Abstract
DNA double-strand breaks (DSBs) arise through both replication errors and from exogenous events such as exposure to ionizing radiation. DSBs are potentially lethal, and cells have evolved a highly conserved mechanism to detect and repair these lesions. This mechanism involves phosphorylation of histone H2AX (γH2AX) and the loading of DNA repair proteins onto the chromatin adjacent to the DSB. It is now clear that the chromatin architecture in the region surrounding the DSB has a critical impact on the ability of cells to mount an effective DNA damage response. DSBs promote the formation of open, relaxed chromatin domains which are spatially confined to the area surrounding the break. These relaxed chromatin structures are created through the coupled action of the p400 SWI/SNF ATPase and histone acetylation by the Tip60 acetyltransferase. The resulting destabilization of nucleosomes at the DSB by Tip60 and p400 is required for ubiquitination of the chromatin by the RNF8 ubiquitin ligase, and for the subsequent recruitment of the brca1 complex. Chromatin dynamics at DSBs can therefore exert a powerful influence on the process of DSB repair. Further, there is emerging evidence that the different chromatin structures in the cell, such as heterochromatin and euchromatin, utilize distinct remodeling complexes and pathways to facilitate DSB. The processing and repair of DSB is therefore critically influenced by the nuclear architecture in which the lesion arises.
Key words: p400, chromatin remodeling, DNA repair, NuA4, H2AX, acetylation, nucleosome, tip60
Damage to cellular DNA can occur through multiple pathways, including exposure to genotoxic agents, the production of endogenous reactive oxygen species or errors which arise during DNA replication. To combat this continuous assault on the genome, mammalian cells have evolved multiple DNA repair pathways. The most challenging lesions to repair are DSBs, which physically cleave the DNA strand. DSBs can occur through exposure to IR, the collapse of replication forks or during the processing of certain types of DNA damage. Over the last 20 years, a clear picture of how the cell detects and repairs DSBs has emerged.1,2 The earliest event in the cell's response to DSBs is the rapid recruitment of the ATM kinase, followed by the phosphorylation of histone H2AX (termed γH2AX) on large chromatin domains which extend for 100's of kilobases on either side of the DSB.3 The mdc1 scaffold protein is then recruited to γH2AX,4 providing a docking platform for the recruitment and retention of additional DNA repair proteins, including the MRN complex, the RNF8 ubiquitin ligase and the brca1 and 53BP1 proteins, onto the chromatin at DSBs.5–7 Eventually, this spreading of DNA repair proteins along the chromatin from the DSB leads to the formation of IRIF, which can be visualized by immunofluorescent techniques. DSBs are then repaired by NHEJ, in which broken DNA ends are directly religated, or by HR, using the undamaged sister chromatid (present during S-phase) as a template. A defining characteristic of DSB repair is the dominant role that chromatin structure plays in the detection and repair of these lesions. In this review, we will examine recent work exploring how remodeling of the chromatin structure adjacent to DSBs plays a key role in the repair of DSBs.
Chromatin Structure
The basic packing unit of chromatin is the nucleosome, which contains 146 bp of DNA wrapped in approximately 1.7 turns around the surface of the nucleosome.8,9 Each nucleosome is composed of a histone octamer, constructed from a central core containing two H3-H4 dimers which are surrounded by two H2A-H2B dimers.8 The n-terminal tails of histones extend outwards from the nucleosome particle, and are the sites for regulatory modification by acetylation, methylation and phosphorylation. Nucleosomes form linear 10 nm strings which can be stacked together to form packed 30 nm arrays and other higher order structures.8,9 The packaging of chromatin is variable, with euchromatin representing open, gene rich, transcriptionally active regions. Typically, histones within euchromatin are highly acetylated and are methylated on lysines 4 and 36 of histone H3.10,11 In contrast, heterochromatin, which constitutes 15–25% of mammalian chromatin, represents condensed regions with low gene density but high levels of repetitive sequences.12,13 Histones within heterochromatin have low levels of acetylation but high levels of histone H3 methylated on lysines 9 and 36.10,13–15
Alterations in chromatin structure are carried out by two linked processes—large motor ATPases, which directly alter chromatin structure, and dynamic regulation of histone post-translational modifications. Motor ATPases are the key functional components of chromatin remodeling complexes. Chromatin remodeling complexes have distinct functional activities, and can: (1) evict nucleosomes from the chromatin, creating free DNA sequences; (2) carry out nucleosome sliding, in which the remodeling ATPase shifts the nucleosome position relative to the DNA, exposing (or burying) a given DNA sequence; or (3) promote histone exchange, in which specific histone variants are exchanged onto the chromatin (reviewed in ref. 16). Post-translational modification of histones (e.g., by acetylation, methylation, phosphorylation or ubiquitination) can create binding sites for the recruitment of chromatin modifying proteins and alter the stability of the interaction between DNA and histones.8,17 By combining chromatin remodeling with histone modification, cells can regulate the dynamic architecture of the chromatin.
Chromatin Structure and DSB Repair
DSBs are associated with changes in chromatin architecture which operate at several layers of chromatin organization. These include changes occurring directly at the DSB, alterations in nucleosome organization on chromatin domains adjacent to the DSB, as well as structural changes which propagate across the entire genome. Here, we will briefly discuss the major types of changes in chromatin organization, and then review recent work on the alterations in nucleosome organization at DSBs.
Nucleosome eviction from DSBs.
A key step in DSB repair is the creation of nucleosome free regions on the DNA at the DSB. These regions were detected using ChIP analysis, and demonstrated a rapid loss of histones from the chromatin within a few 100s of base pairs of the DSB.18–20 This creates short stretches of nucleosome free DNA, extending for several nucleosomes lengths on either side of break. Nucleosome eviction is an active process which requires both the MRN complex and the Ino80 chromatin remodelor.18–20 The MRN complex contains both nuclease (mre11) and ATPase (rad50) activity, and is rapidly recruited to and binds DNA ends at the DSB.21 Whether MRN directly evicts nucleosomes from the DSB or recruits a separate remodeling complex to achieve this is not currently known. However, the localized loss of histones adjacent to the DSB is thought to play a critical role in processing the DNA ends for NHEJ, or to create ssDNA intermediates for HR repair of the DSB.22 The active displacement of intact nucleosomes from the vicinity of the DSB is therefore essential to promote repair.
Global relaxation and heterochromatin structure.
Some of the earliest work indicating alterations in chromatin structure after DNA damage stems from the observation that chromatin is hypersensitive to nuclease digestion following exposure to IR,23–26 indicating increased accessibility of the nuclease to the linker DNA between nucleosomes. The extent of the increased sensitivity to nuclease digestion indicates that the decrease in nucleosome compaction in response to DSBs affects a significant fraction of the chromatin. Subsequent work demonstrated that the ATM-dependent phosphorylation of the kap1 heterochromatin binding protein was required for this global chromatin relaxation.25 Phosphorylated kap-1 is located at DSBs, supporting a key role for phosphorylated kap-1 in the repair of DSBs.25,27 Kap-1 is a transcriptional repressor which forms complexes with HDACs, histone methyltransferases and HP1, promoting the formation of repressed, heterochromatic domains.28,29 Recent studies demonstrated that phosphorylation of kap-1 by ATM is critical for DSB repair within heterochromatin.27,30 These workers demonstrated that DSB repair was slower in heterochromatic regions, and that unrepaired DSBs persisted at the heterochromatin boundaries in the absence of either ATM or kap-1 phosphorylation.27,30,31 This implies that phosphorylation of kap-1 relaxes heterochromatin structure and promotes efficient DSB repair. Currently, it is unclear how phosphorylation of kap1 by ATM alters heterochromatin structure, since phospho-kap1 remains associated with the chromatin.27 One potential mechanism is that phosphorylation of kap-1 alters its interaction with other repressive proteins, such as HDACs and methyltransferases,28,29,32 shifting the balance towards a less repressed chromatin structure. Further, other heterochromatin binding proteins, including HP1 family members, exhibit changes in phosphorylation and chromatin association following DSB production.33–35 These results are consistent with the idea that the compacted, repressive structure of heterochromatin requires a unique pathway, involving phosphorylation of kap-1 and changes in HP1 distribution, to alter heterochromatin and promote efficient DSB repair within this compartment. Further work will be required to address how kap-1 phosphorylation alters heterochromatin structure, and to examine if these changes propagate across the entire heterochromatin structure, or are localized to heterochromatic regions adjacent to the initial DSB.
Localized chromatin destabilization at DSBs.
In addition to the above described chromatin remodeling events, it is now clear that there is a specific relaxation of the local chromatin structure on chromatin domains which are contiguous with the DSB. As discussed above, the sensitivity of DNA to nuclease digestion is increased after DNA damage,23–25 consistent with increased accessibility of the linker DNA between nucleosomes to nucleases. Further work has demonstrated that depletion of linker histones, which promotes decompaction of the chromatin, amplifies DNA damage signaling and increases the ability of cells to repair DSBs.36 A recent biophysical study, which analyzed the incorporation of GFP labeled histone H2B, demonstrated that chromatin undergoes a localized expansion at DSBs.37 Further, this chromatin expansion was an active, ATP-dependent process.37 Together, these results indicate that both chromatin compaction and nucleosome stacking are actively decreased within domains which correspond to the regions containing DSBs.
Studies from the gene transcription and chromatin structure fields have demonstrated that open, actively transcribed, euchromatic domains are associated with high levels of histone acetylation.8,12,38,39 Lysine acetylation promotes the formation of relaxed chromatin structures by neutralizing the negative charge on lysines, and therefore decreasing both histone-DNA and histone-histone interactions within nucleosomes.40,41 This led several groups to examine if increased histone acetylation was associated with altered chromatin compaction at DSBs. Both histones H2A and H4 show increased levels of acetylation following DSB generation,42–47 and the acetylation of histone H4 is specifically increased on chromatin domains extending for several kilobases on either side of the DSB.42,43 Tip60, which acetylates histones H2A,42 and H4,44 has been identified as the acetyltransferase involved in DNA-damage induced chromatin acetylation. Tip60 is a ubiquitously expressed acetyltransferase which plays at least two distinct roles in the repair of DSBs. First, Tip60 is recruited to DSBs were it directly acetylates and activates the ATM kinase.35,48–50 Second, and most important for the current discussion, Tip60 is required for the acetylation of histones H2A and H4 at DSBs after IR exposure.42,43,46 The recruitment of the Tip60 to DSBs may therefore lead to histone hyperacetylation42,45,46 and the generation the open, relaxed chromatin structures previously reported at DSBs.25,26,37,51 However, chromatin remodeling frequently combines histone acetylation with the use of large motor ATPases to modify the chromatin architecture. Although several chromatin remodeling complexes have been implicated in DSB repair, the NuA4 complex plays a pivotal role in histone acetylation and DSB repair.42,43,52–54 Mammalian NuA453 contains at least 16 sub-units, of which 3 possess catalytic activity. These include the Tip60 acetyltransferase, the p400 motor ATPase55 and the Ruvbl1 and Ruvbl2 helicase-like proteins.45 p400 was originally identified as an E1A binding protein and loss of p400 leads to elevated p21 levels and senescence.56 p400 is a SWI/SNF DNA-dependent ATPase56 which functions to alter DNA-histone interactions and facilitates the insertion of histone variants, including H2A.Z, into gene promoters.57 Several sub-units of NuA4 are recruited to DSBs, including Tip60,35 Trrap,42,43,58 p400,59 and ruvbl1 and ruvbl2.45 Inactivation of either Tip60 or Trrap, a scaffold protein which mediates NuA4 formation, leads to loss of histone acetylation after DNA damage and failure to load either 53BP1 or brca1 onto the chromatin.42 Further, p400 and Tip60 function in a common pathway to regulate apoptotic responses to DNA damage,60 implying that the p400 SWI/SNF ATPase and the Tip60 acetyltransferase function together to regulate chromatin structure during DSB repair. These results have led to the proposal that hyperacetylation of histones by the NuA4-Tip60 complex at DSBs reduces both the stability of the histone-DNA interaction within nucleosomes as well as facilitating the unpacking of higher order nucleosome arrays.42,46,61,62
Several key questions remain to be addressed concerning this hypothesis. These include demonstrating that acetylation alters chromatin structure at the DSB, determining the role of NuA4 in this process and, most importantly, determining how changes in nucleosome structure at DSBs impact DSB repair. A key barrier has been the lack of available methodology to monitor nucleosome stability in vivo at mammalian DSBs. Recent work has directly addressed this issue by developing a new approach to monitor changes in chromatin structure at DSBs.59 This is based on the observation that histone-DNA interactions are extremely stable, such that histones are only extracted from chromatin by NaCl concentrations in excess of 1.5 M.63,64 However, if DSBs create domains in which the stability of the histone-DNA interaction is reduced, these regions should exhibit increased sensitivity to NaCl fractionation. Using a biochemical approach, Xu et al.59 have now demonstrated that histones can be preferentially eluted from damaged chromatin by biochemical fractionation with NaCl. This observation is consistent with a decrease in the stability of histone-DNA and histone-histone interactions within nucleosomes following generation of DSBs on the chromatin. The authors refer to this process as a reduce in nucleosome stability.59 Importantly, Xu et al. demonstrated that the domains of decreased nucleosome stability were preferentially located within the γH2AX domains. This indicates that the observed decrease in nucleosome stability after DNA damage was localized to chromatin domains adjacent to the DSB, rather than being propagated across the entire chromatin. This is consistent with previous work indicating that DSBs can alter the local chromatin structure,18,19,25,37 in contrast to global changes in chromatin structure controlled by the ATM/phospho-kap1 pathway.25,27 Overall, this implies that DNA repair foci (defined as γH2AX domains) correspond to regions in which the stability of nucleosomes, which is controlled by histone-histone interactions within the nucleosome core, are significantly decreased, creating regions of open, relaxed chromatin structures.
Xu et al.59 also demonstrated that 3 components of the NuA4 complex—the p400 motor ATPase, the Tip60 acetyltransferase and Trrap, a scaffold protein—were required to decrease nucleosome stability at DSBs. Previous work indicated that several subunits of NuA4 were recruited to DSBs and were required for DSB repair.42,43,45,46 Taken together, this implies that p400 and Tip60 are recruited to DSBs as components of the NuA4 complex, rather than functioning independently. Importantly, histone acetylation in the absence of p400 activity did not lead to nucleosome destabilization,59 indicating that acetylation on its own is insufficient to alter nucleosome structure at DSBs. This implies that recruiting NuA4 to the DSB brings together the remodeling activity of p400 and the acetyltransferase activity of Tip60, which work together to decrease nucleosome stability at the break site. Additional insight into how NuA4 destabilizes nucleosomes is provided by studies of nucleosome structure. The crystal structure of the nucleosome indicate that the n-terminal of histone H4 interacts with an acidic patch on the H2A/H2B dimer.65,66 Further, acetylation of histone H4 on lysine 16 (H4K16Ac) specifically weakens this interaction between H4 and the acidic patch on H2A,67,68 and inhibits the packing of nucleosomes into 30 nm fibers. Histone H4 acetylation therefore inhibits histone-histone interactions both within and between adjacent nucleosomes, preventing the formation of stacked 30 nm nucleosomal arrays and favoring the formation of linear, open nucleosome structures. The Tip60 sub-unit of NuA4 may therefore increase histone H4 acetylation,42,43,59,69 which, in turn, leads to destabilization of histone-histone interactions both within and between adjacent nucleosomes. This favors the unpacking of higher order nucleosome arrays and promotes the formation of localized domains of open, relaxed chromatin at the break site.59,67,68 The role of p400 in this process is less clear. Although the process of nucleosome destabilization required the ATPase activity of the p400 motor protein,59 how p400 alters chromatin structure at DSBs is currently unclear. p400 has histone exchange activity, and can exchange H2A for the histone variant H2A.Z in gene promoters,46,55,57 but p400 does not appear to have either nucleosome sliding activity or the ability to evict nucleosomes from the chromatin. The exact changes in chromatin structure promoted by p400 will need to be clarified to fully understand p400's role in this process. In conclusion, the recruitment of NuA4 to DSBs promotes H4 acetylation, which, in combination with the ATPase activity of p400, decreases histone-histone interactions both within and between nucleosomes, switching the chromatin into a more open, relaxed structure. This leads to the generation of open, relaxed chromatin domains which extend for tens of kilobases on either side of the DSB.
Nucleosome Destabilization Promotes Chromatin Ubiquitination
Although previous studies have described changes in chromatin structure in response to DNA damage,23–25,36,37 it was not clear how these processes impacted DSB repair. Previous work had shown that Trrap, a component of NuA4, was required to recruit brca1 to DSBs,42 implying that chromatin remodeling at DSBs was important for loading repair proteins onto the chromatin. However, the mechanism by which this occurred remained unclear. Xu et al. have now demonstrated that nucleosome destabilization by NuA4 is required for chromatin ubiquitination and loading of the brca1 and 53BP1 proteins onto the chromatin.59 The RNF8 ubiquitin ligase is recruited to DSBs through a direct interaction with the mdc1 scaffold protein,7,70,71 where it ubiquitinates H2A and H2AX, as well as other, unknown chromatin targets. Subsequently, the RNF168 ubiquitin ligase binds to these ubiquitinated sites, promoting polyubiquitination of the chromatin.72,73 These ubiquitin polymers then provide a binding site for the ubiquitin interacting motif of RAP80, facilitating the recruitment of the RAP80/abraxas/brca1 complex to DSBs.74–76 Xu et al.59 have analyzed the impact of nucleosome destabilization on chromatin ubiquitination by the RNF8 ubiquitin ligase. In a novel approach, they used a designer Zinc Finger Nuclease (ZFN) to create a single DSB on chromosome 19. ZFNs are custom engineered nucleases in which the catalytic domain of the Fok1 endonuclease is linked to an engineered zinc finger protein, creating a sequence-specific nuclease which is targeted to a unique sequence within the genome.77 ZFNs were developed for both targeted gene insertion and for correction of inherited mutations through homology directed repair with donor DNA.77,78 By combining ZFNs with Chromatin Immunoprecipitation (ChIP) techniques, it is possible to monitor changes in histone modifications at the break site.59 Using this approach, it was shown that DSBs create domains of ubiquitinated chromatin which extend for at least 10 kb on either side of the DSB.59 Importantly, in the absence of p400-dependent nucleosome destabilization, RNF8 was still recruited to the DSB, but failed to ubiquitinate the chromatin. Consequently, brca1 did not accumulate at the DSB and the cells exhibited increased radiosensitivity and increased numbers of chromosomes aberrations.59 This demonstrates that decreased nucleosome stability mediated by the NuA4 complex is required for RNF8 to ubiquitinate target proteins on the chromatin.42,59 Although RNF8 ubiquitinates histone H2A,7,70,71 it is likely that there are other, unidentified chromatin associated proteins which are also ubiquitinated by RNF8 and RNF168. How the decrease in nucleosome stability facilitates chromatin by ubiquitination by RNF8 is unclear. One potential mechanism is that chromatin relaxation alters the nucleosome structure to expose previously buried histone domains which can then be ubiquitinated by RNF8. This process could be facilitated by the prior acetylation of the histones by Tip60.79 Further, the altered nucleosome structure may promote the recruitment of new proteins to the chromatin which are then ubiquitinated by RNF8. Continued study of the molecular mechanisms involved will provide new insight into the mechanism.
The concept that nucleosome destabilization is required to promote chromatin ubiquitination also has implications for other chromatin modifications which occur at DSBs. For example, although RNF8 is required to recruit 53BP1 to the chromatin,7,70,71 53BP1 does not contain a ubiquitin interacting motif. However, 53BP1 does contain a tudor domain, which can bind to H4K20me2.80,81 The destabilization of nucleosome structure by p400 and RNF8 may expose buried H4K20me2 sites for 53BP1 to associate with.82 However, the restricted distribution of H4K20me2 within mammalian cells10,14 implies that H2K20me2 would only be present at a small fraction of DSBs. An alternative mechanism is that chromatin ubiquitination by RNF8 is required for histone H4K20 methylation (and possibly other histone modifications) at DSBs. This could occur through the ubiquitin-dependent recruitment of methyltransferases to DSBs or through promotion of H4K20 methylation in response to histone ubiquitination. Additional chromatin modifications, including the sumoylation of histones by PIAS1 and PIAS4, also promote ubiquitination by RNF8,83,84 and may be required for recruitment of brca1 to DSBs. The p400-mediated decrease in nucleosome stability may therefore function to decrease histone-histone interactions both within and between nucleosomes, leading to open, flexible nucleosome structures. This destabilization of nucleosomes then facilitates the ubiquitination, sumoylation and methylation of histones along the chromatin and promotes the recruitment of brca1 and 53BP1 to the DSB. In addition, p400 mediated changes in nucleosome structure may also recruit (or evict) proteins from the chromatin, including novel proteins which are targets for either ubiquitination or sumoylation.
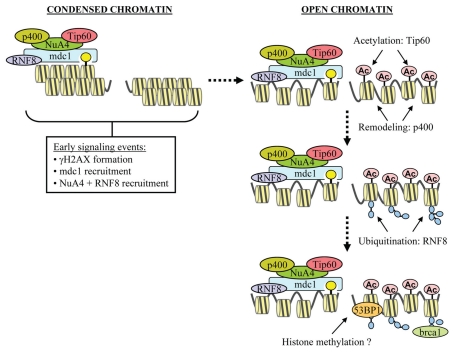
Figure 1 outlines a potential model for how the NuA4 complex regulates nucleosome stability. Previous work has indicated that inactivation of the unique components of NuA4 complex does not affect the early events in DSB repair, such as activation of ATM, binding of the MRN complex to the break, phosphorylation of H2AX and the recruitment of mdc1 or RNF8 to the chromatin.42,48,59 The early events in DNA damage signaling, including loading of mdc1 and RNF8 onto the chromatin, are independent of NuA4-mediated changes in nucleosome stability. Subsequent to these early events, p400, Trrap and Tip60 are recruited to the DSB, most likely as sub-units of the NuA4 complex, through direct interaction with mdc1.59 MRN6,85 and the RNF8 ubiquitin ligase7,70–73 associate with mdc1 by binding to specific phosphorylation sites on mdc1; however, it is currently unclear if NuA4 requires phosphorylation for recruitment, or which sub-unit of NuA4 binds to mdc1. Understanding the mechanism by which NuA4 is recruited to DSBs is therefore a key area for future research. Once the NuA4 complex is positioned at the DSB, the Tip60 sub-unit acetylates adjacent histones, while the p400's ATPase activity remodels both the local histone-DNA interactions as well as histone-histone interactions between adjacent nucleosomes. This promotes unpacking of stacked nucleosome arrays and shifts the local chromatin structure into a relaxed, open conformation. Creation of these relaxed, open chromatin structures then exposes RNF8 ubiquitination substrates on nucleosomes,7,70,71 as well as either exposing potential H4K20me2 sites, or promoting methylation of H4K20.80 Together, these chromatin modifications promote the recruitment of the brca1 and 53BP1 proteins to the DSB. The overall outcome is to facilitate DNA repair by increasing the mobility of the nucleosomes adjacent to the DSBs, promoting the post-translational modification of histones, and directing the recruitment and retention of protein factors such as 53BP1 and brca1 at DSBs.
Figure 1.
Nucleosome destabilization at DSBs by the NuA4 complex. p400 and Tip60 sub-units of NuA4 shown. RAP80/abraxas/brca1 complex is shown as brca1. Potential histone methylation changes which may influence 53BP1 recruitment outlined.
Conclusions and Implications
Alterations in chromatin structure are emerging as key control points during DSB. The demonstration that the Tip60, p400 and Trrap sub-units of NuA4 are required for DSB repair defines these proteins as DNA damage response proteins which function to regulate genomic stability. In fact, several sub-units of NuA4 have been implicated in human cancer. For example, loss of p400 is associated with p21-dependent senescence,56 increased sensitivity to ionizing radiation and increased genomic instability.59 The E1A protein of adenovirus targets p400, and this interaction is essential for the tumor promoting function of adenovirus.85 Further, haploinsufficiency for Tip60 is associated with breast86 and colon cancer87 and disruption of the p400-Tip60 ratio in colorectal cancer cells contributes to loss of the oncogene-induced DNA damage response.59 Mutations or inactivation of NuA4 sub-units may therefore contribute to the etiology and progression of cancer by impacting both chromatin structure and DSB repair at sites of DNA damage.
Cells contain many types of chromatin structure, from compacted heterochromatin to open euchromatin domains. An important issue is whether NuA4 is required to alter nucleosome stability at all chromatin locations, or is restricted to particular types of chromatin structures. NuA4 recruitment to the chromatin requires the prior binding of mdc1 to γH2AX (Fig. 1: reviewed in ref. 58). However, γH2AX does not spread uniformly along the chromosome, suggesting regions of low/absent H2AX.89,90 Similarly, γH2AX foci are preferentially formed in euchromatin27,30,91,92 and γH2AX does not spread through heterochromatin.91 Heterochromatin domains which lack H2AX would not, therefore, recruit either mdc1 or the NuA4 complex, implying that these regions do not require NuA4-mediated decreases in nucleosome stability. However, DSB repair within heterochromatin does require phosphorylation of the kap-1 heterochromatin binding protein,27,30,31 and altered binding of additional heterochromatin proteins (including HP1 family; reviewed in ref. 33–35) at DSBs. This raises the possibility that there are distinct chromatin remodeling mechanisms for DSB repair within heterochromatin (involving kap-1 phosphorylation) and euchromatin (involving NuA4 mediated nucleosome destabilization).
In conclusion, NuA4-mediated decreases in nucleosome stability at DSBs play a crucial role in regulating the post-translational modification of the chromatin, the formation of DNA repair foci and in the mechanism of DSB repair. Understanding the link between chromatin structure and DSB repair, and identifying the unique chromatin remodeling events associated with DSB repair in different chromatin domains, will be an important area for future research.
Acknowledgements
We thank members of the Price laboratory for critical discussions and reading of the manuscript. This work was supported by grants from the NCI (CA64585 and CA93602) and the DOD Breast Cancer Program to B.D.P.
Abbreviations
- HP1α
heterochromatin protein 1α
- MRN
mre11-rad50-nbs1 complex
- DSB
DNA double-strand break
- IR
ionizing radiation
- bp
base-pairs
- IRIF
ionizing radiation induced foci
- NHEJ
non-homologous end-joining
- HR
homologous recombination
- ChIP
chromatin immunoprecipitation
References
- 1.Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, et al. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 5.Melander F, Bekker-Jensen S, Falck J, Bartek J, Mailand N, Lukas J. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol. 2008;181:213–226. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, et al. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J Cell Biol. 2008;181:227–240. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 9.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 10.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Wit E, van Steensel B. Chromatin domains in higher eukaryotes: insights from genome-wide mapping studies. Chromosoma. 2009;118:25–36. doi: 10.1007/s00412-008-0186-0. [DOI] [PubMed] [Google Scholar]
- 13.Peng JC, Karpen GH. Epigenetic regulation of heterochromatic DNA stability. Curr Opin Genet Dev. 2008;18:204–211. doi: 10.1016/j.gde.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26:9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 16.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 17.Cairns BR. Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr Opin Genet Dev. 2005;15:185–190. doi: 10.1016/j.gde.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, et al. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 21.Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26:7749–7758. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- 22.Sinha M, Peterson CL. Chromatin dynamics during repair of chromosomal DNA double-strand breaks. Epigenomics. 2009;1:371–385. doi: 10.2217/epi.09.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubbi CP, Milner J. p53 is a chromatin accessibility factor for nucleotide excision repair of DNA damage. EMBO J. 2003;22:975–986. doi: 10.1093/emboj/cdg082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carrier F, Georgel PT, Pourquier P, Blake M, Kontny HU, Antinore MJ, et al. Gadd45, a p53-responsive stress protein, modifies DNA accessibility on damaged chromatin. Mol Cell Biol. 1999;19:1673–1685. doi: 10.1128/mcb.19.3.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 26.Telford DJ, Stewart BW. Micrococcal nuclease: its specificity and use for chromatin analysis. Int J Biochem. 1989;21:127–137. doi: 10.1016/0020-711x(89)90100-6. [DOI] [PubMed] [Google Scholar]
- 27.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Lin HH, Chen H, Xu X, Shih HM, Ann DK. SUMOylation of the transcriptional co-repressor KAP1 is regulated by the serine and threonine phosphatase PP1. Sci Signal. 2010;3:32. doi: 10.1126/scisignal.2000781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng L, Yap KL, Ivanov AV, Wang X, Mujtaba S, Plotnikova O, et al. Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing. Nat Struct Mol Biol. 2008;15:626–633. doi: 10.1038/nsmb.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noon AT, Shibata A, Rief N, Lobrich M, Stewart GS, Jeggo PA, et al. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol. 2010;12:177–184. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- 31.Goodarzi AA, Noon AT, Jeggo PA. The impact of heterochromatin on DSB repair. Biochem Soc Trans. 2009;37:569–576. doi: 10.1042/BST0370569. [DOI] [PubMed] [Google Scholar]
- 32.Li X, Lee YK, Jeng JC, Yen Y, Schultz DC, Shih HM, et al. Role for KAP1 serine 824 phosphorylation and sumoylation/desumoylation switch in regulating KAP1-mediated transcriptional repression. J Biol Chem. 2007;282:36177–36189. doi: 10.1074/jbc.M706912200. [DOI] [PubMed] [Google Scholar]
- 33.Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–686. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 34.Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, et al. Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol. 2009;185:577–586. doi: 10.1083/jcb.200810035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, et al. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol. 2009;11:1376–1382. doi: 10.1038/ncb1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, et al. Global chromatin compaction limits the strength of the DNA damage response. J Cell Biol. 2007;178:1101–1108. doi: 10.1083/jcb.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Muller WG, McNally JG, et al. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toth K, Brun N, Langowski J. Chromatin compaction at the mononucleosome level. Biochemistry. 2006;45:1591–1598. doi: 10.1021/bi052110u. [DOI] [PubMed] [Google Scholar]
- 39.Ferreira H, Somers J, Webster R, Flaus A, Owen-Hughes T. Histone tails and the H3 alphaN helix regulate nucleosome mobility and stability. Mol Cell Biol. 2007;27:4037–4048. doi: 10.1128/MCB.02229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Hayes JJ. Acetylation mimics within individual core histone tail domains indicate distinct roles in regulating the stability of higher-order chromatin structure. Mol Cell Biol. 2008;28:227–236. doi: 10.1128/MCB.01245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi JK, Howe LJ. Histone acetylation: truth of consequences? Biochem Cell Biol. 2009;87:139–150. doi: 10.1139/O08-112. [DOI] [PubMed] [Google Scholar]
- 42.Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 43.Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, et al. Binding of Chromatin-Modifying Activities to Phosphorylated Histone H2A at DNA Damage Sites. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 44.Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, et al. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 45.Jha S, Shibata E, Dutta A. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol Cell Biol. 2008;28:2690–2700. doi: 10.1128/MCB.01983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 47.Jiang X, Xu Y, Price BD. Acetylation of H2AX on lysine 36 plays a key role in the DNA double-strand break repair pathway. FEBS Lett. 2010;584:2926–2930. doi: 10.1016/j.febslet.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci USA. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun Y, Xu Y, Roy K, Price BD. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol Cell Biol. 2007;27:8502–8509. doi: 10.1128/MCB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun Y, Jiang X, Price BD. Tip60: Connecting chromatin to DNA damage signaling. Cell Cycle. 2010:9. doi: 10.4161/cc.9.5.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Birger Y, West KL, Postnikov YV, Lim JH, Furusawa T, Wagner JP, et al. Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin. EMBO J. 2003;22:1665–1675. doi: 10.1093/emboj/cdg142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Auger A, Galarneau L, Altaf M, Nourani A, Doyon Y, Utley RT, et al. Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants. Mol Cell Biol. 2008;28:2257–2270. doi: 10.1128/MCB.01755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doyon Y, Cote J. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev. 2004;14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 54.Selleck W, Fortin I, Sermwittayawong D, Cote J, Tan S. The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the Enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes. Mol Cell Biol. 2005;25:5535–5542. doi: 10.1128/MCB.25.13.5535-5542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fuchs M, Gerber J, Drapkin R, Sif S, Ikura T, Ogryzko V, et al. The p400 complex is an essential E1A transformation target. Cell. 2001;106:297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 56.Chan HM, Narita M, Lowe SW, Livingston DM. The p400 E1A-associated protein is a novel component of the p53→p21 senescence pathway. Genes Dev. 2005;19:196–201. doi: 10.1101/gad.1280205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gevry N, Chan HM, Laflamme L, Livingston DM, Gaudreau L. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 2007;21:1869–1881. doi: 10.1101/gad.1545707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 59.Xu Y, Sun Y, Jiang X, Ayrapetov MK, Moskwa P, Yang S, et al. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J Cell Biol. 2010;191:31–43. doi: 10.1083/jcb.201001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mattera L, Escaffit F, Pillaire MJ, Selves J, Tyteca S, Hoffmann JS, et al. The p400/Tip60 ratio is critical for colorectal cancer cell proliferation through DNA damage response pathways. Oncogene. 2009;28:1506–1517. doi: 10.1038/onc.2008.499. [DOI] [PubMed] [Google Scholar]
- 61.Brower-Toland B, Wacker DA, Fulbright RM, Lis JT, Kraus WL, Wang MD. Specific contributions of histone tails and their acetylation to the mechanical stability of nucleosomes. J Mol Biol. 2005;346:135–146. doi: 10.1016/j.jmb.2004.11.056. [DOI] [PubMed] [Google Scholar]
- 62.Altaf M, Auger A, Covic M, Cote J. Connection between histone H2A variants and chromatin remodeling complexes. Biochem Cell Biol. 2009;87:35–50. doi: 10.1139/O08-140. [DOI] [PubMed] [Google Scholar]
- 63.von Holt C, Brandt WF, Greyling HJ, Lindsey GG, Retief JD, Rodrigues JD, et al. Isolation and characterization of histones. Methods Enzymol. 1989;170:431–523. doi: 10.1016/0076-6879(89)70061-6. [DOI] [PubMed] [Google Scholar]
- 64.Ye J, Ai X, Eugeni EE, Zhang L, Carpenter LR, Jelinek MA, et al. Histone H4 lysine 91 acetylation a core domain modification associated with chromatin assembly. Mol Cell. 2005;18:123–130. doi: 10.1016/j.molcel.2005.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 66.Chodaparambil JV, Barbera AJ, Lu X, Kaye KM, Hansen JC, Luger K. A charged and contoured surface on the nucleosome regulates chromatin compaction. Nat Struct Mol Biol. 2007;14:1105–1107. doi: 10.1038/nsmb1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 68.Robinson PJ, An W, Routh A, Martino F, Chapman L, Roeder RG, et al. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J Mol Biol. 2008;381:816–825. doi: 10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 71.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 73.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 74.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 75.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci USA. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 78.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, Dekelver RC, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;9:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol. 2007;27:7028–7040. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 81.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huyen Y, Zgheib O, Ditullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 83.Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 85.Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008;9:795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tworkowski KA, Chakraborty AA, Samuelson AV, Seger YR, Narita M, Hannon GJ, et al. Adenovirus E1A targets p400 to induce the cellular oncoprotein Myc. Proc Natl Acad Sci USA. 2008;105:6103–6108. doi: 10.1073/pnas.0802095105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448:1063–1067. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- 88.Sakuraba K, Yasuda T, Sakata M, Kitamura YH, Shirahata A, Goto T, et al. Downregulation of Tip60 gene as a potential marker for the malignancy of colorectal cancer. Anticancer Res. 2009;29:3953–3955. [PubMed] [Google Scholar]
- 89.Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, et al. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Savic V, Yin B, Maas NL, Bredemeyer AL, Carpenter AC, Helmink BA, et al. Formation of dynamic gamma-H2AX domains along broken DNA strands is distinctly regulated by ATM and MDC1 and dependent upon H2AX densities in chromatin. Mol Cell. 2009;34:298–310. doi: 10.1016/j.molcel.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J Cell Biol. 2007;178:209–218. doi: 10.1083/jcb.200612031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cowell IG, Sunter NJ, Singh PB, Austin CA, Durkacz BW, Tilby MJ. gammaH2AX foci form preferentially in euchromatin after ionising-radiation. PLoS One. 2007;2:1057. doi: 10.1371/journal.pone.0001057. [DOI] [PMC free article] [PubMed] [Google Scholar]