Abstract
Translation of cellular mRNAs via initiation at internal ribosome entry sites (IRESs) has received increased attention during recent years due to its emerging significance for many physiological and pathological stress conditions in eukaryotic cells. Expression of genes bearing IRES elements in their mRNAs is controlled by multiple molecular mechanisms, with IRES-mediated translation favored under conditions when cap-dependent translation is compromised. In this review, we discuss recent advances in the field and future directions that may bring us closer to understanding the complex mechanisms that guide cellular IRES-mediated expression. We present examples in which the competitive action of IRES-transacting factors (ITAFs) plays a pivotal role in IRES-mediated translation and thereby controls cell-fate decisions leading to either pro-survival stress adaptation or cell death.
Key words: translation initiation, IRES, canonical initiation factors, ITAFs, stress response, eIF2, angiogenesis, mitosis, nutrient-signaling, hyperosmolar stress
Introduction
Protein biosynthesis is a complex process consisting of three major phases: initiation, elongation and termination.1,2 In prokaryotes, the initiation step leading to assembly of elongation-competent ribosomes at the start codon of the mRNA involves interaction between the 3′-end of a 16S rRNA and a Shine-Dalgarno sequence upstream of the start codon.3–5 In contrast, in eukaryotes, initiation is the most complex phase of the translation process and may proceed via multiple routes.3,6,7 As discussed in detail below, these routes include the cap-dependent and/or scanning mode of initiation as well as cap-independent initiation directed by Internal Ribosome Entry Sites (IRES). The later mode of initiation represents the major focus of this review.
Canonical Cap-Dependent Translation Initiation
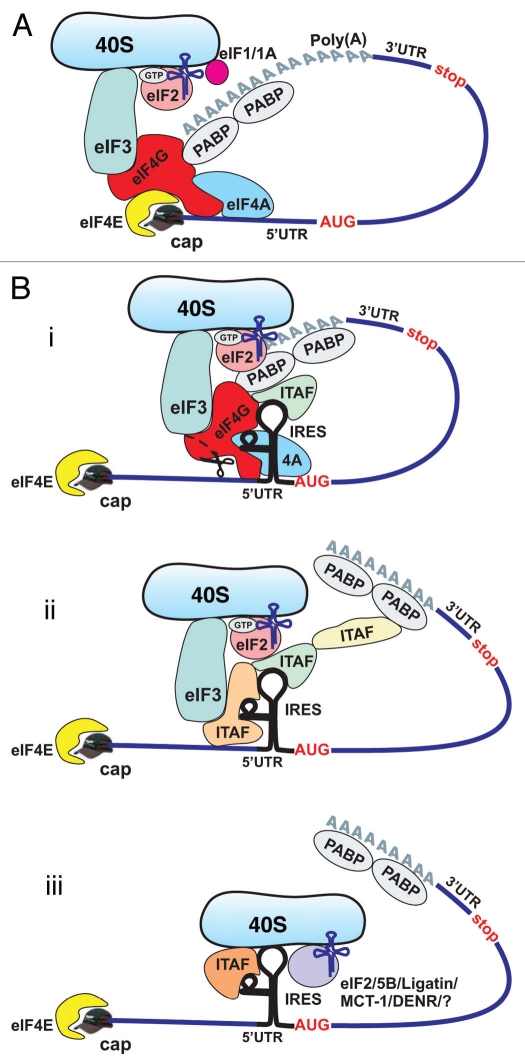
For most eukaryotic mRNAs, translation initiation involves binding of the mRNA 5′- m7G cap-structure to a protein complex referred to as the cap-binding complex or eIF4F.3,6,7 This complex consists of the cap-binding protein eIF4E, the scaffolding protein eIF4G which recruits and links together many other initiation factors, and the ATP-dependent RNA helicase eIF4A, which is thought to unwind the secondary structure in the 5′-untranslated region (5′-UTR) of the mRNA.3,6,7 Binding of eIF4F to the mRNA is followed by recruitment of the 40S ribosomal subunit with its associated initiation factors, resulting in the 43S initiation complex.3,6,7 This complex is comprised of: (1) the 40S subunit, (2) a ternary complex containing an initiator tRNA molecule (eIF2•GTP•Met-tRNAiMet), (3) the multi-subunit initiation factor eIF3, which has multiple functions,8 but also serves as a bridge between the 40S ribosome and mRNA-bound eIF4G and (4) the initiation factors eIF1 and eIF1A, which facilitate recognition of the start codon.3,6,7 It is widely believed that after its assembly at the 5′-end, the 43S initiation complex moves along the 5′-UTR in search of the initiation codon.3,6,7 When the complex encounters and recognizes the initiation AUG codon (usually this is the first AUG encountered), the 40S subunit is joined by a 60S subunit to form an elongation-competent 80S ribosome.3,6,7 It has also been demonstrated that 5′- and 3′-ends of the mRNA cooperate during cap-dependent initiation in eukaryotes (reviewed in refs. 3, 6 and 7). This, in particular, involves interaction between poly(A)-binding protein (PABP) bound at the 3′ end of the message and eIF4G bound at the 5′ end, resulting in circularization of the mRNA (Fig. 1A). The latter, has been suggested to facilitate recycling of ribosomes by allowing 40S subunits to enter the next initiation phase almost immediately after termination of protein synthesis, thus enhancing the overall rate of initiation.6,7,9 In addition, the eIF4G-PABP interaction was suggested to stabilize the interaction of eIF4F with the mRNA (reviewed in ref. 7).
Figure 1.
Cap-dependent and IRES-mediated mechanisms of translation initiation in eukaryotic cells. (A) Cap-dependent initiation is believed to require all canonical initiation factors and involve circularization of the mRNA via interaction of PABP with eIF4G. (B) Cellular IRES-mediated translation generally does not require the cap-binding protein eIF4E and/or intact eIF4G, but may involve circularization of the mRNA. The requirement for canonical initiation factors and ITAFs can vary between different IRES-containing mRNAs. Potential mechanisms of cellular IRES-mediated translation: (i) most, if not all, canonical initiation factors and many ITAFs are required (top part); (ii) a limited number of canonical factors and ITAFs are required (middle); and (iii) canonical factors are dispensable, but some ITAFs may be required (bottom). Delivery of Met-tRNAiMet to the 40S ribosomal subunit may be performed by eIF5B/Ligatin/MCT-1/DENR55,56,58,59 and perhaps some other, yet unidentified proteins, acting in place of eIF2.
The scheme of cap-dependent translation presented here (Fig. 1A) is a simplified representation that omits many mechanistic aspects of the process, including the roles of additional initiation factors (reviewed in refs. 3, 6 and 7).
Alternative Internal Route of Translation Initiation
For a long time the “cap-dependent” or “scanning” mode of initiation was considered the only possible route through which translation of eukaryotic mRNAs could be initiated. However, studies of viral gene expression in the late 1980s led to the discovery of an alternative mode of translation initiation in eukaryotic cells that bypasses the requirement for cap-dependent scanning and allows the 40S ribosome to be directly recruited to the vicinity of the initiation codon (reviewed in refs. 7 and 10–12). The mRNA regions required for this direct recruitment of the 40S ribosomal subunit were termed Internal Ribosome Entry Sites (IRESs) to emphasize that the process is independent of 5′-end recognition.7,10–12
It has been shown that viral IRES-driven translation initiation is typically utilized when cap-dependent initiation is compromised.7,10–12 As might be thus expected, viral IRES-driven translation has a generally reduced requirement for canonical translation initiation factors, particularly members of the eIF4F complex (initiation factors eIF4E and eIF4G). Several other initiation factors also appeared to be dispensable for the internal initiation pathway (reviewed in refs. 7 and 10–13). The involvement of canonical initiation factors in IRES-driven translation initiation appears to vary for IRESs in different mRNAs. Analysis of the structures of viral IRES elements has shown that they possess complex secondary and tertiary structures that are believed to direct non-canonical interactions between the IRESs and components of the canonical translational apparatus, thus allowing for 5′-end-independent initiation.12,13 In some cases, these non-canonical interactions reduce the requirements for canonical initiation factors almost completely.12–14 Moreover, in certain “extreme” cases, initiation can proceed without involvement of any of the canonical initiation factors,14 relying exclusively on direct interactions between the IRES and the 40S ribosome.15 In addition, a number of proteins have been identified that are capable of modulating (typically enhancing) internal initiation. These so-called IRES trans-acting factors (ITAFs) (reviewed in refs. 11 and 16) are cellular RNA-binding proteins that have a variety of cellular functions in addition to promoting internal initiation; however, they do not appear to be involved in cap-dependent translation initiation. The role of ITAFs in IRES-driven initiation is discussed in detail below.
Cellular IRES-Mediated Translation
IRES-mediated translation of cellular transcripts was not widely recognized or extensively studied until recently (reviewed in ref. 16). This delay in attention is due to a number of reasons. First, cellular IRES-mediated translation is typically less efficient than the best studied cases of viral IRES-mediated translation.16 Second, substantial concerns about the validity of cellular internal initiation have been raised in some cases,17,18 prompting investigators to perform thorough analyses of the integrity of the mRNA in order to support their claims of internal initiation.19 Third, cellular IRES-mediated translation appears to be regulated by multiple sophisticated control mechanisms (see below), guided by their physiological significance. The IRESite database20 presents evidence of many eukaryotic cellular internal ribosome entry site elements and the list is growing.20 An increasing body of evidence indicate that these cellular IRESs have two major physiological functions: (1) they support low levels of translation initiation for cellular IRES-containing mRNAs with highly structured 5′-UTRs (incompatible with efficient scanning) under normal physiological conditions when cap-dependent translation is fully active and (2) they support robust translation of cellular mRNAs under a variety of physiological conditions such as mitosis, when cap-dependent translation is compromised. All cellular mRNAs are presumed to be capped and should be capable of binding the eIF4F complex. However, it is generally believed that conventional scanning from the 5′-end is not efficient for most IRES-containing cellular mRNAs because their 5′UTRs are typically long, GC-rich, highly structured and may contain several upstream translation initiation codons.21 Nevertheless, cases when both mechanisms operate on the same message do exist. The mRNA for neurogranin, a neuronal calmodulin-binding protein, is an example of a message that is translated by both 5′-cap-dependent and internal initiation mechanisms.22
It is now apparent that under conditions of decreased cap-dependent initiation, cellular (like viral) IRES-mediated initiation takes over.23,24 It has been demonstrated that many physiological, pathophysiological and stress conditions that lead to inhibition of cap-dependent translation cause a substantial increase in cellular IRES-mediated translation (reviewed in refs. 16 and 25). Such conditions include, but are not limited to, endoplasmic reticulum (ER) stress, hypoxia, nutrient limitation, mitosis and cell differentiation. Since cap-dependent translation is suppressed under these conditions, it is believed that IRES-containing mRNAs become more competitive for the available pool of ribosomes and translation initiation factors, including both canonical initiation factors and ITAFs (Fig. 1B). It is striking that many of the cellular mRNAs that contain IRES elements20 encode proteins that are involved in protection of cells from stress or, alternatively, induction of programmed cell death (apoptosis). Therefore, it is currently believed that cellular IRES-mediated translation plays an important role in cell-fate decisions under a variety of conditions.
It should be noted, however, that in contrast to viral IRES elements whose mechanism of action is becoming better understood,12,13,26 very little is currently known about the mechanism underlying cellular IRES function. In this review article, we will focus on the emerging mechanisms of the cellular IRES structure and function, as well as the many questions that remain to be answered.
Structure of Cellular IRES Elements
No common sequence and/or structural motifs have been identified to allow prediction of cellular IRES elements from an mRNA sequence. Therefore, the existence of an IRES in a particular mRNA must be experimentally determined in each and every case following a set of defined tests (reviewed in refs. 16 and 27). The vast majority of cellular IRES elements are located within the 5′-UTRs immediately upstream of the initiation codon. Nevertheless, there are cases in which the IRES is downstream of the initiation codon or located in the coding regions of the message, thereby triggering synthesis of a truncated protein.28,29 Chemical and enzymatic probing of the structure of a variety of cellular IRESs revealed (as found for viral IRES elements) complex structures that often include stem loops and pseudoknots.16,30 However, no common structural motifs have been detected among cellular IRES elements.31,32 In general, as compared to their viral counterparts, cellular IRES elements appear to be much more diverse in their structures and surprisingly less stable in terms of the Gibbs free energy of the folded mRNA.32
Mechanism of Cellular IRES-Directed Translation Initiation: Involvement of Canonical Initiation Factors
Similar to viral IRESs, cellular IRES elements likely participate in multiple interactions with components of the translational machinery (canonical initiation factors, ITAFs and 40S ribosomal subunits). Together, these interactions are believed to provide (in most of the cases) a means for proper positioning of the initiation codon at the ribosomal P-site without ribosomal scanning from the 5′ end of the message (reviewed in ref. 16). However, this remains largely hypothetical, since there have not been extensive systematic studies of the ability of cellular IRES elements to bind the 40S ribosomal subunit or of the requirements for canonical translation initiation factors in cellular internal initiation. It also remains possible that certain cellular IRESs may utilize the so-called “land” (in the vicinity of initiation codon) “and scan” mechanism typical for picornavirus IRES elements.33 IRESs found in c-myc, L-myc and N-myc mRNAs were suggested to utilize this mechanism of internal initiation.34 In addition, it has been postulated that some cellular IRESs, e.g., a short nine nt IRES from the mRNA of the human homeodomain protein Gtx35 and a 90 nucleotide IRES found in the human proto-oncogene IGF1R mRNA may operate through a Shine-Dalgarno-like interaction between the IRES and the 18S rRNA.36 Yet, it is unclear, how many other cellular IRESs may utilize a prokaryotic-like mechanism of initiation. Clarification of these issues is an important goal since several lines of evidence suggest that IRES-driven initiation appears to account for a significant proportion of cellular translation initiation: (1) a large number of cellular IRESs have been experimentally verified,20 (2) under stress conditions, 3–5% of the mRNAs are translated efficiently as determined by their association with polyribosomes,37 (3) 10–15% of cellular mRNAs were suggested to rely on cap-independent mechanisms of translation initiation, independently of stress38 and (4) several recent reports highlight the in vivo significance of IRES-mediated translation of specific mRNAs.39–41
Given the prevalence of IRES-mediated initiation under stress conditions, it is notable that in most cases of cellular internal initiation, the cap-binding protein eIF4E and the scaffolding protein eIF4G (which undergoes caspase-mediated cleavage during stress, reviewed in ref. 16) seem not to be required.42 Studies using siRNAs and specific chemical inhibitors (such as, for example, hippuristanol, a potent inhibitor of eIF4A43 revealed that the c-myc and N-myc IRESs do not require eIF4E or intact eIF4G for their activity, but do require eIF4A and eIF3).34 In this regard, these IRESs are similar to many viral IRES elements, e.g., EMCV.44 A strong requirement for the RNA helicase eIF4A suggests that the c- and N-myc IRESs likely need to be remodeled in order to be competent for initiation. Interestingly, the same study showed that the L-myc IRES requires both eIF4E and full-length eIF4G for its activity,34 which is reminiscent of the factor requirements of the hepatitis A virus (HAV) IRES.45,46 Recently, the first case of a cellular IRES element that seem to be capable of direct binding to 40S ribosomal subunits was reported for the c-Src kinase mRNA.47 This feature of the c-Src kinase IRES element makes it similar to hepatitis C virus-like IRESs.48 However, it is currently unclear how many other cellular elements utilize the same pathway.
The role of eIF2, the major eukaryotic initiation factor that delivers an initiator methionine-charged tRNA (Met-tRNAiMet) to the ribosome during canonical cap-dependent translation, has also been investigated for cellular internal initiation. Many cellular IRES-containing mRNAs (e.g., cat-1, N-myc, s-Src, etc.,) were shown to be insensitive, or much less sensitive than mRNAs without IRESs, to the inhibition of protein synthesis caused by eIF2 phosphorylation.16,49–52 Phosphorylation of the alpha subunit of eIF2 (eIF2α) is a common consequence of many stress conditions (e.g., ER stress, nutrient limitation and many others).53 This reduces the activity of the eIF2•GTP•Met-tRNAiMet ternary complex and thereby suppresses the overall rate of cap-dependent protein synthesis (reviewed in ref. 5). Some viral IRESs (SCFV, HCV),54–56 as well as some cellular IRESs16,49–52 are insensitive to this mode of translation inhibition. These observations suggest that different mRNAs and, in particular, different IRES-containing mRNAs might differ in their requirements for the active ternary complex and/or might utilize different pathway(s) to deliver Met-tRNAiMet to the ribosome.57 The latter pathway(s) might involve initiation factor eIF5B and/or Ligatin or perhaps some other proteins.55,56,58,59 Eukaryotic translation initiation factor 5B is an ortholog of prokaryotic IF2, which delivers the initiator tRNAi to the ribosome in prokaryotes and also participates in subunit joining (reviewed in ref. 5). Two recent reports clearly showed that two structurally similar viral IRES elements from mRNAs of the Flaviviridae family of viruses (classical swine fever virus (CSFV) and HCV) utilize the “eIF5B pathway” to promote Met-tRNAiMet binding to ribosomes.55,56 Therefore, it is possible that some cellular IRESs might utilize the same pathway, although this has yet to be determined. Another explanation for eIF2-independent internal initiation might be utilization of Ligatin58,59 and/or the oncogene MCT-1 and density-regulated protein (DENR) (together),59 in the place of eIF2. It was recently shown that Ligatin (alone) and MCT-1/DENR (in combination) can promote attachment of Met-tRNAiMet to ribosomal complexes assembled on mRNAs that place their initiation codon directly in the P site; e.g., HCV-like IRESs.59 The exact mechanism of action of Ligatin and MCT-1/DENR in promoting recruitment of tRNA to the 40S ribosomal subunit is not known, yet.59 It can not be excluded that some other/additional proteins can promote Met-tRNAiMet binding to ribosomes in eukaryotes.
We should also mention that in contrast to the wealth of positive views, as presented here, negative considerations regarding the existence and function of cellular IRES elements have also been published.17,18,27,60 These negative considerations17,18,27,60 are mostly influenced by findings showing that in certain cases not all the necessary tests have been performed for careful validation of IRES activity.27 In almost all such controversial cases, bicistronic vectors were initially used to test IRES activities in normal non-stress conditions, when cap-dependent translation prevails. It has recently become clear that these vector systems may not be the most appropriate27 for evaluating the physiological significance of a given IRES, which functions at its best during conditions of inhibition of cap-dependent translation. An example of such controversial cellular IRES element is the XIAP IRES,61 which has been studied by several groups and in particular has been suggested to be an invalid one due to the presence of splicing and/or cryptic promoter activity within the 5′UTR mRNA fragment comprising the IRES.60,61 However, it has recently become clear that alternative splicing within the 5′-UTR generates two alternative XIAP mRNA transcripts, one which is translated in a cap-dependent manner and another which is translated via an IRES.61 The IRES-containing XIAP mRNA was associated with polyribosomes (efficient translation) under conditions of nutritional stress (serum starvation).61 This was in contrast to the non-IRES-containing XIAP mRNA transcript which was translated inefficiently during stress.61
Complexity of the control mechanisms regulating gene expression in eukaryotes makes the study of the cellular IRES-mediated translation challenging. We believe, however, that evaluation of the efficiency of translation (i.e., association with polyribosomes) of mRNAs under stress conditions in which the encoded proteins have biological significance can represent an appropriate validation test for the presence of an IRES in the mRNA of interest. This type of evaluation can then be followed by the delineation/identification of the 5′-UTRs of such efficiently translated mRNAs and further testing of these UTRs under stress conditions using monocistronic reporter expression vector systems. We propose that the physiological significance of an IRES should drive its discovery and validation of its existence, followed by studies on the mechanism of ribosome recruitment.
Examples of IRES-mediated translation operating in physiological and pathological states (discussed below) may open the path for more important findings on the existence and function of additional cellular IRES elements.
The Role of ITAFs in Cellular IRES-Mediated Translation
The complex nature of regulation of cellular mRNA translation under different pathophysiological conditions suggests that there may be multiple diverse pathways leading to cellular IRES-mediated initiation (Fig. 1B). In addition, it has been shown that different cellular IRESs reveal different responses to various stress conditions that inhibit cap-dependent translation. For example, during mitosis, the Unr, c-myc and PITSLREp58 kinase IRESs become more active, while others do not.62 Furthermore, during apoptosis, the Apaf-1 IRES is active (reviewed in ref. 63). However the XIAP IRES is inhibited.64 Studies aimed at explaining these types of differential responses indicated that ITAFs are responsible for sensing changes in cellular metabolism and influence IRES activity.16,24,38
A striking feature of many ITAFs is that they belong to the group of heterogeneous nuclear ribonucleoproteins (HnRNP A1, C1/C2, I, E1/E2, K and L) known to shuttle between the nucleus and the cytoplasm.16,24,38 In addition to their participation in a variety of cellular activities (e.g., RNA splicing and/or export), ITAFs are generally believed to be able to increase (or, in certain cases, decrease) the affinity of binding between IRESs and components of the translational apparatus (canonical initiation factors and ribosomes). Although the exact mechanism(s) underlying ITAF function is unknown, hypotheses include: (1) they remodel IRES spatial structures to produce conformations with higher or lower affinity for components of the translation apparatus, (2) they build or abolish bridges between the mRNA and the ribosome in addition to those provided by canonical initiation factors and (3) they take the place of canonical factors in building bridges between the mRNA and the ribosome.16,24,38
Overexpression and/or depletion of specific ITAFs in normal cells can affect the activity of the cellular IRESs that normally utilize those ITAFs without altering cap-dependent translation (reviewed in ref. 24). Thus, it is clear that the intracellular concentration of ITAFs plays an important role in modulating the activity of IRESs, but the mechanism(s) responsible for regulating ITAF concentration have not been fully defined. Several studies have suggested that the subcellular (nuclear/cytoplasmic) distribution of ITAFs is an important determinant of IRES activity.24,38 Two alternative mechanisms have been proposed to explain the effects of ITAF compartmentalization. In one model, nuclear localized ITAFs were suggested to associate with their target IRES-containing mRNAs and sequester them in the nucleus away from the translational machinery.65 Alternatively, ITAFs in the nucleus were suggested to be primarily in an mRNA-unbound form, separated from their target IRES-containing messages residing in the cytoplasm. Following the appropriate signals (caused by stress or other physiological conditions), either the ITAF-bound mRNAs (in the first model) or the unbound ITAFs themselves (in the second model) were proposed to translocate from the nucleus to the cytoplasm, allowing translation of the mRNAs to proceed.24
Although these two models are sufficient to explain some cases of IRES activation via corresponding ITAFs, they do not explain others. It has recently been shown that the ITAFs PTB and HnRNP L associate with the cat-1 mRNA in both the nucleus and the cytoplasm during amino acid starvation following the kinetics of cat-1 mRNA accumulation and cat-1 IRES activation.66 This observation suggests that additional mechanism(s) may govern increased association of these ITAFs with the cat-1 mRNA both in the nucleus and in the cytoplasm. In some cases, posttranslational modifications of ITAFs, triggered by stress, have been shown to affect both their subcellular localization and binding affinity for IRESs. For example, phosphorylation of HnRNP A1 was shown to affect both its subcellular distribution and its ability to modulate the activity of target IRESs, such as cyclin D1, c-myc, FGF2, VEGF, XIAP, Apaf-1 and unr, respectively.67–71
Despite advancements in our understanding of the requirements of ITAFs in cellular IRES-mediated initiation, the mechanism of ITAF-mediated translation initiation remains unclear. The recently discovered family of A-rich yeast IRESs72 provides an interesting case in this regard since they rely on poly(A) binding protein (PABP) for their activity. PABP is known to interact with two scaffolding translation initiation factors, eIF4G and eIF3.9,73 Therefore, the strong requirement for PABP in the case of the A-rich yeast IRES elements may be explained by the necessity to build a bridge that can bring the ribosome to the IRES. PABP, eIF4G and eIF3 are all considered canonical initiation factors. However, it is unclear whether additional non-canonical factors (e.g., ITAFs) might be involved in this particular pathway. For example, in mammalian cells, PABP is known to interact with many other RNA binding proteins, including the ITAFs Unr and NSAP1, which may contribute to building missing bridges between the RNA and the ribosome.74,75 Interestingly, in apparent contrast to the yeast situation, A-rich stretches upstream of the initiation codon in mammalian IRESs were shown to have a negative regulatory effect on the activities of some IRESs. For the p27(kip1) mRNA (encoding a cyclin-dependent kinase inhibitor that regulates cell cycle progression), binding of the ITAF HuR to a A-rich region was shown to inhibit the activity of the p27(kip1) IRES element, resulting in decreased p27(kip1) expression and enhanced tumor cell proliferation.76,77 One possible explanation for the differential effect of the A-rich elements found in IRES-containing mRNAs is that these A-rich elements might act as competitive targets for both canonical initiation factors and ITAFs and thus be capable of either enhancing or suppressing IRES-mediated translation of mRNAs.
Ultimately, a complete understanding of the requirements for canonical initiation factors and ITAFs for every cellular IRES could be obtained by using purified factors in a reconstituted in vitro translation system(s) and toe-printing assays (reviewed in ref. 78). This has been done for a number of “classic” viral IRES elements, including HCV, FMDV, EMCV, etc.48,79 It is generally believed that different cellular IRESs have very different requirements for canonical initiation factors, as well as ITAFs. However, at present, the data are scarce and many more studies will be required to generalize this conclusion with confidence. Further, kinetic analysis of the factor(s) binding to the IRES and the binding of the factor(s)-IRES complexes to the 40S ribosomal subunit may be extremely helpful in understanding the mechanisms of intracellular competition between mRNAs possessing different IRES elements. As can be seen from the discussion below, these mechanisms play an important role in various cell fate decisions.
Physiological Significance of IRES-Mediated Translation
There are a number of studies that highlight the significance of IRES-mediated translation in physiological and pathological/stress conditions. Below, we review representative examples of IRES-mediated translation in the cellular response to growth, nutritional, environmental and proliferation signals. A major obstacle in addressing the significance of IRES-mediated translation in different cellular pathways arises from the complex nature of many of the proteins translated via internal initiation and the multiple mechanisms that control their expression. As mentioned above, many of these proteins (e.g., c-myc, Apaf-1, FGF, XIAP, p53, VEGF and others) are master regulators of cell survival, proliferation or death. Expression of these proteins is usually controlled by multiple mechanisms operating at different levels, including transcription, RNA splicing, translation and protein localization and stability. Although it is common for many of these mechanisms to be switched on simultaneously, studies have been successful in discerning the importance of IRES-mediated translation in several cases.
IRES-mediated Translation in Angiogenesis
Angiogenesis is the physiological process of new blood vessel formation/growth from pre-existing vessels.80 In addition to its role in “normal” situations (e.g., embryonic development, etc.) angiogenesis also acts as an important stress-response mechanism that promotes repair of damaged tissues requiring an intensive supply of nutrients and oxygen.81 Angiogenesis also plays a key role in the transition of tumors from dormancy to malignancy.81 The vascular endothelial growth factor (VEGF) family of factors plays a pivotal role in angiogenesis by stimulating the proliferation, migration and proteolytic activity of endothelial cells.82 VEGF-A is the key member of the family and its expression is regulated by multiple mechanisms.83 The importance of tight control of VEGF expression is illustrated by data showing that either depletion (by 50%) or moderate overexpression (2–3-fold) of VEGF results in embryonic lethality due to improper vascularization (reviewed in ref. 83). Alternative splicing gives rise to at least nine different VEGF-A transcript variants, the functions of which have not yet been fully defined.83 Although five protein isoforms have been described, three isoforms are the most abundant (VEGF 121, VEGF 165 and VEGF 189). VEGF 121 (as compared to VEGF 165 and VEGF 189) possesses higher angiogenic activity since it is not able to bind to the extracellular matrix. Interestingly, in many pathological conditions (e.g., prostate tumors), expression of VEGF 121 predominates over all other VEGF-A isoforms.83 The mechanisms responsible for the high level of expression of VEGF 121 under these conditions are not fully understood; however, as discussed below, recent experiments indicate a role for IRES-mediated translational control.
The VEGF-A mRNA harbors two IRES-elements termed IRES-A and IRES-B. IRES-A directs translation starting from the AUG located 1,038 nt downstream of the 5′-end of the mRNA. IRES-B triggers translation starting at an upstream CUG codon (located 499 nt downstream of the 5′-end of the mRNA).83 Translation from the upstream CUG codon produces a 180 amino acid longer VEGF isoform called L-VEGF. L-VEGF is further proteolytically processed to an N-terminal fragment, named N-VEGF and a C-terminal fragment of a size similar to that of the AUG-initiated VEGF.83 N-terminal N-VEGF is intracellular, while C-terminal VEGF fragment of L-VEGF is a secreted VEGF isoform.83 Translation from the AUG codon generates secreted VEGF isoforms only. It appeared that a 121 amino acids long VEGF 121 isoform is mainly produced from the spliced mRNA via internal initiation events utilizing the CUG start codon (IRES-B) followed by proteolytic cleavage of the polypeptide precursor.83
Recent experiments have shown the expression from the VEGF-A IRES-B is negatively regulated by microRNA-16 (miR-16). Although the miR-16 target site is located in the 3′UTR of the VEGF-A mRNA, this microRNA appeared to preferentially suppress the activity of IRES-B, but not IRES-A. It should be noted that miR-16 was shown to act specifically on VEGF IRES-B-mediated translation and not on VEGF transcription or VEGF mRNA stability.84 Therefore, miR-16 can be viewed as the prototype of a new class of negative-regulatory cellular ITAFs. These findings suggest that the enhanced expression of VEGF 121 observed in prostate tumor cells might be due to loss of miR-16 translational control. Although the exact mechanism underlying the effect of miR-16 on VEGF-A translation remains unknown, it may involve the hnRNP L protein, which binds within the VEGF-A 3′-UTR at a site adjacent to the miR-16 target site85 and may thereby alter interaction of miR-16 with the mRNA. HnRNP L is a known ITAF66 and was recently shown to affect VEGF mRNA translation during hypoxia.86 Thus, miR-16 may function as an anti-angiogenic factor that represses IRES-mediated translation under hypoxic conditions. As has been mentioned the loss of miR-16 control may well explain increased VEGF-121 levels in prostate tumors in agreement with the hypoxic environment that will also favor internal initiation of VEGF mRNA translation at a CUG initiation codon, due to increased eIF2α phosphorylation (which compromises AUG initiation codon recognition). Overall, the example of IRES-regulated VEGF expression illustrates the significance of controlled internal initiation as a key element in the decision between two cellular fates—normal growth and malignant transformation.
The significance of IRES-mediated translation in angiogenesis during cancer development and progression is also highlighted by findings demonstrating that increased levels of the translation initiation factors eIF4G1 and 4E-BP-1 in inflammatory breast cancer (IBC) sensitizes the tumor cells to induce a hypoxic-stress response at higher oxygen levels than normal.87 Factor 4E-BP-1 is known to sequester eIF4E and thereby inhibit cap-dependent translation. Thus, increased levels of eIF4G1 and 4E-BP-1 cause a switch from cap-dependent to IRES-mediated translation, thus favoring expression of IRES-containing pro-angeogenic mRNAs.88 Among these mRNAs is the VEGF-A mRNA.39 Translation of VEGF-A under these conditions was shown to be induced in an eIF4G1-dependent manner.39 The exact mechanism(s) leading to the enhanced VEGF mRNA translation and/or particular VEGF isoform production regulated by eIF4G1 has yet to be determined. Although eIF4G1 is a canonical initiation factor, in this example it can be considered as an ITAF, since it enhances IRES-mediated translation in a manner independent of its function in the eIF4F-mediated cap-dependent translation pathway.
IRES-mediated Translation in Mitosis: Positive- and Negative-Regulatory ITAFs Control Cell Fate
Regulated IRES-mediated translation events have also been shown to play important roles in controlling the progression of cells through mitosis. Here, we discuss IRES-mediated synthesis of the CDK11/PITSLREp58 kinase, which is involved in spindle formation, and of the anti-apoptotic proteins BCL2 and CDK1.62,89
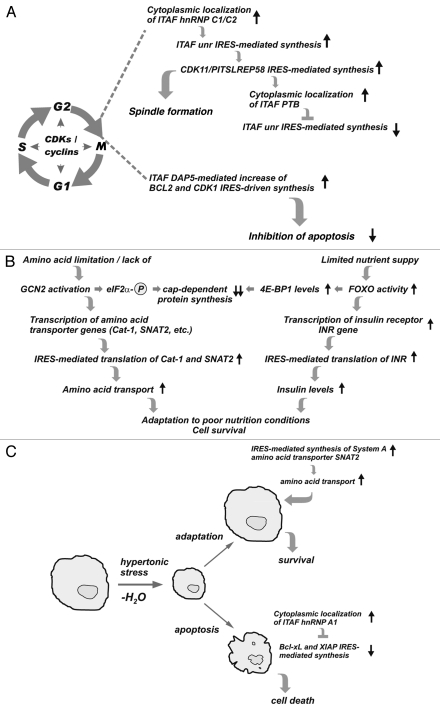
The central role of cyclin-dependent kinases (CDKs) in the regulation of mitosis is well established. However the role of IRES-mediated translation in this process is still emerging. Specific modulations of IRES activities were found to be central (Fig. 2A) for the progression of cells through the cell cycle, specifically at the G2/M transition when cap-dependent initiation is severely compromised. IRES-mediated translation of PITSLREp58, which is involved in spindle formation, was found to be under the control of several IRES-mediated events. First, cytoplasmic localization of the ITAF hnRNP C1/C2 stimulates IRES-mediated translation of another ITAF, unr. Unr, in turn, is required for enhanced IRES-mediated translation of PITSLREp58.62 Termination of this stimulatory cascade occurs through inhibition of unr synthesis by a feedback loop involving unr itself and cytoplasmic accumulation of a third ITAF, PTB.62 Therefore, progression through mitosis occurs via regulation of IRES-mediated translation involving changes in the relative cytoplasmic levels/activities of positive- and negative-regulatory ITAFs (Fig. 2A). The exact mechanism leading to these changes is unknown. However, it is likely to involve associated intracellular signaling events that may lead to post-translational modifications of ITAFs, thus altering their levels, subcellular localization, activities and/or interactions. In this regard, it was reported that AKT-mediated phosphorylation of hnRNP A1, an ITAF for the cyclin D1 IRES, represses its activity.69 In contrast, the unphosphorylated form of HnRNP A1 acts as a cyclin D1 IRES activator.69 These findings support the possibility that cell cycle progression signaling events influence ITAF activity and function. Future studies aimed at isolation of different ITAF cytoplasmic complexes and analysis of their modifications would likely help elucidate the mechanism(s) of IRES-mediated translational control in mitosis. The molecular mechanism(s) leading to repression of IRES-mediated translation might also involve sequestration of the corresponding IRES-containing mRNAs in stress granules (SGs), dynamic cytoplasmic structures comprised of translationally-arrested mRNAs.90,91 This possibility will be discussed further below with respect to IRES-mediated translation under hyperosmolar conditions.
Figure 2.
Adaptation to physiological or pathological stress involves regulation of IRES-mediated translation: model pathways where the competitive action of ITAFs determines cell fate. (A) Coordinated changes in the subcellular localization of ITAFs controls IRES-mediated translation of specific mRNAs during mitosis. (B) Adaptation to limited amino acid or nutrient availability involves transcriptional and translational mechanisms that amplify insulin signaling and amino acid transporter levels. (C) Adaptive (pro-survival) and apoptotic cellular responses to hyperosmolar stress involve regulation of IRES-mediated translation. Increased expression of the System A amino acid transporter SNAT2,109 restores cell volume and promotes survival. Cytoplasmic localization of the ITAF HnRNP A1 during hyperosmolar stress represses IRES-mediated translation of anti-apoptotic mRNAs and promotes apoptosis.
Another example of IRES-mediated control of protein synthesis with relevance to mitosis is the modulation of BCL-2 and CDK1 expression by the ITAF DAP5 (p96), a member of the eIF4G family of proteins. Synthesis of BCL-2 and CDK1 during mitosis protects cells from undergoing apoptosis.89 Interestingly, during apoptosis, DAP5 is cleaved to a shorter protein (p86) which is most likely a positive-acting ITAF for IRES-mediated translation of death-promoting mRNAs. In fact, cleaved DAP5 has been suggested to increase IRES-mediated synthesis of DAP5 itself.92 This provides an example of regulation of ITAF levels via proteolytic cleavage resulting in an impact on cell fate.
Nutritional Control of IRES-mediated Translation: Nutrient-Signaling is Amplified via Transcriptional and Translational Mechanisms
Nutrient availability is crucial for the growth and function of all organisms. Amino acid and insulin signaling are important pathways for nutrient sensing.93–95 It is well established that an excess supply of nutrients can cause obesity, insulin resistance and development of diabetes.96 On the other hand, shortage or imbalance of dietary amino acids can influence an organism's life span via specific nutrient-sensing mechanisms.97–99 At the molecular level, regulation of the levels and activity of the insulin receptor and amino acid transporters are important cellular mechanisms that respond to nutrient availability95 and studies have shown that controlled IRES activity plays a role in this regulation100 (Fig. 2B). For example, in Drosophila under poor nutritional conditions, FOXO-mediated transcriptional activation of 4E-BP causes a decrease in cap-dependent translation, while IRES-mediated translation is sustained.101 Interestingly, FOXO also induces transcription of the insulin receptor (INR) gene, resulting in accumulation of INR via IRES-mediated translation of the INR mRNA. These results suggest that transcriptional mechanisms together with IRES-mediated translation of a nutrient sensor (INR) may contribute to cell survival under poor nutritional conditions (Fig. 2B).
A recent study demonstrated that the human INR mRNA also contains an IRES in its 5′ UTR and provided some insight into the mechanisms that control the activity of this element.100 The human INR IRES element was shown to be positively regulated by insulin treatment of sub-confluent cultured cells.100 This suggests that IRES-mediated translation of INR in vivo might be regulated in a manner dependent on blood glucose levels. If true, this would imply that the plasma glucose levels in diabetic individuals could serve to sustain or increase IRES-mediated INR synthesis as a mechanism to compensate for peripheral insulin resistance. This mode of regulation could fit nicely at early stages of type II diabetes, when insulin resistance of peripheral tissues causes increased plasma glucose levels and the subsequent increase of insulin levels. In addition, the INR IRES was shown to be more active in the brain and cell lines of neuronal origin than other tissues and cell lines.100 This suggests that IRES-directed translation is important for maintaining INR expression and insulin sensitivity in tissues with low-efficiency cap-dependent translation, such as the brain. The work of Spriggs et al. also demonstrated that the ITAF PTB is required for function of the human INR IRES both in vitro and in vivo. It remains to be determined, however, if the expression or activity of PTB is affected by physiological changes in blood glucose levels. It was previously reported that PTB stabilizes the insulin mRNA in response to glucose and at the same time is phosphorylated by cAMP-dependent Protein Kinase A (PKA) and translocated from the nucleus to the cytoplasm of cells.102 It is therefore possible that insulin signaling in response to glucose involves coordinated regulation of both insulin and INR synthesis via mechanisms with shared elements. For example, glucose-induced PKA-mediated modification of PTB might contribute to INR synthesis by increasing the level of PTB in the cytoplasm and thereby stimulating IRES-dependent translation initiation. Additional studies will be required to test this and other hypotheses in order to fully decipher how hormone-nutrient interactions impact the regulation of ITAFs and IRES-directed translation.
Another example of nutritional control of transcription/translation via modulation of IRES activity is provided by the cellular response to limited amino acid availability.93,103 Amino acid depletion induces GCN2 kinase-mediated phosphorylation of eIF2α, leading to a global decrease in protein synthesis and induction of an adaptive survival program.103,104 This mechanism involves ATF4-mediated (downstream of GCN2 activation) induction of transcription of amino acid transporter genes105,106 and IRES-mediated translation of the resulting mRNAs,107,108 thus preparing cells to transport amino acids once they become available (Fig. 2B). This, as well as the example of the insulin system discussed above, highlights the importance of coordinated transcriptional and translational mechanisms in cellular responses to nutrient availability.
Regulation of IRES-mediated Translation by Hyperosmolar Conditions: eIF2α Phosphorylation at the Crossroads of Survival and Death
Acute hypertonic conditions cause cell shrinkage and induce an adaptive response leading to volume recovery.109 In contrast, severe and prolonged hypertonic stress induces apoptosis. This cell fate decision in response to hyperosmolar stress involves a translation “war” between IRES-containing pro-survival and pro-apoptotic mRNAs67,68,110 (Fig. 2C). We have recently shown that phosphorylation of the translation initiation factor eIF2α leads to apoptosis prevailing over cell survival in this “war.”71 Phosphorylation of eIF2α induces accumulation of hnRNP A1, an inhibitory ITAF for the anti-apoptotic XIAP and Bcl-xL IRESs, in cytoplasmic SGs.71 Interestingly, hyperosmolar stress decreased protein synthesis and induced SG formation independently of eIF2α phosphorylation.71 Therefore, HnRNP A1 may associate with a specific group of translationally repressed IRES-containing mRNAs during osmotic stress. We have proposed a model in which hnRNP A1 binds to the IRESs of anti-apoptotic mRNAs and sequesters them in SGs, thereby suppressing their translation71 (Fig. 2C). This would be expected to tip the balance towards activation of execution caspases and subsequent cleavage of survival factors. However, HnRNP A1 may also bind and inhibit translation of additional mRNAs that promote cell survival. This possibility is currently under investigation. Although the precise mechanisms of phospho-eIF2α signaling and HnRNP A1-mediated inhibition of IRES activity are not known, they are likely to involve post-translational modifications of HnRNP A1 and/or its interacting proteins. In future studies it will be interesting to identify ITAFs with positive or negative effects on IRES-mediated translation during osmotic stress and determine their contribution to cell fate decisions. By experimentally modulating the balance of pro-survival and pro-apoptotic ITAFs in the cytoplasm, it should be possible to study their target mRNAs and their contribution to cell fate decisions.
There are many more interesting examples of regulated IRES-mediated translation that we were not able to cover in this review. These include, (1) the IGF1 receptor IRES which is positively regulated by HnRNP C and negatively regulated by HuR;111 (2) the GATA 4 IRES which is active during cardiac hypertrophy;112 and (3) the PcG (Polycomb) IRES which is 91 bp long and requires intact eIF4Ffor its activity,113 suggesting that a structural component of the IRES might mimic the 5′-cap structure. As mentioned above, the cellular IRES found in the L-myc mRNA has also been recently shown to require eIF4F.34 It is therefore possible that an experimental system could be established to identify the eIF4F-dependent element in these two IRESs and determine potential structural/functional similarities within this element. This type of study could be assisted by the use of guanosine analogues such as ribavirin that can compete with the 5′-cap structure for the eIF4E binding site within the eIF4F complex.114
Conclusions and Future Directions
It is clear that IRES-mediated translation, while still considered an “alternative” pathway, is used relatively frequently under both normal physiological and pathological conditions. In this review, we have discussed the role of regulated IRES-mediated translation in angiogenesis, mitosis, nutritional and osmotic control. Despite the recent increase in recognition and study of these and other systems impacted by IRES-mediated translation, there remain many questions to be addressed: (1) Is the mechanism of cap-independent cellular mRNA translation different from viral IRES-mediated cap-independent translation? (2) Is the term internal ribosome entry site (IRES)-mediated cellular mRNA translation synonymous to cellular cap-independent mRNA translation? (3) Is there cellular cap-independent non-IRES-mediated mRNA translation60 which is regulated by factors similar to IRES Trans Acting Factors (ITAFs)? (4) Is the term bona fide cellular or viral IRES correct?
The original term “ribosome landing pad” which was suggested by Nahum Sonenberg and colleagues,115 adequately described the nature of the novel mechanism of the cap-independent viral IRES-mediated translation. Recent conflicting reports27,60 urge us to rethink on the cap-independent mechanisms of cellular mRNA translation. We believe, however, that one should follow the Ockham's razor principle and among several hypotheses, chose the one that is supported the most by experimental data, without the necessity of multiplying entities. We thus believe that the term cellular IRES-mediated translation may yet provide an adequate description of the complex cap-independent translation events taking place under the conditions of reduced cap-dependent translation.
In this review article, we suggest that the focus of the future studies in the field should be on the physiological significance of the cellular IRES-mediated translation. We believe that this may help to clarify many issues related to the mechanism(s) of the cellular cap-independent translation and may lead us to the discovery of new candidate mRNAs, whose cap-independent IRES-mediated translation may be facilitated/driven by group(s) of common regulatory ITAFs. Identification of common positive- or negative-regulatory ITAFs can further lead to isolation of groups of mRNAs that are regulated via IRES elements and allow determination of the composition of IRES-protein complexes. Identification of these proteins will facilitate studies of reconstituted cell-free experimental systems for determining the mechanism of ribosome recruitment (ITAF-mediated) to cellular IRESs.
The requirement for canonical factors in IRES-mediated translation is another key question that should be systematically addressed. The availability of techniques for isolation of polysome-associated mRNAs116 and stable isotope labeling of cells117 should enable identification of mRNAs that are translated under conditions of prevailing IRES-mediated translation and relative quantification of the encoded proteins. A similar approach could be followed for IRES-containing mRNAs that are translationally repressed. An appealing idea is that inhibitory ITAFs may bind their corresponding IRESs, or IRES-containing mRNAs and sequester them in SGs. Because SGs are dynamic structures, increased cytoplasmic levels of positive-acting ITAFs might reverse this inhibition by interacting with the IRES-containing mRNAs.
Overall, we envision that future studies of the mechanism of IRES-mediated translation will focus on specific pathways in vivo with parallel in vitro analysis of identified factors. Understanding the role of the IRES pathway and upstream and downstream regulatory mechanisms under different cellular conditions has the potential to identify new therapeutic targets.
Acknowledgements
This work was supported by National Institute of Health grants DK060596 and DK053307 (to M.H.) and in part by National American Heart Association grant 0730120N and the Human Frontier Science Program (HFSP) RGP0024 (to A.A.K.). Drs. Martin Snider, Dawid Krokowski, Elena Bevilacqua and Mithu Majumder are gratefully acknowledged for helpful discussions and many valuable suggestions. We are also grateful to Dr. Patricia Stanhope Baker for help in preparation of the manuscript.
References
- 1.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 2.Rodnina MV, Wintermeyer W. Recent mechanistic insights into eukaryotic ribosomes. Curr Opin Cell Biol. 2009;21:435–443. doi: 10.1016/j.ceb.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 3.Hershey JWB, Merrick WC. The pathway and mechanism of initiation of protein synthesis. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 33–38. [Google Scholar]
- 4.Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev. 2005;69:101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Myasnikov AG, Simonetti A, Marzi S, Klaholz BP. Structure-function insights into prokaryotic and eukaryotic translation initiation. Curr Opin Struct Biol. 2009;19:300–309. doi: 10.1016/j.sbi.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinnebusch AG. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Derry MC, Yanagiya A, Martineau Y, Sonenberg N. Regulation of poly(A)-binding protein through PABP-interacting proteins. Cold Spring Harb Symp Quant Biol. 2006;71:537–543. doi: 10.1101/sqb.2006.71.061. [DOI] [PubMed] [Google Scholar]
- 10.Jackson RJ. A comparative view of initiation site selection mechanisms. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 127–183. Translational Control of Gene Expression. [Google Scholar]
- 11.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 12.Balvay L, Soto Rifo R, Ricci EP, Decimo D, Ohlmann T. Structural and functional diversity of viral IRESes. Biochim Biophys Acta. 2009;1789:542–557. doi: 10.1016/j.bbagrm.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Hellen CU. IRES-induced conformational changes in the ribosome and the mechanism of translation initiation by internal ribosomal entry. Biochim Biophys Acta. 2009;1789:558–570. doi: 10.1016/j.bbagrm.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pisarev AV, Shirokikh NE, Hellen CU. Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites. C R Biol. 2005;328:589–605. doi: 10.1016/j.crvi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Kieft JS. Viral IRES RNA structures and ribosome interactions. Trends Biochem Sci. 2008;33:274–283. doi: 10.1016/j.tibs.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komar AA, Hatzoglou M. Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J Biol Chem. 2005;280:23425–23428. doi: 10.1074/jbc.R400041200. [DOI] [PubMed] [Google Scholar]
- 17.Kozak M. Alternative ways to think about mRNA sequences and proteins that appear to promote internal initiation of translation. Gene. 2003;30:1–23. doi: 10.1016/s0378-1119(03)00774-1. [DOI] [PubMed] [Google Scholar]
- 18.Kozak M. A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 2005;33:6593–6602. doi: 10.1093/nar/gki958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Eden ME, Byrd MP, Sherrill KW, Lloyd RE. Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA. 2004;10:720–730. doi: 10.1261/rna.5225204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mokrejs M, Masek T, Vopálensky V, Hlubucek P, Delbos P, Pospísek M. IRESite—a tool for the examination of viral and cellular internal ribosome entry sites. Nucleic Acids Res. 2010;38:131–136. doi: 10.1093/nar/gkp981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park EH, Lee JM, Blais JD, Bell JC, Pelletier J. Internal translation initiation mediated by the angiogenic factor Tie2. J Biol Chem. 2005;280:20945–20953. doi: 10.1074/jbc.M412744200. [DOI] [PubMed] [Google Scholar]
- 22.Pinkstaff JK, Chappell SA, Mauro VP, Edelman GM, Krushel LA. Internal initiation of translation of five dendritically localized neuronal mRNAs. Proc Natl Acad Sci USA. 2001;98:2770–2775. doi: 10.1073/pnas.051623398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spriggs KA, Stoneley M, Bushell M, Willis AE. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol Cell. 2008;100:27–38. doi: 10.1042/BC20070098. [DOI] [PubMed] [Google Scholar]
- 24.Lewis SM, Holcik M. For IRES trans-acting factors, it is all about location. Oncogene. 2008;27:1033–1035. doi: 10.1038/sj.onc.1210777. [DOI] [PubMed] [Google Scholar]
- 25.Graber TE, Holcik M. Cap-independent regulation of gene expression in apoptosis. Mol Biosyst. 2007;3:825–834. doi: 10.1039/b708867a. [DOI] [PubMed] [Google Scholar]
- 26.Filbin ME, Kieft JS. Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol. 2009;19:267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreev DE, Dmitriev SE, Terenin IM, Prassolov VS, Merrick WC, Shatsky IN. Differential contribution of the m7G-cap to the 5′ end-dependent translation initiation of mammalian mRNAs. Nucleic Acids Res. 2009;37:6135–6147. doi: 10.1093/nar/gkp665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komar AA, Lesnik T, Cullin C, Merrick WC, Trachsel H, Altmann M. Internal initiation drives the synthesis of Ure2 protein lacking the prion domain and affects [URE3] propagation in yeast cells. EMBO J. 2003;22:1199–1209. doi: 10.1093/emboj/cdg103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grover R, Candeias MM, Fähraeus R, Das S. p53 and little brother p53/47: linking IRES activities with protein functions. Oncogene. 2009;28:2766–2772. doi: 10.1038/onc.2009.138. [DOI] [PubMed] [Google Scholar]
- 30.Baird SD, Lewis SM, Turcotte M, Holcik M. A search for structurally similar cellular internal ribosome entry sites. Nucleic Acids Res. 2007;35:4664–4677. doi: 10.1093/nar/gkm483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le SY, Chen JH, Konings D, Maizel JV., Jr Discovering well-ordered folding patterns in nucleotide sequences. Bioinformatics. 2003;19:354–361. doi: 10.1093/bioinformatics/btf826. [DOI] [PubMed] [Google Scholar]
- 32.Xia X, Holcik M. Strong eukaryotic IRESs have weak secondary structure. PLoS One. 2009;4:4136. doi: 10.1371/journal.pone.0004136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belsham GJ, Jackson RJ. Translation initiation on picornavirus RNA. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 869–900. [Google Scholar]
- 34.Spriggs KA, Cobbold LC, Jopling CL, Cooper RE, Wilson LA, Stoneley M, et al. Canonical initiation factor requirements of the Myc family of internal ribosome entry segments. Mol Cell Biol. 2009;29:1565–1574. doi: 10.1128/MCB.01283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chappell SA, Edelman GM, Mauro VP. Biochemical and functional analysis of a 9-nt RNA sequence that affects translation efficiency in eukaryotic cells. Proc Natl Acad Sci USA. 2000;97:1536–1541. doi: 10.1073/pnas.0308759101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meng Z, Jackson NL, Shcherbakov OD, Choi H, Blume SW. The human IGF1R IRES likely operates through a Shine-Dalgarno-like interaction with the G961 loop (E-site) of the 18S rRNA and is kinetically modulated by a naturally polymorphic polyU loop. J Cell Biochem. 2010;110:531–544. doi: 10.1002/jcb.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johannes G, Carter MS, Eisen MB, Brown PO, Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc Natl Acad Sci USA. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spriggs KA, Bushell M, Mitchell SA, Willis AE. Internal ribosome entry segment-mediated translation during apoptosis: the role of IRES-trans-acting factors. Cell Death Differ. 2005;12:585–591. doi: 10.1038/sj.cdd.4401642. [DOI] [PubMed] [Google Scholar]
- 39.Silvera D, Arju R, Darvishian F, Levine PH, Zolfaghari L, Goldberg J, et al. Essentail role for eIF4G1 overexpression in the pathigenesis of inflammatory breast cancer. Nat Cell Biol. 2009;11:903–908. doi: 10.1038/ncb1900. [DOI] [PubMed] [Google Scholar]
- 40.Bellodi C, Kopmar N, Ruggero D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. EMBO J. 2010;29:1865–1876. doi: 10.1038/emboj.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schrufer TL, Antonetti DA, Sonenberg N, Kimball SR, Gardner TW, Jefferson LS. Ablation of 4E-BP1/2 prevents hyperglycemia-mediated induction of VEGF expression in the rodent retina and in Muller cells in culture. Diabetes. 2010;59:2107–2116. doi: 10.2337/db10-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prevot D, Darlix JL, Ohlmann T. Conducting the initiation of protein synthesis: the role of eIF4G. Biol Cell. 2003;95:141–156. doi: 10.1016/s0248-4900(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 43.Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, et al. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol. 2006;2:213–220. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- 44.Kolupaeva VG, Lomakin IB, Pestova TV, Hellen CU. Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosomal entry site. Mol Cell Biol. 2003;23:687–698. doi: 10.1128/MCB.23.2.687-698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borman AM, Michel YM, Kean KM. Detailed analysis of the requirements of hepatitis A virus internal ribosome entry segment for the eukaryotic initiation factor complex eIF4F. J Virol. 2001;75:7864–7871. doi: 10.1128/JVI.75.17.7864-7871.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ali IK, McKendrick L, Morley SJ, Jackson RJ. Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J Virol. 2001;75:7854–7863. doi: 10.1128/JVI.75.17.7854-7863.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allam H, Ali M. Initiation factor eIF2-independent mode of c-Src mRNA translation occurs via an internal ribosome entry site. J Biol Chem. 2010;285:5713–5725. doi: 10.1074/jbc.M109.029462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clemens MJ. Initiation factor eIF2alpha phosphorylation in stress responses and apoptosis. Prog Mol Subcell Biol. 2001;27:57–89. doi: 10.1007/978-3-662-09889-9_3. [DOI] [PubMed] [Google Scholar]
- 50.Gerlitz G, Jagus R, Elroy-Stein O. Phosphorylation of initiation factor-2 is required for activation of internal translation initiation during cell differentiation. Eur J Biochem. 2002;269:2810–2819. doi: 10.1046/j.1432-1033.2002.02974.x. [DOI] [PubMed] [Google Scholar]
- 51.Sonenberg N, Dever TE. Eukaryotic translation initiation factors and regulators. Curr Opin Struct Biol. 2003;13:56–63. doi: 10.1016/s0959-440x(03)00009-5. [DOI] [PubMed] [Google Scholar]
- 52.Tinton SA, Schepens B, Bruynooghe Y, Beyaert R, Cornelis S. Regulation of the cell cycle dependent internal ribosome entry site of the PITSLRE protein kinase: roles of Unr (upstream of N ras) protein and phosphorylated translation initiation factor eIF-2alpha. Biochem J. 2005;385:155–163. doi: 10.1042/BJ20040963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 54.Vyas J, Elia A, Clemens MJ. Inhibition of the protein kinase PKR by the internal ribosome entry site of hepatitis C virus genomic RNA. RNA. 2003;9:858–870. doi: 10.1261/rna.5330503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CU. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J. 2008;27:1060–1072. doi: 10.1038/emboj.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat Struct Mol Biol. 2008;15:836–841. doi: 10.1038/nsmb.1445. [DOI] [PubMed] [Google Scholar]
- 57.Adams SL, Safer B, Anderson WF, Merrick WC. Eukaryotic initiation complex formation. Evidence for two distinct pathways. J Biol Chem. 1975;250:9083–9089. [PubMed] [Google Scholar]
- 58.Dmitriev SE, Terenin IM, Andreev DE, Ivanov PA, Dunaevsky JE, Merrick WC, et al. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol Chem. 2010;285:26779–26787. doi: 10.1074/jbc.M110.119693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shatsky IN, Dmitriev SE, Terenin IM, Andreev DE. Cap- and IRES-independent scanning mechanism of translation initiation as an alternative to the concept of cellular IRESs. Mol Cells. 2010;30:285–293. doi: 10.1007/s10059-010-0149-1. [DOI] [PubMed] [Google Scholar]
- 61.Riley A, Jordan LE, Holcik M. Distinct 5′ UTRs regulate XIAP expression under normal growth conditions and during cellular stress. Nucleic Acids Res. 2010;38:4665–4674. doi: 10.1093/nar/gkq241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schepens B, Tinton SA, Bruynooghe Y, Parthoens E, Haegman M, Beyaert R, et al. A role for hnRNP C1/C2 and Unr in internal initiation of translation during mitosis. EMBO J. 2007;26:158–169. doi: 10.1038/sj.emboj.7601468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 64.Ungureanu NH, Cloutier M, Lewis SM, de Silva N, Blais JD, Bell JC, et al. Internal ribosome entry site-mediated translation of Apaf-1, but not XIAP, is regulated during UV-induced cell death. J Biol Chem. 2006;281:15155–15163. doi: 10.1074/jbc.M511319200. [DOI] [PubMed] [Google Scholar]
- 65.Semler BL, Waterman ML. IRES-mediated pathways to polysomes: nuclear versus cytoplasmic routes. Trends Microbiol. 2008;16:1–5. doi: 10.1016/j.tim.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 66.Majumder M, Yaman I, Gaccioli F, Zeenko VV, Wang C, Caprara MG, et al. The hnRNA-binding proteins hnRNP L and PTB are required for efficient translation of the cat-1 arginine/lysine transporter mRNA during amino acid starvation. Mol Cell Biol. 2009;29:2899–2912. doi: 10.1128/MCB.01774-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cammas A, Pileur F, Bonnal S, Lewis SM, Lévêque N, Holcik M, et al. Cytoplasmic relocalization of heterogeneous nuclear ribonucleoprotein A1 controls translation initiation of specific mRNAs. Mol Biol Cell. 2007;18:5048–5059. doi: 10.1091/mbc.E07-06-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lewis SM, Veyrier A, Hosszu Ungureanu N, Bonnal S, Vagner S, Holcik M. Subcellular relocalization of a trans-acting factor regulates XIAP IRES-dependent translation. Mol Biol Cell. 2007;18:1302–1311. doi: 10.1091/mbc.E06-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jo OD, Martin J, Bernath A, Masri J, Lichtenstein A, Gera J. Heterogeneous nuclear ribonucleoprotein A1 regulates cyclin D1 and c-myc internal ribosome entry site function through Akt signaling. J Biol Chem. 2008;283:23274–23287. doi: 10.1074/jbc.M801185200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi Y, Frost PJ, Hoang BQ, Benavides A, Sharma S, Gera JF, et al. IL-6-induced stimulation of c-myc translation in multiple myeloma cells is mediated by myc internal ribosome entry site function and the RNA-binding protein, hnRNP A1. Cancer Res. 2008;68:10215–10222. doi: 10.1158/0008-5472.CAN-08-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bevilacqua E, Wang X, Majumder M, Gaccioli F, Yuan CL, Wang C, et al. eIF2α phosphorylation tips the balance to apoptosis during osmotic stress. J Biol Chem. 2010;285:17098–17111. doi: 10.1074/jbc.M110.109439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gilbert WV, Zhou K, Butler TK, Doudna JA. Cap-independent translation is required for starvation-induced differentiation in yeast. Science. 2007;317:1224–1227. doi: 10.1126/science.1144467. [DOI] [PubMed] [Google Scholar]
- 73.Smith RW, Gray NK. Poly(A)-binding protein (PABP): a common viral target. Biochem J. 2010;426:1–12. doi: 10.1042/BJ20091571. [DOI] [PubMed] [Google Scholar]
- 74.Grosset C, Chen CY, Xu N, Sonenberg N, Jacquemin-Sablon H, Shyu AB. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell. 2000;103:29–40. doi: 10.1016/s0092-8674(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 75.Chang TC, Yamashita A, Chen CY, Yamashita Y, Zhu W, Durdan S, et al. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes Dev. 2004;18:2010–2023. doi: 10.1101/gad.1219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, et al. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 77.Coleman J, Miskimins WK. Structure and activity of the internal ribosome entry site within the human p27Kip1 5′-untranslated region. RNA Biol. 2009;6:84–89. doi: 10.4161/rna.6.1.7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pisarev AV, Unbehaun A, Hellen CU, Pestova TV. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 2007;430:147–177. doi: 10.1016/S0076-6879(07)30007-4. [DOI] [PubMed] [Google Scholar]
- 79.Kolupaeva VG, de Breyne S, Pestova TV, Hellen CU. In vitro reconstitution and biochemical characterization of translation initiation by internal ribosomal entry. Methods Enzymol. 2007;430:409–439. doi: 10.1016/S0076-6879(07)30016-5. [DOI] [PubMed] [Google Scholar]
- 80.Costa C, Soares R, Schmitt F. Angiogenesis: now and then. APMIS. 2004;112:402–412. doi: 10.1111/j.1600-0463.2004.apm11207-0802.x. [DOI] [PubMed] [Google Scholar]
- 81.Walsh DA. Pathophysiological mechanisms of angiogenesis. Adv Clin Chem. 2007;44:187–221. doi: 10.1016/s0065-2423(07)44006-9. [DOI] [PubMed] [Google Scholar]
- 82.Ferrara N. Vascular endothelial growth factor. Arterioscler Thromb Vasc Biol. 2009;29:789–791. doi: 10.1161/ATVBAHA.108.179663. [DOI] [PubMed] [Google Scholar]
- 83.Bastide A, Karaa Z, Bornes S, Hieblot C, Lacazette E, Prats H, et al. An upstream open reading frame within an IRES controls expression of a specific VEGF-A isoform. Nucleic Acids Res. 2008;36:2434–2445. doi: 10.1093/nar/gkn093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Karaa ZS, Iacovoni JS, Bastide A, Lacazette E, Touriol C, Prats H. The VEGF IRESes are differentially susceptible to translation inhibition by miR-16. RNA. 2009;15:249–254. doi: 10.1261/rna.1301109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Du M, Roy KM, Zhong L, Shen Z, Meyers HE, Nichols RC. VEGF gene expression is regulated post-transcriptionally in macrophages. FEBS J. 2006;273:732–745. doi: 10.1111/j.1742-4658.2006.05106.x. [DOI] [PubMed] [Google Scholar]
- 86.Ray PS, Jia J, Yao P, Majumder M, Hatzoglou M, Fox PL. A stress-responseive RNA switch regulates VEGFA expression. Nature. 2009;457:915–919. doi: 10.1038/nature07598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Silvera D, Schneider RJ. Inflammatory breast cancer cells are constitutively adapted to hypoxia. Cell Cycle. 2009;8:3091–3096. doi: 10.4161/cc.8.19.9637. [DOI] [PubMed] [Google Scholar]
- 88.Braunstein S, Karpisheva K, Pola C, Goldberg J, Hochman T, Yee H, et al. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol Cell. 2007;28:501–512. doi: 10.1016/j.molcel.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 89.Marash L, Liberman N, Henis-Korenblit S, Sivan G, Reem E, Elroy-Stein O, et al. DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Mol Cell. 2008;30:447–459. doi: 10.1016/j.molcel.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 90.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 92.Henis-Korenblit S, Shani G, Sines T, Marash L, Shohat G, Kimchi A. The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc Natl Acad Sci USA. 2002;99:5400–5405. doi: 10.1073/pnas.082102499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Harding HP, Zhang Y, Zeng Y, Novoa I, Lu P, Calfon M, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 94.Reynolds B, Laynes R, Ogmundsdottir MH, Boyd CAR, Goberdhan DCI. Amino acid transporters and nutrient-sensing mechanisms: new targets for treating insulin-linked disorders? Biochem Soc Trans. 2007;35:1215–1217. doi: 10.1042/BST0351215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hietakangas V, Cohen SM. Regulation of tissue growth through nutrient sensing. Annu Rev Genet. 2009;43:389–410. doi: 10.1146/annurev-genet-102108-134815. [DOI] [PubMed] [Google Scholar]
- 96.Boden G. Endoplasmic reticulum stress: another link between obesity and insulin resistance/inflammation? Dabetes. 2009;58:518–519. doi: 10.2337/db08-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hao S, Sharp JW, Ross-Inta CM, McDaniel BJ, Anthony TG, Wek RC, et al. Uncharged tRNA and sensing of amino acid deficiency in mammalian piriform cortex. Science. 2005;307:1776–1778. doi: 10.1126/science.1104882. [DOI] [PubMed] [Google Scholar]
- 98.Grandison CR, Piper MDW, Partridge L. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature. 2009;462:1061–1064. doi: 10.1038/nature08619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab. 2009;20:436–443. doi: 10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Spriggs KA, Cobbold LC, Ridley SH, Coldwell M, Bottley A, Bushell M, et al. The human insulin receptor mRNA contains a functional internal ribosome entry segment. Nucleic Acids Res. 2009;37:5881–5893. doi: 10.1093/nar/gkp623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Marr MT, 2nd, D'Alessio JA, Puig O, Tjian R. IRES-mediated functional coupling of transcription and translation amplifies insulin receptor feedback. Genes Dev. 2007;21:175–183. doi: 10.1101/gad.1506407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fred RG, Tillmar L, Welsh N. The role of PTB in insulin mRNA stability control. Curr Diabetes Rev. 2006;2:363–366. doi: 10.2174/157339906777950570. [DOI] [PubMed] [Google Scholar]
- 103.Bruhat A, Cherasse Y, Chaveroux C, Maurin AC, Jousse C, Fafournoux P. Amino acids as regulators of gene expression in mammals: molecular mechanisms. Biofactors. 2009;35:249–257. doi: 10.1002/biof.40. [DOI] [PubMed] [Google Scholar]
- 104.Zhang P, McGrath BC, Reinert J, Olsen DS, Lei L, Gill S, et al. The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol. 2002;22:6681–6688. doi: 10.1128/MCB.22.19.6681-6688.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hatzoglou M, Fernandez J, Yaman I, Closs E. Regulation of cationic amino acid transport: the story of the CAT-1 transporter. Annu Rev Nutr. 2004;24:377–399. doi: 10.1146/annurev.nutr.23.011702.073120. [DOI] [PubMed] [Google Scholar]
- 106.Palii SS, Thiaville MM, Pan YX, Zhong C, Kilberg MS. Characterization of the amino acid response element within the human sodium-coupled neutral amino acid transporter 2 (SNAT2) System A transporter gene. Biochem J. 2006;395:517–527. doi: 10.1042/BJ20051867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yaman I, Fernandez J, Liu H, Caprara M, Komar AA, Koromilas AE, et al. The zipper model of translational control: a small upstream OFR is the switch that controls structural remodeling of an mRNA leader. Cell. 2004;113:519–531. doi: 10.1016/s0092-8674(03)00345-3. [DOI] [PubMed] [Google Scholar]
- 108.Gaccioli F, Huang CC, Wang C, Bevilacqua E, Franchi-Gazzola R, Gazzola GC, et al. Amino acid starvation induces the SNAT2 neutral amino acid transporter by a mechanism that involves eukaryotic initiation factor 2alpha phosphorylation and cap-independent translation. J Biol Chem. 2006;281:17929–17940. doi: 10.1074/jbc.M600341200. [DOI] [PubMed] [Google Scholar]
- 109.Franchi-Gazzola R, Dall'Asta V, Sala R, Visigalli R, Bevilacqua E, Gaccioli F, et al. The role of the neutral amino acid transporter SNAT2 in cell volume regulation. Acta Physiol. 2006;187:273–283. doi: 10.1111/j.1748-1716.2006.01552.x. [DOI] [PubMed] [Google Scholar]
- 110.Cammas A, Lewis SM, Vagner S, Holcik M. Post-transcriptional control of gene expression through subcellular relocalization of mRNA binding proteins. Biochem Pharmacol. 2008;76:1395–1403. doi: 10.1016/j.bcp.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 111.Meng Z, Jackson NL, Choi H, King PH, Emanuel PD, Blume SW. Alterations in RNA-binding activities of IRES-regulatory proteins as a mechanism for physiological variability and pathological dysregulation of IGF-IR translational control in human breast tumor cells. J Cell Physiol. 2008;217:172–183. doi: 10.1002/jcp.21486. [DOI] [PubMed] [Google Scholar]
- 112.Sharma A, Masri J, Jo OD, Bernath A, Martin J, Funk A, et al. Protein kinase C regulates internal initiation of translation of the GATA-4 mRNA following vasopressin-induced hypertrophy of cardiac myocytes. J Biol Chem. 2007;282:9505–9516. doi: 10.1074/jbc.M608874200. [DOI] [PubMed] [Google Scholar]
- 113.Boutsma E, Noback S, van Lohuizen M. The Polycomb protein and E3 ubiquitin ligase Ring1B harbors an IRES in its highly conserved 5′ UTR. PLoS One. 2008;3:2322. doi: 10.1371/journal.pone.0002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kentsis A, Topisirovic I, Culjkovic B, Shao L, Borden KL. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc Natl Acad Sci USA. 2004;101:18105–18110. doi: 10.1073/pnas.0406927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 116.Thomas JD, Johannes GJ. Identification of mRNAs that continue to associate with polysomes during hypoxia. RNA. 2007;13:1116–1131. doi: 10.1261/rna.534807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Doherty MK, Hammond DE, Clague MJ, Gaskell SJ, Beynon RJ. Turnover of the human proteome: determination of protein intracellular stability by dynamic SILAC. J Proteome Res. 2009;8:104–112. doi: 10.1021/pr800641v. [DOI] [PubMed] [Google Scholar]