Abstract
Collagens contain a unique triple helical structure with a repeating sequence -G-X-Y-, where proline and hydroxyproline are major constituents in X and Y positions, respectively. Folding of the collagen triple helix requires trimerization domains. Once trimerized, collagen chains are correctly aligned and the folding of the triple helix proceeds in a zipper-like fashion. Here we report the isolation, characterization and crystal structure of the trimerization domain of human type XVIII collagen, a member of the multiplexin family. This domain differs from all other known trimerization domains in other collagens and exhibits a high trimerization potential at picomolar concentrations. Strong chain association and high specificity of binding are needed for multiplexins, which are present at very low levels.
Keywords: collagen XVIII and XV, crystal structure, trimerization domain, non-collagenous domain, endostatin
Introduction
A self-organizing step, common to all collagen types, is trimerization that selects, binds and registers specific chains which subsequently oligomerize into specific suprastructures. The mechanism of chain selection and initiation of the triple helix formation is generally attributed to terminal non-collagenous domains.1 Early studies on fibril-forming collagen I and III suggested that the carboxyl-terminal non-collagenous domains (NC) play a crucial role in the trimerization step.2–6 The carboxyl-terminal NC domains govern chain selection and trimerization for all classes except for the transmembrane collagens XIII and XVII which are thought to be governed by the amino-terminal NC domains,7,8 and the fibril-associated collagens IX, XII, XIV, XVI, XIX, XX and XXI which are most probably governed by the NC2 domains (the second non-collagenous domain counted from the carboxyl-terminal end).9,10 The trimerization domains of network-forming collagens IV, VIII and X were crystallized and their atomic structures were recently solved,11–14 which allowed a detailed atomic analysis of chain selectivity for those collagen types. Many important features remain to be elucidated about the NC trimerization domains in other collagen types.
Collagens type XV and XVIII, identified as a chondroitin sulfate and heparan sulfate proteoglycan, respectively,15,16 are closely related non-fibrillar collagens that define the multiplexin subfamily (multiple triple helix domains with interruptions).17–20 α1 (XV) and α1 (XVIII) chains form homotrimers. Each chain is a composition of 10 collagenous (COL) repeats alternating with 11 non-collagenous (NC) repeats.21
Collagen XVIII came into the focus of medical interest when an 18-kDa anti-angiogenic peptide with tumor-suppressing activity, named endostatin, turned out to be the carboxyl-terminal part of collagen XVIII.22
The carboxyl-terminal NC domains (NC1 domains) of collagens XV and XVIII have been isolated from invertebrate and vertebrate basement membranes and identified as circulating fragments in serum.23 They share similar primary structures, which are organized into three different subdomains (Fig. 1).24,25 An amino-terminal region potentially responsible for trimerization is followed by a protease-labile segment and the endostatin domain.
Figure 1.
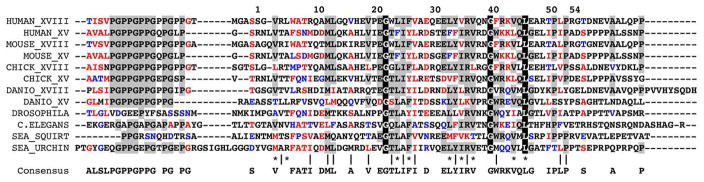
Schematic drawing of collagen XVIII carboxyl-terminus including a part of the COL1 domain and the full-length NC1 domain. The NC1 domain compromised of a trimerization domain, a hinge region and the endostatin domain.
Preliminary data indicates the presence of a trimerization domain at the beginning of the NC1 domain. Initially, it was shown by sieve chromatography that the recombinantly expressed full-length murine NC1 domain had an apparent molecular weight of ~100kDa which corresponds to a trimer (a single chain molecular weight is 38kDa), although the endostatin domain was found to be a monomer for collagens XV and XVIII.24 Moreover, the NC1 domain of collagen XVIII was sensitive to endogenous proteolysis which caused the appearance of electrophoretic bands of 30–32 and 5 kDa in significant amounts. The 5kDa fragment showed the original amino-terminus and was eluted at a position of ~12 kDa in molecular sieve chromatography which indicated its trimeric organization.24 Based on these findings and also on the sequence alignment between types XV and XVIII collagens and the exon positions, it was concluded that the trimerization domain spans residues 10–60 of the NC1 domain.24 Later, the NC1 domain of collagen XV was also found to form a trimer according to sieve chromatography.25 Independently, chemical cross-linking experiments on the recombinantly expressed human collagen XVIII NC1 domain demonstrated its trimeric nature.26 Recently, the proposed trimerization domain of murine collagen XVIII was successfully used to trimerize a single-chain antibody as part of a fusion molecule.27 Endostatin is probably the most well-studied endogenous angiogenesis inhibitor that potently inhibits the growth of various tumors in mice. Its anti-tumor activity is the consequence of an inhibition of endothelial proliferation and migration and an induction of endothelial apoptosis.22,28–31 Both mouse and human studies demonstrate that endostatin produces virtually no toxicity after long-term delivery.32 Many published reports show that endostatin mainly suppresses pathological angiogenesis and does not affect wound healing or reproduction.33 However, its antitumor activity remains a controversial issue, which is probably due to differences in expression systems, protein folding and solubility, heparin-binding affinities, zinc binding, dosages, bolus versus sustained delivery, and gene therapy versus protein therapy.32 Another function of the collagen XVIII NC1 domain is its ability to regulate ECM-dependent motility and morphogenesis of both endothelial and nonendothelial cells in a manner strictly requiring the endostatin domain oligomerization. These properties are distinct from and in fact antagonized by its physiologic cleavage product an endostatin domain monomer, and the NC1 trimer is inhibited by the endostatin monomer in a potential negative autoregulatory loop.26
We confirm in this study the presence of the trimerization domain at the beginning of the NC1 domain of both type XV and XVIII collagens by analytical ultracentrifugation and present the crystal structure of the type XVIII collagen trimerization domain with excellent association properties.
RESULTS
NC1 domain contains a trimerization domain at the amino-terminus
A set of the full-length NC1 domains and the endostatin domains of murine collagens XV and XVIII were expressed in 293 cells, purified and analyzed on an analytical ultracentrufuge to determine their oligomeric states. This was done to verify preliminary indications of a trimeric amino-terminal part of the NC1 domain, obtained by a sieve chromatography. Data are summarized in Table 1. The isolated endostatin domains of both collagen XV and XVIII are monomers, while the full-length NC1 domains are trimers.
TABLE 1.
Molecular mass determinations of the full-length NC1 domain and the endostatin domain of both murine type XV and XVIII collagens by sedimentation equilibrium. Samples were run in PBS at 4°C.
| Protein | <U> (cm3 g−1) | RPM | Calculated mass (kDa) | AUC mass (kDa) | Oligomeric state | Concentration (mg/ml) |
|---|---|---|---|---|---|---|
| XV-ES | 0.7326 | 20,000 | 20.0 | 17.4±0.1/16.6±0.1 | 0.87/0.83 | 0.5/1.0 |
| XVIII-ES | 0.7253 | 20,000 | 20.7 | 18.5±0.1 | 0.89 | 0.5 |
| XV-NC1 | 0.7378 | 12,000 | 28.9 | 90.1±0.7/104.1±1. | 3.11/3.6 | 0.27/0.55 |
| XVIII-NC1 | 0.7299 | 12,000 | 38.0 | 97.6±0.3 | 2.56 | 0.5 |
Defining a stable trimeric domain
The first three constructs listed in Table 2 were cloned and expressed in E. coli as fusion proteins with the mini-fibritin 34,35 which as obligatory trimer induces homotrimerization. Cleavage of this homotrimerizing mini-fibritin allows then to identify homotrimers even in absence of this domain. The constructs include adjacent COL1 and NC1 sequences of varying length. The cleaved and isolated proteins were then subjected to trypsin proteolysis and a common stable domain was identified (Table 2). There is only one potential trypsin cleavage site in construct (43)COL1-NC1(69), and two sites in constructs (66)COL1-NC1(69) and (66)COL1-NC1(136) within the COL1 region, and 5 and 11 sites in (43)COL1-NC1(69)/(66)COL1-NC1(69) and (66)COL1-NC1(136) within the NC1 region, respectively. The major cleavage product observed on SDS-PAGE for all three constructs had an apparent molecular weight of about 8kDa. Three tryptic fragments of (43)COL1-NC1(69) were purified on HPLC C18 column and analyzed by amino-terminal sequencing and mass-spectrometry. These are fragments 1–19, 20–97 and 98–112, based on the sequence numbering of construct (43)COL1-NC1(69). Thus the fragment 20–97 (or (24)COL1-NC1(54)) is the major cleavage product which presumably includes a trimerization domain and a short sequence of the collagenous domain with six residues of a triple helix interruption and six tripeptide units -G-X-Y- with no potential trypsin cleavage sites. Since E. coli expressed collagen sequences do not contain hydroxyprolines in the Y position the ability of such a short collagenous sequence to form a stable triple helix is questionable. The melting transition curves monitored at 225nm (a characteristic wavelength for monitoring a triple helix) by CD in a range of temperature 4–70°C showed no triple helix formation (data not shown). A new construct, NC1(54), was made based on the trypsin stable fragment with all the triple helical residues deleted. The last amino acid residue of the new construct is arginine, exactly as after trypsin digestion of longer constructs. NC1(54) was successfully crystallized and the structure showed a compact trimeric organization.
TABLE 2.
Sequences of peptides expressed as fusion proteins with the mini-fibritin. First and last numbers in peptide naming indicate the number of residues included from the carboxyl-terminal part of COL1 and the amino-terminal part of NC1, respectively. Numbering is given for the NC1(54) peptide only, where residues −1 and 0 do not belong to the NC1 domain. Underlined sequences are potential triple helix. Italics are non-natural sequences at the amino termini where GS were a part of the thrombin cleavage site (in all constructs) and P was added artificially for the continuous triple helix in constructs (66)COL1-NC1(69) and (66)COL1-NC1(136). The NC1(54) peptide contains only a single non-natural amino acid, G(−1) instead of A. Highlighted with gray are trypsin resistant region containing the trimerization domain.
 |
Crystallization
The NC1(54) protein was crystallized in two crystal forms, cubic at neutral pH and tetragonal at acidic pH. Crystallization was highly dependent on peptide preparation batch, varying from nice crystals to small disordered crystals to none at all. The cubic crystal form was exceptionally sensitive to the purity of the peptide. Reproducibility was solved by eliminating contamination by a tryptic fragment lacking eleven amino-terminal residues whose cleavage site mapped to the starred location in H2N-GSSGVRLWATR*QAML… (by mass-spectrometry). The cleavage site is located at the beginning of an α-helix and is solvent exposed.
Solving the crystal structure
No protein of known atomic structure with significant sequence homology to NC1(54) was identified for use as an initial phasing model. To obtain the phase information necessary for crystal structure determination we produced a seleno-methionine derivative which was crystallized under the same conditions as the native protein. The structure of two crystal forms was independently determined by SeMet MAD phasing after determining the positions of six Se atoms per asymmetric unit of the tetragonal crystal form and one Se atom per asymmetric unit of the cubic form. For details see the Materials and Methods section.
Crystal structure
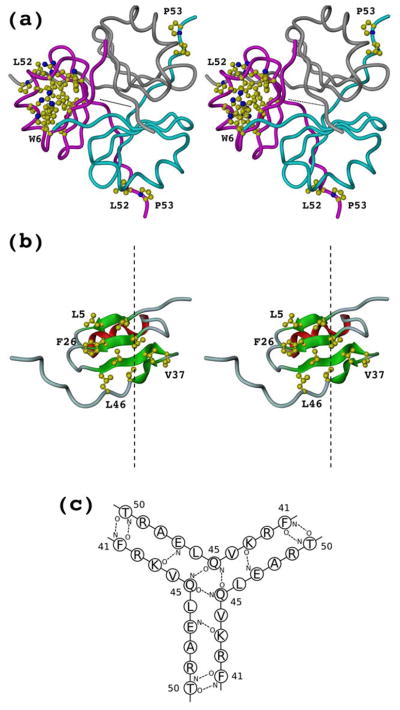
The tetragonal crystal form of NC1(54) has six chains per asymmetric unit which form two trimers. There is a non-crystallographic two-fold rotation symmetry relating those trimers and each trimer has a three-fold non-crystallographic rotation axis. The cubic form has a single chain per asymmetric unit. A trimer, the biological unit of NC1(54), is formed by a crystal three-fold rotation symmetry.
Although two crystal forms were grown at two different pH levels, one highly acidic (pH 3–4), no significant structural differences were observed, the r.m.s.d. of the least-square fit of the single chain Cα atoms of the cubic form and corresponding atoms of any chain of the tetragonal form varies from 0.65 to 0.75 Å. The detailed analysis of the biological form, the trimer, is based on chains A, B and C of the tetragonal form, which is at much higher resolution. The choice of chains A, B and C is favored over chains D, E and F because the latter have significantly higher B-factor of ~40 Å2 (Fig. 2). The same difference in B-factor values within asymmetric unit was observed both in native and seleno-methionine crystals which reflects the nature of the crystal packing. Each chain has four β-strands, one α-helix and a short 310 helix (Figs. 2 and 3). Four β-strands of each chain form a mixed parallel and antiparallel β-sheet which faces β-sheets of two adjacent chains. Side chains of all three β-sheets form a hydrophobic interior of the trimer. The α-helix and 310 helix are solvent exposed and interact with all four β-strands of the same chain as well as with the carboxyl-terminal end of the adjacent chain (Fig. 3). The overall shape of the trimer might be approximated by a triangular prism with a side of ~40Å and a height of ~20Å. The amino-terminal end is fully structured up to the very first unnatural Gly (it substitutes for Ala). The protein numbering starts at −1 so that residue 1 is the first residue of the NC1 domain. The carboxyl-terminal end is structured to the last R54, although in some chains it is partially or fully disordered, depending on crystal contacts. We anticipate that R54 is not important for the trimer formation since it is fully exposed to solvent.
Figure 2.
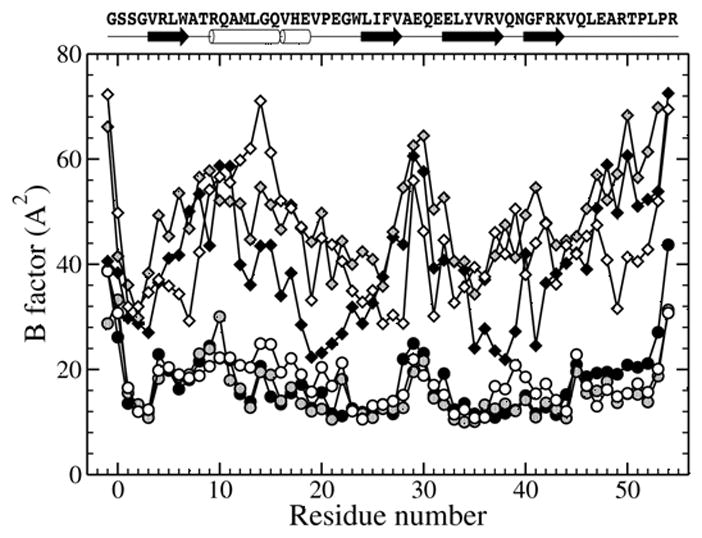
Secondary structure elements and B-factor values for chains A to F of the tetragonal crystal form. Chains A, B and C are depicted with black, gray and open circles, while chains D, E and F with black, gray and open diamonds, respectively.
Figure 3.
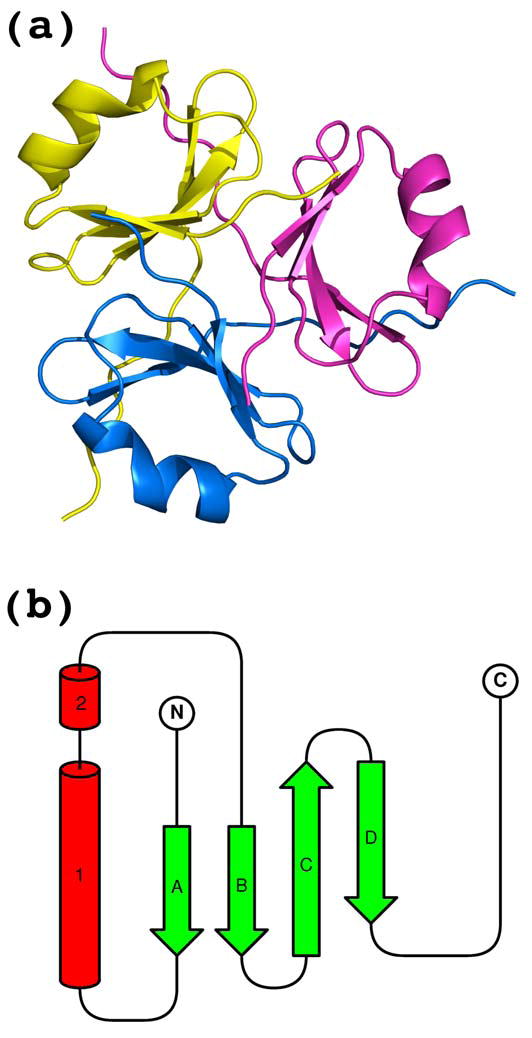
(a) The trimeric biological unit with secondary structure elements and (b) the topology diagram of the single chain. α-Helix (labeled with 1) and 310-helix (labeled with 2) are shown as red cylinders, β-strands (labeled with A through D) are shown as green arrows.
Two types of hydrophobic core are found in the structure. One forms an interface between β-sheet and helices of each chain which stabilizes the monomeric subunit (Fig. 4a) and another forms the central interior, which must be the driving force for trimer formation (Fig. 4b). The hydrophobic core of each subunit is mainly formed by side chains of residues, M12, L13, V16, V19, I25, V27, F41, and is additionally stabilized by hydrophobic parts of charged and polar residues R4, W6, R9, W23, Y34 and R36 (Fig. 4a). The trimer interface is exclusively formed by hydrophobic residues V3, L5, L24, F26, L33, V35, V37, V44 and L46 (Fig. 4b). Two residues L52 and P53 contribute to the hydrophobic core of the adjacent monomeric subunit (Fig. 4a) and thus stabilize both the monomeric and trimeric cores. No solvent molecules are observed in either hydrophobic core.
Figure 4.
Hydrophobic cores and intersubunit hydrogen bonding. (a) Residues forming the hydrophobic core of the monomeric subunit (shown for one chain only). Residues L52 and P53 are shown for all chains to emphasize their dual role in stabilizing the monomer and the trimer. (b) Residues forming the hydrophobic interior of the trimer (shown for one chain). Non-crystallographic three-fold axes are shown in dashed line. (c) Main chain to main chain intersubunit hydrogen bonding diagram.
Interchain hydrogen bonds are also involved in establishing the trimer interface. The most interesting main chain interactions involve hydrogen bonds F41(N)-T50(O), Q45(N,O)-Q45(O,N), E47(N)-K43(O), T50(N)-F41(O) (Fig. 4c). Additional main chain-to-main chain hydrogen bonds S1(N)-V3(O), as well as side chain-to-main chain hydrogen bond between R42 and E47(O) and side chain-to-side chain between E31 and R42, E47 and R42 also contribute to the trimer specificity.
An interesting substructure might be observed in the crystal packing of the tetragonal form (but not in the cubic crystal form). Two chains, C and D, within one asymmetric unit but belonging to two different trimers have a crystal contact mediated by a sulfate ion (Fig. 5). The ion is located on the non-crystallographic two-fold rotation symmetry axis. The negatively charged sulfate ion is coordinated by positively charged side chains of R36 and additionally stabilized by hydrogen bonds with the amino groups of N39 and protonated side chains of E21 (pKa=4.4 for glutamate side chain, while the crystals were grown at pH 3.0–4.0) (Fig. 5b). The same type of crystal contact is observed between trimers of different asymmetric units. Twelve such sulfate ions are involved in organizing eight trimers of four symmetry related asymmetric units to form an ideal cube, where trimers occupy corners in a way that their central axes point to the center of the cube (Fig. 5a). Although this fascinating cubic structure observed in the tetragonal crystal form is not biologically relevant (non physiological pH range), it might serve as a model for generating self-assembling nanostructures with defined composition and geometry by adding sulfate ions and lowering pH, but further experiments are required for testing.
Figure 5.
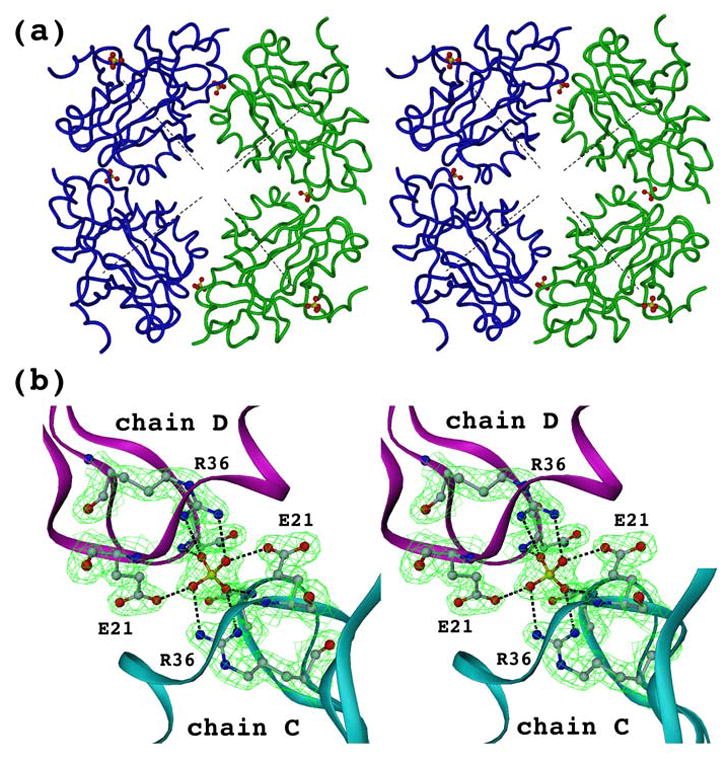
One type of crystal contact of the tetragonal crystal form. (a) Crystal contacts between two trimers within one asymmetric unit (shown in green or blue) as well as within different units (green and blue) are mediated by sulfate ions. These two symmetry related asymmetric units constitute only a half of a cube which requires another pair of symmetry related asymmetric units to fulfill it (not shown for clarity). Central axes of all eight trimers, which constitute the cube, intersects at the same point (shown only four of them as dashed lines). (b) Closer view of the crystal contact between chains C and D within one asymmetric unit. Contour map (2Fo−Fc) is shown at a sigma level of 1.5.
Denaturant-induced equilibrium transitions
To test whether the isolated trimeric form of the NC1(54) peptide observed in the crystal structure is able to trimerize itself without the need of a helper trimerization molecule, which has been used for the expression, the equilibrium unfolding-refolding transitions were measured. Figure 6a shows the CD spectrum of the NC1(54) peptide in 50mM sodium phosphate buffer, pH 8.0, with two prominent peaks centered at 205 nm (negative) and 233 nm (positive). Intrestingly, a similar positive peak was also observed for T4 page fibritin trimerization domain.36 The maximal positive CD signal at 233 nm was chosen for monitoring (GdmCl)-induced transitions due to spectroscopic limitations of high concentrations of the denaturant at the wavelength range below 210nm. The unfolding of the initial sample used for refolding curve was performed at monomer concentration of 300 μM in 7 M GdmCl for 10 minutes and its completeness was verified by CD signal at 233 nm (data not shown). Figure 6b shows unfolding (open circles) and refolding (black circles) curves at a monomer concentration of 30 μM at 20 °C. To confirm that these transitions are a trimer to monomer conversion, the samples were also chemically cross-linked with glutaraldehyde and analyzed on SDS-PAGE (Fig.6d). Since the NC1(54) peptide has only two potential sites for glutaraldehyde cross-linking (the amino terminus and a single lysine) the appearance of a trimeric band was rather limited. However, the presence and disappearance of the trimeric (as well as dimeric) band follows exactly the CD monitored unfolding/refolding with most of the transition located between 1.5 and 2.5M GdmCl (Fig.6d, lanes 5–7). The coincidence of unfolding and refolding curves demonstrates the reversible folding of the trimeric form of the NC1(54) peptide and thus confirms its trimerization properties (Fig.6b). Assuming that the trimer unfolds in a cooperative two-state transition of a native trimer (N) to unfolded monomers (U):
the transition curve can be used for calculating the equilibrium constant (Keq) for every GdmCl concentration:
| (1) |
where [M]0 indicates the total monomer concentration ([M]0=[U]+3[N]) and fU is the fraction of unfolded monomer (fu=[U]/[M]0). The standard free energy ΔG0 is related to Keq by:
Figure 6.
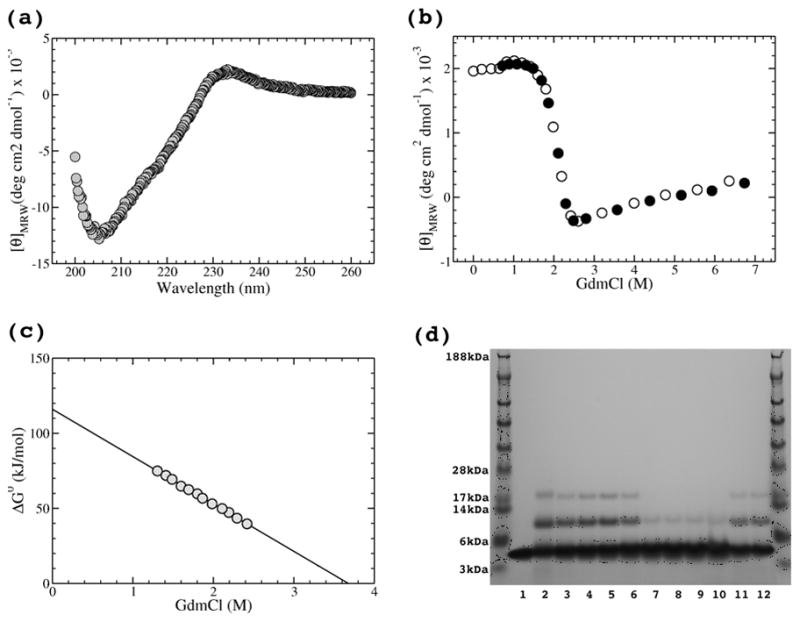
CD spectrum and GdmCl-induced unfolding-refolding transition of the NC1(54) peptide at 30μM chain concentration in 50mM sodium phosphate buffer, pH 8.0, recorded using a 1mm path-length quartz cuvette equilibrated at 20°C. (a) CD spectrum of the native form. (b) GdmCl induced unfolding (open circles) and refolding (circles) transitions monitored at 233nm. (c) ΔG0 dependence on GdmCl concentration with the linear fit. (d) GdmCl induced transitions observed on 4–12% SDS-PAGE. Equilibrated samples (the same conditions as for CD monitored transitions) were chemically cross-linked with 0.3% glutarldehyde for 15 minutes. Lane 1 shows non-cross linked NC1(54). Unfolding trace is shown on lanes 2–10, where GdmCl concentrations were 0, 0.5, 1.0, 1.5, 2.0, 2.5, 3.0, 5.0 and 7.0M, respectively. Refolding is shown on lanes 11,12, where GdmCl concentrations were 0.7 and 1.4M, respectively.
A denaturant dependence of ΔG0 shown in Figure 6c appears to be linear:37,38
where ΔG0(H2O) is the free energy for unfolding in water and m is the change in free energy with GdmCl. The linear fit yields a free energy of unfolding of ΔG0(H2O) = 116.0(±0.7) kJ/mol and the change in free energy with GdmCl (m-value) of −31.5 (kJ/mol)/M. ΔG0(H2O) is noticeably higher compared to the stability of another small trimerization domain from bacteriophage T4 fibritin, the foldon domain (27-residue long), for which ΔG0(H2O) = 89.2(±0.6) kJ/mol.39 A small difference in absolute values of ΔG0(H2O) leads to an impressive difference in Keq being 2.4×10−21 mol2 for the NC1(54) peptide and 1.4×10−16 mol2 for the fibritin foldon domain. From Keq follows the midpoint total chain concentration at which a half of it is incorporated into a trimer, while another half stays in monomeric form, i.e. 3[N]1/2=[U]1/2. From Equation (1):
follows
The midpoint total chain concentrations are then twice [U]1/2 and equals 56.6 pM for NC1(54) and 13.7nM for the foldon, which demonstrates remarkable trimerization abilities of the NC1(54) peptide.
For globular proteins, an m-value was found to correlate with ΔASA (ASA=accessible surface area) between native and unfolded state.40 A good correlation of m-value was also found for the foldon domain assuming its trimeric composition.39 This is not the case for the collagen XVIII trimerization domain. For unfolding of the NC1(54) peptide a ΔASA of ~20,000 Å2 is expected (see Materials and Methods). Based on the correlations found in globular proteins a ΔASA of 20,000 Å2 should result in an m-value of ~−22 (kJ/mol)/M,40 which significantly deviates from the value found for the NC1(54) peptide and possibly indicates the necessity for special consideration of multimeric proteins.
Preliminary experiments on refolding of the NC1(54) peptide show that the reaction is in a subsecond interval. The unfolded peptide at monomer concentration of 300 μM in 7M GdmCl was manually diluted ten times with 50mM sodium phosphate, pH 8, to a final GdmCl concentration of 0.7M and a final peptide concentration of 30 μM as fast as possible. The change in the CD signal was then monitored. The dead time of mixing was about 10 seconds. Only a very small change (less than 5% of the difference between denatured and native signals of the corresponding protein concentration) was observed within 5–10 seconds in the CD signal, which reached the value of the native trimeric form. The observed slow rate trace of refolding was probably observed due to a number of prolines in the protein sequence and their slow cis-trans isomerization.
Attempts to study thermal unfolding of the trimerization domain were unsuccessful. In 50mM sodium phosphate, pH 8, the protein retains its native structure upon heating to about 70°C, but rapidly and irreversibly precipitates upon heating above 75°C.
Homology modeling of type XV trimerization domain
Sequence identity between type XVIII and XV collagen is rather small, being only 31% (Fig. 8). The crystal structure of the collagen XVIII trimerization domain was used as an initial model for three-dimensional homology modeling of the collagen XV trimerization domain. After changing the non-identical residues the model was subject to several rounds of an energy minimization in vacuo and in a water shell using a program Insight II (Accelrys Software Inc.). The total energy substantially dropped to a value of −43,459 kJ/M/trimer. The final model was inspected manually and did not reveal any clashes or severe problems with the backbone traces and side-chain packing. The least-square fit of Cα atoms of two structures, the NC1(54) and the modeled collagen XV domain, had the r.m.s.d. of only 0.47 Å. A set of interesting results were observed in the modeled structure. Two leucine residues (24 and 33) of every chain of the NC1(54) forming a central hydrophobic core are replaced by two phenylalanine residues which leads to an increased volume of the central core (Fig. 7). In addition V35 and V44 of the hydrophobic interior are substituted by I35 and L44 with bulkier side chains. Other residues of the trimer core L5 and F26 are A5 and Y26 in the modeled collagen XV trimerization domain (Fig. 7). The rest of the hydrophobic interior, residues V3, V37, L46, is conserved. Solvent exposed W23 which has a hydrophobic contact with G2 is replaced by T23 which allows a substitution of G2 to L2. All differences in amino acid sequences between collagen XVIII and XV trimerization domain are reasonably accommodated in the three-dimensional model of collagen XV.
Figure 8.
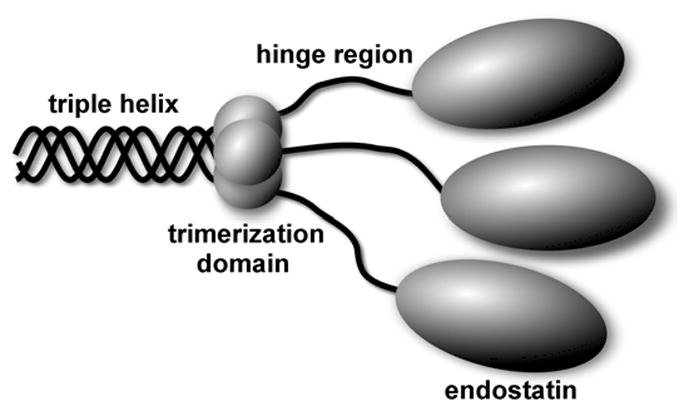
Sequence alignment of a potential trimerization region of different multiplexins. Sequence alignment of the trimerization domain of different multiplexins. Multiplexins of human, mouse, chicken, zebrafish, drosophila, nematode, sea squirt and sea urchin were aligned using Vector NTI software. Residue numbering is given for the human collagen XVIII NC1 domain. Identical residues across all sequences are shown in white with black shading, conservative residues in black with gray shading, blocks of similar residues are depicted in red, and weakly similar residues in blue. GenBank™ accession numbers for the sequences are as follows: human collagen XVIII, P39060; human collagen XV, P39059; mouse collagen XVIII, P39061; mouse collagen XV, O35206; chicken collagen XVIII, NP_989495; chicken collagen XV, XP_418896; zebrafish collagen XVIII, Q2PBM7; zebrafish collagen XV, Q05H57; drosophila multiplexin, ACD03747; C. elegans CLE-1 protein, Q7JL30; sea squirt (Ciona intestinalis) collagen XVIII homologue, Q86SC8; and sea urchin (Strongylocentrotus purpuratis) collagen XVIII homologue, XP_781637. Residues forming hydrophobic interior of the trimerization domain are indicated with asterisk, whereas residues forming hydrophobic core of each subunit are marked with a vertical line.
Figure 7.
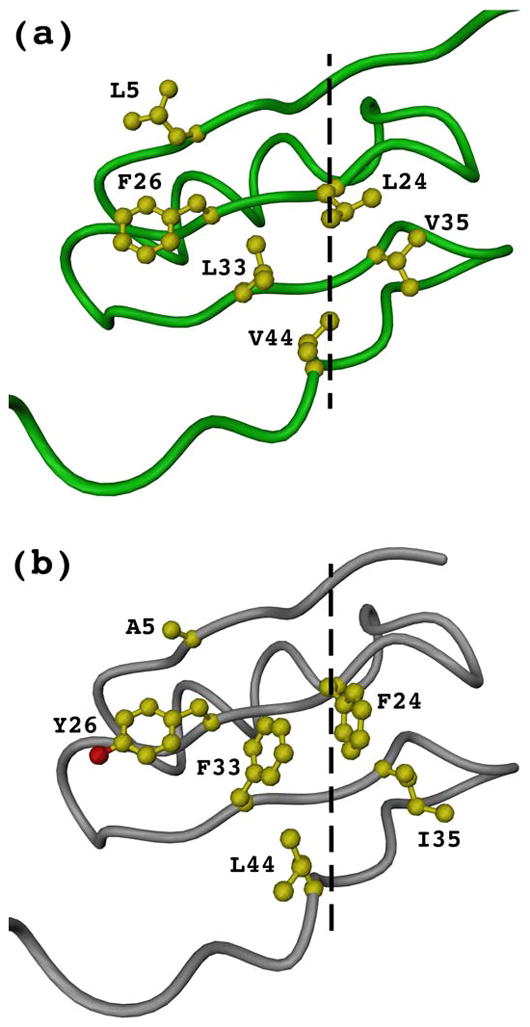
Human collagen XVIII trimerization domain crystal structure (a) and the modeled analog of human collagen XV (b). Shown are traces of single chain backbones and side chains of those hydrophobic interior residues that differ between XVIII and XV trimerization domains. The central axes of trimers are shown as dashed lines.
Sequence pattern of trimerization domain of other known multiplexins
Multiplexins of different organisms were analyzed in order to identify their trimerization domain. Interestingly, although there is no significant sequence homology among them in the region where the trimerization domain is located in human type XVIII collagen, this region has a defined length, it has no deletions or insertions. Only three residues are identical throughout all aligned sequences, i.e. G22, G40 (involved in formation of β-turns) and L46 (a part of the trimer hydrophobic interior). The most striking thing is a conserved pattern of key residues involved in formation of both monomer and trimer hydrophobic cores (Fig. 8).
Comparison to structures of other collagen trimerization domains
There are only three known atomic structures of trimerization domains in collagens, the NC1 domain of collagen IV11,12 and the homologous NC1 domain of collagens VIII14 and X.13 All of them including the newly solved collagen XVIII trimerization domain have a high content of β-structure, but share no structural homology. The trimerization domains of collagen IV and collagens VIII and X are ~230 and ~160 residues long, respectively, compared to 54 residues of collagen XVIII. The NC1 domains of collagens IV, VIII and X are also involved in network formation which can possibly explain their bigger size, whereas the trimerization domain of multiplexins is only a small part of the NC1 domain and it is probably optimized for chain association only. It is worth mentioning that the amino-terminal part (about 30 residues) of the NC1 domain in collagens VIII and X was unexpectedly unstructured, although that part should provide the alignment of collagen chains and initiate folding of the triple helix.
The trimeric structure of NC1(54) is mainly stabilized by a central hydrophobic core (Fig. 4b), where no solvent molecules are observed. That is not the case for crystal structures of trimerization domains in collagens VIII and X, where solvent molecules as well as polar residues were found in the hydrophobic core that is important for trimeric interface.13,14 Trimer formation in collagen IV is not governed by a general hydrophobic core at all and is primarily based on exchanges of β-strands between subunits, local hydrophobic contacts and hydrogen bonding, moreover an internal axial tunnel with the diameters varying from 4 to 15Å has mainly negatively charged surface.13
Three-dimensional homologous folds
Searching matches of NC1(54) to all structures in the PDB using the DaliLite server41 results in a list of hits with the highest Z-score (a representative value of similarity) being only 4.1. The threshold for any reasonable similarity is about 2. A more convenient analysis is provided by comparison with PDB90, which is a representative subset of PDB chains, where no two chains contain more than 90% sequence identity. Although about 70 hits have z-scores above 2 the majority of them represents structures where similarity is even not relevant to trimeric folds. Nevertheless, the first two hits with maximal z-scores and two more with much lower scores reveals structures with the similarities in their trimeric domains. These include Streptococcus pyogenes bacteriophage-associated hyaluronidase (z-score: 4.1, PDB accession: 2dp5), Bordetella phage Bmp-1 major tropism determinant (z-score: 3.6, PDB: 1yu2), Salmonella enterica phage DET7 tailspike protein (z-score: 2.4, PDB: 2v5i)42 and another tailspike protein of Enterobacteria phage P22 (z-score: 2.2, PDB: 2vfp)43. All hits are large trimeric proteins from bacteriophages. The first two structures share the structural homology with NC1(54) at their very amino-terminal domain, whereas two others share homology at the very carboxyl-terminal domain. Since the two last proteins share ~50% overall sequence identity and exhibit very similar overall three-dimensional structures,42 they should be considered basically as a single hit. To the best of our knowledge, the amino-terminal domains of neither the phage hyaluronidases nor the major tropism determinants were reported as trimerization domains. The hyaluronidase genes found in the bacteriophage genomes show a high degree of similarity to each other with a major difference being the presence or absence of a 102-bp fragment that encodes a collagen-like motif, -G-X-Y- repeating units.44 The collagen-like fragment is located just after the amino-terminal domain that structurally resembles the collagen XVIII trimerization domain. A role and structural organization of this collagen-like fragment remains unclear, as well as the proximity to the amino-terminal domain.
The P22 tailspike carboxyl-terminal domain (residues 544–666) was shown to be involved in stabilization of the trimeric structure.45 Very recently, the trimerization ability of the isolated carboxyl-terminal domain of P22 tailspike was demonstrated in a fusion chimera with the monomeric maltose binding protein.46 The carboxyl-terminal domain can be visually separated into the β-prism and caudal fin subdomains, although their trimeric structure is stabilizied by the continuous central hydrophobic core.45 It is the caudal fin subdomain which share the three-dimensional homology with the collagen XVIII trimerization domain. It is unclear whether the two subdomains pertain the trimerization ability only as a single domain or the caudal fin part might be an independent trimerization unit.
The monomeric core of all homologues includes both hydrophobic and polar/charged residues involved in a set of hydrogen bonds. Strikingly, all structural homologues reveal a strong hydrophobic core at the trimer interface, we thus anticipate the potential trimerization role of these regions in the folding of full-length proteins. It might be the case that the caudal fin subdomain of the tailspikes is already sufficient to drive the trimerization of the whole molecule.
DISCUSSION
Collagens are the major components of the extracellular matrix of multicellular animals. At present, 28 types of collagen are classified for vertebrate species. The characteristic feature of collagen and collagen-like proteins is a formation of a triple helix with the G-X-Y repeated sequence. Although short collagen model peptides are able to form the triple helix without any additional trimerization domain47 it becomes increasingly clear that full-length collagen molecules require specialized domains for chain selection, trimerization and subsequent triple helix formation.1 Moreover, the stability of the triple helix as well as folding kinetics is greatly affected by the presence of such domains.36 In vitro studies of the collagen triple helix stabilized by a specific trimerization domain is then more biologically relevant. Available methods to mimic the natural trimerization of the collagen triple helix are limited to either use of disulfide cross-links48 or an exogenous trimerization domain of bacteriophage T4.36
We have recently identified an α-helical trimerization domain of human type XIX collagen and demonstrated its stabilizing effect on the triple helix.10 Type XIX collagen is a member of fibril-associated collagens with interrupted triple helix (FACITs) which include type IX, XII, XIV, XVI, XIX, XX, XXI and XXII collagens. The discovery of the trimerization domain of collagen XVIII further extends our knowledge on the folding of collagens as well as provide us with another small, efficient and collagen-specific association domain for better understanding of collagen folding, stability and influence of mutations.
The multiplexins have a sparse presentation in tissues, type XV collagen in one of its most abundant tissue sources (an umbilical cord) was estimated at only (1–2)×10−4% of dry weight.49 Upon expression in a cell, a local concentration of multiplexins in the ER, where the folding of collagens takes place, is probably much higher, although it is difficult to expect that it might be comparable to that of fibrillar collagens. To initiate chain association into a trimer, the multiplexin trimerization domain has to have a sufficient association constant, much higher than that of any fibrillar collagen, and a fast kinetics of association. Being relatively small (less than 60 residues, compared to ~250 residues for fibrillar collagens) this domain can form a trimer at picomolar concentrations. Small size and lack of disulfide bonds also facilitate fast folding. Moreover, despite the predicted structure identity between the trimerization domains of collagen types XVIII and XV, differences in hydrophobic residues which constitute the trimer association interface should allow for co-expression of these collagen types in the cell without being interfered and mixed.
In earlier studies, the beginning of the multiplexin trimerization domain was localized at about residue 10 of the NC1 domain. Analysis of the crystal structure of the NC1(54) peptide, which includes the very beginning of the NC1, reveals that those ten residues participate in a number of interactions and are thus presumably important for the folding of the monomeric unit as well as for the trimer association. The amino-terminal residues are adjacent to the collagen triple helical part of the full-length collagen molecule and have to accommodate the three-fold rotational symmetry of the trimerization domain and the collagen three chain-staggering. So far, the only example with known atomic structure of such accommodation is an artificial fusion of the collagen model peptide, (GPP)10, and the fibritin foldon domain.50 The structure reveals a dramatic kink with an angle of 62.5° and a distortion of a few residues in both domains. As observed in the crystal structure of the collagen XVIII trimerization domain, the amino-terminal residues that should just follow the collagen triple helix are spatially distant (even more distant than in the foldon!) and stabilized by interactions with the surface of the molecule. Possibly, detaching or pulling a different number of residues for each chain (one, two, and three, respectively?) should align them with the necessary stagger of the triple helix. Future experiments to crystallize the collagen XVIII trimerization domain with stable triple helices will show whether nature has succeeded in optimizing such an interface in collagens.
In 2003 McAlinden et al.9 had performed an extensive analysis for potential α-helical coiled-coil regions among all collagen and collagen-like proteins known at that time. An α-helical coiled-coil is a versatile motif which is able to form a variety of monomeric, dimeric, trimeric, tetrameric and pentameric parallel or antiparallel ensembles.51 Three-dimensional structures of the three-stranded coiled-coil domains were determined for the collectin family, i.e. for the mannose-binding protein52,53 and the lung surfactant protein.54 Collectins are trimeric molecules with a collagen-like domain followed by an α-helical coiled-coil region and then a carbohydrate recognition domain. Although the coiled-coil region of lung surfactant protein D forms a parallel triple helix as determined by NMR,55 the coiled-coil peptide of mannose-binding protein A does not form stable triple helices and requires the carbohydrate recognition domain to stabilize the trimer.53 Surprisingly, McAlinden et al.9 were able to find potential coiled-coil sequences in most of the collagens and collagen-like molecules. These sequences might play a role in chain selection and trimerization of collagens, as was initially suggested for collagens XIII and XVII,7,8 but experimental proof is needed in each case. Potential coiled-coil sequences were also found in the oligomerization domains of the multiplexins, they were located to residues 27–48 of the NC1 domain. All the residues are in the crystal structure of the collagen XVIII trimerization domain, where no coiled coil is present, moreover these residues form β-strands and loops only (Fig. 2). All predictions of coiled coil structures must be considered with caution and proven experimentally.
The small size, high expression level and solubility of the type XVIII trimerization domain makes it an attractive choice for engineering different kinds of homotrimeric collagen molecules with an inherited propensity to form a triple helix. Although the foldon domain of phage T4 fibritin was successfully applied to design a set of different collagen peptides, including the full-length type I and III collagen,56 the use of human collagen XVIII trimerization domain looks more appropriate due to several reasons: no potential antigenicity, stronger association and presumably better arrangement of staggered triple helix.
Materials and Methods
Cloning, expression and purification
Recombinant full-lenght NC1 domains and endostatin domains of mouse collagen types XV and XVIII were expressed in HEK 293 cells and purified as described previously.24,25 To facilitate expression and purification of deletion mutants containing the putative trimerization domain of human collagen XVIII they were cloned as part of a fusion molecule with a His-tagged mini-fibritin with a thrombin cleavage site (HT-mf-thr). Mini-fibritin is an obligatory trimer with both amino- and carboxyl-termini exposed to the solvent.34 Recently the modified version of mini-fibritin (i.e. HT-mf-thr) has been successfully exploited to express a set of collagen fragments35,57 and a set of hantavirus nucleocapsid coiled coil fragments.58 The plasmid pET23-HisMf58 has multiple cloning sites just after the HT-mf-thr gene.
Human α1(XVIII) cDNA clone pNF18-218 was used as a template to amplify the sequences encoding COL1 and NC1 regions by PCR. Constructs (43)COL1-NC1(69), (66)COL1-NC1(69), (66)COL1-NC1(136) (Table 2) were PCR amplified using a corresponding pair of oligonucleotides: either 5′-TGCAGATCTCCCGGCCCTCCGGGCCCCCCTGGGCCC-3′ ((43)COL1, the BglII site is underlined) or 5′-TGCAGATCTCCGGGCCAGCCCGGCCCACCTGGACCT-3′ ((66)COL1, the BglII site is underlined) as a forward primer and either 5′-GTCAGTCGACTTACTGCACCACGGGGGGCTGCAAGGC-3′ (NC1(69), the SalI site is underlined) or 5′-GTCAGTCGACTTACTGGAAGTCGCGGTGGCTGTGGGC-3′ (NC1(136), the SalI site is underlined) as a reversed primer. The construct NC1(54) (Table 2) was PCR amplified using two oligonucleotides: 5′-TGCAGATCTTCAGGGGTGAGGCTCTGGGCT-3′ (forward, the BglII site is underlined) and 5′-GTCAGTCGACTTATCGTGGGAGTGGTGTCCGGG (reversed, the SalI site is underlined).
The PCR fragments were cut with BglII and SalI restriction enzymes and cloned into the pET23-HisMf vector using restriction sites BamHI and SalI. The DNA inserts were verified by Sanger dideoxy DNA sequencing. The recombinant constructs (43)COL1-NC1(69), (66)COL1-NC1(69), (66)COL1-NC1(136) and NC1(54) (Table 2), each as part of a fusion molecule with a His-tagged mini-fibritin were expressed at 30°C in the E. coli BL21(DE3) host strain (Novagen) after IPTG induction (final concentration 1mM) for 6 hours. Purification of the 6xHis-taged fusion proteins by immobilized metal affinity chromatography on HisTrap™ HP column (Amersham Biosciences) and separation of the fragments (43)COL1-NC1(69), (66)COL1-NC1(69), (66)COL1-NC1(136) and NC1(54) after thrombin/trypsin cleavage were carried out as described in the manufacturer’s instructions. Thrombin cleavage for all but NC1(54) was performed at 4°C for 24 hours with thrombin protease (ICN) in 50mM Tris buffer, pH 8.3, supplemented with 150mM NaCl. NC1(54) was cleaved from the fusion molecule by trypsin (at a final trypsin concentration of 0.05 mg/ml) under the same conditions. The resulting fragments still had the residues GS at the amino termini as a part of the thrombin cleavage site (LVPRGS). An additional residue P was added during the cloning after GS in constructs (66)COL1-NC1(69), (66)COL1-NC1(136) to maintain a continuous triple helix (Table 2). Correct molecular masses were confirmed by mass-spectroscopy.
Analytical ultracentrifugation
Sedimentation equilibrium measurements were performed on a Beckman model XLA analytical ultracentrifuge. Absorbance was measured at 280 nm. Runs were carried out at 20°C in an An60-Ti rotor using 12 mm cells and Epon, 2 channels, centerpieces. Data analysis was done using Ultrascan II (Demeler, B. UltraScan version 9.3. A Comprehensive Data Analysis Software Package for Analytical Ultracentrifugation Experiments. The University of Texas Health Science Center at San Antonio, Department of Biochemistry. http:/www.ultrascan.uthscsa.edu).
Crystallization and data collection
The NC1(54) peptide of human collagen XVIII was crystallized in two forms, cubic and tetragonal. The best crystals of the cubic form were obtained by the hanging drop vapor diffusion method using a reservoir solution of 0.2–0.3M MgCl2, 0.1M BisTris (pH 6.0–7.0), 18–22% (w/v) polyethylene glycol 8000 which was mixed for the drop solution in 1:1 proportion with protein solution at 15 mg/mL. The crystals grew to a final size of 0.4 mm×0.4 mm×0.4 mm after about two-seven days at 20°C. The crystals were briefly dipped into a cryo-protectant solution containing the reservoir solution and 20%(v/v) ethylene glycol, and then frozen in a liquid nitrogen.
The best crystals of the tetragonal form were obtained by the same method using a reservoir solution of 1.5–1.8M (NH4)2SO4, 0.1M citric acid (pH3.0–4.0). The crystals grew to a final size of 1.5 mm×0.7 mm×0.3 mm after about two-seven days at 20 °C. The crystals were briefly dipped into a cryo-protectant solution containing the reservoir solution and 20%(v/v) glycerol, and then frozen in a liquid nitrogen.
Data collection was performed on crystals cryocooled to 100K on the “NOIR-1” detector system at the Molecular Biology Consortium Beamline 4.2.2 of the Advanced Light Source, Lawrence Berkeley National Laboratory.
Crystal structure determination
SeMet crystals were used for a three-wavelength, MAD data collection (Table 3) procedure.59 The programs MOSFLM and SCALA from CCP4 package60 were used to index, integrate and scale the diffraction data sets. The program PHENIX61 was used for automated structure solution: determination of Se atom positions, phasing, density modifications and automatic model building. Atomic models were checked and if needed manually corrected with the help of the program COOT.62 The programs CNS63 and PHENIX61 were used in refinement of cubic and tetragonal crystal forms, respectively (Table 3). In the case of the tetragonal crystal form, two TLS groups (chains A,B,C in one and chains D,E,F in another) were used for combined TLS and individual isotropic B-factor refinement at final stages.
TABLE 3.
Crystallographic data statistics Crystal type
| Crystal type | Native cubic | SeMet cubic | Native tetragonal | SeMet tetragonal |
|---|---|---|---|---|
| Data collectiona | ||||
| Wavelength (Å) | 1.106 | 0.9795, 0.9798, 0.995 | 1.106 | 0.9795, 0.9798, 0.9640 |
| Resolution range (Å) | 38.7–3.0(3.2–3.0) | 48.0–2.8(2.95–2.8) | 40.5–1.8(1.9–1.8) | 47.4–2.0(2.1–2.0) |
| Spacegroup | I4132 | I4132 | P4212 | P4212 |
| Cell dimensions | a=b=c=94.9 | a=b=c=95.9 | a=b=71.7, c=134.7 | a=b=71.5, c=135.3 |
| Rmerge (%) | 7.4 (65.5) | 6.1(48.8), 6.2(55.0), 5.6(45.6) | 6.3 (44.0) | 8.4(49.6), 8.4(51.0), 7.8(45.5) |
| I/sigmaI | 8.8 (1.4) | 8.0(1.5), 8.2(1.4), 8.6(1.7) | 8.4 (1.6) | 6.9(1.4), 6.9(1.3), 7.2(1.5) |
| Completeness (%) | 100 (100) | 99.9 (99.9) | 100 (100) | 99.7(98.8), 99.8(99.0), 99.6(98.4) |
| Multiplicity | 12.6 (13.2) | 38.7(40.4), 19.2(20.1), 19.3(20.1) | 11.9 (9.7) | 10.2(7.5), 10.3(7.6), 10.2(8.2) |
| Wilson plot B factor (Å2) | 100.6 | 105.9, 107.9, 106.5 | 22.6 | 23.0, 23.3, 23.7 |
| Matthews coefficient (Å3Da−1) | 2.8 | 2.8 | 2.3 | 2.3 |
| Contents of asymmetric unit | Single chain | Single chain | Two trimers | Two trimers |
| Solvent content (%) | 51 | 52 | 38 | 38 |
| Phasing | ||||
| PHENIX BAYES-CC/FOM | 67.73/0.69 | 71.15/0.71 | ||
| Refinementa | ||||
| Number of reflections used in refinement/for Rfree | 1625/80 | 33423/3336 | ||
| R-factor/Rfree (%) | 27.9/28.8 (43.7/43.4) |
17.5/22.4 (20.0/26.4) |
||
| Number of protein/solvent atoms | 444/0 | 2789/395 | ||
| RMSD of bonds(Å)/angles from idealized values | 0.008/1.356 | 0.009/1.113 | ||
Data in parentheses show the results in the highest resolution range
Circular dichroism analysis
CD spectra were recorded on an AVIV model 202 spectropolarimeter (AVIV Instruments, Inc.) with thermostated quartz cells of 0.1–1 mm path length. The spectra were normalized for concentration and path length to obtain the mean molar residue ellipticity after subtraction of the buffer contribution. Peptide concentrations were determined by amino acid analysis.
Gdm-induced transitions
Two forms of the peptide were used to measure GdmCl transitions: the native form in 50mM sodium phosphate buffer (pH 8.0) and the unfolded form in 7M GdmCl, 50mM sodium phosphate buffer (pH 8.0), both at a chain concentration of 300μM. Complete loss of the secondary structure of the unfolded form was verified by the CD spectrum. The native and unfolded peptides were diluted with varying denaturant concentrations in 50 mM sodium phosphate buffer (pH 8.0) to a final peptide concentration of 30μM. Final GdmCl concentrations were determined using refractive indexes. CD measurements were performed at 233 nm with 1 mm path-length. The coincidence of unfolding and refolding curves was achieved by the equilibration time of ~20 hours at 20°C.
The fraction of unfolded monomer fu was calculated as:
where Θn and Θu are the extrapolated native and unfolded ellipticities to the transition region and Θ the observed ellipticity at the given denaturant concentration.
In addition to CD monitoring of the transitions, equilibrated samples at different GdmCl concentrations were subjected to chemical cross-linking with 0.3% glutaraldehyde for 15 minutes and quenched with 0.2M glycine. Samples were then desalted on desalting spin-columns (Thermo Fisher Scientific, Inc) and ran on 4–12% SDS-PAGE to detect oligomers.
Calculation of ΔASA
The difference in accessible surface area (ΔASA) was calculated by using the crystal structure of the trimer (chains A, B and C) and the program MOLMOL.64 For the unfolded state an extended conformation was assumed.
Homology modeling
The crystal structure of the NC1(54) peptide of human collagen XVIII was used as an initial model for human type XV collagen trimerization domain. Residues GS from NC1(54) were omitted and the others were changed according to the sequence of human collagen XV (see Fig. 8) with the help of the program COOT.62 The model was subject to several rounds of an energy minimization in vacuo and in a water shell using a program Insight II (Accelrys Software Inc.) with the CVFF force field.
Figure preparation
The following programs were used in preparation of figures: Fig. 1, INKSCAPE: Open source scalable vector graphics editor (http://www.inkscape.org); Fig. 3, PYMOL: DeLano, W.L. The PyMol Molecular Graphics System (2002) (http://www.pymol.org); Figs. 4,5,7, DINO: Visualizing Structural Biology (2002) (http://www.dino3d.org).
Data deposition
The refined atomic models and the observed structure factors were deposited into the RCSB Protein Data Bank (PDB) with the accession numbers 3HSH and 3HON for the tetragonal and cubic crystal forms, respectively.
Acknowledgments
This work was supported by a grant from Shriners Hospital for Children.
The authors would like to thank Dr. Kerry Maddox, Jessica Hacker and Jesse Vance for mass-spectrometry, amino-terminal peptide sequencing and DNA sequencing.
Part of this research was performed at the Advanced Light Source, which is supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences Division, US Department of Energy, under contract no. DE-AC03-76SF00098, at Lawrence Berkeley National Laboratory.
Abbreviations used
- COL
collagenous
- NC
non-collagenous
- NC1
carboxyl-terminal non-collagenous domain
References
- 1.Khoshnoodi J, Cartailler J, Alvares K, Veis A, Hudson BG. Molecular recognition in the assembly of collagens: terminal noncollagenous domains are key recognition modules in the formation of triple helical protomers. J Biol Chem. 2006;281:38117–38121. doi: 10.1074/jbc.R600025200. [DOI] [PubMed] [Google Scholar]
- 2.Fessler LI, Fessler JH. Protein assembly of procollagen and effects of hydroxylation. J Biol Chem. 1974;249:7637–7646. [PubMed] [Google Scholar]
- 3.Bächinger HP, Fessler LI, Timpl R, Fessler JH. Chain assembly intermediate in the biosynthesis of type III procollagen in chick embryo blood vessels. J Biol Chem. 1981;256:13193–13199. [PubMed] [Google Scholar]
- 4.Doege KJ, Fessler JH. Folding of carboxyl domain and assembly of procollagen I. J Biol Chem. 1986;261:8924–8935. [PubMed] [Google Scholar]
- 5.Dion AS, Myers JC. COOH-terminal propeptides of the major human procollagens. Structural, functional and genetic comparisons. J Mol Biol. 1987;193:127–143. doi: 10.1016/0022-2836(87)90632-2. [DOI] [PubMed] [Google Scholar]
- 6.Lees JF, Bulleid NJ. The role of cysteine residues in the folding and association of the COOH-terminal propeptide of types I and III procollagen. J Biol Chem. 1994;269:24354–24360. [PubMed] [Google Scholar]
- 7.Snellman A, Tu H, Väisänen T, Kvist AP, Huhtala P, Pihlajaniemi T. A short sequence in the N-terminal region is required for the trimerization of type XIII collagen and is conserved in other collagenous transmembrane proteins. EMBO J. 2000;19:5051–5059. doi: 10.1093/emboj/19.19.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Areida SK, Reinhardt DP, Muller PK, Fietzek PP, Kowitz J, Marinkovich MP, Notbohm H. Properties of the collagen type XVII ectodomain. Evidence for n- to c-terminal triple helix folding. J Biol Chem. 2001;276:1594–1601. doi: 10.1074/jbc.M008709200. [DOI] [PubMed] [Google Scholar]
- 9.McAlinden A, Smith TA, Sandell LJ, Ficheux D, Parry DAD, Hulmes DJS. Alpha-helical coiled-coil oligomerization domains are almost ubiquitous in the collagen superfamily. J Biol Chem. 2003;278:42200–42207. doi: 10.1074/jbc.M302429200. [DOI] [PubMed] [Google Scholar]
- 10.Boudko SP, Engel J, Bächinger HP. Trimerization and triple helix stabilization of the collagen XIX NC2 domain. J Biol Chem. 2008;283:34345–34351. doi: 10.1074/jbc.M806352200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sundaramoorthy M, Meiyappan M, Todd P, Hudson BG. Crystal structure of NC1 domains. Structural basis for type IV collagen assembly in basement membranes. J Biol Chem. 2002;277:31142–31153. doi: 10.1074/jbc.M201740200. [DOI] [PubMed] [Google Scholar]
- 12.Than ME, Henrich S, Huber R, Ries A, Mann K, Kühn K, Timpl R, Bourenkov GP, Bartunik HD, Bode W. The 1.9-A crystal structure of the noncollagenous (NC1) domain of human placenta collagen IV shows stabilization via a novel type of covalent Met-Lys cross-link. Proc Natl Acad Sci USA. 2002;99:6607–6612. doi: 10.1073/pnas.062183499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bogin O, Kvansakul M, Rom E, Singer J, Yayon A, Hohenester E. Insight into Schmid metaphyseal chondrodysplasia from the crystal structure of the collagen X NC1 domain trimer. Structure. 2002;10:165–173. doi: 10.1016/s0969-2126(02)00697-4. [DOI] [PubMed] [Google Scholar]
- 14.Kvansakul M, Bogin O, Hohenester E, Yayon A. Crystal structure of the collagen alpha1(VIII) NC1 trimer. Matrix Biol. 2003;22:145–152. doi: 10.1016/s0945-053x(02)00119-1. [DOI] [PubMed] [Google Scholar]
- 15.Halfter W, Dong S, Schurer B, Cole GJ. Collagen XVIII is a basement membrane heparan sulfate proteoglycan. J Biol Chem. 1998;273:25404–25412. doi: 10.1074/jbc.273.39.25404. [DOI] [PubMed] [Google Scholar]
- 16.Li D, Clark CC, Myers JC. Basement membrane zone type XV collagen is a disulfide-bonded chondroitin sulfate proteoglycan in human tissues and cultured cells. J Biol Chem. 2000;275:22339–22347. doi: 10.1074/jbc.M000519200. [DOI] [PubMed] [Google Scholar]
- 17.Abe N, Muragaki Y, Yoshioka H, Inoue H, Ninomiya Y. Identification of a novel collagen chain represented by extensive interruptions in the triple-helical region. Biochem Biophys Res Commun. 1993;196:576–582. doi: 10.1006/bbrc.1993.2288. [DOI] [PubMed] [Google Scholar]
- 18.Oh SP, Kamagata Y, Muragaki Y, Timmons S, Ooshima A, Olsen BR. Isolation and sequencing of cDNAs for proteins with multiple domains of Gly-Xaa-Yaa repeats identify a distinct family of collagenous proteins. Proc Natl Acad Sci USA. 1994;91:4229–4233. doi: 10.1073/pnas.91.10.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rehn M, Pihlajaniemi T. Alpha 1(XVIII), a collagen chain with frequent interruptions in the collagenous sequence, a distinct tissue distribution, and homology with type XV collagen. Proc Natl Acad Sci USA. 1994;91:4234–4238. doi: 10.1073/pnas.91.10.4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rehn M, Hintikka E, Pihlajaniemi T. Primary structure of the alpha 1 chain of mouse type XVIII collagen, partial structure of the corresponding gene, and comparison of the alpha 1(XVIII) chain with its homologue, the alpha 1(XV) collagen chain. J Biol Chem. 1994;269:13929–13935. [PubMed] [Google Scholar]
- 21.Saarela J, Ylikärppä R, Rehn M, Purmonen S, Pihlajaniemi T. Complete primary structure of two variant forms of human type XVIII collagen and tissue-specific differences in the expression of the corresponding transcripts. Matrix Biol. 1998;16:319–328. doi: 10.1016/s0945-053x(98)90003-8. [DOI] [PubMed] [Google Scholar]
- 22.O’Reilly MS, Boehm T, Shing Y, Fukai N, Vasios G, Lane WS, Flynn E, Birkhead JR, Olsen BR, Folkman J. Endostatin: an endogenous inhibitor of angiogenesis and tumor growth. Cell. 1997;88:277–285. doi: 10.1016/s0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- 23.Ständker L, Schrader M, Kanse SM, Jürgens M, Forssmann WG, Preissner KT. Isolation and characterization of the circulating form of human endostatin. FEBS Lett. 1997;420:129–133. doi: 10.1016/s0014-5793(97)01503-2. [DOI] [PubMed] [Google Scholar]
- 24.Sasaki T, Fukai N, Mann K, Göhring W, Olsen BR, Timpl R. Structure, function and tissue forms of the C-terminal globular domain of collagen XVIII containing the angiogenesis inhibitor endostatin. EMBO J. 1998;17:4249–4256. doi: 10.1093/emboj/17.15.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasaki T, Larsson H, Tisi D, Claesson-Welsh L, Hohenester E, Timpl R. Endostatins derived from collagens XV and XVIII differ in structural and binding properties, tissue distribution and anti-angiogenic activity. J Mol Biol. 2000;301:1179–1190. doi: 10.1006/jmbi.2000.3996. [DOI] [PubMed] [Google Scholar]
- 26.Kuo CJ, LaMontagne KRJ, Garcia-Cardeña G, Ackley BD, Kalman D, Park S, Christofferson R, Kamihara J, Ding YH, Lo KM, Gillies S, Folkman J, Mulligan RC, Javaherian K. Oligomerization-dependent regulation of motility and morphogenesis by the collagen XVIII NC1/endostatin domain. J Cell Biol. 2001;152:1233–1246. doi: 10.1083/jcb.152.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sánchez-Arévalo Lobo VJ, Cuesta AM, Sanz L, Compte M, García P, Prieto J, Blanco FJ, Alvarez-Vallina L. Enhanced antiangiogenic therapy with antibody-collagen XVIII NC1 domain fusion proteins engineered to exploit matrix remodeling events. Int J Cancer. 2006;119:455–462. doi: 10.1002/ijc.21851. [DOI] [PubMed] [Google Scholar]
- 28.Dhanabal M, Ramchandran R, Waterman MJ, Lu H, Knebelmann B, Segal M, Sukhatme VP. Endostatin induces endothelial cell apoptosis. J Biol Chem. 1999;274:11721–11726. doi: 10.1074/jbc.274.17.11721. [DOI] [PubMed] [Google Scholar]
- 29.Dhanabal M, Ramchandran R, Volk R, Stillman IE, Lombardo M, Iruela-Arispe ML, Simons M, Sukhatme VP. Endostatin: yeast production, mutants, and antitumor effect in renal cell carcinoma. Cancer Res. 1999;59:189–197. [PubMed] [Google Scholar]
- 30.Yamaguchi N, Anand-Apte B, Lee M, Sasaki T, Fukai N, Shapiro R, Que I, Lowik C, Timpl R, Olsen BR. Endostatin inhibits VEGF-induced endothelial cell migration and tumor growth independently of zinc binding. EMBO J. 1999;18:4414–4423. doi: 10.1093/emboj/18.16.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dixelius J, Larsson H, Sasaki T, Holmqvist K, Lu L, Engström A, Timpl R, Welsh M, Claesson-Welsh L. Endostatin-induced tyrosine kinase signaling through the Shb adaptor protein regulates endothelial cell apoptosis. Blood. 2000;95:3403–3411. [PubMed] [Google Scholar]
- 32.Cao Y. Molecular mechanisms and therapeutic development of angiogenesis inhibitors. Adv Cancer Res. 2008;100:113–131. doi: 10.1016/S0065-230X(08)00004-3. [DOI] [PubMed] [Google Scholar]
- 33.Folkman J. Antiangiogenesis in cancer therapy--endostatin and its mechanisms of action. Exp Cell Res. 2006;312:594–607. doi: 10.1016/j.yexcr.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 34.Boudko SP, Strelkov SV, Engel J, Stetefeld J. Design and crystal structure of bacteriophage T4 mini-fibritin NCCF. J Mol Biol. 2004;339:927–935. doi: 10.1016/j.jmb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 35.Boudko SP, Engel J. Structure formation in the C terminus of type III collagen guides disulfide cross-linking. J Mol Biol. 2004;335:1289–1297. doi: 10.1016/j.jmb.2003.11.054. [DOI] [PubMed] [Google Scholar]
- 36.Frank S, Kammerer RA, Mechling D, Schulthess T, Landwehr R, Bann J, Guo Y, Lustig A, Bächinger HP, Engel J. Stabilization of short collagen-like triple helices by protein engineering. J Mol Biol. 2001;308:1081–1089. doi: 10.1006/jmbi.2001.4644. [DOI] [PubMed] [Google Scholar]
- 37.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Meth Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 38.Greene RFJ, Pace CN. Urea and guanidine hydrochloride denaturation of ribonuclease, lysozyme, alpha-chymotrypsin, and beta-lactoglobulin. J Biol Chem. 1974;249:5388–5393. [PubMed] [Google Scholar]
- 39.Güthe S, Kapinos L, Möglich A, Meier S, Grzesiek S, Kiefhaber T. Very fast folding and association of a trimerization domain from bacteriophage T4 fibritin. J Mol Biol. 2004;337:905–915. doi: 10.1016/j.jmb.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 40.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holm L, Kääriäinen S, Rosenström P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walter M, Fiedler C, Grassl R, Biebl M, Rachel R, Hermo-Parrado XL, Llamas-Saiz AL, Seckler R, Miller S, van Raaij MJ. Structure of the receptor-binding protein of bacteriophage det7: a podoviral tail spike in a myovirus. J Virol. 2008;82:2265–2273. doi: 10.1128/JVI.01641-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steinbacher S, Seckler R, Miller S, Steipe B, Huber R, Reinemer P. Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer. Science. 1994;265:383–386. doi: 10.1126/science.8023158. [DOI] [PubMed] [Google Scholar]
- 44.Hynes WL, Hancock L, Ferretti JJ. Analysis of a second bacteriophage hyaluronidase gene from Streptococcus pyogenes: evidence for a third hyaluronidase involved in extracellular enzymatic activity. Infect Immun. 1995;63:3015–3020. doi: 10.1128/iai.63.8.3015-3020.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gage MJ, Robinson AS. C-terminal hydrophobic interactions play a critical role in oligomeric assembly of the P22 tailspike trimer. Protein Sci. 2003;12:2732–2747. doi: 10.1110/ps.03150303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webber T, Gurung S, Saul J, Baker T, Spatara M, Freyer M, Robinson AS, Gage MJ. The C-terminus of the P22 tailspike protein acts as an independent oligomerization domain for monomeric proteins. Biochem J. 2009 doi: 10.1042/BJ20081449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brodsky B, Thiagarajan G, Madhan B, Kar K. Triple-helical peptides: an approach to collagen conformation, stability, and self-association. Biopolymers. 2008;89:345–353. doi: 10.1002/bip.20958. [DOI] [PubMed] [Google Scholar]
- 48.Boulègue C, Musiol H, Götz MG, Renner C, Moroder L. Natural and artificial cystine knots for assembly of homo- and heterotrimeric collagen models. Antioxid Redox Signal. 2008;10:113–125. doi: 10.1089/ars.2007.1868. [DOI] [PubMed] [Google Scholar]
- 49.Myers JC, Amenta PS, Dion AS, Sciancalepore JP, Nagaswami C, Weisel JW, Yurchenco PD. The molecular structure of human tissue type XV presents a unique conformation among the collagens. Biochem J. 2007;404:535–544. doi: 10.1042/BJ20070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stetefeld J, Frank S, Jenny M, Schulthess T, Kammerer RA, Boudko S, Landwehr R, Okuyama K, Engel J. Collagen stabilization at atomic level: crystal structure of designed (GlyProPro)10foldon. Structure. 2003;11:339–346. doi: 10.1016/s0969-2126(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 51.Parry DAD, Fraser RDB, Squire JM. Fifty years of coiled-coils and alpha-helical bundles: a close relationship between sequence and structure. J Struct Biol. 2008;163:258–269. doi: 10.1016/j.jsb.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 52.Sheriff S, Chang CY, Ezekowitz RA. Human mannose-binding protein carbohydrate recognition domain trimerizes through a triple alpha-helical coiled-coil. Nat Struct Biol. 1994;1:789–794. doi: 10.1038/nsb1194-789. [DOI] [PubMed] [Google Scholar]
- 53.Weis WI, Drickamer K. Trimeric structure of a C-type mannose-binding protein. Structure. 1994;2:1227–1240. doi: 10.1016/S0969-2126(94)00124-3. [DOI] [PubMed] [Google Scholar]
- 54.Håkansson K, Lim NK, Hoppe HJ, Reid KB. Crystal structure of the trimeric alpha-helical coiled-coil and the three lectin domains of human lung surfactant protein D. Structure. 1999;7:255–264. doi: 10.1016/s0969-2126(99)80036-7. [DOI] [PubMed] [Google Scholar]
- 55.Hoppe HJ, Barlow PN, Reid KB. A parallel three stranded alpha-helical bundle at the nucleation site of collagen triple-helix formation. FEBS Lett. 1994;344:191–195. doi: 10.1016/0014-5793(94)00383-1. [DOI] [PubMed] [Google Scholar]
- 56.Pakkanen O, Hämäläinen E, Kivirikko KI, Myllyharju J. Assembly of stable human type I and III collagen molecules from hydroxylated recombinant chains in the yeast Pichia pastoris. Effect of an engineered C-terminal oligomerization domain foldon. J Biol Chem. 2003;278:32478–32483. doi: 10.1074/jbc.M304405200. [DOI] [PubMed] [Google Scholar]
- 57.Bachmann A, Kiefhaber T, Boudko S, Engel J, Bächinger HP. Collagen triple-helix formation in all-trans chains proceeds by a nucleation/growth mechanism with a purely entropic barrier. Proc Natl Acad Sci USA. 2005;102:13897–13902. doi: 10.1073/pnas.0505141102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boudko SP, Kuhn RJ, Rossmann MG. The coiled-coil domain structure of the Sin Nombre virus nucleocapsid protein. J Mol Biol. 2007;366:1538–1544. doi: 10.1016/j.jmb.2006.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hendrickson WA, Ogata CM. Phase determination from multiwavelength anomalous diffraction measurements. Methods Enzymol. 1997;276:494–523. doi: 10.1016/S0076-6879(97)76074-9. [DOI] [PubMed] [Google Scholar]
- 60.Collaborative Computational Project N4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 61.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 62.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 63.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 64.Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–5. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]