-coupled beads in vitro (Fig. 6D). Thus, although we cannot rule out the possibility that CK1 is acting elsewhere in the assembly of the preinitiation complex, our data are most consistent with a model in which CK1 modulates cap binding through direct phosphorylation of the rpS6 C terminus (Fig. 7).
FIGURE 7.
A model for dynamic phosphorylation and dephosphorylation of rpS6 C terminus. A, initiation and maintenance of rpS6 phosphorylation. Upon mitogenic stimulation, p90 RSK and p70 S6K kinase families phosphorylate Ser-235/236. Ser-235/236 phosphorylation (circled P) promotes p70 S6K-dependent phosphorylation of Ser-240 and Ser-244. Ser-244 phosphorylation then promotes CK1-dependent phosphorylation of Ser-247. Phospho-Ser-247 triggers feedback phosphorylation of Ser-240/244, which sustains rpS6 phosphorylation and its association with the 5′-m7GpppG cap complex. B, dephosphorylation of rpS6 by PP1. After cessation of mitogenic stimuli and inactivation of rpS6 kinases, PP1 rapidly dephosphorylates rpS6 beginning with Ser-247. Unphosphorylated rpS6 then dissociates from the 5′-m7GpppG cap complex. A and B, circled P indicates phosphorylation. Phosphorylation sites are underlined.
Finally, our results confirm a previously established link between rpS6 phosphorylation and recruitment to the mRNA cap complex. This is consistent with a number of reports that have found that polysomes contain a greater percentage of phosphorylated rpS6 than subpolysomal fractions (21–23). However, cells derived from rpS6P−/− mice, which express a mutant form of rpS6 that cannot be phosphorylated, have higher rates of global protein synthesis, suggestive of an inhibitory role for rpS6 phosphorylation (9). Thus, how site-specific rpS6 phosphorylation contributes to each step of translation requires further investigation. It is likely that rpS6 phosphorylation may affect protein synthesis differently throughout translation initiation, elongation, and termination. Phosphorylation of rpS6 may serve as a tuning mechanism, promoting translation initiation while inhibiting other critical steps of translation.
This work was supported, in whole or in part, by National Institutes of Health Grant CA124722 (to R. S. T.) and National Institutes of Health Grant T32ES007015 through the NIEHS (to J. A. H.). This work was also supported by grants from the American Cancer Society and a Shaw Scientist Award from the Greater Milwaukee Foundation (to R. S. T.) and an American Heart Association Predoctoral Fellowship (to N. P. S.).
N. P. Shanware, J. A. Hutchinson, S. H. Kim, L. Zhan, M. J. Bowler, and R. S. Tibbetts, manuscript submitted.
- CK
- casein kinase
- rpS6
- ribosomal protein S6
- S6K
- p70 S6 kinase
- RSK
- p90 ribosomal S6 kinase
- PP1
- protein phosphatase type 1
- PER2
- PERIOD2
- mTOR
- mammalian target of rapamycin
- PMA
- phorbol-12-myristate-13-acetate
- h
- human
- P
- phospho.
REFERENCES
- 1. Knippschild U., Gocht A., Wolff S., Huber N., Löhler J., Stöter M. (2005) Cell. Signal. 17, 675–689 [DOI] [PubMed] [Google Scholar]
- 2. Meggio F., Pinna L. A. (2003) FASEB J. 17, 349–368 [DOI] [PubMed] [Google Scholar]
- 3. Gao Z. H., Metherall J., Virshup D. M. (2000) Biochem. Biophys. Res. Commun. 268, 562–566 [DOI] [PubMed] [Google Scholar]
- 4. Meyuhas O. (2008) Int. Rev. Cell Mol. Biol. 268, 1–37 [DOI] [PubMed] [Google Scholar]
- 5. Ruvinsky I., Meyuhas O. (2006) Trends Biochem. Sci. 31, 342–348 [DOI] [PubMed] [Google Scholar]
- 6. Krieg J., Hofsteenge J., Thomas G. (1988) J. Biol. Chem. 263, 11473–11477 [PubMed] [Google Scholar]
- 7. Wettenhall R. E., Erikson E., Maller J. L. (1992) J. Biol. Chem. 267, 9021–9027 [PubMed] [Google Scholar]
- 8. Martin-Pérez J., Thomas G. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 926–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ruvinsky I., Sharon N., Lerer T., Cohen H., Stolovich-Rain M., Nir T., Dor Y., Zisman P., Meyuhas O. (2005) Genes Dev. 19, 2199–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ruvinsky I., Katz M., Dreazen A., Gielchinsky Y., Saada A., Freedman N., Mishani E., Zimmerman G., Kasir J., Meyuhas O. (2009) PLoS One. 4, e5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shima H., Pende M., Chen Y., Fumagalli S., Thomas G., Kozma S. C. (1998) EMBO J. 17, 6649–6659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pende M., Um S. H., Mieulet V., Sticker M., Goss V. L., Mestan J., Mueller M., Fumagalli S., Kozma S. C., Thomas G. (2004) Mol. Cell. Biol. 24, 3112–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roux P. P., Shahbazian D., Vu H., Holz M. K., Cohen M. S., Taunton J., Sonenberg N., Blenis J. (2007) J. Biol. Chem. 282, 14056–14064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chung J., Kuo C. J., Crabtree G. R., Blenis J. (1992) Cell 69, 1227–1236 [DOI] [PubMed] [Google Scholar]
- 15. Belandia B., Brautigan D., Martín-Pérez J. (1994) Mol. Cell Biol. 14, 200–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Olivier A. R., Ballou L. M., Thomas G. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 4720–4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rena G., Bain J., Elliott M., Cohen P. (2004) EMBO Rep. 5, 60–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wojda I., Cytryńska M., Frajnt M., Jakubowicz T. (1999) Acta Biochim. Pol. 46, 211–215 [PubMed] [Google Scholar]
- 19. Haas D. W., Hagedorn C. H. (1991) Arch. Biochem. Biophys. 284, 84–89 [DOI] [PubMed] [Google Scholar]
- 20. Cegielska A., Gietzen K. F., Rivers A., Virshup D. M. (1998) J. Biol. Chem. 273, 1357–1364 [DOI] [PubMed] [Google Scholar]
- 21. Thomas G., Martin-Pérez J., Siegmann M., Otto A. M. (1982) Cell 30, 235–242 [DOI] [PubMed] [Google Scholar]
- 22. Duncan R., McConkey E. H. (1982) Eur. J. Biochem. 123, 535–538 [DOI] [PubMed] [Google Scholar]
- 23. Burkhard S. J., Traugh J. A. (1983) J. Biol. Chem. 258, 14003–14008 [PubMed] [Google Scholar]