Abstract
Heart failure (HF) is a modern epidemic and a heterogeneous disorder with many therapeutic options. While the average response to each individual treatment is favorable, significant interindividual variation exists in the response to HF therapeutics. As a result, the optimal regimen for an individual patient or subgroup of patients is elusive, with current treatment being mainly empirical. Pharmacogenetic customization of HF therapy may provide an important opportunity to improve the treatment of HF. Common genetic variations exist in genes related to most classes of HF drugs, many of which have known functional consequences for or established relationships with drug response. This review summarizes the current understanding of the pharmacogenetics of HF therapeutics, including angiotensin-converting enzyme inhibitors and β-blockers, and focuses on recent advances and medium-term expectations for the field.
Keywords: Aldosterone antagonist, angiotensin-converting enzyme inhibitor, angiotensin receptor blocker, β-blocker, heart failure, personalized medicine, pharmacogenetics
Introduction
Heart failure (HF) is a modern epidemic and an increasing public health concern. As mortality from coronary artery disease and stroke decline, the prevalence of HF increases, with approximately 5 million individuals affected and more than 500,000 new cases annually in the US. HF is lethal, with 1-year mortality rate estimates ranging from 25 to 45% [1,2]. The management of HF is also costly because of the chronic, progressive nature and frequent exacerbations of the disease, resulting in 3.4 million hospital visits in 2006 in the US [2]. Furthermore, with an incidence of approximately 10% in patients over 65 years of age and an aging population, this disease is likely to increase in importance [2].
HF is a heterogeneous disorder arising from a variety of etiologies that result in inadequate cardiac performance. After an initial (and potentially ongoing) insult, the disease is characterized by cascades of adverse physiological responses that lead to vasoconstriction, fluid retention and further compromise of cardiac function. These sympotoms are often accompanied by adverse cardiac remodeling and dilation, with reduced ejection fraction, although half of all patients with HF maintain an ejection fraction that is almost normal [3]. Maladaptive physiological responses in HF are well described, including the upregulation of the renin-angiotensin-aldosterone system (RAAS) and sympathetic nervous system, as well as other responses that are less understood, such as the involvement of inflammatory and apoptotic pathways in this disease [1,4–6].
Treatment strategies that are proven to reduce mortality are limited to use in patients with HF with reduced ejection fraction; therefore, this review focuses mainly on this patient group. Most therapeutic agents with established benefit act by interrupting the neurohormonal pathways discussed previously (ie, RAAS and the sympathetic nervous system), such as β-blockers (BBs), angiotensin-converting enzyme (ACE) inhibitors (ACE-Is), angiotensin receptor blockers (ARBs) and aldosterone antagonists [7]. However, other therapies such as hydralazine-isosorbide dinitrate (HN) combination therapy have mechanisms of action that are less understood. Other important drugs that are commonly used in HF include drugs targeting disease symptoms, such as diuretics and digoxin, or complications, such as warfarin.
As the number of drug classes that are indicated for use in HF has increased (currently at least seven drug classes), it has become more difficult to determine whether each additional therapy has incremental benefit that outweighs the added risks and cost for specific individuals or subgroups of patients. While the average population response to HF therapeutics is favorable, significant interindividual variation exists in the response (Figure 1). This variability should not be surprising given the diverse etiologies and genetic backgrounds upon which the HF phenotype can occur. Common genetic variation exists in genes related to most classes of HF drugs, and many of these variants have known functional consequences. As more is understood about the interplay of drugs and genes, genetic sequence variants may help to explain some of the variation in patient responses, and therefore may help physicians to provide more rational, efficient and targeted treatments for HF [8].
Figure 1. Variation in drug response within a disease population.
The different coloring of figures represents underlying genetic heterogeneity between individuals with the same medical diagnosis. Pharmacogenomics seeks to understand this heterogeneity, use it to have better estimates of the risks and benefits of medical intervention, and then utilize it to provide improved care for individual patients.
This review discusses the current understanding of the pharmacogenetics of HF therapies, focusing on recent advances and expectations for the near future. Therapies discussed include ACE-Is, ARBs, aldosterone receptor antagonists, BBs, natriuretic peptides (NPs), HN, diuretics and warfarin. These examples vary in terms of the amount of data available and, therefore, are illustrative of the range of advances in studying pharmacogenetics from an early stage of understanding (eg, HN) to more advanced stages (eg, warfarin). A better understanding of the pharmacogenetics of HF therapies, its advances and limitations, and how this field is advancing toward clinical use is important to optimize how physicians incorporate this emerging clinical science.
Antagonizing the renin-angiotensin-aldosterone system
Inhibition of the RAAS is a major focus of therapy in HF. There have been a large number of pharmacogenetic studies related to agents targeting the RAAS. The most interesting genetic variants and associated phenotypes for response to HF therapy, categorized by relevant therapeutic agent, are summarized in Table 1.
Table 1.
Summary of variants with pharmacogenetic interactions for RAAS-modulating agents in HF.
| Drug | Gene | Polymorphism | rs number | Molecular phenotype | Clinical phenotype | Reference |
|---|---|---|---|---|---|---|
| ACE inhibitors | ACE | Intron 16 I/D | rs4646994 | Increased ACE level associated with D-allele | D-allele homozygotes with HF had lower event-free survival on low-dose ACE inhibitor therapy. | [14, 91] |
| TACR2 | Gly231Glu | Coding SNP of unknown function | In patients (n = 91) receiving ACE inhibitor therapy, only Gly231 carriers developed chronic cough. | [22] | ||
| Angiotensin receptor blockers | ACE | Intron 16 I/D | rs4646994 | Increased ACE level associated with D-allele | Faster reduction of LVH in D-allele carriers with angiotensin receptor blocker therapy. | [26,91] |
| AGTR1 | A1166C | rs5186 | 3′ Untranslated region SNP located in a microRNA binding site | C-allele carriers had better BP and NT-proBNP responses with the addition of candesartan to treatment including an ACE inhibitor. | [27,28] | |
| AGTR1 | A-480G | rs1492078 | Located 5′ to gene in the promoter region | A significant interaction between irbesartan treatment and BP reduction was observed. | [29] | |
| CYP2C9 | Arg144Cys (CYP2C9* 2) | rs1799853 | Reduced catalytic capacity for certain angiotensin receptor blockers compared with CYP2C9*1 | CYP2C9*2 genotype increased BP response to irbesartan treatment. | [30,31] | |
| Aldosterone antagonists | ACE | Intron 16 I/D | rs4646994 | Increased ACE level associated with D-allele | Among patients with HF, only carriers of the I-allele demonstrated improvements in EF, EDV and ESV with spironolactone treatment. | [37,91] |
ACE Angiotensin 1-converting enzyme, AGTR1 angiotensin II receptor type 1, BP blood pressure, CYP2C9 cytochrome P450 family 2 subfamily C polypeptide 9, EDV end diastolic volume, EF ejection fraction, ESV end systolic volume, HF heart failure, I/D insertion/deletion, LVH left ventricular hypertrophy, NT-proBNP N-terminal proB-type natriuretic peptide, RAAS renin-angiotensin-aldosterone system, TACR2 tachykinin receptor 2
Angiotensin-converting enzyme inhibitors
Treatment with an ACE-I is a cornerstone of HF therapy, and is recommended to all patients with HF without contraindication [7]. ACE-Is have demonstrated survival benefit in multiple randomized clinical trials for HF with reduced ejection fraction [9]. These agents block the production of angiotensin II by antagonizing ACE (encoded by the ACE gene), reducing the adverse effects of angiotensin II, including vasoconstriction, aldosterone production and ventricular remodeling [5].
Many studies have been conducted to identify pharmacogenetic interactions of ACE-I, but a clear understanding of these interactions remains elusive [10–19]. One polymorphism that has been studied extensively is a 287-bp insertion/deletion (I/D) in intron 16 of the ACE gene (rs4646994). While one study suggested that the ACE I/D genotype affects ACE-I efficacy in heart failure [14], this variant remains of uncertain importance, and has been reviewed elsewhere recently (see references [20,21]). In terms of adverse drug effects, a polymorphism in the gene encoding the neurokinin-2 receptor (TACR2) has been associated with ACE-I-induced cough, a frequent side effect that often leads to discontinuation of the drug [22]. A large, ongoing clinical trial, the PERindopril GENEtic association study (PERGENE), may provide new insights into ACE-I pharmacogenetics [23]. This cohort trial aims to genotype polymorphisms from 11 candidate genes in patients (n = 12,218) treated with ACE-Is for stable coronary artery disease. Although HF is not an endpoint of this study, new pharmacogenetic interactions that can be tested in patients with HF may be identified.
Angiotensin receptor blockers
Clinically, ARBs are primarily useful in HF as a substitute for ACE-Is and have been demonstrated to have comparable survival benefit in this setting [24,25]. ARBs inhibit the binding of angiotensin II to its primary receptor, angiotensin receptor type 1, which is encoded by AGTR1. While there are relatively few studies of ARB pharmacogenetics specifically in patients with HF, several studies in hypertension have been conducted and many of the response phenotypes (eg, blood pressure lowering and reversal of hypertrophy) are relevant for HF. One study revealed significantly faster reduction of left ventricular hypertrophy after the initiation of ARB therapy in ACE D-allele carriers [26]. Another study examining the efficacy of ARBs as add-on therapy to ACE-Is demonstrated that carriers of the C-allele at the AGTR1 A1166C polymorphism (rs5186), which is located in a microRNA binding site in the 3′ untranslated region [27], had greater blood pressure and N-terminal proB-type NP (NT-proBNP) responses to treatment [28]. The study was provocative, but was underpowered, necessitating validation studies before further inferences can be made. Additional AGTR1 variants may also be related to blood pressure reductions resulting from ARB treatment. A small study of irbesartan in patients with hypertension identified a significant relationship between irbesartan concentration and AGTR1 genotype for blood pressure reduction. The associated SNP is in the promoter of AGTR1 (rs1492078) [29], suggesting a potential role via transcriptional regulation. ARBs are metabolized via the cytochrome P450 (CYP) enzymes, and genetic variations in CYP enzymes have been implicated in affecting the response to some ARBs [30]. Specifically, the CYP2C9*2 variant was demonstrated to increase the blood pressure reduction observed with irbesartan in patients with hypertension [31], with some evidence for an impact on losartan efficacy [32–34].
Aldosterone receptor antagonists
Aldosterone receptor antagonists have demonstrated reductions in mortality in two clinical trials: one in patients with severe HF [35] and one in patients with HF after acute myocardial infarction [36]. Thus, these agents are Class I indicated in suitable patients [7]. Pharmacogenetic data for the effect of aldosterone antagonists is limited, but one small study has investigated the pharmacogenetics of these agents in patients with HF [37]. Patients receiving standard HF therapy (n = 93) were randomly assigned to receive spironolactone or placebo. Among patients receiving spironolactone, only ACE I/D insertion carriers had significant improvement in ejection fraction, compared with baseline values. Conversely, when comparing changes in ejection fraction between the spironolactone and placebo groups, ACE D/D homozygotes trended toward a stronger effect (3.0 in D/D vs 1.7; p = not significant) [37]. Additional studies to further elucidate the role of pharmacogenetics in response to aldosterone receptor antagonists are needed.
β-adrenergic antagonists
BBs have been demonstrated to reduce HF mortality in multiple randomized clinical trials and are recommended for the treatment of all patients with HF without contraindications [7]. BBs antagonize the β-adrenergic receptors (β-ARs), a family of GPCRs that increase heart rate and cardiac contractility, and stimulate renin release in the kidneys. Despite the efficacy of these compounds in clinical trials, response to BBs varies significantly, with inconsistent recovery of ejection fraction, and many patients continuing to experience disease progression [38]. These agents also have potential adverse effects, particularly during dose titration, such as reduced contractility, bradycardia and the potential to cause or worsen HF exacerbations [39]. Increasing evidence suggests that genetic factors may explain some of this variability. The pharmacogenetic factors associated with BBs have been reviewed in detail elsewhere (see reference [40]), and the discussion in this review summarizes the key points and focuses on current and future directions. The key genetic variants relevant to BB therapy, their molecular phenotypes and the associated clinical phenotypes are summarized in Table 2.
Table 2.
Summary of variants with pharmacogenetic interactions for β-blocker therapy in heart failure.
| Gene | Polymorphism/mutation | rs number | Molecular phenotype | Clinical phenotype | Reference |
|---|---|---|---|---|---|
| ADRB1 | Arg389Gly | rs1801253 | Arg389 demonstrated differential effects on adenylyl levels in normal and heart failure tissues. | Increased survival or LVEF β-blocker response with Arg389Arg. Gly389 associated with greater survival response in a separate study. Possibly involved in gene-gene interactions. | [44,47,51,56,57,92–94] |
| Ser49Gly | rs1801252 | Gly49 allele leads to greater receptor downregulation on stimulation. | Lower 5-year mortality in Gly49 carriers on low-dose β-blockers, but no difference with high-doses. | [95,96] | |
| ADRB2 | Glu27Gln | rs1042714 | Glu27 allele has decreased receptor downregulation. | Glu27 allele associated with better response to carvedilol. | [43,97] |
| Gly16Arg | rs1042713 | Gly16 allele has greater receptor downregulation with receptor stimulation. | Increased risk of death or transplantation detected for Arg16/Gln27 double homozygotes. | [45,97] | |
| ADRA2C | Exon 1 I/D | rs61767072 | D-allele results in loss of autoinhibition. | Arg389 homozygotes carrying D-allele exhibited greatest LVEF improvements on β-blocker therapy. In bucindolol-treated patients, only WT-homozygotes exhibited reductions in mortality and transplantation. | [53,57,98,99] |
| ACE | Intron 16 I/D | rs4646994 | Increased ACE level associated with D-allele. | D-allele associated with risk of death or transplantation in patients not receiving β-blockers. | [56,83] |
| EDN1 | rs5370 rs2071942 |
rs5370 rs2071942 |
SNPs of unknown functionality in tight LD. | Study of placebo- and bucindolol- treated patients (n = 159 and 150, respectively) demonstrated significant interaction between treatment and genotype at both loci. | [59] |
| GRK5 | Gln41Leu | rs17098707 | Leu41 enhances β-adrenoreceptor desensitization. | Leu41 leads to increased survival in African American patients in the absence of β-blockers. | [47,55] |
ACE angiotensin 1-converting enzyme, ADRA2C adrenergic receptor α 2c, ADRB1 adrenergic receptor β 1, ADRB2 adrenergic receptor β 2, EDN1 endothelin 1, GRK5 G-protein coupled receptor kinase 5, I/D insertion/deletion, LVEF left ventricular ejection fraction, WT wild-type
Adrenergic receptor polymorphisms
The adrenergic receptor genes are highly polymorphic, and many of the variants have functional consequences. Much attention has focused on ADRB1 and ADRB2, which encode the β-AR1 and β-AR2, respectively; these receptors are the molecular targets for BBs. The variants in these genes and their functional consequences have been well researched (see references [41,42]). Variants in both ADRB1 and ADRB2 have been associated with improvements in ejection fraction with BB treatment [43,44]. Subsequently, several large cohort studies have been conducted to examine the relationship between β-AR polymorphisms, mortality rates and treatment with carvedilol or metoprolol in patients with HF [45–47]; one study has been conducted in patients with acute coronary syndrome [46]. These studies have revealed varying results, with two studies indicating an important association for the ADRB2 haplotype, but not ADRB1 variants [45,46], one indicating that the ADRB1 Arg389Gly variant is significant, but not the ADRB2 variants [47], and two studies demonstrating no significant association with any of these variants [48,49]. Taken together, these data do not support a definitive conclusion. It is important to note that all of these studies are limited because they included few or no patients who were BB naive, making inference as to the effects of the drug more difficult.
Conversely, the randomized, controlled β-blocker Evaluation of Survival Trial (BEST) investigated the effects of bucindolol (ARCA biopharma Inc) [50]. The clinical trial was terminated prematurely, failed to meet its primary endpoint and demonstrated little overall benefit with bucindolol therapy, in contrast with other published trials of BB therapy in HF. Subsequent pharmacogenetic analyses of these data revealed that there was enhanced benefit for bucindolol among ADRB1 Arg389 homozygotes [51]. Following the discovery of this pharmacogenetic association, the developers of bucindolol submitted an NDA to the FDA for bucindolol to be used in conjunction with a genetic test for the ADRB1 Arg389Gly genotype; bucindolol would have been the first genetic-guided therapy to be approved for HF. Recently, the FDA review panel examined this issue, but did not recommend approval [52]. While clinical use of pharmacogenetics to guide BB therapy awaits further data, the experience with bucindolol is indicative of how close pharmacogenetics may be to being applied in the clinic for HF.
α-AR variants may also have an impact on the response to BB therapy. ADRA2C encodes the α2c-AR, which regulates presynaptic norepinephrine release. Another substudy of the BEST trial examined the effects of a frame-shift mutation caused by a 12-nucleotide deletion in exon 1 of ADRA2C (rs61767072) [53]. A previous analysis suggested that patients with a strong sympatholytic response to bucindolol were at an increased risk of adverse events, indicating that changes in the ability of ADRA2C to regulate sympathetic activity might be involved [54]. While no difference in sympathetic activity by genotype was observed in the placebo group, patients with the ADRA2C deletion demonstrated a greater sympatholytic response than wild-type homozygotes to bucindolol treatment; only wild-type homozygotes exhibited reductions in mortality and in need for transplantation with bucindolol treatment [53].
Within the adrenergic system, some important multilocus or epigenetic effects have recently been discovered. Pharmacogenetic analysis of a large cohort of patients with hypertension demonstrated that the ADRB1 Ser49/Arg389 haplotype was associated with increased mortality at baseline and significant improvement with atenolol therapy, but no response to treatment with verapamil [55]. However, allelic associations were not tested, making the contribution of each allele and the extent of any interaction unclear. The ADRB1 Arg389 allele may also interact with polymorphisms in ADRA2C. Kardia et al recently identified significant gene-gene interactions between ADRB1 and ADRA2C [56], confirming results from a previous study demonstrating that Arg389 homozygotes carrying a deletion in ADRA2C experienced the greatest improvement in left ventricle ejection fraction (LVEF) with BB treatment [57].
Other associated genes
Downstream of the gene encodingβ-AR, polymorphisms in proteins related to signal transduction may also affect individual response to BBs. For example, G-protein receptor kinase 5 (GRK5) phosphorylates β-ARs in the myocardium, resulting in uncoupling of the receptor from adenyl cyclase [58]. A non-synonymous Glu41Leu substitution in the GRK5 gene identified by Liggett et al results in enhanced uncoupling in vitro and in animal models [59]. A pharmacogenetic interaction between the Gln41Leu polymorphism and BB use was also identified in humans. In a prospective cohort of African American patients (n = 375), the authors also showed that Leu41 carriers had better survival than Gln41 homozygotes in the absence of BB therapy, while there was no difference between genotype groups when treated with BB, indicating selective benefit of BB for Gln homozygotes [59]. These findings were confirmed by Cresci et al in African Americans; however, no association was observed in Caucasians, in whom the variant is approximately one-tenth as frequent compared with African Americans [47].
Given the far-ranging effects of manipulating the adrenergic system, polymorphisms in even more distantly associated genes or pathways may impact variability in response to BB. For example, one of the early associations of the ACE I/D polymorphism was BB response [60], although this finding has not been confirmed by further studies [61]. More recently, a study of patients (n = 309) with idiopathic dilated cardiomyopathy from the BEST trial was conducted examining the polymorphisms in the gene encoding endothelin 1 (EDN1), based upon previous research demonstrating decreases in endothelin levels with bucindolol treatment [62]. A pharmacogenetic interaction was identified between bucindolol treatment and EDN1 genotype. Two SNPs, rs5370 and rs2071942 (loci are in linkage disequilibrium (LD)), were associated with the rate of HF hospitalization and all-cause mortality in bucindolol-treated patients, but not in the placebo group [63].
β-blocker metabolism
Polymorphisms affecting drug metabolism may also be important in BB selection and dosing. CYP2D6 metabolizes metoprolol and, to a lesser extent, carvedilol. A study by Bijl et al determined that CYP2D6*4 homozygotes (ie, poor metabolizers) had significantly lower heart rate and diastolic blood pressure with metoprolol treatment than either CYP2D6*1 homozygotes (ie, extensive metabolizers) or heterozygotes (ie, intermediate metabolizers) [64]. No differences were observed by genotype in patients treated with atenolol. In patients with HF receiving metoprolol, a small study by Sharp et al demonstrated that the CYP2D6 genotype influenced metoprolol blood concentration significantly, but had no effect on metoprolol dosing or clinical outcomes [65]. However, the study was underpowered with respect to poor metabolizers (n = 3), suggesting that further research is required [65].
Other agents of interest in heart failure
Natriuretic peptide system
The importance of the NP system in cardiovascular homeostasis and HF has been recognized increasingly [66]. The NP system is understood to be an important counter-regulatory system that reduces blood pressure and has general salutary effects in HF. There are three naturally occurring NPs: atrial NP (ANP); B-type NP (BNP); and C-type NP (CNP). BNP has applications in HF diagnosis and prognosis, but this molecule and the other NPs are also useful as therapeutics. Recombinant BNP (ie, nesiritide) and ANP (ie, carperitide) have been approved for the treatment of HF in the US and Japan, respectively, and other ‘designer’ peptides are being investigated [67,68].
Improved targeting of NP therapies is highly desirable because these compounds are currently expensive, parenteral and associated with adverse effects despite being efficacious. Pharmacogenetics may help to improve targeting, as substantial evidence suggests the importance of genetic variation in the NP system (reviewed in reference [66]). An analysis from the Framingham Study identified three SNPs in NPPA and NPPB that were significantly associated with both circulating NP levels and blood pressure [69]. From a strictly pharmacogenetic perspective, there have been limited published studies related to NPs and HF, but ongoing studies promise important results within the next few years. An NIH-funded pharmacogenetic assessment of the effect of recombinant BNP in patients with HF, measuring pharmacokinetic (ie, drug levels and elimination) and pharmacodynamic (ie, serum and urine cyclic guanosine monophosphate) endpoints, is ongoing. In addition, an ongoing, large (n = 7000), randomized clinical trial of BNP in acutely decompensated HF (ASCEND-HF) is also collecting genetic samples and should have adequate power to investigate the effects of drug and genotype interactions on important clinical outcomes such as dyspnea and clinical events (ie, death or hospitalization) [70].
Isosorbide-dinitrate/hydralazine
Combination therapy with HN is, to our knowledge, the first drug that is approved and marketed based on race, having been tested for the treatment of HF in self-identified African-Americans. This therapy has resulted in many interesting questions regarding medical care, race and genetics. Racial differences in response to HN treatment were first identified in a post-hoc analysis of the V-HeFT-II trial that compared HN with enalapril in patients with HF [71]. As a result, the benefit of HN in addition to standard therapy (ie, BBs and ACE-Is) was then tested in self-identified African-Americans in the African-American Heart Failure Trial (AA-HeFT) [72]. The randomized trial demonstrated a 40% reduction in the relative risk for death, leading to FDA approval of the drug. This approach highlights the question of whether it is desirable to use race to assign medical therapy, and underscores the need to better understand the genetic and biological differences that belie such race-based differences in efficacy. Race is a social construct that is at best an approximate proxy for biological differences, and includes several other components such as shared cultural and environmental factors that must be separated from genetic/biological factors. Pharmacogenetics may be able to more precisely identify the genetic differences in such cases, and may nullify the motivation to use race as a classifier for medical treatment. Thus, it is a high research priority to identify the genetic underpinnings of race-based difference in drug efficacy, such as those observed with HN.
With respect to HN therapy, the genetic determinants of response remain an active area of investigation, but some data indicate that polymorphisms in the gene NOS3, encoding endothelial nitric oxide synthase, may contribute to the race-specific benefit. Because HN is thought to act as a nitric oxide donor, individuals with lower NOS3 activity might be expected to benefit more from HN therapy. A substudy of the AA-HeFT trial identified three NOS3 polymorphisms with significant differences in frequency in African-Americans and Caucasians [73]. A pharmacogenetic interaction was identified between a non-synonymous polymorphism (rs1799983) that results in an Asp298Glu mutation. Glu298 homozygotes benefited significantly from HN treatment, whereas Asp298 carriers did not [73]. However, because this effect was mostly in quality-of-life scores and given the limited sample size (n = 352), this study should be interpreted as hypothesis-generating.
Diuretics
Diuretics are recommended in patients with HF who have evidence of fluid retention [7]. Several classes of diuretics are administered; however, loop diuretics are the most frequently used agents. Although there are relatively few pharmacogenetic studies examining diuretics, some results relevant to HF exist.
Specific to diuretic therapy in HF, a small study examining the interaction of polymorphisms in CYP2C9 and SLCO1B1 (solute carrier organic anion transporter family, member 1B1) genes and torsemide, a long-acting loop diuretic used in patients with HF, demonstrated that CYP2C9*3 and SLCO1B1 C521T (rs4149056) had a significant effect on the plasma concentration and half-life of torsemide [74]. Other diuretic classes have also been investigated. For example, Lynch et al studied two SNPs in the NPPA/NPPB gene regions, rs5063 and rs5065, in patients (approximate n = 38,000) in a subgroup of the Antihypertensive and Lipid-Lowering Treatment to Prevent Heart Attack Trial (ALLHAT) [75]. In patients carrying at least one C-allele at rs5065, treatment with chlorthalidone resulted in a significantly lower incidence of coronary heart disease, stroke and combined cardiovascular disease, as well as significantly lower all-cause mortality, when compared with amplodipine treatment; these effects appeared to be independent of changes in blood pressure.
Warfarin
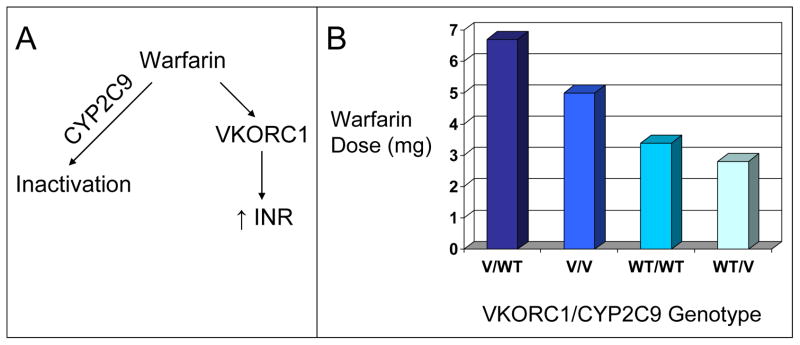
Anticoagulation therapy with warfarin is commonly indicated in patients with HF because of comorbid conditions that result in an increased risk of thromboembolism, such as atrial fibrillation or severely impaired left ventricular function. Warfarin acts by binding to the vitamin K 2,3-epoxide reductase complex (VKORC1), the enzyme responsible for reducing vitamin K to its active form [76]. Warfarin is primarily metabolized to its inactive form by CYP2C9 [77]. By interfering with vitamin K recycling, warfarin prevents the vitamin K-dependent carboxylation of the clotting Factors II, VII, IX and X, thus exerting its anticoagulant effect [78]. Warfarin is one of the best understood examples of the role of pharmacogenetic interactions, with excellent reviews recently published on this topic [79,80]. The two key genes in warfarin pharmacogenetics VKORC1 and CYP2C9 account for approximately 40% of variation in warfarin dosing (Figure 2) [81]. Several developments that are illustrative of the advancement of pharmacogenetics in HF are discussed in this section.
Figure 2. Pharmacogenetic influences on warfarin dosing.
(A) The warfarin drug pathway. (B) The average maintenance dose of warfarin is dependent on VKORC1 (vitamin K 2,3-epoxide reductase complex) and CYP2C9 (cytochrome P450 family 2 subfamily C polypeptide 9) genotypes (data from reference [81]).
INR international normalized ratio, V variant, WT wild-type
The use of pharmacogenetics to determine the dosing of warfarin is at advanced stages of development, serving as a good example to illustrate the requirements for bringing pharmacogenetics to the clinic, including practical genotype-based dosing guide and randomized interventional pharmacogenetic trials Warfarin is one of the only drugs to have been investigated for pharmacogenetic interactions using a genome-wide approach. Two studies by Cooper et al [82] and Takeuchi et al [83] examined patients (n = 379 and 1053, respectively) beginning warfarin therapy. While neither study identified any new associations, the feasibility of a genome-wide approach was confirmed, as were the effects from VKORC1 and CYP2C9 variants.. Genetically driven dosing algorithms have been derived and published for warfarin [84,85]. In addition, a large, NIH-sponsored interventional trial (Clarification of Optimal Anticoagulation through Genetics [COAG]) is ongoing to assess the benefit of genotyping prior to the initiation of treatment on warfarin dosing and on adverse effects. The COAG trial is expected to be completed in 2012 (ClinicalTrials.gov identifier: NCT00839657).
Warfarin also serves as a good test case for examining the cost-effectiveness of genetic testing prior to drug initiation. Although genotyping can help guide warfarin dosing [86–88], it has not been demonstrated adequately that genotyping helps to avoid adverse effects. A recent meta-analysis did not find sufficient evidence to support the use of genetic information for warfarin dosing [89]. Another study suggested that there was a 10% chance that genotype-guided warfarin dosing would be cost-effective, even with fairly optimistic assumptions for genotyping cost and processing time [90]. However, with the results of studies such as the COAG trial or with future advances in genotyping technologies, genotyping may become an important clinical strategy for determining warfarin dose.
Conclusion
HF should be considered a prime target for pharmacogenetics and personalized medicine given the great burden of disease and multiplicity of therapeutic options. Research progress in pharmacogenetics is broadly accelerating as a result of improving research technologies with lower costs, larger study cohorts, and increased awareness and acceptance in the wider medical community. Cardiovascular therapies are no exception to the increase in interest in pharmacogenetics, as illustrated by the examples described in this review, such as BBs and warfarin, which are at an advanced stage. However, even in these advanced cases, as well as in newer examples such as NPs, much research remains before clinical pharmacogenetics will be commonplace. First, and most importantly, is the need for emphasis to be placed on investigating the clinical implementation of genetically guided therapy. Warfarin is an instructive example, demonstrating that the creation of usable pharmacogenetic tools rely not only on associating a genotype with a drug-response phenotype, but also requires creating usable decision guidelines, establishing superiority to empirical therapy and demonstrating cost-effectiveness. These requirements necessitate more interventional pharmacogenetic studies (ie, assigning patients to genetic-based therapy compared with empirical therapy) to meet this goal. The second main challenge is to apply pharmacogenetics earlier in the drug development process such that future clinical applications of PGs do not require as long to develop as the current generation. If early-phase trials included broad genotyping and association to surrogate endpoints, phase III pivotal trials could simultaneously include candidate gene studies, greatly accelerating progress toward clinically useful genetic markers and genetically guided therapy. While there has been relatively slow clinical adaptation of pharmacogenetic findings thus far, this situation is changing, and is likely to accelerate. If successful, pharmacogenetics will define a new era of advancement, producing many new tools to improve the treatment of patients using genetics.
References
•• of outstanding interest
• of special interest
- 1.Jessup M, Brozena S. Heart failure. N Engl J Med. 2003;348(20):2007–2018. doi: 10.1056/NEJMra021498. [DOI] [PubMed] [Google Scholar]
- 2.Lloyd-Jones D, Adams R, Carnethon M, De Simone G, Ferguson TB, Flegal K, Ford E, Furie K, Go A, Greenlund K, Haase N, et al. Heart disease and stroke statistics – 2009 update: A report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2009;119(3):480–486. doi: 10.1161/CIRCULATIONAHA.108.191259. [DOI] [PubMed] [Google Scholar]
- 3.Senni M, Redfield MM. Heart failure with preserved systolic function. A different natural history? J Am Coll Cardiol. 2001;38(5):1277–1282. doi: 10.1016/s0735-1097(01)01567-4. [DOI] [PubMed] [Google Scholar]
- 4.Adams KF., Jr Pathophysiologic role of the renin-angiotensin-aldosterone and sympathetic nervous systems in heart failure. Am J Health Syst Pharm. 2004;61(Suppl 2):S4–S13. doi: 10.1093/ajhp/61.suppl_2.S4. [DOI] [PubMed] [Google Scholar]
- 5.Cody RJ. The sympathetic nervous system and the renin-angiotensin-aldosterone system in cardiovascular disease. Am J Cardiol. 1997;80(9B):9J–14J. doi: 10.1016/s0002-9149(97)00832-1. [DOI] [PubMed] [Google Scholar]
- 6.Kjaer A, Hesse B. Heart failure and neuroendocrine activation: Diagnostic, prognostic and therapeutic perspectives. Clin Physiol. 2001;21(6):661–672. doi: 10.1046/j.1365-2281.2001.00371.x. [DOI] [PubMed] [Google Scholar]
- 7.Hunt SA, Abraham WT, Chin MH, Feldman AM, Francis GS, Ganiats TG, Jessup M, Konstam MA, Mancini DM, Michl K, Oates JA, et al. 2009 focused update incorporated into the ACC/AHA 2005 guidelines for the diagnosis and management of heart failure in adults: A report of the American College of Cardiology Foundation/American Heart Association task force on practice guidelines: Developed in collaboration with the International Society for Heart and Lung Transplantation. Circulation. 2009;119(14):e391–e479. doi: 10.1161/CIRCULATIONAHA.109.192065. [DOI] [PubMed] [Google Scholar]
- 8.Lanfear DE, McLeod HL. Pharmacogenetics: Using DNA to optimize drug therapy. Am Fam Physician. 2007;76(8):1179–1182. [PubMed] [Google Scholar]
- 9.Demers C, Mody A, Teo KK, McKelvie RS. ACE inhibitors in heart failure: What more do we need to know? Am J Cardiovasc Drugs. 2005;5(6):351–359. doi: 10.2165/00129784-200505060-00002. [DOI] [PubMed] [Google Scholar]
- 10.Bleumink GS, Schut AF, Sturkenboom MC, van Duijn CM, Deckers JW, Hofman A, Kingma JH, Witteman JC, Stricker BH. Mortality in patients with hypertension on angiotensin-I converting enzyme (ACE)-inhibitor treatment is influenced by the ACE insertion/deletion polymorphism. Pharmacogenet Genomics. 2005;15(2):75–81. doi: 10.1097/01213011-200502000-00003. [DOI] [PubMed] [Google Scholar]
- 11.Bozkurt O, de Boer A, Grobbee DE, Kroon AA, Schiffers P, de Leeuw P, Klungel OH. Renin-angiotensin system polymorphisms and the association between use of angiotensin II receptor blockers or angiotensin-converting enzyme inhibitors and the risk of diabetes. J Renin Angiotensin Aldosterone Syst. 2009;10(2):101–108. doi: 10.1177/1470320309104877. [DOI] [PubMed] [Google Scholar]
- 12.Bozkurt O, Verschuren WM, van Wieren-de Wijer BM, Knol MJ, de Boer A, Grobbee DE, Geerlings MI, Heerdink ER, Klungel OH. Genetic variation in the renin-angiotensin system modifies the beneficial effects of ACE inhibitors on the risk of diabetes mellitus among hypertensives. J Hum Hypertens. 2008;22(11):774–780. doi: 10.1038/jhh.2008.62. [DOI] [PubMed] [Google Scholar]
- 13.Hingorani AD, Jia H, Stevens PA, Hopper R, Dickerson JE, Brown MJ. Renin-angiotensin system gene polymorphisms influence blood pressure and the response to angiotensin converting enzyme inhibition. J Hypertens. 1995;13(12 Pt 2):1602–1609. [PubMed] [Google Scholar]
- 14.McNamara DM, Holubkov R, Postava L, Janosko K, MacGowan GA, Mathier M, Murali S, Feldman AM, London B. Pharmacogenetic interactions between angiotensin-converting enzyme inhibitor therapy and the angiotensin-converting enzyme deletion polymorphism in patients with congestive heart failure. J Am Coll Cardiol. 2004;44(10):2019–2026. doi: 10.1016/j.jacc.2004.08.048. [DOI] [PubMed] [Google Scholar]
- 15.Schelleman H, Klungel OH, van Duijn CM, Witteman JC, Hofman A, de Boer A, Stricker BH. Insertion/deletion polymorphism of the ACE gene and adherence to ACE inhibitors. Br J Clin Pharmacol. 2005;59(4):483–485. doi: 10.1111/j.1365-2125.2004.02332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schelleman H, Klungel OH, Witteman JC, Breteler MM, Yazdanpanah M, Danser AH, Hofman A, van Duijn CM, de Boer A, Stricker BH. Angiotensinogen M235T polymorphism and the risk of myocardial infarction and stroke among hypertensive patients on ACE-inhibitors or β-blockers. Eur J Hum Genet. 2007;15(4):478–484. doi: 10.1038/sj.ejhg.5201789. [DOI] [PubMed] [Google Scholar]
- 17.Schelleman H, Klungel OH, Witteman JC, Hofman A, van Duijn CM, de Boer A, Stricker BH. The influence of the α-adducin G460W polymorphism and angiotensinogen M235T polymorphism on antihypertensive medication and blood pressure. Eur J Hum Genet. 2006;14(7):860–866. doi: 10.1038/sj.ejhg.5201632. [DOI] [PubMed] [Google Scholar]
- 18.Schelleman H, Klungel OH, Witteman JC, Hofman A, van Duijn CM, de Boer A, Stricker BH. Pharmacogenetic interactions of three candidate gene polymorphisms with ACE-inhibitors or β-blockers and the risk of atherosclerosis. Br J Clin Pharmacol. 2007;64(1):57–66. doi: 10.1111/j.1365-2125.2007.02848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Knaap R, Siemes C, Coebergh JW, van Duijn CM, Hofman A, Stricker BH. Renin-angiotensin system inhibitors, angiotensin I-converting enzyme gene insertion/deletion polymorphism, and cancer: The Rotterdam Study. Cancer. 2008;112(4):748–757. doi: 10.1002/cncr.23215. [DOI] [PubMed] [Google Scholar]
- 20.Sayed-Tabatabaei FA, Oostra BA, Isaacs A, van Duijn CM, Witteman JC. ACE polymorphisms. Circ Res. 2006;98(9):1123–1133. doi: 10.1161/01.RES.0000223145.74217.e7. [DOI] [PubMed] [Google Scholar]
- 21.Beitelshees AL, Zineh I. Renin-angiotensin-aldosterone system (RAAS) pharmacogenomics: Implications in heart failure management. Heart Fail Rev. 2010;15(3):209–217. doi: 10.1007/s10741-008-9092-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim TB, Oh SY, Park HK, Jeon SG, Chang YS, Lee KY, Cho YS, Chae IH, Kim YK, Cho SH, Moon HB, et al. Polymorphisms in the neurokinin-2 receptor gene are associated with angiotensin-converting enzyme inhibitor-induced cough. J Clin Pharm Ther. 2009;34(4):457–464. doi: 10.1111/j.1365-2710.2008.01018.x. [DOI] [PubMed] [Google Scholar]
- 23.Brugts JJ, de Maat MP, Boersma E, Witteman JC, van Duijn C, Uitterlinden AG, Bertrand M, Remme W, Fox K, Ferrari R, Danser AH, et al. The rationale and design of the PERindopril GENEtic association study (PERGENE): A pharmacogenetic analysis of angiotensin-converting enzyme inhibitor therapy in patients with stable coronary artery disease. Cardiovasc Drugs Ther. 2009;23(2):171–181. doi: 10.1007/s10557-008-6156-1. [DOI] [PubMed] [Google Scholar]
- 24.Pfeffer MA, McMurray JJ, Velazquez EJ, Rouleau JL, Kober L, Maggioni AP, Solomon SD, Van de Werf M, White H, Leimberger JD, Henis M, et al. Valsartan, captopril, or both in myocardial infarction complicated by heart failure, left ventricular dysfunction, or both. N Engl J Med. 2003;349(20):1893–1906. doi: 10.1056/NEJMoa032292. [DOI] [PubMed] [Google Scholar]
- 25.Pfeffer MA, Swedberg K, Granger CB, Held P, McMurray JJ, Michelson EL, Olofsson B, Ostergren J, Yusuf S, Pocock S. Effects of candesartan on mortality and morbidity in patients with chronic heart failure: The CHARM-overall programme. Lancet. 2003;362(9386):759–766. doi: 10.1016/s0140-6736(03)14282-1. [DOI] [PubMed] [Google Scholar]
- 26.Nakayama M, Nakano H, Tsuboi N, Kurosawa T, Tsuruta Y, Iwasaki Y, Yokoyama K, Hosoya T, Fukagawa M. The effect of angiotensin receptor blockade ARB on the regression of left ventricular hypertrophy in hemodialysis patients: Comparison between patients with D allele and non-D allele ACE gene polymorphism. Clin Nephrol. 2005;64(5):358–363. doi: 10.5414/cnp64358. [DOI] [PubMed] [Google Scholar]
- 27.Martin MM, Buckenberger JA, Jiang J, Malana GE, Nuovo GJ, Chotani M, Feldman DS, Schmittgen TD, Elton TS. The human angiotensin II type 1 receptor +1166 A/C polymorphism attenuates microRNA-155 binding. J Biol Chem. 2007;282(33):24262–24269. doi: 10.1074/jbc.M701050200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.de Denus S, Zakrzewski-Jakubiak M, Dubé MP, Bélanger F, Lepage S, Leblanc MH, Gossard D, Ducharme A, Racine N, Whittom L, Lavoie J, et al. Effects of AGTR1 A1166C gene polymorphism in patients with heart failure treated with candesartan. Ann Pharmacother. 2008;42(7):925–932. doi: 10.1345/aph.1K657. [DOI] [PubMed] [Google Scholar]
- 29.Kurland L, Hallberg P, Melhus H, Liljedahl U, Hashemi N, Syvänen AC, Lind L, Kahan T. The relationship between the plasma concentration of irbesartan and the antihypertensive response is disclosed by an angiotensin II type 1 receptor polymorphism: Results from the Swedish Irbesartan Left Ventricular Hypertrophy Investigation vs. Atenolol (SILVHIA) trial. Am J Hypertens. 2008;21(7):836–839. doi: 10.1038/ajh.2008.190. [DOI] [PubMed] [Google Scholar]
- 30.Zhou SF, Zhou ZW, Huang M. Polymorphisms of human cytochrome P450 2C9 and the functional relevance. Toxicology. 2009 doi: 10.1016/j.tox.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 31.Hallberg P, Karlsson J, Kurland L, Lind L, Kahan T, Malmqvist K, Ohman KP, Nystrom F, Melhus H. The CYP2C9 genotype predicts the blood pressure response to irbesartan: Results from the Swedish Irbesartan Left Ventricular Hypertrophy Investigation vs Atenolol (SILVHIA) trial. J Hypertens. 2002;20(10):2089–2093. doi: 10.1097/00004872-200210000-00030. [DOI] [PubMed] [Google Scholar]
- 32.Yin T, Maekawa K, Kamide K, Saito Y, Hanada H, Miyashita K, Kokubo Y, Akaiwa Y, Otsubo R, Nagatsuka K, Otsuki T, et al. Genetic variations of CYP2C9 in 724 Japanese individuals and their impact on the antihypertensive effects of losartan. Hypertens Res. 2008;31(8):1549–1557. doi: 10.1291/hypres.31.1549. [DOI] [PubMed] [Google Scholar]
- 33.Joy MS, Dornbrook-Lavender K, Blaisdell J, Hilliard T, Boyette T, Hu Y, Hogan SL, Candiani C, Falk RJ, Goldstein JA. CYP2C9 genotype and pharmacodynamic responses to losartan in patients with primary and secondary kidney diseases. Eur J Clin Pharmacol. 2009;65(9):947–953. doi: 10.1007/s00228-009-0707-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donner KM, Hiltunen TP, Suonsyrja T, Hannila-Handelberg T, Tikkanen I, Antikainen M, Hirvonen A, Kontula K. CYP2C9 genotype modifies activity of the renin-angiotensin-aldosterone system in hypertensive men. J Hypertens. 2009;27(10):2001–2009. doi: 10.1097/HJH.0b013e32832f4fae. [DOI] [PubMed] [Google Scholar]
- 35.Pitt B, Zannad F, Remme WJ, Cody R, Castaigne A, Perez A, Palensky J, Wittes J. The effect of spironolactone on morbidity and mortality in patients with severe heart failure. Randomized Aldactone Evaluation Study Investigators. N Engl J Med. 1999;341(10):709–717. doi: 10.1056/NEJM199909023411001. [DOI] [PubMed] [Google Scholar]
- 36.Pitt B, Remme W, Zannad F, Neaton J, Martinez F, Roniker B, Bittman R, Hurley S, Kleiman J, Gatlin M. Eplerenone, a selective aldosterone blocker, in patients with left ventricular dysfunction after myocardial infarction. N Engl J Med. 2003;348(14):1309–1321. doi: 10.1056/NEJMoa030207. [DOI] [PubMed] [Google Scholar]
- 37.Cicoira M, Rossi A, Bonapace S, Zanolla L, Perrot A, Francis DP, Golia G, Franceschini L, Osterziel KJ, Zardini P. Effects of ACE gene insertion/deletion polymorphism on response to spironolactone in patients with chronic heart failure. Am J Med. 2004;116(10):657–661. doi: 10.1016/j.amjmed.2003.12.033. [DOI] [PubMed] [Google Scholar]
- 38.Khand A, Gemmel I, Clark AL, Cleland JG. Is the prognosis of heart failure improving? J Am Coll Cardiol. 2000;36(7):2284–2286. doi: 10.1016/s0735-1097(00)00995-5. [DOI] [PubMed] [Google Scholar]
- 39.Ko DT, Hebert PR, Coffey CS, Curtis JP, Foody JM, Sedrakyan A, Krumholz HM. Adverse effects of β-blocker therapy for patients with heart failure: A quantitative overview of randomized trials. Arch Intern Med. 2004;164(13):1389–1394. doi: 10.1001/archinte.164.13.1389. [DOI] [PubMed] [Google Scholar]
- 40.Shin J, Johnson JA. β-Blocker pharmacogenetics in heart failure. Heart Fail Rev. 2010;15(3):187–196. doi: 10.1007/s10741-008-9094-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liggett SB. Polymorphisms of β-adrenergic receptors in heart failure. Am J Med. 2004;117(7):525–527. doi: 10.1016/j.amjmed.2004.07.039. [DOI] [PubMed] [Google Scholar]
- 42.Dorn GW, 2nd, Liggett SB. Mechanisms of pharmacogenomic effects of genetic variation within the cardiac adrenergic network in heart failure. Mol Pharmacol. 2009;76(3):466–480. doi: 10.1124/mol.109.056572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaye DM, Smirk B, Williams C, Jennings G, Esler M, Holst D. β-Adrenoceptor genotype influences the response to carvedilol in patients with congestive heart failure. Pharmacogenetics. 2003;13(7):379–382. doi: 10.1097/00008571-200307000-00002. [DOI] [PubMed] [Google Scholar]
- 44.Terra SG, Hamilton KK, Pauly DF, Lee CR, Patterson JH, Adams KF, Schofield RS, Belgado BS, Hill JA, Aranda JM, Yarandi HN, et al. β1-adrenergic receptor polymorphisms and left ventricular remodeling changes in response to β-blocker therapy. Pharmacogenet Genomics. 2005;15(4):227–234. doi: 10.1097/01213011-200504000-00006. [DOI] [PubMed] [Google Scholar]
- 45.Shin J, Lobmeyer MT, Gong Y, Zineh I, Langaee TY, Yarandi H, Schofield RS, Aranda JM, Jr, Hill JA, Pauly DF, Johnson JA. Relation of β2-adrenoceptor haplotype to risk of death and heart transplantation in patients with heart failure. Am J Cardiol. 2007;99(2):250–255. doi: 10.1016/j.amjcard.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 46.Lanfear DE, Jones PG, Marsh S, Cresci S, McLeod HL, Spertus JA. β2-Adrenergic receptor genotype and survival among patients receiving β-blocker therapy after an acute coronary syndrome. JAMA. 2005;294(12):1526–1533. doi: 10.1001/jama.294.12.1526. [DOI] [PubMed] [Google Scholar]
- 47•.Cresci S, Kelly RJ, Cappola TP, Diwan A, Dries D, Kardia SL, Dorn GW., 2nd Clinical and genetic modifiers of long-term survival in heart failure. J Am Coll Cardiol. 2009;54(5):432–444. doi: 10.1016/j.jacc.2009.05.009. Describes the largest cohort study in patients with HF examining BB response and adrenergic pathway polymorphisms. Important associations were identified with ADRB1 and GRK5 variants, offseting any potential race difference in BB benefit. The main limitation of this study was the low number of BB-naive patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Groote P, Helbecque N, Lamblin N, Hermant X, McFadden E, Foucher-Hossein C, Amouyel P, Dallongeville J, Bauters C. Association between β-1 and β-2 adrenergic receptor gene polymorphisms and the response toβ-blockade in patients with stable congestive heart failure. Pharmacogenet Genomics. 2005;15(3):137–142. doi: 10.1097/01213011-200503000-00001. [DOI] [PubMed] [Google Scholar]
- 49.Sehnert AJ, Daniels SE, Elashoff M, Wingrove JA, Burrow CR, Horne B, Muhlestein JB, Donahue M, Liggett SB, Anderson JL, Kraus WE. Lack of association between adrenergic receptor genotypes and survival in heart failure patients treated with carvedilol or metoprolol. J Am Coll Cardiol. 2008;52(8):644–651. doi: 10.1016/j.jacc.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 50.A trial of the β-blocker bucindolol in patients with advanced chronic heart failure. N Engl J Med. 2001;344(22):1659–1667. doi: 10.1056/NEJM200105313442202. No authors listed. [DOI] [PubMed] [Google Scholar]
- 51••.Liggett SB, Mialet-Perez J, Thaneemit-Chen S, Weber SA, Greene SM, Hodne D, Nelson B, Morrison J, Domanski MJ, Wagoner LE, Abraham WT, et al. A polymorphism within a conserved β1-adrenergic receptor motif alters cardiac function and β-blocker response in human heart failure. Proc Natl Acad Sci USA. 2006;103:30–11293. doi: 10.1073/pnas.0509937103. Describes the most definitive pharmacogenetic interaction that has been demonstrated in HF therapuetics. The major limitation is that the agent studied is not approved for the treatment of HF, and agents that are approved for HF do not clearly have a similar association to the agent studied. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.ARCA Biopharma Inc. ARCA Biopharma receives complete response letter from FDA on the Gencaro™ NDA. Press Release. 2009 June 02; [Google Scholar]
- 53.Bristow MR, Murphy GA, Krause-Steinrauf H, Anderson JL, Carlquist JF, Thaneemit-Chen S, Krishnan V, Abraham WT, Lowes BD, Port JD, Davis GW, et al. An α2C-adrenergic receptor polymorphism alters the norepinephrine lowering effects and therapeutic response of the β blocker bucindolol in chronic heart failure. Circ Heart Fail. 2010;3(1):21–28. doi: 10.1161/CIRCHEARTFAILURE.109.885962. [DOI] [PubMed] [Google Scholar]
- 54.Bristow MR, Krause-Steinrauf H, Nuzzo R, Liang CS, Lindenfeld J, Lowes BD, Hattler B, Abraham WT, Olson L, Krueger S, Thaneemit-Chen S, et al. Effect of baseline or changes in adrenergic activity on clinical outcomes in the β-blocker evaluation of survival trial. Circulation. 2004;110(11):1437–1442. doi: 10.1161/01.CIR.0000141297.50027.A4. [DOI] [PubMed] [Google Scholar]
- 55.Pacanowski MA, Gong Y, Cooper-Dehoff RM, Schork NJ, Shriver MD, Langaee TY, Pepine CJ, Johnson JA. β-adrenergic receptor gene polymorphisms and β-blocker treatment outcomes in hypertension. Clin Pharmacol Ther. 2008;84(6):715–721. doi: 10.1038/clpt.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kardia SL, Kelly RJ, Keddache MA, Aronow BJ, Grabowski GA, Hahn HS, Case KL, Wagoner LE, Dorn GW, 2nd, Liggett SB. Multiple interactions between the α2C- and β1-adrenergic receptors influence heart failure survival. BMC Med Genet. 2008;9:93. doi: 10.1186/1471-2350-9-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lobmeyer MT, Gong Y, Terra SG, Beitelshees AL, Langaee TY, Pauly DF, Schofield RS, Hamilton KK, Herbert Patterson J, Adams KF, Jr, Hill JA, et al. Synergistic polymorphisms of β1 and α2C-adrenergic receptors and the influence on left ventricular ejection fraction response to β-blocker therapy in heart failure. Pharmacogenet Genomics. 2007;17(4):277–282. doi: 10.1097/FPC.0b013e3280105245. [DOI] [PubMed] [Google Scholar]
- 58.Premont RT, Koch WJ, Inglese J, Lefkowitz RJ. Identification, purification, and characterization of GRK5, a member of the family of G protein-coupled receptor kinases. J Biol Chem. 1994;269(9):6832–6841. [PubMed] [Google Scholar]
- 59.Liggett SB, Cresci S, Kelly RJ, Syed FM, Matkovich SJ, Hahn HS, Diwan A, Martini JS, Sparks L, Parekh RR, Spertus JA, et al. A GRK5 polymorphism that inhibits β-adrenergic receptor signaling is protective in heart failure. Nat Med. 2008;14(5):510–517. doi: 10.1038/nm1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60•.McNamara DM, Holubkov R, Janosko K, Palmer A, Wang JJ, MacGowan GA, Murali S, Rosenblum WD, London B, Feldman AM. Pharmacogenetic interactions between β-blocker therapy and the angiotensin-converting enzyme deletion polymorphism in patients with congestive heart failure. Circulation. 2001;103(12):1644–1648. doi: 10.1161/01.cir.103.12.1644. Suggested that ACE D/D patients had an enhanced response to BBs. This was the first pharmacogenetic finding in HF and helped advance the field of pharmacogenetics for HF. [DOI] [PubMed] [Google Scholar]
- 61.de Groote P, Helbecque N, Lamblin N, Hermant X, Amouyel P, Bauters C, Dallongeville J. β-Adrenergic receptor blockade and the angiotensin-converting enzyme deletion polymorphism in patients with chronic heart failure. Eur J Heart Fail. 2004;6(1):17–21. doi: 10.1016/j.ejheart.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 62.Frantz RP, Lowes BD, Grayburn PA, White M, Krause-Steinrauf H, Krishnan V, Uyeda L, Burnett JC. Baseline and serial neurohormones in patients with congestive heart failure treated with and without bucindolol: Results of the neurohumoral substudy of the β-Blocker Evaluation of Survival Study (BEST) J Card Fail. 2007;13(6):437–444. doi: 10.1016/j.cardfail.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 63.Taylor MR, Slavov D, Humphrey K, Zhao L, Cockroft J, Zhu X, Lavori P, Bristow MR, Mestroni L, Lazzeroni LC. Pharmacogenetic effect of an endothelin-1 haplotype on response to bucindolol therapy in chronic heart failure. Pharmacogenet Genomics. 2009;19(1):35–43. doi: 10.1097/FPC.0b013e328317cc57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bijl MJ, Visser LE, van Schaik RH, Kors JA, Witteman JC, Hofman A, Vulto AG, van Gelder T, Stricker BH. Genetic variation in the CYP2D6 gene is associated with a lower heart rate and blood pressure in β-blocker users. Clin Pharmacol Ther. 2009;85(1):45–50. doi: 10.1038/clpt.2008.172. [DOI] [PubMed] [Google Scholar]
- 65.Sharp CF, Gardiner SJ, Jensen BP, Roberts RL, Troughton RW, Lainchbury JG, Begg EJ. CYP2D6 genotype and its relationship with metoprolol dose, concentrations and effect in patients with systolic heart failure. Pharmacogenomics J. 2009;9(3):175–184. doi: 10.1038/tpj.2009.9. [DOI] [PubMed] [Google Scholar]
- 66.Lanfear DE. Genetic variation in the natriuretic peptide system and heart failure. Heart Fail Rev. 2010;15(3):219–228. doi: 10.1007/s10741-008-9113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee CY, Chen HH, Lisy O, Swan S, Cannon C, Lieu HD, Burnett JC., Jr Pharmacodynamics of a novel designer natriuretic peptide, CD-NP, in a first-in-human clinical trial in healthy subjects. J Clin Pharmacol. 2009;49(6):668–673. doi: 10.1177/0091270009336233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee CY, Lieu H, Burnett JC., Jr Designer natriuretic peptides. J Investig Med. 2009;57(1):18–21. doi: 10.231/JIM.0b013e3181946fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Newton-Cheh C, Larson MG, Vasan RS, Levy D, Bloch KD, Surti A, Guiducci C, Kathiresan S, Benjamin EJ, Struck J, Morgenthaler NG, et al. Association of common variants in NPPA and NPPB with circulating natriuretic peptides and blood pressure. Nat Genet. 2009;41(3):348–353. doi: 10.1038/ng.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hernandez AF, O’Connor CM, Starling RC, Reist CJ, Armstrong PW, Dickstein K, Lorenz TJ, Gibler WB, Hasselblad V, Komajda M, Massie B, et al. Rationale and design of the Acute Study of Clinical Effectiveness of Nesiritide in Decompensated Heart Failure Trial (ASCEND-HF) Am Heart J. 2009;157(2):271–277. doi: 10.1016/j.ahj.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 71.Carson P, Ziesche S, Johnson G, Cohn JN. Racial differences in response to therapy for heart failure: Analysis of the vasodilator-heart failure trials. Vasodilator-Heart Failure Trial Study Group. J Card Fail. 1999;5(3):178–187. doi: 10.1016/s1071-9164(99)90001-5. [DOI] [PubMed] [Google Scholar]
- 72••.Taylor AL, Ziesche S, Yancy C, Carson P, D’Agostino R Jr, Ferdinand K, Taylor M, Adams K, Sabolinski M, Worcel M, Cohn JN. Combination of isosorbide dinitrate and hydralazine in blacks with heart failure. N Engl J Med. 2004;351(20):2049–2057. doi: 10.1056/NEJMoa042934. Describes a successful clinical trial of HN combination therapy in HF. The trial is most notable because it is the first and only trial of a racially targeted cardiovascular therapy that was subsequently approved by the FDA. [DOI] [PubMed] [Google Scholar]
- 73.McNamara DM, Tam SW, Sabolinski ML, Tobelmann P, Janosko K, Venkitachalam L, Ofili E, Yancy C, Feldman AM, Ghali JK, Taylor AL, et al. Endothelial nitric oxide synthase (NOS3) polymorphisms in African Americans with heart failure: Results from the A-HeFT trial. J Card Fail. 2009;15(3):191–198. doi: 10.1016/j.cardfail.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 74.Werner D, Werner U, Meybaum A, Schmidt B, Umbreen S, Grosch A, Lestin HG, Graf B, Zolk O, Fromm MF. Determinants of steady-state torasemide pharmacokinetics: Impact of pharmacogenetic factors, gender and angiotensin II receptor blockers. Clin Pharmacokinet. 2008;47(5):323–332. doi: 10.2165/00003088-200847050-00003. [DOI] [PubMed] [Google Scholar]
- 75.Lynch AI, Boerwinkle E, Davis BR, Ford CE, Eckfeldt JH, Leiendecker-Foster C, Arnett DK. Pharmacogenetic association of the NPPA T2238C genetic variant with cardiovascular disease outcomes in patients with hypertension. JAMA. 2008;299(3):296–307. doi: 10.1001/jama.299.3.296. [DOI] [PubMed] [Google Scholar]
- 76.Li T, Chang CY, Jin DY, Lin PJ, Khvorova A, Stafford DW. Identification of the gene for vitamin K epoxide reductase. Nature. 2004;427(6974):541–544. doi: 10.1038/nature02254. [DOI] [PubMed] [Google Scholar]
- 77.Higashi MK, Veenstra DL, Kondo LM, Wittkowsky AK, Srinouanprachanh SL, Farin FM, Rettie AE. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002;287(13):1690–1698. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 78.Holbrook AM, Pereira JA, Labiris R, McDonald H, Douketis JD, Crowther M, Wells PS. Systematic overview of warfarin and its drug and food interactions. Arch Intern Med. 2005;165(10):1095–1106. doi: 10.1001/archinte.165.10.1095. [DOI] [PubMed] [Google Scholar]
- 79.Cavallari LH, Limdi NA. Warfarin pharmacogenomics. Curr Opin Mol Ther. 2009;11(3):243–251. [PubMed] [Google Scholar]
- 80.Glurich I, Burmester JK, Caldwell MD. Understanding the pharmacogenetic approach to warfarin dosing. Heart Fail Rev. 2008;15(3):239–248. doi: 10.1007/s10741-008-9115-9. [DOI] [PubMed] [Google Scholar]
- 81.Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, Blough DK, Thummel KE, Veenstra DL, Rettie AE. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352(22):2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 82•.Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, Ritchie MD, Stein CM, Roden DM, Smith JD, Veenstra DL, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112(4):1022–1027. doi: 10.1182/blood-2008-01-134247. Describes one of the first whole genome approaches to drug response in cardiovascular therapy that is notable not for the findings of the study, but for the approach used. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, Whittaker P, Ranganath V, Kumanduri V, McLaren W, Holm L, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5(3):e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sconce EA, Khan TI, Wynne HA, Avery P, Monkhouse L, King BP, Wood P, Kesteven P, Daly AK, Kamali F. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: Proposal for a new dosing regimen. Blood. 2005;106(7):2329–2333. doi: 10.1182/blood-2005-03-1108. [DOI] [PubMed] [Google Scholar]
- 85.Wu AH, Wang P, Smith A, Haller C, Drake K, Linder M, Valdes R., Jr Dosing algorithm for warfarin using CYP2C9 and VKORC1 genotyping from a multi-ethnic population: Comparison with other equations. Pharmacogenomics. 2008;9(2):169–178. doi: 10.2217/14622416.9.2.169. [DOI] [PubMed] [Google Scholar]
- 86.Millican EA, Lenzini PA, Milligan PE, Grosso L, Eby C, Deych E, Grice G, Clohisy JC, Barrack RL, Burnett RS, Voora D, et al. Genetic-based dosing in orthopedic patients beginning warfarin therapy. Blood. 2007;110(5):1511–1515. doi: 10.1182/blood-2007-01-069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Anderson JL, Horne BD, Stevens SM, Grove AS, Barton S, Nicholas ZP, Kahn SF, May HT, Samuelson KM, Muhlestein JB, Carlquist JF. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation. 2007;116(22):2563–2570. doi: 10.1161/CIRCULATIONAHA.107.737312. [DOI] [PubMed] [Google Scholar]
- 88.Caraco Y, Blotnick S, Muszkat M. CYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: A prospective randomized controlled study. Clin Pharmacol Ther. 2008;83(3):460–470. doi: 10.1038/sj.clpt.6100316. [DOI] [PubMed] [Google Scholar]
- 89.Kangelaris KN, Bent S, Nussbaum RL, Garcia DA, Tice JA. Genetic testing before anticoagulation? A systematic review of pharmacogenetic dosing of warfarin. J Gen Intern Med. 2009;24(5):656–664. doi: 10.1007/s11606-009-0949-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eckman MH, Rosand J, Greenberg SM, Gage BF. Cost-effectiveness of using pharmacogenetic information in warfarin dosing for patients with nonvalvular atrial fibrillation. Ann Intern Med. 2009;150(2):73–83. doi: 10.7326/0003-4819-150-2-200901200-00005. [DOI] [PubMed] [Google Scholar]
- 91.Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86(4):1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mason DA, Moore JD, Green SA, Liggett SB. A gain-of-function polymorphism in a G-protein coupling domain of the human beta1-adrenergic receptor. J Biol Chem. 1999;274(18):12670–12674. doi: 10.1074/jbc.274.18.12670. [DOI] [PubMed] [Google Scholar]
- 93.Mialet Perez J, Rathz DA, Petrashevskaya NN, Hahn HS, Wagoner LE, Schwartz A, Dorn GW, Liggett SB. Beta 1-adrenergic receptor polymorphisms confer differential function and predisposition to heart failure. Nat Med. 2003;9(10):1300–1305. doi: 10.1038/nm930. [DOI] [PubMed] [Google Scholar]
- 94.Biolo A, Clausell N, Santos KG, Salvaro R, Ashton-Prolla P, Borges A, Rohde LE. Impact of beta1-adrenergic receptor polymorphisms on susceptibility to heart failure, arrhythmogenesis, prognosis, and response to beta-blocker therapy. Am J Cardiol. 2008;102(6):726–732. doi: 10.1016/j.amjcard.2008.04.070. [DOI] [PubMed] [Google Scholar]
- 95.Levin MC, Marullo S, Muntaner O, Andersson B, Magnusson Y. The myocardium-protective Gly-49 variant of the beta 1-adrenergic receptor exhibits constitutive activity and increased desensitization and downregulation. J Biol Chem. 2002;277(34):30429–30435. doi: 10.1074/jbc.M200681200. [DOI] [PubMed] [Google Scholar]
- 96.Magnusson Y, Levin MC, Eggertsen R, Nystrom E, Mobini R, Schaufelberger M, Andersson B. Ser49Gly of beta1-adrenergic receptor is associated with effective beta-blocker dose in dilated cardiomyopathy. Clin Pharmacol Ther. 2005;78(3):221–231. doi: 10.1016/j.clpt.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 97.Green SA, Turki J, Innis M, Liggett SB. Amino-terminal polymorphisms of the human beta 2-adrenergic receptor impart distinct agonist-promoted regulatory properties. Biochemistry. 1994;33(32):9414–9419. doi: 10.1021/bi00198a006. [DOI] [PubMed] [Google Scholar]
- 98.Neumeister A, Charney DS, Belfer I, Geraci M, Holmes C, Sharabi Y, Alim T, Bonne O, Luckenbaugh DA, Manji H, Goldman D, et al. Sympathoneural and adrenomedullary functional effects of alpha2C-adrenoreceptor gene polymorphism in healthy humans. Pharmacogenet Genomics. 2005;15(3):143–149. doi: 10.1097/01213011-200503000-00002. [DOI] [PubMed] [Google Scholar]
- 99.Small KM, Forbes SL, Rahman FF, Bridges KM, Liggett SB. A four amino acid deletion polymorphism in the third intracellular loop of the human alpha 2C-adrenergic receptor confers impaired coupling to multiple effectors. J Biol Chem. 2000;275(30):23059–23064. doi: 10.1074/jbc.M000796200. [DOI] [PubMed] [Google Scholar]