Stem cells are central to developing new treatment options for tissue regeneration and constructing controllable models for biological research. Bioengineered cell culture environments that combine microenvironmental control with tissue-specific transport and signaling are critical tools in our efforts to study tissue development, regeneration, and disease under conditions that predict the human in vivo context. We propose that experimentation at the interfaces of biology, engineering and medical sciences is critical for unlocking the full potential of stem cells. Here, we focus on the design and utilization of in vitro platforms that recapitulate the environments associated with tissue development, disease and regeneration.
The regeneration of worn and diseased tissues using some form of cell therapy is becoming increasingly plausible. The need is obvious. Medical advances have extended the functional life of our organs, and enabled us to live longer and better. It has been estimated that roughly one in five people reaching age of 65 would benefit from some kind of tissue replacement therapy during their lifetime (Lysaght and Reyes, 2001). At this time, tissues that are failing beyond repair, or missing due to surgical resection or congenital abnormalities are being replaced by transplantation, an ultimate measure limited by the scarcity of matching donor organs. Recent advances in stem cell biology and tissue engineering are enabling us to “instruct” multipotent cells – the ultimate “tissue engineers” - to differentiate into the right phenotypes in the right place and at the right time, in order to assemble functional tissue structures. It is a true integration of biology and engineering that makes it possible to design “biomimetic” environments that subject the cell to the combinations of factors known to guide tissue development and regeneration in vivo.
We are just starting to fully realize the importance of the entire context of a cell’s microenvironment, including the presence of other cells, three-dimensional matrix, and sequences of molecular and physical morphogens. The premise behind the design of biomimetic models is that to unlock the full potential of stem cells at least some aspects of the dynamic in vivo environments need to be reconstructed in experimental systems used in vitro. It has been argued that we now need a new generation of 3D culture systems that would offer a middle ground between the bare bones approach of a standard Petri dish and a live organism model, such as a rat, or mouse (Lutolf, 2009; Burdick and Vunjak-Novakovic, 2009). Advanced culture systems that combine high biological fidelity with tight control over the cellular environment are in active development, and have begun to take center stage in efforts to observe stem cell responses that predict their behavior in vivo. By definition, the development of biomimetic environments depends on replicating the physiological context, based on an existing knowledge of the in vivo conditions present in a target organ or tissue. However, productive, functional artificial environments may also be constructed according to lessons learned from the in vivo settings, without aiming to fully recapitulate those conditions. Indeed, it is increasingly possible, and sometimes may be necessary, to generate engineered tissues by going beyond a purely biomimetic context towards a set of conditions that promote organ and tissue regeneration to the extent achievable without external manipulation.
Overall, research at the interface between stem cell science and tissue engineering is currently driving important advances into the regeneration of functional tissue structures. The two communities – biologists and engineers –have been disconnected for a very long time, but are now starting to effectively communicate in order to establish an entirely new interdisciplinary field of stem cell bioengineering. In fact, some of the most exciting recent breakthroughs in regenerative medicine have been achieved by integrating stem cell science with the application of bioengineering methods (Peterson et al., 2010; Ott et al., 2010; Grayson et al., 2009; Macchiarini et al., 2009; Uygun et al., 2010; Zimmermann et al., 2006). At the same time, bioengineering research is becoming focused on fundamental biological questions that cannot be addressed using the traditional cell culture plates (Altman et al., 2002; Au et al., 2007; Connelly et al., 2010; Discher et al., 2009; Freytes et al., 2009; Gilbert et al., 2010; Hui and Bhatia 2007; Lucchetta et al., 2005; Lutolf et al., 2009; Lutolf, 2009; Nelson et al., 2005; Park et al., 2009; Skelley et al., 2009; Terraciano et al., 2007). The bioengineered stem cell “niche” – comprising multiple cell types, extracellular matrix, cytokines and physical factors – has emerged as a new paradigm for stem cell research that brings together the two communities in a most effective way.
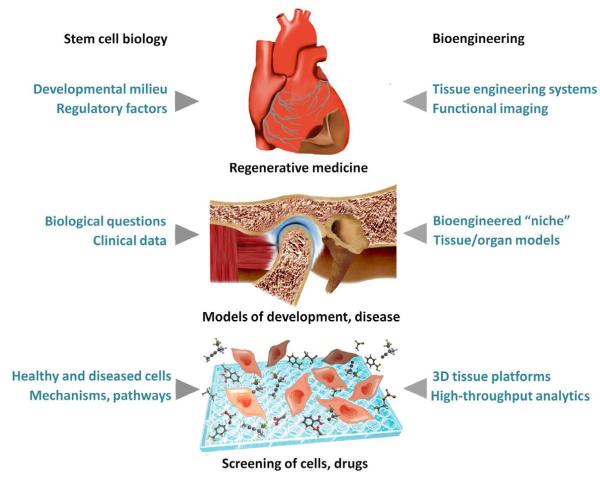
The implications of this collaborative approach are yet to be seen, but are likely to extend beyond the current goal of answering complex biological questions using new bioengineering tools, to the derivation of entirely new, as yet unforeseen concepts that will shape future advances in regenerative medicine. In this Perspective article, we focus on the development and use of biomimetic platforms that provide the interface between biological questions and engineering tools, towards (i) new insights into environmental regulation of stem cells, (ii) study of disease, and (iii) developing new treatment modalities for regenerative medicine (Figure 1).
Figure 1. Bioengineering platforms.
The work at the interface between stem cell science and bioengineering is now resulting in controllable models of high biological fidelity that are driving progress in three major areas: (i) new treatment modalities for regenerative medicine, (ii) study of development and disease, and (iii) fundamental biological research.
Cells
Access to cells, both in number and with the appropriate developmental potential, for use in bioengineering has historically been limiting. However, two parallel developments indicate that this issue may be becoming less problematic. There have been numerous advances in understanding the hierarchy of cells comprising particular tissues. Tissues are typically organized such that mature cells are generated and replaced by a proliferative pool of less differentiated progenitors, which in turn arise from a reserve set of stem cells that is relatively small is size. In the past, the presence of tissue specific stem/progenitors had been hypothesized, but the phenotype of these cells was often not well defined, making their isolation challenging. The last decade has seen an explosion in the identification of stem/progenitor populations in multiple somatic tissues such that it is now possible to define with precision some key stem/progenitors in the skeleton, muscle, brain, intestine, skin and blood (Barker et al., 2007; Cotsarelis et al., 1990; Frederiksen et al., 1988; Sacchetti et al., 2007; Sherwood et al., 2004; Snippert et al., 2010; Kiel et al., 2005; Osawa et al., 1996; Rietze et al., 2001). Many of these advances have been achieved using animal models, and human correlates are still being sought. Nonetheless, that work is ongoing and likely to be productive.
Identification of adult, multipotent stem/progenitor cells for use as a source of lineage committed mature cells may foster the creation of engineered tissue constructs, at least for situations in which a pool of precursors is both accessible and can be grown in a robust fashion. This concept is perhaps best demonstrated by the isolation and expansion of mesenchymal stem cells, which can be cultured in order to produce sufficiently large numbers of cells capable of giving rise to multiple mesenchymal lineages. These mature, long lived progeny can then be incorporated into engineered constructs (Pereira et al., 1995; Jaiswal et al., 1997), or even into some tissues. However, for some tissues, the existence of an adult stem cell pool is still under debate (such as in pancreatic islets, (Dor et al., 2004; Smukler et al., 2011) or the tissue-specific stem cells may be in a relatively inaccessible location, such as in brain, which makes them either impossible, or at least challenging to isolate for ex vivo expansion purposes. For these tissues, pluripotent stem cells remain the best option.
Access to pluripotent stem cells represents the second recent development that has had a significant impact on the growing potential of bioengineering applications. Pluripotent stem cells (which are capable of making any intraembryonic cell type) can now be obtained not only from blastocysts as embryonic stem cells (ESCs), but also in the form of induced pluripotent stem cells (iPSCs) following the reprogramming of adult somatic cells. The ability to convert a mature skin fibroblast or blood cell to a pluripotent cell represents an extraordinary achievement that has opened the possibility of generating cells from the individual who is the intended recipient of a bioengineered construct (Takahashi et al., 2007). The reprogrammed pluripotent cells appear to be no longer constrained by the ‘Hayflick limit’ (Hayflick, 1979). That is, reprogrammed cells are not subject to the same constraints of senescence that a more mature cell population encounters, and their final numbers are essentially unlimited. The generation of large numbers of cells for use in engineered constructs is therefore made possible. Those cells may now be generated from the patient so that either engineered models of disease or engineered therapeutics with reduced risk of immunologic rejection can be constructed (Saha and Jaenisch, 2009).
The ability to take full advantage of the inherent potential of iPSCs depends upon two important issues. First, reprogramming methods that do not result in permanent genetic alteration of the resulting cells must be developed in order to mitigate the risk of tumor formation. Recent studies using RNA based strategies have moved the field markedly closer to that goal (Warren et al., 2010). Second, it will be necessary to achieve high fidelity means for directing the differentiation of iPSCs to tissue specific stem/progenitor and mature cells. Mapping of the molecular signatures of individual cell lineages during their maturation process is an ongoing area of investigation that will provide better guidance in selecting the desired features of target cell populations (Guenther et al., 2010; Bernstein et al., 2010; Novershtern et al., 2010). By combining molecular signatures of the various cell states within a lineage hierarchy with larger scale screening strategies, it is becoming possible to define methods for differentiation, or even reprogramming to a particular cell state that offers potential for therapeutic applications (Yuan et al., 2011; Borowiak and Melton, 2009).
Scaffolds
In vivo, cells are surrounded by an extracellular matrix (ECM) that is responsible for the multidimensional and long-range ordering of highly organized tissues, and which interacts with the local cell populations and their secreted factors. Disease processes are often characterized by inappropriate cell-mediated remodeling that leads to unbalanced turnover of the ECM and negatively affects local cell function. 3D scaffolds present an engineered, in vitro alternative to the native extracellular matrix for the expansion and organization of cultured cells. A scaffold can be considered as a structural and “cell-instructive” template for cells and the forming tissue (Burdick and Vunjak-Novakovic, 2009). Scaffold materials –in most cases biodegradable and custom-designed to mimic the matrix of a specific tissue – can be processed into a range of 3D architectures suitable for cell seeding and cultivation (Dawson et al., 2007; Nair and Laurencin, 2007).
The specific choice of biomaterial for any given application is guided by the need to restore cell-matrix interactions, direct cell alignment and migration, and apply physical signals (such as flow-induced shear, mechanical stretch or electrical pacing). The Discher lab demonstrated that stem cells specify lineage and commit to phenotypes with extreme sensitivity to elasticity of the substrate (Engler et al., 2006), such that soft matrices that mimic brain are neurogenic, stiffer matrices that mimic muscle are myogenic, and the stiffest matrices that mimic bone are osteogenic. The results of their studies significantly improved our understanding of how physical factors influence stem cell differentiation.
Advanced scaffold designs are now being developed to implement patterning, binding of ligands, sustained release of cytokines, and the structural and mechanical anisotropy intrinsic to specific tissues, such as heart muscle, or bone (Engelmayr et al., 2008; Kloxin et al., 2009; Zhao et al., 2011). A “biomimetic” scaffold would mimic the properties of a native tissue, dynamically interact with the cells by generation and transmission of biophysical signals, and undergo gradual replacement by newly synthesized tissue matrix.
Bioreactors
In a Petri dish, the cell culture environment is determined by the concentrations of oxygen, nutrients and metabolites surrounding the cells that all change between one exchange or refresh of media solution to another. In cultures of 3D tissues, these concentrations will also change in space, because of diffusion gradients across the thickness of the tissue. In particular, oxygen penetration depth can be as little as 100 μm for dense tissues such as heart or bone. To overcome these limitations of static culture plates, bioreactors can be developed to provide control over the cell environment (through enhanced mass transport to and from the cells), and physical signals (hydrodynamic, mechanical, electrical), and also to enable insight into cellular behavior (through imaging and on line measurements). Design of a tissue engineering bioreactor should ideally support cell viability and 3D organization by mechanisms similar to those present in the native cell environment (Griffith and Swartz, 2006). When designing a bioreactor, we aim at mimicking an in vivo cell niche (Burdick and Vunjak-Novakovic, 2010). In reality, bioreactors provide an opportunity to manipulate and control only certain aspects of a given niche, but do allow for quantitative studies of cellular interactions with their environment.
Biosynthetically active cells are central to any of our efforts to grow tissue grafts, to construct models of disease, or to develop in vitro platforms for therapeutic screening. In order to mobilize their full biological potential, the scaffold-bioreactor system should serve as an in vitro mimic of the milieu of the development, regeneration or disease under investigation. Such a biologically inspired approach is behind the design of highly specialized, tightly controlled culture systems that are replacing the conventional “one size fits all” Petri dishes. With the capability to generate spatial gradients of regulatory signals, to subject cells to dynamic changes in their environment, and to offer insight into cellular responses in real time, these new technologies are setting a stage for an entirely new approach to stem cell research. The examples that follow illustrate some of the recent work at the interface between biology and engineering.
Biomimetic paradigm
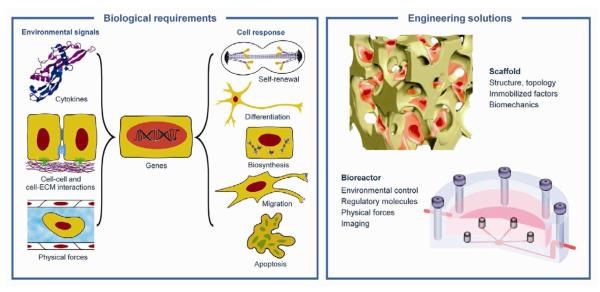
During development and regeneration, tissues emerge from coordinated sequences of stem cell renewal, specialization and assembly that are orchestrated by cascades of environmental factors. In vitro and in vivo, stem cell fate and function are regulated by a combination of the intrinsic genetic (and epigenetic) program of the cell, and the cellular microenvironment, also termed a “niche”. Cells interact with the entire context of their environment, rather than with one single dominating factor (Figure 2). The four main groups of key factors: (i) regulatory molecules (oxygen, nutrients, cytokines), (ii) other cells (3D context, cell-cell contacts, autocrine and paracrine signals), (iii) extracellular matrix (immobilized and released factors, structure, topology, stiffness), and (iv) physical factors (flow shear, compression, stretch, electrical signals) act in concert, with synergistic and competing effects.
Figure 2. Biomimetic paradigm.
Stem cell fate and function are regulated by the entire context of the cellular microenvironment (niche), through dynamic interactions of the cells with cascades of multiple factors: molecular, structural and physical. Native-like (biomimetic) cell environments can be engineered by a combined use of a scaffold (providing a structural and logistic template for cell differentiation and functional assembly) and a bioreactor (providing environmental control, molecular and physical signaling).
The overall complexity of cell regulation is further increased by the dynamic nature of regulatory signals, which change in space and time, and in ways that are not entirely known. Also, the interactions between the cells and their environment occur in both directions. Cells both respond to and actively modify the properties of their environment by synthesizing or degrading the extracellular matrix, secreting cytokines, and communicating with other cells and matrix by molecular and physical signals. The “dynamic reciprocity” (Nelson and Bissell, 2005) of cell-cell and cell-matrix signaling takes place at multiple hierarchical levels – from the scale of cell membrane molecules, to tissues, and whole organs. At each level, there are specific readouts that change from one level to another, and from one cell or tissue type to another.
The study of the individual and combined effects of regulatory signals, via precise spatiotemporal control of signal type and magnitude, is not a trivial task and certainly not achievable by using traditional well plates. Recent developments in cell culture technology offer the opportunities of singling out one factor of interest from other systemic signals, and superimposing this factor with other, well-defined signals. Clearly, in vitro systems cannot possibly capture the complexity of actual regulatory pathways, but the bioengineered “niche” allows sophisticated and controllable studies of multiple factors regulating developmental processes. A biomimetic approach to the formation of engineered tissues was established to direct the differentiation and functional assembly of stem cells by factors known to regulate cell fate and function during native development and regeneration. Acting in concert, the two components of a tissue engineering system – scaffold and bioreactor - provide a controllable environment for cultured cells, with a multitude of cytokines (diffusing or immobilized) and physical factors (hydrodynamic shear, mechanical stretch, electrical gradients) (Figure 2).
Both in vivo (during development and regeneration) and in vitro (for tissue engineering), the cues presented to cells are principal determinants of their phenotype. Hence, the designs of systems for cell culture, cell delivery and tissue engineering are necessarily inspired by biology (in a developing or adult organism). The complementary engineering principles help recapitulate the combinations of parameters in the native environments of a specific tissue or organ, in order to orchestrate the conversion of ‘collections of cells’ into specific tissue phenotypes.
Microbioreactors
Microtechnologies have been developed to precisely manipulate the cellular microenvironment and study cellular responses in real time and in a quantitative fashion. Such a small scale allows for high-throughput studies within a large experimental space while utilizing minimal amounts of cells and materials. In one set of recent studies (Figallo et al., 2007; Cimetta et al., 2009), a simple and practical device was developed by coupling a microfluidic platform with an array of culture wells, to enable systematic and precise variation of mass transport and hydrodynamic shear in cultures of human ESCs. This microarray bioreactor with twelve culture wells on a standard microscope slide format was designed to accommodate stem cells attached to a 2D substrate, and cells encapsulated in 3D hydrogel. Both culture formats allow for controlled perfusion of medium, and tight control of medium composition and hydrodynamic shear. Using this microfluidic platform, hESCs were systematically studied for their cardiovascular differentiation potential. Cell differentiation correlated with the level of hydrodynamic shear and transport rates of oxygen and growth factors (Cimetta et al., 2009). As this technology is compliant with standard imaging formats, differentiation patterns could be studied with the aid of quantitative image processing (Figallo et al., 2007).
Another microfluidic device was developed to enable cultivation of adherent murine ESCs over a range of flow rates, with concentration gradients applied across the culture space. Medium composition was precisely controlled through mass transport of individual molecular species to and away from the cells. For the first time, mESCs were cultured in continuous, logarithmically scaled perfusion for 4 days, with more than a 3000-fold variation in flow rates across the array (Kamei et al., 2009). The associated hydrodynamic shear was shown to determine the size of cell colonies. Subsequently, another microfluidic platform was developed for semi-automated cultivation of hESCs in a way that allowed parallel study of cell self-renewal and differentiation using a large parameter matrix.
Microfluidic platforms
Another application of “tiny technologies” is for the manipulation of individual cells in culture. Cell fusion is a key event during embryonic development, and this process has been used to study the epigenetic reprogramming of somatic cells to pluripotent stem cells. The use of cell fusion as a research tool is rapidly moving from non-mammalian model organisms (such as Drosophila) into human stem cells. A major challenge in studies of cell fusion is the low efficiency and specificity of cell pairing. Random cell aggregation and fusion result in heterogeneity of cell aggregates, which in turn translates into heterogeneity of the resulting cellular responses. A recent design of a microfluidic device may overcome this problem, by achieving efficiencies of cell pairing of up to 70%, by specifically pairing only two cells, and by performing cell pairing in a high-throughput fashion (Skelley et al., 2009). The design is remarkably simple, and it is based on cell trapping into small “niches” by manipulating flow streams. This approach has great potential for systematic study of cell reprogramming by fusion.
An elegant early example of the use of microfluidic technology is the study of the response of the Drosophila embryo to dynamic perturbation of temperature (Luchetta et al., 2005). Embryos were cultured in a Y-shaped chamber that operates with two fluid streams maintained at different temperatures, and allows on-line imaging of the fluid flow and the embryo. This way, the anterior and posterior halves of the embryo were forced to develop at two different temperatures, resulting in different rates of development. This study exemplifies how microfluidic technology allows controllable “perturb and observe” experiments involving complex biological phenomena.
The use of microfluidic technologies is now extending to the generation of concentration gradients of cytokines in cultures of stem cells. Spatial gradients of diffusible signaling molecules are known to determine cell migration, lineage specification and maturation during development and are of paramount interest to human stem cell research efforts. A microfluidic platform of this kind was designed to expose the cultured cells to stable concentration gradients for more than a week, with only minimal handling and no external power source (Park et al., 2009). The gradient was maintained by a combination of osmotic and capillary action. To demonstrate the utility of the system, hESC-derived neural progenitors were cultured for 8 days with exposure to gradients of Shh, FGF8, and BMP4. Neural progenitors successfully differentiated into neurons, generating a complex neural network. The average numbers of neuronal cell body clusters and neurite bundles were directly proportional to Shh concentrations.
Bioengineering a cell niche
Tissue viability and function depend upon regulated replenishment of differentiated cells, through cell propagation, differentiation and architectural organization. Coordinating these processes occurs at multiple levels, but must begin with stem/progenitor cells. Without preservation of these cells and balance of their self-replenishment versus production of maturing cells, tissue integrity cannot be maintained. Regulation of stem cells has been associated with a stem cell niche. All cells engage with other cells and extracellular environments, but the regulation of cell fate is perhaps most exquisite for the stem/progenitor cells. Therefore, cell niches have generally focused on the stem cell niche, an anatomically definable tissue site where self-renewal and differentiation of stem cells is regulated (Voog and Jones, 2010).
Understanding in detail the components and physiology of stem cell niches is still quite limited. There has been progress for a number of tissues (Hsu et al., 2011; Sato et al. 2011), but the complexity of niche inputs is still best defined for bone marrow hematopoiesis. It is now clear that multiple cell types and extracellular components participate in altering the number, proliferative activity and localization of stem cells. It is also clear that thinking of a niche as a single cell type or single matrix component is too simple and that physiologic regulation depends upon multiple and often countervailing influences. Among these are cues from multiple mesenchymal populations (from undifferentiated cells to osteolineages to adipocytic cells), hematopoietic descendents like osteoclasts and macrophages and sympathetic neurons. (Calvi et al., 2003; Zhang et al., 2003; Kollet et al., 2006; Naveiras et al., 2009; Omatsu et al., 2010; Winkler et al., 2010; Katayama, et al., 2006). In addition, extracellular matrix glycoproteins (like osteopontin), signaling and small molecules (like ionic calcium and oxygen) all appear to play a role (Adams et al., 2006; Nilsson et al., 2005; Parmar et al., 2007; Stier et al., 2005). Recapitulating this system in vitro is extremely challenging. However, progress is being made using constructs of reduced complexity.
It has long been known the hematopoietic stem cell (HSC) preservation and differentiation can occur on adherent ‘stomal’ feeder layers. Refining these methods based on knowledge of the endogenous cell niche has been moving forward, now enabling relatively robust co-culture systems that can be used in intermediate-throughput chemical screens. In addition, highly promising engineered 2-D and 3-D niche models have been developed by the laboratory of Helen Blau (Lutolf et al., 2009). Using micropatterned hydrogels of varying properties containing growth factors, function of hematopoietic and, dramatically, muscle stem cells can be modulated. In the latter case, altering the elasticity of the hydrogel substrate to more closely mimic the conditions found in vivo, resulted in self-renewal of muscle stem cells ex vivo such that they were then effective sources of muscle upon transplantation (Gilbert et al., 2010). These results strongly support the notion that recapitulation of some elements of the in vivo niche can be productively used to increase stem cell number and provide useful cell populations for either transplantation of subsequent participation in bioengineered devices. Micropatterning to mimic architectural relationships in vivo has also been shown to result in tissue morphogenesis ex vivo that may be the basis for the reconstruction of complex tissues (Nelson et al., 2006).
Directed stem cell differentiation
Scaffold-bioreactor systems
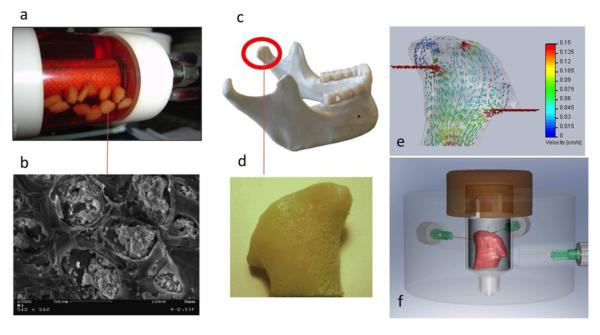
In a pioneering bioengineering study, hESCs were cultured in porous alginate scaffolds using a hydrodynamically active bioreactor, under conditions that promote the formation of embryoid bodies (EBs) and subsequent vascular differentiation (Gerecht et al., 2004). The confining environment of scaffold pores resulted in the formation of small, uniformly sized EBs, each in a scaffold pore (Figure 3 a, b). Once differentiation factors were introduced, these same EBs underwent vascular differentiation. Control of EB size – for example by micropatterning followed by suspension culture - proved beneficial for scaling up cell differentiation to large cell numbers (Niebruegge et al., 2009).
Figure 3. Scaffold-bioreactor systems for human stem cells.
hESCs were cultured in porous alginate scaffolds using a rotating bioreactor (a) to form spatially defined EBs inside scaffold pores (b) An anatomically correct bone graft in the exact size and shape of a human temporomandibular joint condyle (reproduced with permission from Gerecht et al. 2004). (c) has been engineered by using an anatomically shaped decellularized bone scaffold (d) seeded with human mesenchymal stem cells and cultured with medium perfusion (e) in an “anatomical” bioreactor (panels c, d, e reproduced with permission from Grayson et al. 2009)
Various types of hydrogels have been developed over the last decade for propagation and early differentiation of hESCs. The importance of these studies is that animal feeder layers are replaced by cell-friendly macromolecules (such as hyaluronane, Gerecht et al., 2007) that encapsulate viable cells by photopolymerization, have tailored structural and mechanical properties, and can incorporate functional groups or control-release microcarriers (Ferreira et al., 2007). Insights into the dynamics of cell-matrix interactions helped derive design requirements for cell culture scaffolds (Engler et al., 2006 & 2008, Guilak et al., 2009; Discher et al., 2009).
New technologies are emerging in conjunction with the use of these new hydrogels to enable precise and systematic variation of environmental factors in high-throughput settings (Underhill and Bhatia, 2007; Flaim et al., 2005; Figallo et al., 2007). Overall, the hydrogel-bioreactor platforms provide unique ways to study the role of stem cells as mediators of repair. It will be most interesting to see how effectively we can translate these in vitro models into in vivo platforms for tissue repair.
Mechanical conditioning
Subjecting stem cells to electrical stimulation and mechanical loading could be a way to direct their fate and function during various stages of development, in vitro and in vivo. We are only beginning to learn about the effects of physical signals on cell commitment, differentiation and assembly. Electrical and mechanical signals are related – for example, muscle cells are induced to contract by electrical signals – and they both enhance mass transport of nutrients, most critically oxygen, to and from the cells. From the early days of tissue engineering, mechanical conditioning of cells cultured on scaffolds was explored based on the premise that the same forces that govern cell differentiation and tissue development in vivo would also enhance cell differentiation and tissue development in vitro. Human ligaments were engineered by applying dynamic tension and torsion in a specialized bioreactor designed to mimic mechanical forces in human knee. Interestingly, mechanical loading alone, without specific growth factors, induced cell alignment and the accumulation of ligament-specific markers in favor of alternate differentiation paths into cartilage or bone (Altman et al., 2002).
Also using a biomimetic paradigm, functional blood vessels were engineered from adult hMSCs by staged application of morphogens and pulsatile fluid pressure (Gong and Niklason, 2008). The conditions of mechanical stimulation were designed to mimic those associated with native vessels: (i) hydrodynamic shear acting on endothelial cells due to lumen flow and on smooth muscle cells due to interstitial flow, (ii) cyclic pressure, and (iii) circumferential and (iv) longitudinal stretch. An elegant approach was recently proposed that enables geometry-force control of stem cell differentiation, by cell culture on geometrically defined patterns (Ruiz and Chen, 2008; Wan et al., 2010).
Electrical conditioning
Electrical signals play major roles in stem cell differentiation into cardiac, vascular and neural lineages. During early development, direct currents gradually give way to the time-varying currents present in adult tissues. Interestingly, in the case of injury, the body again reverts to direct currents to drive the repair processes. Engineering strategies may well need to follow the same pattern and utilize the developmental and would-healing currents. For example, in hESC-derived EBs, the application of direct current electrical fields enhanced cardiac differentiation, through mechanisms involving the generation of reactive oxygen species (Serena et al., 2009). When subjected to direct currents similar to those encountered during wound healing, human adipose-derived mesenchymal stem cells elongated and aligned in parallel, disassembled gap junctions, and upregulated the expression of genes for connexin-43, thrombomodulin, VEGF, and FGF. The same effects were observed for human epicardial fat-derived stem cells (Tandon et al., 2009).
To enable high-throughput studies of electrical signals at the cell level, a microscale bioreactor has been developed with an interdigitated array of electrodes generated by laser ablation of a conductive coating on a glass slide (Tandon et al., 2010). The culture space consisted of an array of 200 μm wide electrodes positioned at 200 μm distances, and the cells were cultured between each pair of positive and negative electrodes. When subjected to pulsatile electrical fields, adipose derived mesenchymal stem cells oriented and aligned, and increased their proliferation rate and the number of gap junctions. This simple and practical culture system allows the study of interactive effects of surface topography and pulsatile electrical fields on stem cells, a regime shown to greatly affect the behavior of cardiac myocytes and cardiac fibroblasts (Au et al., 2007).
Growing tissues and organs
Repopulation of native tissue matrix
There is a long clinical history of using decellularized heart valves, which exhibit an extremely complex shape and structure that determines its biomechanical function, and can be recellularized in vivo for adequate long-term function (Elkins et al., 2001). About twenty years ago, the tissue engineering community started to seek out various “biological scaffold” candidates, produced by removing cellular material from tissues or whole organs, because of their ability to maintain much of the complexities of the composition, structure and biomechanics of native tissue (Gilbert et al., 2006; Badylak, 2007; Ott et al., 2008; Ott et al., 2010; Petersen et al., 2010; Uygun et al., 2010; Grayson et al., 2009). From blood vessels to bladder, muscle, bone and lung, these scaffolds enabled studies of stem cells in native-like environments and resulted in some recent remarkable examples of engineering complete organs.
In 2008, a patient was implanted with a bioengineered airway (Macchiarini et al., 2008) made from allogeneic human trachea. The donor’s trachea was processed to remove cells and antigens and repopulated by the recipient’s cells, to obtain a graft that was used to replace the whole left bronchus. The same year, another group decellularized rodent hearts by coronary perfusion with detergents, using a method that preserved most of the composition and architecture of the heart matrix. When the biological template was reseeded with cells and cultured in a bioreactor with medium perfusion, the engineered construct started to beat and by one month generated some pumping function (2% of the adult heart, 16% of the fetal heart) (Ott et al., 2008).
Most recently, two studies published within a week of each other reported the engineering of a rodent lung by repopulating fully decellularized lungs (Ott et al., 2010; Peterson et al., 2010). Remarkably, these engineered lungs functioned in vivo, and persisted for several hours. Lessons learned from using such scaffolds supplied by nature are leading into the design of the next generation of synthetic scaffolds with hierarchical architectures that mimic the structure and function of the native extracellular matrix (Moutos et al., 2007; Engelmayr et al., 2008).
Customized tissue grafts
Personalized tissue grafts - engineered from the patient’s own stem cells in the precise anatomical shapes of the defects that need to be treated would revolutionize the way we currently treat large tissue reconstructions. This approach would combine best of the two worlds: the advantages of living bone autografts (the right structure, mechanical and metabolic function, ability to integrate and remodel) and synthetic materials (precise anatomical shape, off-the shelf availability). Engineering of living grafts that would also be anatomically shaped and tailored to the patient critically depends on our ability to direct stem cell differentiation towards functional tissue assembly within clinically sized engineered grafts (Grayson et al., 2008).
A novel tissue engineering system has been established for the in vitro creation of an entire bone condyle containing viable cells at physiologic density surrounded by bone matrix (Grayson et al., 2009). Anatomically-shaped scaffolds with the exact geometry of a temporomandibular joint (TMJ) were generated from fully decellularized bone using digitized clinical images, seeded with human mesenchymal cells, and cultured with interstitial flow of culture medium in an “anatomical” bioreactor (Figure 3 c-f). For the first time for bone grafts of this size and complexity, cells were fully viable and present at a physiologically high density. Flow patterns associated with the complex geometry of the bone graft provided a unique opportunity to correlate the architecture of the forming bone with interstitial flow patterns. In another study, the articular surface of a synovial joint was regenerated in a rabbit model by homing of endogenous cells into a bioactive scaffold (Lee et al., 2010). We expect that these approaches, used individually or in combination, will help provide a variety of anatomically shaped grafts to meet the needs of a specific patient for a given tissue reconstruction.
Summary and challenges ahead
Novel bioengineered culture platforms can provide tight environmental control along with the physiological transport and signaling, and thereby enable study of development, regeneration, and disease under conditions that predict the human in vivo context. In vitro, human stem/progenitor cells are still mostly studied in systems that lack the structural and signaling specification of native tissues, the temporal and spatial sequences of molecular and physical regulatory factors, and the dynamic forces and systemic factors provided by blood circulation. In whole animal models, human cells are studied in an environment not necessarily representative of their native organism, and with limited control of and insight into cellular responses. As a result, in vitro studies of human cells/tissues often fail to predict findings in translational animal models and human clinical studies, increasing the time and cost, and decreasing the effectiveness of any resulting therapeutic strategies.
The path forward will almost certainly require a deeper understanding of how tissues are formed in the body. Lineage tracing models in mice are providing information about how cell types are related to one another during development and under stress conditions. Animal models enable durable fluorescent tagging of particular cell types at particular time points in vivo, serving as an in vivo equivalent of a pulse-chase experiment. As such, they can be leveraged to understand more than lineage relationships. For example, coupling these models with in vivo imaging technologies permits precise definition of 3D architectural relationships of multiple aspects of organ development and repair. Further, these novel approaches permit analysis of cells in particular positions or under particular conditions. As such, they can be the source of high-density data sets of the type needed for reconstructing tissues ex vivo.
Full leverage of the biologic systems to bioengineering contexts requires interaction of biologists and engineers in ways not yet achieved. The methods of analysis and even the language of individuals in these disciplines create a cultural divide that still limits the field. History tells us that such a gap has existed in other settings before and been successfully spanned. Enter any genomics laboratory and the collision between the worlds of computer science and molecular biology is immediately evident. When the technology of high throughput sequencing made the potential of unraveling complex genetic information a reality, visionary individuals in divergent disciplines came together, driven by opportunity. There is a similar change currently happening in the field of regenerative medicine. Tools are becoming available to study with engineering precision the complex biology of tissues. Materials, including cells, are now sufficiently plastic to enable recreation of complex in vivo environments with significant precision. The leaders in the field and trainees motivated to take the leap are now catalyzing truly interdisciplinary approaches to the study of stem cells and their application in regenerative medicine.
In summary, a more profound understanding of the biological requirements by bioengineers, and of the capabilities and limitations of advanced technologies by biologists are among the barriers we need to overcome in order to define the critical questions and devise approaches to address these questions. The interdisciplinary research is now moving the field forward and, for the very first time, the gap between engineering and biology is becoming manageable.
Acknowledgments
The fusion of the fields of cell biology and bioengineering has been driven by the pioneering efforts of many contributors working at the interface of the two disciplines. We acknowledge their collective contributions, and regret that space constraints prevented us from citing many meritorious studies that will continue to inspire future efforts. We also acknowledge research funding by NIH (HL076485, DE16525, HL089913, EB002520 to GVN and HL044851, HL097794, HL100402 and DK050234 to DTS), and thank Dr Nebojsa Mirkovic for his expert help with illustrations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams GB, Chabner KT, Alley IR, Olson DP, Szczepiorkowski ZM, Poznansky MC, Kos CH, Pollak MR, Brown EM, Scadden DT. Stem cell engraftment at the endosteal niche is specified by the calcium-sensing receptor. Nature. 2006;439:599–603. doi: 10.1038/nature04247. [DOI] [PubMed] [Google Scholar]
- Altman G, Horan RL, Martin I, Farhadi J, Stark PRH, Volloch V, Richmond JC, Vunjak-Novakovic G, Kaplan DL. Cell Differentiation by Mechanical Stress. FASEB J. 2002;16:270–272. doi: 10.1096/fj.01-0656fje. [DOI] [PubMed] [Google Scholar]
- Au HT, Cheng I, Chowdhury MF, Radisic M. Interactive effects of surface topography and pulsatile electrical field stimulation on orientation and elongation of fibroblasts and cardiomyocytes. Biomaterials. 2007;28:4277–4293. doi: 10.1016/j.biomaterials.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badylak SF. The extracellular matrix as a biologic scaffold material. Biomaterials. 2007;28:3587–3593. doi: 10.1016/j.biomaterials.2007.04.043. [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nature Biotechnology. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiak M, Melton DA. How to make beta cells? Current opinion in cell biology. 2009;21:727–732. doi: 10.1016/j.ceb.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick JA, Vunjak-Novakovic G. Engineered microenvironments for controlled stem cell differentiation. Tissue Eng Part A. 2009;15:205–219. doi: 10.1089/ten.tea.2008.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- Cimetta E, Cannizzaro C, Elvasore N, Vunjak-Novakovic G. Microarray bioreactors for steady-state and transient studies of stem cells. Methods. 2009;47:81–89. doi: 10.1016/j.ymeth.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JT, Gautrot JE, Trappmann B, Tan DW, Donati G, Huck WT, Watt FM. Actin and serum response factor transduce physical cues from the microenvironment to regulate epidermal stem cell fate decisions. Nat Cell Biol. 2010;12:711–718. doi: 10.1038/ncb2074. [DOI] [PubMed] [Google Scholar]
- Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- Dawson D, Mapili G, Erickson K, Taqvi S, Roy K. Biomaterials for stem cell differentiation. Adv Drug Del Rev. 2008;60:215–228. doi: 10.1016/j.addr.2007.08.037. [DOI] [PubMed] [Google Scholar]
- Discher DE, Mooney DJ, Zandstra PW. Growth factors, matrices, and forces combine and control stem cells. Science. 2009;324:1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- Frederiksen K, Jat PS, Valtz N, Levy D, McKay R. Immortalization of precursor cells from the mammalian CNS. Neuron. 1988;1:439–448. doi: 10.1016/0896-6273(88)90175-4. [DOI] [PubMed] [Google Scholar]
- Engelmayr GC, Cheng M, Bettinger CJ, Borenstein JT, Langer R, Freed LE. Accordion-like honeycombs for tissue engineering of cardiac anisotropy. Nature Materials. 2008;7:1003–1010. doi: 10.1038/nmat2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Engler AJ, Carag-Krieger C, Johnson CP, Raab M, Tang HY, Speicher DW, Sanger JW, Sanger JM, Discher DE. Embryonic cardiomyocytes beat best on a matrix with heart-like elasticity: scar-like rigidity inhibits beating. J Cell Sci. 2008;121:3794–802. doi: 10.1242/jcs.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira LS, Gerecht S, Fuller J, Shieh HF, Vunjak-Novakovic G, Langer R. Bioactive hydrogel scaffolds for controllable vascular differentiation of human embryonic stem cells. Biomaterials. 2007;28:2706–2717. doi: 10.1016/j.biomaterials.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figallo E, Cannizzaro C, Gerecht S, Burdick JA, Langer R, Elvassore N, Vunjak-Novakovic G. Micro-bioreactor array for controlling cellular microenvironments. Lab on a Chip. 2007;7:710–719. doi: 10.1039/b700063d. [DOI] [PubMed] [Google Scholar]
- Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat Methods. 2005;2:119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- Freytes D, Wan L, Vunjak-Novakovic G. Geometry and Force Control of Cell Function. Journal of Cellular Biochemistry. 2009;108:1047–1058. doi: 10.1002/jcb.22355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerecht-Nir S, Cohen S, Ziskind A, Itskovitz-Eldor J. Three-dimensional porous alginate scaffolds provide a conducive environment for generation of well-vascularizedembryoid bodies from human embryonic stem cells. Biotechnology and Bioengineering. 2004;88:313–320. doi: 10.1002/bit.20248. [DOI] [PubMed] [Google Scholar]
- Gerecht S, Burdick JA, Ferreira LS, Townsend SA, Langer R, Vunjak-Novakovic G. Hyaluronic acid hydrogel for controlled self-renewal and differentiation of human embryonic stem cells. Proc Nat Academy Sci. 2007;104:11298–11303. doi: 10.1073/pnas.0703723104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert PM, Havenstrite KL, Magnusson KE, Sacco A, Leonardi NA, Kraft P, Nguyen NK, Thrun S, Lutolf MP, Blau HM. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert TW, Sellaro TL, Badylak SF. Decellularization of tissues and organs. Biomaterials. 2006;27:3675–3683. doi: 10.1016/j.biomaterials.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Gong Z, Niklason LE. Small-diameter human vessel wall engineered from bone marrow-derived mesenchymal stem cells (hMSCs) FASEB J. 2008;22:1635–1648. doi: 10.1096/fj.07-087924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith LG, Swartz MA. Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol. 2006;7:211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- Grayson WG, Fröhlich M, Yeager K, Bhumiratana S, Cannizzaro C, Wan LQ, Chan ME, Liu ME, Guo X. Edward, Vunjak-Novakovic GV. Engineering anatomically shaped human bone grafts. Proceedings of the National Academy of Sciences USA. 2009;107:3299–3304. doi: 10.1073/pnas.0905439106. EX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, Young RA. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7:249–257. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilak F, Cohen DM, Estes BT, Gimble JM, Liedtke W, Chen CS. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 2009;5:17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayflick L. The cell biology of aging. J Invest Dermatol. 1979;73:8–14. doi: 10.1111/1523-1747.ep12532752. [DOI] [PubMed] [Google Scholar]
- Hsu YC, Pasolli HA, Fuchs E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell. 2011;144:92–105. doi: 10.1016/j.cell.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui EE, Bhatia SN. Micromechanical control of cell-cell interactions. Proc Natl Acad Sci U S A. 2007;104:5722–5726. doi: 10.1073/pnas.0608660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal N, Haynesworth SE, Caplan AI, Bruder SP. Osteogenic differentiation of purified, culture-expanded human mesenchymal stem cells in vitro. J Cell Biochem. 1997;64:295–312. [PubMed] [Google Scholar]
- Kamei K, Guo S, Yu ZT, Takahashi H, Gschweng E, Suh C, Wang X, Tang J, McLaughlin J, Witte ON, Lee KB, Tseng HR. An integrated microfluidic culture device for quantitative analysis of human embryonic stem cells. Lab Chip. 2009;9:555–563. doi: 10.1039/b809105f. [DOI] [PubMed] [Google Scholar]
- Katayama Y, Battista M, Kao W-M, Hidalgo A, Peired AJ, Thomas SA, Frenette PS. Signals from the sympathetic nervous system regulate hematopoietic stem cell egress from bone marrow. Cell. 2006;124:407–421. doi: 10.1016/j.cell.2005.10.041. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C, Morrison SJ. SLAM Family Receptors Distinguish Hematopoietic Stem and Progenitor Cells and Reveal Endothelial Niches for Stem Cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollet O, Dar A, Shivtiel S, Kalinkovich A, Lapid K, Sztainberg Y, Tesio M, Samstein RM, Goichberg P, Spiegel A, et al. Osteoclasts degrade endosteal components and promote mobilization of hematopoietic progenitor cells. Nature Medicine. 2006;12:657–664. doi: 10.1038/nm1417. [DOI] [PubMed] [Google Scholar]
- Lee CH, Cook JL, Mendelson A, Moioli EK, Yao H, Mao JJ. Regeneration of the articular surface of the rabbit synovial joint by cell homing: a proof of concept study. Lancet. 2010;376:440–448. doi: 10.1016/S0140-6736(10)60668-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchetta EM, Lee JH, Fu LA, Patel NH, Ismagilov RF. Dynamics of Drosophila embryonic patterning network perturbed in space and time using microfluidics. Nature. 2005;434:1134–1138. doi: 10.1038/nature03509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutolf MP, Gilbert PM, Blau HM. Designing materials to direct stem-cell fate. Nature. 2009;462:433–441. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutolf MP. Integration column: Artificial ECM: expanding the cell biology toolbox in 3D. Integr. Biol. 2009;1:235–241. doi: 10.1039/b902243k. [DOI] [PubMed] [Google Scholar]
- Lysaght MJ, Reyes J. The growth of tissue engineering. Tissue Eng. 2001;7:485–493. doi: 10.1089/107632701753213110. [DOI] [PubMed] [Google Scholar]
- Macchiarini P, Jungebluth P, Go T, Asnaghi MA, Rees LE, Cogan TA, Dodson A, Martorell J, Bellini S, Parnigotto PP, Dickinson SC, Hollander AP, Mantero S, Conconi MT, Birchall MA. Clinical transplantation of a tissue-engineered airway. Lancet. 2008;372:2023–2230. doi: 10.1016/S0140-6736(08)61598-6. [DOI] [PubMed] [Google Scholar]
- Moutos FT, Freed LE, Guilak F. A biomimetic three-dimensional woven composite scaffold for functional tissue engineering of cartilage. Nature materials. 2007;6:162–167. doi: 10.1038/nmat1822. [DOI] [PubMed] [Google Scholar]
- Nair LS, Laurencin CT. Biodegradable polymers as biomaterials. Prog Poly Sci. 2007;32:762–798. [Google Scholar]
- Naveiras O, Nardi V, Wenzel PL, Hauschka PV, Fahey F, Daley GQ. Bone-marrow adipocytes as negative regulators of the haematopoietic microenvironment. Nature. 2009;460:259–263. doi: 10.1038/nature08099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Bissell MJ. Modeling dynamic reciprocity: engineering three-dimensional culture models of breast architecture, function, and neoplastic transformation. Semin Cancer Biol. 2005;15:342–352. doi: 10.1016/j.semcancer.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Jean RP, Tan JL, Liu WF, Sniadecki NJ, Spector AA, Chen CS. Emergent patterns of growth controlled by multicellular form and mechanics. Proc Natl Acad Sci U S A. 2005;102:11594–11599. doi: 10.1073/pnas.0502575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Vanduijn MM, Inman JL, Fletcher DA, Bissell MJ. Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science. 2006;314:298–300. doi: 10.1126/science.1131000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebruegge S, Bauwens CL, Peerani R, Thavandiran N, Masse S, Sevaptisidis E, Nanthakumar K, Woodhouse K, Husain M, Kumacheva E, Zandstra PW. Generation of human embryonic stem cell-derived mesoderm and cardiac cells using size-specified aggregates in an oxygen-controlled bioreactor. BiotechnolBioeng. 2009;102:493–507. doi: 10.1002/bit.22065. [DOI] [PubMed] [Google Scholar]
- Nilsson SK, Johnston HM, Whitty GA, Williams B, Webb RJ, Denhardt DT, Bertoncello I, Bendall LJ, Simmons PJ, Haylock DN. Osteopontin, a key component of the hematopoietic stem cell niche and regulator of primitive hematopoietic progenitor cells. Blood. 2005;106:1232–1239. doi: 10.1182/blood-2004-11-4422. [DOI] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2010;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omatsu Y, Sugiyama T, Kohara H, Kondoh G, Fujii N, Kohno K, Nagasawa T. The essential functions of adipo-osteogenic progenitors as the hematopoietic stem and progenitor cell niche. Immunity. 2010;33:387–399. doi: 10.1016/j.immuni.2010.08.017. [DOI] [PubMed] [Google Scholar]
- Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- Ott HC, Matthiesen TS, Goh S-K, Black LD, Kren SM, Netoff TI, Taylor DA. Perfusion-decellularized matrix: using nature’s platform to engineer a bioartificial heart. Nat Med. 2008;14:213–221. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- Ott HC, Clippinger B, Conrad C, Schuetz C, Pomerantseva I, Ikonomou L, Kotton D, Vacanti JP. Regeneration and orthotopic transplantation of a bioartificial lung. Nat Med. 2010;16:927–933. doi: 10.1038/nm.2193. [DOI] [PubMed] [Google Scholar]
- Park JY, Kim S-K, Woo D-H, Lee E-J, Kim J-H, Lee S-H. Differentiation of neural progenitor celles in amicrofluidic chip-generated cytokine gradient. Stem Cells. 2009;27:2646–2654. doi: 10.1002/stem.202. [DOI] [PubMed] [Google Scholar]
- Parmar K, Mauch P, Vergilio JA, Sackstein R, Down JD. Distribution of hematopoietic stem cells in the bone marrow according to regional hypoxia. Proc Natl Acad Sci U S A. 2007;104:5431–5436. doi: 10.1073/pnas.0701152104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira RF, Halford KW, O’Hara MD, Leeper DB, Sokolov BP, Pollard MD, Bagasra O, Prockop DJ. Cultured adherent cells from marrow can serve as long-lasting precursor cells for bone, cartilage, and lung in irradiated mice. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:4857–4861. doi: 10.1073/pnas.92.11.4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TH, Calle EA, Zhao L, Lee EJ, Gui L, Raredon MB, Gavrilov K, Yi T, Zhuang ZW, Breuer C, Herzog E, Niklason LE. Tissue-engineered lungs for in vivo implantation. Science. 2010;329:538–541. doi: 10.1126/science.1189345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietze RL, Valcanis H, Brooker GF, Thomas T, Voss AK, Bartlett PF. Purification of a pluripotent neural stem cell from the adult mouse brain. Nature. 2001;412:736–739. doi: 10.1038/35089085. [DOI] [PubMed] [Google Scholar]
- Ruiz SA, Chen CS. Emergence of patterned stem cell differentiation within multicellular structures. Stem Cells. 2008;26:2921–2927. doi: 10.1634/stemcells.2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchetti B, Funari A, Michienzi S, Di Cesare S, Piersanti S, Saggio I, Tagliafico E, Ferrari S, Robey PG, Riminucci M, et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- Saha K, Jaenisch R. Technical Challenges in Using Human Induced Pluripotent Stem Cells to Model Disease. Cell Stem Cell. 2009;5:584–595. doi: 10.1016/j.stem.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RI, Christensen JL, Conboy IM, Conboy MJ, Rando TA, Weissman IL, Wagers AJ. Isolation of adult mouse myogenic progenitors: functional heterogeneity of cells within and engrafting skeletal muscle. Cell. 2004;119:543–554. doi: 10.1016/j.cell.2004.10.021. [DOI] [PubMed] [Google Scholar]
- Skelley AM, Kirak O, Suh H, Jaenisch R, Voldman J. Microfluidic control of cell pairing and fusion. Nat Methods. 2009;6:147–152. doi: 10.1038/nmeth.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smukler, et al. Cell Stem Cell. 2011;(this issue) xxx-xxx. [Google Scholar]
- Snippert HJ, Haegebarth A, Kasper M, Jaks V, van Es JH, Barker N, van de Wetering M, van den Born M, Begthel H, Vries RG, et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010;327:1385–1389. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- Stier S, Ko Y, Forkert R, Lutz C, Neuhaus T, Grunewald E, Cheng T, Dombkowski D, Calvi LM, Rittling SR, et al. Osteopontin is a hematopoietic stem cell niche component that negatively regulates stem cell pool size. J Exp Med. 2005;201:1781–1791. doi: 10.1084/jem.20041992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tandon N, Cannizzaro C, Chao PH, Maidhof R, Marsano A, Au HT, Radisic M, Vunjak-Novakovic G. Electrical stimulation systems for cardiac tissue engineering. Nat Protoc. 2009;4:155–173. doi: 10.1038/nprot.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandon N, Marsano A, Maidhof R, Numata K, Montori-Sorentino C, Cannizzaro C, Voldman J, Vunjak-Novakovic G. Surface-patterned indium tin oxide electrodes for electrical stimulation of cardiac cells. Lab on a Chip. 2010;10:692–700. doi: 10.1039/b917743d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terraciano V, Hwang N, Moroni L, Park HB, Zhang Z, Mizrahi J, Seliktar D, Elisseeff J. Differential response of adult and embryonic mesenchymal progenitor cells to mechanical compression in hydrogels. Stem Cells. 2007;25:2730–2738. doi: 10.1634/stemcells.2007-0228. [DOI] [PubMed] [Google Scholar]
- Underhill GH, Bhatia SN. High-throughput analysis of signals regulating stem cell fate and function. Curr Opin Chem Biol. 2007;11:357–366. doi: 10.1016/j.cbpa.2007.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uygun BE, Soto-Gutierrez A, Yagi H, Izamis ML, Guzzardi MA, Shulman C, Milwid J, Kobayashi N, Tilles A, Berthiaume F, Hertl M, Nahmias Y, Yarmush ML, Uygun K. Organ reengineering through development of a transplantable recellularized liver graft using decellularized liver matrix. Nat Med. 2010;16:814–820. doi: 10.1038/nm.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voog J, Jones DL. Stem Cells and the Niche: A Dynamic Duo. Cell Stem Cell. 2010;6:103–115. doi: 10.1016/j.stem.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan LQ, Kang SM, Eng G, Grayson WL, Lu XL, Huo B, Gimble J, Guop XE, Mow VC, Vunjak-Novakovic G. Geometric Control of Adult Human Stem Cell Morphology and Differentiation. Integrative Biology. 2010;2:346–353. doi: 10.1039/c0ib00016g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler IG, Sims NA, Pettit AR, Barbier V, Nowlan B, Helwani F, Poulton IJ, van Rooijen N, Alexander KA, Raggatt LJ, et al. Bone marrow macrophages maintain hematopoietic stem cell (HSC) niches and their depletion mobilizes HSCs. Blood. 2010;116:4815–4828. doi: 10.1182/blood-2009-11-253534. [DOI] [PubMed] [Google Scholar]
- Yuan X, Li W, Ding S. Small molecules in cellular reprogramming and differentiation. Progress in drug research. 2011;67:253–266. doi: 10.1007/978-3-7643-8989-5_13. [DOI] [PubMed] [Google Scholar]
- Zhang J, Niu C, Ye L, Huang H, He X, Tong W-G, Ross J, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- Zhao X, Kim J, Cezar CA, Huebsch N, Lee K, Bouhadir K, Mooney DJ. Active scaffolds for on-demand drug and cell delivery. Proc Natl Acad Sci U S A. 2011;108:67–72. doi: 10.1073/pnas.1007862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann W-H, Melnychenko I, Wasmeier G, Didie M, Naito H, Nixdorff U, Hess A, Budinsky L, Brune K, Michaelis B, Dhein S, Schwoerer A, Ehmke H, Eschenhagen T. Engineered heart tissue grafts improve systolic and diastolic function in infarcted rat hearts. Nat Med. 2006;12:452–458. doi: 10.1038/nm1394. [DOI] [PubMed] [Google Scholar]