Abstract
For the first time the genetic diversity among the uniformed personnel in Kinshasa, the capital city of the Democratic Republic of Congo (DRC), a country that has experienced military conflicts since 1998 and in which the global HIV-1/M pandemic started, has now been documented. A total of 94 HIV-1-positive samples, collected in 2007 in Kinshasa garrison settings from informed consenting volunteers, were genetically characterized in the pol region (protease and RT). An extensive diversity was observed, with 51% of the strains corresponding to six pure subtypes (A 23%, C 13.8%, D, G, H, J, and untypable), 15% corresponding to nine different CRFs (01, 02, 11, 13, 25, 26, 37, 43, and 45), and 34% being unique recombinants with one-third being complex mosaic viruses involving three or more different subtypes/CRFs. Only one strain harbored a single mutation, I54V, associated with drug resistance to protease inhibitors. Due to their high mobility and potential risk behavior, HIV infections in military personnel can lead to an even more complex epidemic in the DRC and to a possible increase of subtype C.
The global HIV/AIDS epidemic is characterized by a high diversity of human immunodeficiency viruses (HIV). Based on phylogenetic analyses of numerous isolates obtained from diverse geographic origins, HIV-1 is classified into four groups, M, N, O, and P and each HIV-1 group corresponds to an independent cross-species transmission from chimpanzees (Pan troglodytes troglodytes) and gorillas (Gorilla gorilla) in West Central Africa.1,2 Group M (for Major) represents the majority of HIV-1 strains found worldwide and is responsible for the global pandemic. HIV-1 group M can be further subdivided into nine subtypes (A–D, F–H, J, K), denoted with letters, and subtypes A and F are subdivided into subsubtypes, A1 to A4 and F1 and F2. Numerous intersubtype recombinant viruses are also observed and today at least 45 CRFs and numerous URFs are recognized (http://www.hiv.lanl.gov).
The classification of HIV strains has helped in tracking the course of the HIV pandemic. The initial diversification of group M may have occurred within or near the Democratic Republic of Congo(DRC), where the highest diversity of group M strains has been observed and the earliest cases of HIV-1 infection (1959 and 1960) have been documented.3 More precisely, molecular epidemiological studies revealed that the epicenter of the HIV pandemic is situated in the area of Kinshasa, the capital city of the DRC, where the highest genetic diversity of HIV-1 M, in number of cocirculating subtypes, intrasubtype diversity, and recombinants, has been observed and about 5% of sequences reported from DRC are not clustering with any of the known HIV-1 variants.4 Interestingly, the geographic distribution of HIV-1 M variants is also heterogeneous within the country, and between 6 and 11 HIV-1 variants cocirculate in the different regions studied.
The highest genetic diversity has been observed in Kinshasa and subtype A is the most prevalent variant in almost each region, except in the extreme south where subtype C largely predominates. Subtypes D, G, and H were found mostly in the northern and western parts of the country. In Kisangani, in the northeast, subtypes A and D are relatively well represented, as in countries bordering the DRC in the northeast.4–6
Since 1998, the country has experienced an internal armed conflict involving six foreign armed forces, a situation that has led to profound socioeconomic changes and important displacement of population groups. The political instability that prevailed in the country did not result in a substantial change in overall HIV prevalence.7 However, comparison of subtype distribution in Kinshasa between 1997 and 2002 revealed an overall increase of subtype C from 2.1 to 9.7% and more precisely from 0% to 18.9% in female sex workers (FSW) (p = 0.013).6 Military personnel also constitute a very high-risk population for sexually transmitted infections (STI) including HIV, especially during conflicts when their economic and political powers are higher and are exerted away from their normal social control and partners.8,9 In addition, they are often deployed in different areas of the country and can subsequently spread new HIV-1 variants and/or recombinants across the different regions.
HIV-1 variants that circulate among military personnel in the DRC were documented in this project. The study was designed on the basis of HIV sentinel surveillance among the uniformed services in Kinshasa from June to September 2007. It received support from the DRC Ministry of Defense, Ministry of Public Health and was carried out in close collaboration with the National AIDS Control Program, the National Institute for Biomedical Research, and the Kinshasa School of Public Health. Blood samples were collected on EDTA from informed consenting individuals among all members of the military services in Kinshasa, the capital city of the DRC. Samples were collected between 21 June and 11 September 2007 inside the Kinshasa garrison. HIV screening was done through the national algorithm that included a rapid test (Determine HIV1/2; Abbott, Tokyo, Japan), an ELISA test (Enzygnost HIV1/2 integral, Dade Behring, Dresden, Germany), and Western blot assay.
Plasma samples from 94 HIV-positive participants were shipped on dry ice for further characterization of the HIV strains. The majority of the study population was male (92.5%), although seven women were also present (7.5%). The median age was 35 years and ranged between 20 and 63 years. The main route of HIV transmission appeared to be heterosexual contact. The majority reported being married or living together with a regular partner; 17 individuals reported no regular partner. At the time of the study all were based in Kinshasa, but the time they spent in the garrison away from Kinshasa varied from 1 month to more than 20 years and participants also originated from different regions in the country: 40% were from the extreme south of the country; 19% were from Kinshasa; 16% originated from the Equateur province, principally from the extreme northwest; 10% were from the Oriental province, mainly from the extreme northeast; 7% came each from the Bas-Congo and Kivu regions; and 4–5% were from the Bandundu and Occidental Kasaï provinces. In addition, they have been assigned to military missions in several regions of the country between 2002 and 2007. However, the period of the different missions was quite variable, ranging from less than 1 month to 5 years. Also, the mission destinations corresponded to numerous regions in the country that were located at or near well-known internal armed conflict areas, mainly along the eastern frontiers from north to south, but also in the northwest and the southwest of the country, and to a lesser extent in the center of the DRC.
The plasma samples from the 94 HIV-positive participants were first subjected to an indirect ELISA assay using synthetic peptides derived from V3 loop consensus sequences to discriminate between HIV-1 group M, N, O and HIV-2 infection.10 The serological assays revealed that all samples were HIV-1 and no HIV-1 group O or N infections were identified. HIV-1 group M variants and genotypic drug resistance mutations were then studied in the pol region as previously described.11 Briefly, RNA was extracted using the QIAamp Viral RNA extraction kit (Qiagen SA, Courtabeauf, France) and processed for reverse transcription polymerase chain reaction (RT-PCR) with the integrase-specific primer IN3 5′-TCTATBCCATCTAAAAATAGTACTTTCCTGATTCC-3′ using the Expand reverse transcriptase (Roche Diagnostics, Meylan, France) according to the manufacturer's instructions. The resulting cDNA served as a template in the subsequent nested PCR reaction during which a 1865-base pairs fragment, corresponding to the protease and the first 440 amino acids of the reverse transcriptase region of the pol gene, was amplified with previously described primers and cycling conditions11 using the Expand Long Template PCR system (Roche Diagnostics, Meylan, France).
The amplified HIV-1 nucleic acid fragments were purified using the Geneclean Turbo Kit (Q-Biogen, MPbiomedicals, France) and directly sequenced with primers encompassing the pol region using BigDye Terminator version 3.1 (Applied Biosystems, Courtaboeuf, France) according to the manufacturer's instructions. Electrophoresis and data collection were done on an Applied Biosystems 3130XL Genetic Analyzer. The sequenced fragments from both strands were reconstituted using Seqman II from the DNAstar package v5.08 (Lasergene, Madison, WI).
The 94 HIV-1 samples were successfully amplified and sequenced and were subsequently aligned with known representatives of the different groups, subtypes, subsubtype and CRFs. We paid particular attention to include all CRFs that circulate in central and west-central Africa, including the most recently characterized CRF26_A5U12 and CRF45_AK.13 Positions with any gap between the sequences and areas of uncertain alignment were excluded from the analysis. Pairwise evolutionary distances were estimated with Kimura's two-parameter method. Phylogenetic trees were constructed by the neighbor-joining method, and the reliability of the tree topology was assessed by bootstrap analysis as implemented in the Clustal X software.14 All 94 sequences were systematically investigated for recombination using SimPlot 3.2 beta software (Stuart Ray, http://www.med.jhu.edu/deptmed/sray/). SimPlot performs similarity plots that determined the percent similarity between a newly characterized sequence and selected groups of references, by moving a window of 350–400 base pairs (bp) with 10–20 bp increments along the sequence alignment; similarity values are plotted at the midpoint of the 350–400 bp fragments. SimPlot also performs bootscanning on neighbor-joining (NJ) trees from the same sliding windows by using Seqboot, DNAdist (with Kimura two-parameter method), Neighbor, and Consensus from the Phylip package. One hundred bootstrap replicates are evaluated for each phylogeny and the bootstrap values are plotted at the midpoint of each sliding window. In these two sets of analyses, the new sequences were aligned with consensus sequences (50% threshold) representing all the references from the same alignment used for the phylogenetic analyses. Individual phylogenetic trees were then processed for each segment constituting a mosaic pattern, to better define the mosaic structure of each strain.
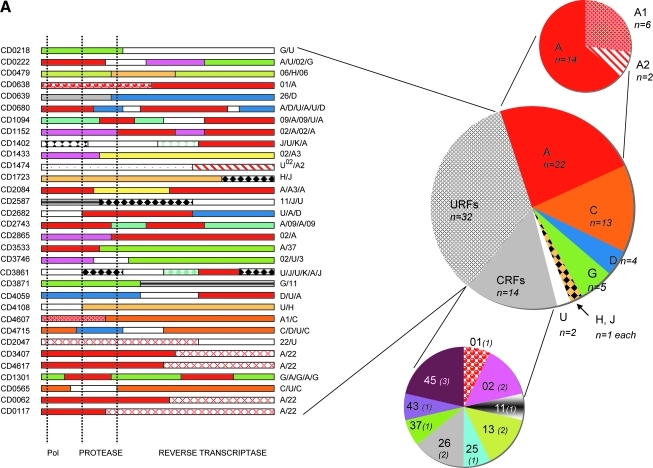
As shown in Fig. 1, numerous subtypes and CRFs were identified. At least six different pure subtypes were seen: 22 (23%) subtype A comprising two representatives of subsubtype A2 and 6 A1; 13 (13.8%) subtype C; four (4.3%) subtype D; five (5.3%) subtype G from which two sequences were very close to the subtype G fragment from CRF14_BG; one representative for subtype H and J each; and two strains that remained untypable and were then assigned as U. In addition, representatives of nine different circulating recombinant forms (CRFs) were identified: CRF01_AE, CRF02_AG, CRF11_cpx, CRF13_cpx, CRF25_cpx, CRF26_A5U, CRF37_cpx, CRF43_02G, and CRF45_cpx, each represented by one to three sequences, and globally representing 15% of all the strains from the study population. Subsequent to the detailed analysis of each individual sequence for recombination, we found a remarkable number of 32 (34%) unique recombinant strains (URFs). The detailed mosaic structures are shown in Fig. 1A.
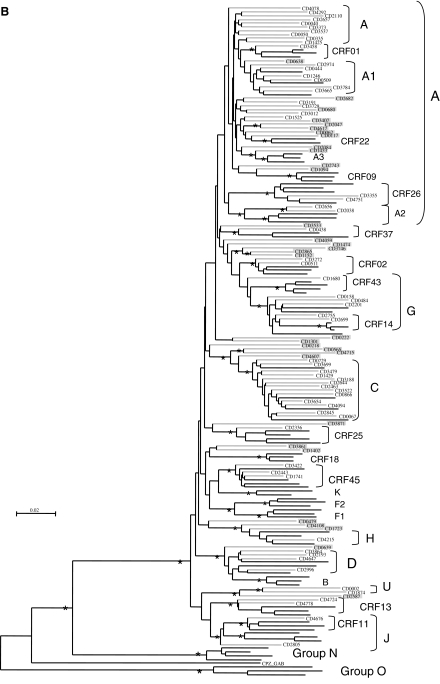
FIG. 1.
(A) Detailed distribution of the HIV-1 variants in the military population from the DRC and schematized structure of the unique recombinant forms. The start of the pol gene and the protease and RT regions are indicated at the bottom of the figure. (B) Phylogenetic tree analysis of the 94 newly obtained pol sequences from the military population. The phylogenetic analysis was done as indicated in the text using the neighbor-joining method with 100 bootstrap resamplings on 1455 aligned bases from the gap-stripped alignment. For better clarity, the tree was drawn with the minimal number of references, i.e., without some CRFs and unique sequences not represented among the samples of the study. The bootstrap values above 80 were indicated with an asterisk at each node. The branches for the reference sequences are in black and those for the sequences of the study population are in gray. Each newly sequenced URF was highlighted in gray and the corresponding mosaic structure is shown in (A).
The observed mosaic structures mainly involved subtypes and CRFs already known to circulate in the DRC and in the study population, but also with HIV-1 variants that were untypable or described in countries other than the DRC, such as subsubtype A3 and CRF22_01A1. Four strains were recombinants between CRF22_01A1 and subtype A and one strain was a mosaic of CRF22_01A1 and an untypable segment. CRF02_AG fragments were observed in five URFs in addition to the two pure CRF02 pol sequences. Finally, we could note that 22 of the 32 URFs (68.75%) were mosaics of two different subtypes/CRFs and 10 (31.25%) were complex recombinants in which three or more different subtypes/CRFs were involved.
We also analyzed the amino acid sequences for the presence of drug resistance mutations according to the last update of the WHO list for the surveillance of drug resistance mutations in antiretroviral treatment-naive patients (SDRM version 2009, http://hivdb.stanford.edu/pages/WHOResistanceList.html).15 Despite important genetic differences with the consensus B sequence, results indicate a single (1.2%) mutation I54IV in one patient, inducing low level resistance to protease inhibitors such as saquinavir/ritonavir (SQV/r), nelfinavir (NFV), and tipranavir/ritonavir (TPV/r). This mutation was detected in an HIV-1 strain obtained from a 39-year-old man infected with CRF01_AE. No mutation inducing potentially high level resistance to any class of reverse transcriptase inhibitors was found.
In this study we investigated the genetic diversity of HIV-1 group M among the military personnel in Kinshasa, DRC. We observed a high genetic diversity, with the cocirculation of at least six subtypes and nine CRFs; 34% of the samples were URFs. This high genetic diversity is in line with and is even more complex than what has been observed in previous reports, although results cannot be easily compared, because we studied the pol gene here whereas previous studies in the DRC focused mainly on env or gag sequences.4–6,16,17 In addition to the high numbers of cocirculating subtypes and CRFs, a high intrasubtype diversity was also seen, especially for subtype A, which predominated in our study population and represented 23% of the samples followed by subtype C, representing 13.8%. Previous reports also revealed that subtype A predominates in Kinshasa, Mbuyi-Mayi, and northern cities of the DRC.4,6 A high genetic diversity with the predominance of subtype A has also been reported in the Republic of Congo (Brazzaville) and Angola,18,19 two countries that share similar HIV molecular epidemiological features with the DRC, probably because of frequent population movements through these areas.
In this study we reported more CRFs than in previous reports, but five of the nine CRFs observed have only recently been described. CRF26_A5U and CRF45_AKU have been recently been described in samples from the DRC,12,13 whereas CRF25_cpx and CRF37_cpx were described in neighboring Cameroon and CRF43_02G was found in Saudi Arabia.20,21 The CRF43_02G sequence from our study clustered at a basal position of the CRF43_02G radiation suggesting a more divergent virus; characterization of other genomic regions from this sample might provide more information on the strains from Saudi Arabia and Central Africa and provide insights into the origin of this variant in Saudi Arabia. Unique recombinant pol sequences accounted for 34% of all the sequences, which is apparently in concordance with a previous study among 70 samples collected in 2002 in four provinces in the DRC and in different population groups, which reported 32.9% of recombinant pol sequences with often complex structures.6 Similarly, a study on a limited number of samples (n = 24) from Likasi (southeast DRC) reported a high recombination rate of 62% based on the pol-integrase and env-C2V3 regions.11
In general, the prevalence of CRF02_AG is lower in the DRC as compared to other west central African countries such as Cameroon, Gabon, and Equatorial Guinea.22 However, we observed CRF02_AG segments in 7% of the samples analyzed in the present study. A recent study also revealed that CRF02_AG sequences were involved in several recombination patterns in Angola, a country that shares borders with the DRC and Congo.19 It is worthwhile to study these CRF02_AG sequences in more detail to determine whether this variant originated in this part of Africa or whether this was more recently introduced. Similarly, we observed CRF22_01A1 segments in pol sequences of five (5.3%) individuals. CRF22_01A1 was previously mainly documented in Cameroon and recently also in Equatorial Guinea.23,24 Also, we document for the first time the presence of subsubtype A3 fragments in the DRC.
The high prevalence of URFs and their complex structure suggest that dual infections are not rare events. In general, the proportion of dual infections and URFs is higher in population groups with high-risk behavior, such as commercial sex workers.25 Also, in our previous studies in the DRC the prevalence of mosaic and/or intersubtype recombinant strains seems to be higher in commercial sex workers when compared to the general population. More than half (57.2%) of FSWs harbored interregion recombinant or mosaic strains in the DRC, versus less than one-third (28.6%) for the general population (N. Vidal and M. Peeters, personal observation).
The military population of this study is at high risk of infection with different strains, not only because of their potential risk behavior but also due to their high mobility across the country and to areas in which numerous variants cocirculate. These factors can therefore favor the intermixing of HIV-1 strains across the country. Although it was not possible to know when and where each participant from our study became infected, we traced back the region of origin and military missions. For example, for subtype C, which represented 13.8% of the HIV-1 strains in our study, we tried to see whether the region of origin and/or missions were located in the south of the country, where this subtype is more prevalent and represents 50% of the HIV-1 infections.6 Among the 13 subtype C-infected individuals, seven performed missions or originated from the south of the country and six individuals performed missions and/or originated in the northwest, northeast, west, and east of the country. Overall, no significant linkage could be found between observed subtypes and regions of origin and military missions, although the sample size was limited, especially per HIV-1 variant.
The epidemic in Central Africa is different from that in other parts of the world, where subtypes and CRFs are clearly defined in most cases and where certain variants have been introduced due to founder effects with the subsequent intermixing of recently introduced strains leading to new CRFs and URFs derived from the initial epidemic strains. In the DRC, the viral diversity is so high that what we found is most probably a continuum of evolution initiated since the beginning of the twentieth century.3 This high genetic diversity implies that it is not easy to detect changes in subtype distribution over time or among different population groups. Interestingly, in another study performed in 2007 in Kinshasa, only 3 (4.4%) of 67 patients, recently diagnosed with HIV infection, were infected with subtype C (J. Muwonga and M. Peeters, personal observations). In general, the subtype C prevalence in the military personnel from Kinshasa seemed higher than in the general population in 2007 (13.3% versus 4.4%), which is in line with our previous observations concerning the higher prevalence and the increase of subtype C in FSW and in other population groups at high risk for HIV infection.6
Finally, we have also reported only one drug resistance mutation to protease inhibitors in this putatively antiretroviral-naive population. The I54V mutation was seen in one CRF01_AE strain. The prevalence of circulating drug-resistant strains is in accordance with that reported (0–5%) among antiretroviral-naive populations in other sub-Saharan countries in which routine use of antiretroviral treatment is now a reality.26 Because the use of protease inhibitors in the DRC is limited, it is also possible that the observed mutation is a naturally occurring polymorphism. However, as access to antiretroviral treatment in the DRC continues to increase, care must be taken to ensure total compliance in order to prevent further development of drug resistance.
In a country in which so many subtypes and complex recombinants already cocirculate, the introduction of new strains could lead to even more complex recombinants or CRFs with possible different biological properties. The mobility of military personnel as well as displaced people related to political instability can lead to even more complex epidemics. The genetic diversity of HIV has an impact on serological and nucleic acid-based diagnostics as well as on antiretroviral therapy and the development of drug resistance.2 This ever increasing genetic diversity of HIV can thus continue to be a challenge for treatment, biological monitoring, and drug resistance in the DRC.
Accession Numbers
The sequences described in this chapter were submitted to the EMBL Nucleotide Sequence Database under accession numbers FR666603 to FR666696.
Acknowledgments
We thank the Ministry of Public Health and the Ministry of National Defense of the Democratic Republic of Congo for permission to undertake this study and the U.S. Embassy in the DRC for their continued support. C.F.D. was supported, in part, by funds from the National Institutes of Health (NIH) Fogarty International Center (FIC) AIDS International Training and Research Program (2 D 43 TW000010-16/17). N.D.W. is supported by the NIH Director's Pioneer Award (DP1-OD000370). G.V.F. is supported by the DoD HIV/AIDS Prevention Program (DHAPP), the Henry M. Jackson Foundation for the Advancement of Military Medicine, google.org, The Skoll Foundation, and the Global Emerging Infections Surveillance and Response System (GEIS)—a Division of the United States Armed Forces Health Surveillance Center. Additional support was provided by The Institut de Recherche pour le Dévelopement (IRD), France. We owe sincere gratitude to the staff at the Global Viral Forecasting Initiative (GVF) Cameroon for their technical support.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Keele BF. Van Heuverswyn F. Li Y, et al. Chimpanzee reservoirs of pandemic and non-pandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Heuverswyn F. Li Y. Neel C, et al. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature. 2006;444:164. doi: 10.1038/444164a. [DOI] [PubMed] [Google Scholar]
- 3.Worobey M. Gemmel M. Teuwen DE, et al. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature. 2008;455:661–664. doi: 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vidal N. Peeters M. Mulanga-Kabeya C, et al. Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J Virol. 2000;74:10498–10507. doi: 10.1128/jvi.74.22.10498-10507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang C. Li M. Mokili JL, et al. Genetic diversification and recombination of HIV type 1 group M in Kinshasa, Democratic Republic of Congo. AIDS Res Hum Retroviruses. 2005;21:661–666. doi: 10.1089/aid.2005.21.661. [DOI] [PubMed] [Google Scholar]
- 6.Vidal N. Mulanga C. Bazepeo SE, et al. Distribution of HIV-1 variants in the Democratic Republic of Congo suggests increase of subtype C in Kinshasa between 1997 and 2002. J Acquir Immune Defic Syndr. 2005;40:456–462. doi: 10.1097/01.qai.0000159670.18326.94. [DOI] [PubMed] [Google Scholar]
- 7.Behets F. Edmonds A. Kitenge F. Crabbé F. Laga M for the PTME Group. Heterogeneous and decreasing HIV prevalence among women seeking antenatal care in Kinshasa, Democratic Republic of Congo. Int J Epidemiol. 2010;39(4):1066–1073. doi: 10.1093/ije/dyq060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ba O. O'Regan C. Nachega J, et al. HIV/AIDS in African militaries: An ecological analysis. Med Confl Surviv. 2008;24:88–100. doi: 10.1080/13623690801950260. [DOI] [PubMed] [Google Scholar]
- 9.Omba Kalonda JC. Sexual violence in the Democratic Republic of Congo: Impact on public health? Med Trop (Mars) 2008;68:576–578. [PubMed] [Google Scholar]
- 10.Vergne L. Bourgeois A. Mpoudi-Ngole E, et al. Biological and genetic characteristics of HIV infections in Cameroon reveals dual group M and O infections and a correlation between SI-inducing phenotype of the predominant CRF02_AG variant and disease stage. Virology. 2003;17:851–855. doi: 10.1016/s0042-6822(03)00167-3. [DOI] [PubMed] [Google Scholar]
- 11.Vidal N. Mulanga C. Bazepeo ES. Kasali MJ, et al. HIV type 1 pol gene diversity and antiretroviral drug resistance mutations in the Democratic Republic of Congo (DRC) AIDS Res Hum Retroviruses. 2006;22:202–206. doi: 10.1089/aid.2006.22.202. [DOI] [PubMed] [Google Scholar]
- 12.Vidal N. Bazepeo SE. Mulanga C. Delaporte E. Peeters M. Genetic characterization of eight full-length HIV type 1 genomes from the Democratic Republic of Congo (DRC) reveal a new subsubtype, A5, in the A radiation that predominates in the recombinant structure of CRF26_A5U. AIDS Res Hum Retroviruses. 2009;25:823–832. doi: 10.1089/aid.2008.0283. [DOI] [PubMed] [Google Scholar]
- 13.Niama FR. Vidal N. Bazepeo SE, et al. CRF45_AKU, a circulating recombinant from Central Africa, is probably the common ancestor of HIV type 1 MAL and HIV type 1 NOGIL. AIDS Res Hum Retroviruses. 2009;25:1345–1353. doi: 10.1089/aid.2009.0169. [DOI] [PubMed] [Google Scholar]
- 14.Thompson JD. Gibson TJ. Plewniak F, et al. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett DE. Camacho RJ. Otelea D, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug resistance: 2009 update. Plos One. 2009;4(3):e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalish ML. Robbins KE. Pieniazek D, et al. Recombinant viruses and early global HIV-1 epidemic. Emerg Infect Dis. 2004;10:1227–1234. doi: 10.3201/eid1007.030904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kita K. Ndembi N. Ekwalanga M, et al. Genetic diversity of HIV type 1 in Likasi, Southeast of the Democratic Republic of Congo. AIDS Res Hum Retroviruses. 2004;20:1352–1357. doi: 10.1089/aid.2004.20.1352. [DOI] [PubMed] [Google Scholar]
- 18.Niama FR. Toure-Kane C. Vidal N, et al. HIV-1 subtypes and recombinants in the Republic of Congo. Infect Genet Evol. 2006;6:337–343. doi: 10.1016/j.meegid.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Bártolo I. Rocha C. Bartolomeu J, et al. Highly divergent subtypes and new recombinant forms prevail in the HIV/AIDS epidemic in Angola: New insights into the origins of the AIDS pandemic. Infect Genet Evol. 2009;9:672–682. doi: 10.1016/j.meegid.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Yamaguchi J. Badreddine S. Swanson P, et al. Identification of new CRF43_02G and CRF25_cpx in Saudi Arabia based on full genome sequence analysis of six HIV type 1 isolates. AIDS Res Hum Retroviruses. 2008;24:1327–1335. doi: 10.1089/aid.2008.0101. [DOI] [PubMed] [Google Scholar]
- 21.Powell RL. Zhao J. Konings FA, et al. Circulating recombinant form (CRF) 37_cpx: An old strain in Cameroon composed of diverse, genetically distant lineages of subtypes A and G. AIDS Res Hum Retroviruses. 2007;23:923–933. doi: 10.1089/aid.2007.0040. [DOI] [PubMed] [Google Scholar]
- 22.Peeters M. Toure-Kane C. Nkengasong JN. Genetic diversity of HIV in Africa: Impact on diagnosis, treatment, vaccine development and trials. AIDS. 2003;17:2547–2560. doi: 10.1097/01.aids.0000096895.73209.89. [DOI] [PubMed] [Google Scholar]
- 23.Carr JK. Wolfe ND. Torimiro JN, et al. HIV-1 recombinants with multiple parental strains in low-prevalence, remote regions of Cameroon: Evolutionary relics? Retrovirology. 2010;7:39. doi: 10.1186/1742-4690-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Djoko CF. Wolfe ND. Vidal N, et al. HIV-1 pol gene diversity and genotypic antiretroviral drug resistance mutations in Malabo, Equatorial Guinea. AIDS Res Hum Retroviruses. 2010;26(9):1027–1031. doi: 10.1089/aid.2010.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herbinger KH. Gerhardt M. Piyasirisilp S, et al. Frequency of HIV type 1 dual infection and HIV diversity: Analysis of low- and high-risk populations in Mbeya Region, Tanzania. AIDS Res Hum Retroviruses. 2006;22:599–606. doi: 10.1089/aid.2006.22.599. [DOI] [PubMed] [Google Scholar]
- 26.Hamers RL. Derdelinckx I. van Vugt M, et al. The status of HIV-1 resistance to antiretroviral drugs in sub-Saharan Africa. Antivir Ther. 2008;13:625–639. [PubMed] [Google Scholar]