Abstract
Correct folding of the collagen triple helix requires a self-association step which selects and binds α-chains into trimers. Here we report the crystal structure of the trimerization domain of human type XV collagen. The trimerization domain of type XV collagen contains three monomers each composed of four β-sheets and an α-helix. The hydrophobic core of the trimer is devoid of solvent molecules and is shaped by β-sheet planes from each monomer. The trimerization domain is extremely stable and forms at picomolar concentrations. It is found that the trimerization domain of type XV collagen is structurally similar to that of type XVIII, despite only 32 % sequence identity. High structural conservation indicates that the multiplexin trimerization domain represents a three dimensional fold that allows for sequence variability while retaining structural integrity necessary for tight and efficient trimerization.
Keywords: collagen XV, collagen XVIII, trimerization domain, non-collagenous domain, multiplexin
1. Introduction
Collagens are a diverse family of proteins that constitute the major structural component of the extracellular matrix. Although collagen proteins are identified by the presence of one or more triple-helical domains, each α-chain also contains two or more non-collagenous domains. To date, 28 different types of collagen have been identified (Gordon and Hahn, 2010; Myllyharju and Kivirikko, 2004). Further classification according to supramolecular structure assigns collagens to fibril, fibril-associated containing interrupted triple helicies (FACIT), beaded filament, anchoring fibril, network-forming, transmembrane or multiple triple helicies with interruptions (Multiplexin) families (Shoulders and Raines, 2009). Each type of collagen is composed of one, two or three different α chains that can assemble into hetero- or homo-trimers. Assembly of all collagens begins with a self-association step that selects, aligns and trimerizes α-chains.
The mechanism of trimerization is attributed to non-collagenous (NC) domains (Khoshnoodi et al., 2006). Studies on classic fibril-forming collagens I and III found that the carboxy-terminal NC (NC1) domains were essential to trimerization (Doege and Fessler, 1986; McLaughlin and Bulleid, 1998). C-terminal NC domains govern chain selection for all collagen families except the transmembrane collagens, which are believed to trimerize via amino-terminal NC domains (Snellman et al., 2000). Fibril associated-collagens IX and XIX have recently been shown to trimerize via their NC2 domains (the second NC domain from the carboxy-terminal end) (Boudko et al., 2008; Boudko et al., 2010). The NC2 domains of the other FACIT family members are also most likely responsible for self association. For all remaining collagens, the NC1 domain is believed to be responsible for chain selection and trimerization.
Crystal structures for the NC1 domains of the network-forming collagens IV, VIII and X have been solved (Bogin et al., 2002; Kvansakul et al., 2003; Sundaramoorthy et al., 2002; Than et al., 2002). Along with the NC1 domain of the multiplexin type XVIII collagen (Boudko et al., 2009), these represent the only known atomic structures for collagen trimerization domains. Although both the multiplexin and network-forming collagen trimerization domains are composed primarily of β-sheets, they share no overall structural homology.
Collagens XV and XVIII are the only known members of the multiplexin family (Pihlajaniemi and Rehn, 1995). Both collagens are homo-trimers composed of a single α-chain that contains a central highly interrupted collagenous domain flanked by N- and C-terminal non collagenous domains (Myers et al., 1992; Rehn et al., 1994; Rehn and Pihlajaniemi, 1994). The collagenous domain of human type XV is divided into nine collagenous domains with eight non-collagenous repeats. Human type XVIII exists as three variants with differing N-terminal NC domains, but have identical interrupted collagenous and NC1 domains. There are ten collagenous sequences alternating with nine non-collagenous repeats in the type XVIII interrupted collagenous domain (Saarela et al., 1998). Type XVIII is a heparan sulfate proteoglycan (Halfter et al., 1998) and type XV can carry chondroitin/dermatan sulfate alone or chondroitin/dermatan sulfate and heparan sulfate chains (Amenta et al., 2005). Type XVIII collagen localizes in epidermal and vascular basement membrane, similar to other heparan sulfate proteoglycans found in basement membranes (Marneros and Olsen, 2005). Type XV is not an integral basement membrane component; rather, it localizes to areas peripheral to basement membranes and associates directly with collagen fibers/fibrils in a manner consistent with other chondroitin sulfate proteoglycans (Amenta et al., 2005; Iozzo, 1999).
Collagen XV null mice develop collapsed capillaries and degenerating endothelial cells in the heart and skeletal muscles and exhibit increased sensitivity to exercise-induced muscle damage (Eklund et al., 2001). However, there are currently no known diseases states that are associated with type XV collagen. Although COL15 was shown to be upregulated in patients with systemic sclerosis, polymorphisms in the COL15 gene were shown to not be associated with the disease (Pushpakom et al., 2008). In contrast, the human autosomal recessive Knobloch syndrome has been mapped to the COL18A1 gene, which leads to an early stop codon. Knobloch syndrome manifests with severe myopia, retinal degeneration and retinal detachment leading to blindness. Col18a1−/− mice display impaired blood vessel development in the eye (Fukai et al., 2002), as well as hydrocephalus and structurally altered basement membranes (Utriainen et al., 2004). Double knockout mice for Col18a1 and Col15a1did not demonstrate any functional compensation between collagens XV and XVIII.
The highest level of sequence homology between type XV and XVIII lies in their NC1 domain. The NC1 domains of collagens XV and XVIII are organized into a trimerization domain, a hinge region and an endostatin domain (Sasaki et al., 1998; Sasaki et al., 2000). The trimerization domain of type XVIII collagen has been isolated, characterized and crystallized (Boudko et al., 2009). In this study we present the crystal structure of the trimerization domain of type XV collagen along with biochemical analysis of the protein stability and oliogmeric state. Despite having only 32 % sequence identity, the type XV structure is remarkably similar to and displays comparable biochemical properties to the type XVIII trimerization domain. The multiplexin trimerization domain (TD) is therefore a structurally conserved domain that trimerizes at very low concentrations and may thus be a useful tool in the engineering of protein trimers.
2. Results
2.1 Crystallization and Structure Determination
The trimerization domain of human type XV collagen, designated XV(TD), was designed to cover the homologous sequence to the NC1(54) peptide used in the crystal structure of the collagen XVIII trimerization domain (Boudko et al., 2009). The XV(TD) construct covers amino acids 587 through 643 of the full length protein (Myers et al., 1992) and is numbered such that the first visible residue is labeled as residue 1 (Fig. 1).
Fig 1.
Sequence alignments. Alignment of type XV and XVIII trimerization domain sequences used to solve their respective crystal structures. Sequences are derived from GenBank™ accession numbers P39059 and P39060 for human type XV and XVIII collagens, respectively. Residues common to formation of the trimeric hydrophobic core are indicated with an asterisk (*) and the residue involved in trimeric hydrophobic core of type XVIII collagen but not in type XV is indicated with a backslash (\). Residues common to the formation of the monomeric hydrophobic core are marked with dots (●). Italics indicate residues not visible in the crystal structure of type XV. Underlining represents non-natural sequence introduced during cloning in type XVIII. The first visible residue in the type XV structure (N) is labeled as residue 1.
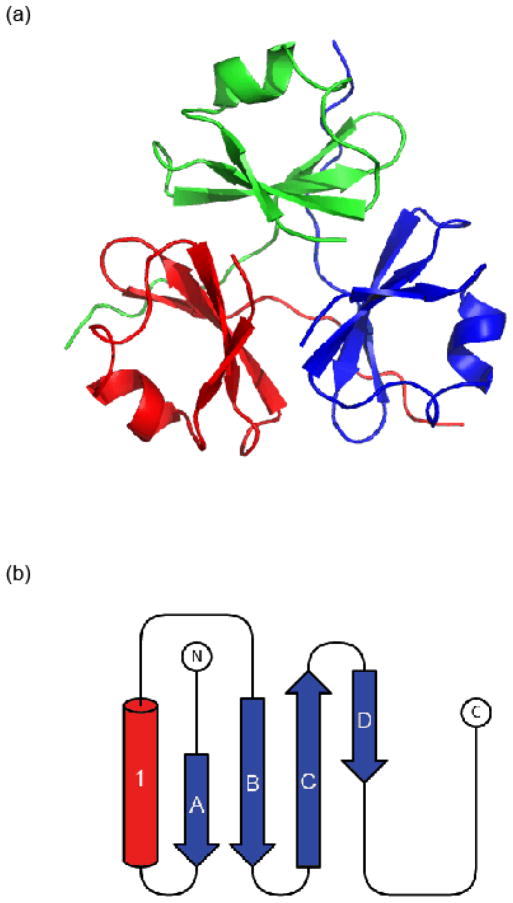
The XV(TD) protein crystallized in a hexagonal crystal form (Table 1). The trimerization domain of type XVIII (PDB 3HSH) was used for molecular replacement (see Experimental Procedures for details). Two single chains were found per asymmetric unit; two trimers were related by the 3-fold crystal symmetry. The crystal packing was very tight (Table 1) with a solvent content of only 30 %. Each monomer is comprised of one α-helix and four β-strands in a mixed parallel and antiparallel β-sheet (Fig. 2). The α-helix interacts with all four strands of the β-sheet to shape a hydrophobic core within each monomer. The core is comprised of residues F6, M9, M12, L13, A16, V19, I25, L27, F34 and is stabilized by the hydrophobic parts of residues T4, T23, R36 and W41 (Fig. 3).
Table 1.
Crystallographic Statistics
|
Data Collectiona | |
| Wavelength (Å) | 1.54 |
| Resolution range (Å) | 52.5-2.0 (2.11-2.01) |
| Space group | P63 |
| Cell dimensions |
a = b = 49.12 c = 65.6 |
| Rmerge (%) | 3.4 (11.0) |
| I/σI | 35.4 (15.6) |
| Completeness (%) | 100 (100) |
| Multiplicity | 6.6 (6.5) |
| Wilson plot B (Å2) | 20.8 |
| Matthews Coefficient (Å3 Da−1) | 1.74 |
| Contents of asymmetric unit | Two monomers |
| Solvent content (%) | 29.5 |
| Refinement | |
| No. of reflections used in refinement | 6079 |
| No. of reflections used for Rfree | 343 |
| R-factor/Rfree (%) | 21.6/26.5 |
| RMSD bonds (Å)/angles(°) | 0.008/1.4 |
| No. of protein/solvent atoms | 874/40 |
| Ramachandran plot (%)b | 97.1/1.9/1.0 |
Data in parentheses show the results in the highest resolution range.
Percentage of residues in favored/allowed/outlier regions of the Ramachandran plot.
Fig 2.
Structure of the type XV trimerization domain, (a) Cartoon representation of the trimeric biological unit. (b) Topology diagram for a single chain: α-helix shown as a red cylinder labeled 1 and beta sheets (labeled A through D) are shown as blue arrows.
Fig 3.
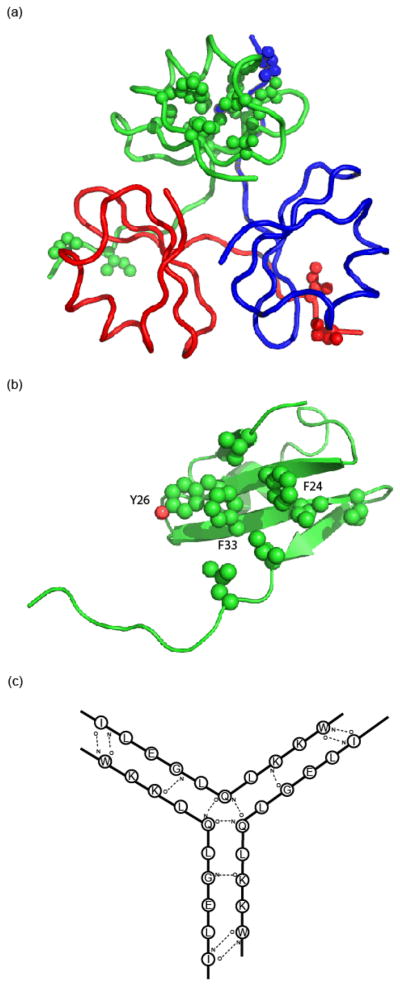
Hydrophobic cores and intersubunit hydrogen bonding. (a) Residues forming the hydrophobic core of a monomeric subunit are shown in green. Additionally, residues I50 and I52 which form hydrophobic contacts between subunits are shown for all three monomers. (b) Residues forming the hydrophobic interior of the trimer are shown for a monomeric subunit. (c) Main chain to main chain intersubunit hydrogen bonding diagram.
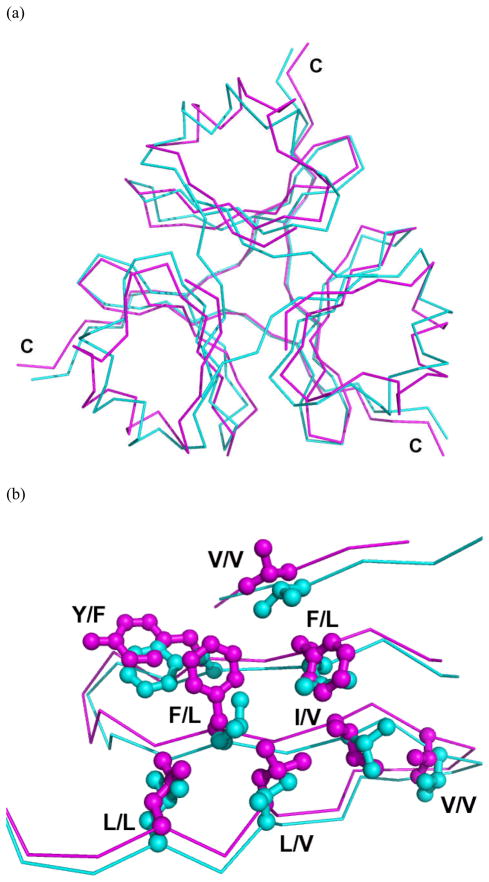
The trimer measures approximately 40 Å in length and 20 Å in height. The trimer is stabilized by a central hydrophobic core, interchain hydrophobic interactions between adjacent monomers, and an interchain hydrogen bond network (Fig. 3). The trimer core is comprised of residues V3, F24, F33, I35, V37, L44, L46 and the hydrophobic face of Y26 (Figs. 2 and 3). No solvent molecules are observed in the trimer interior. Residues I50 and I52 interact with the monomeric hydrophobic core of the adjacent subunit. Interchain hydrogen bonds also stabilize trimerization, forming a net of bonds that cover the carboxyl-terminal end of the trimer. The majority of the hydrogen bonds connect two adjacent chains, specifically the hydrogen bonds between the backbones of W4 to T50 and K45 to G47; however, at the core of the hydrogen bond network, the Q45 backbone forms hydrogen bonds with both adjacent Q45 backbones. A comparison between the structures of the trimerization domains of type XV and type XVIII is shown in Figure 4.
Fig 4.
Structure comparison between type XV (magenta) and XVIII (cyan) trimerization domains. (a) Least square fit of Cα traces. The carboxy-terminal end is labeled with C. (b) Superimposition of residues forming the hydrophobic trimer core (shown for one subunit; residues labeled as XV/XVIII).
2.2 Oligomeric State
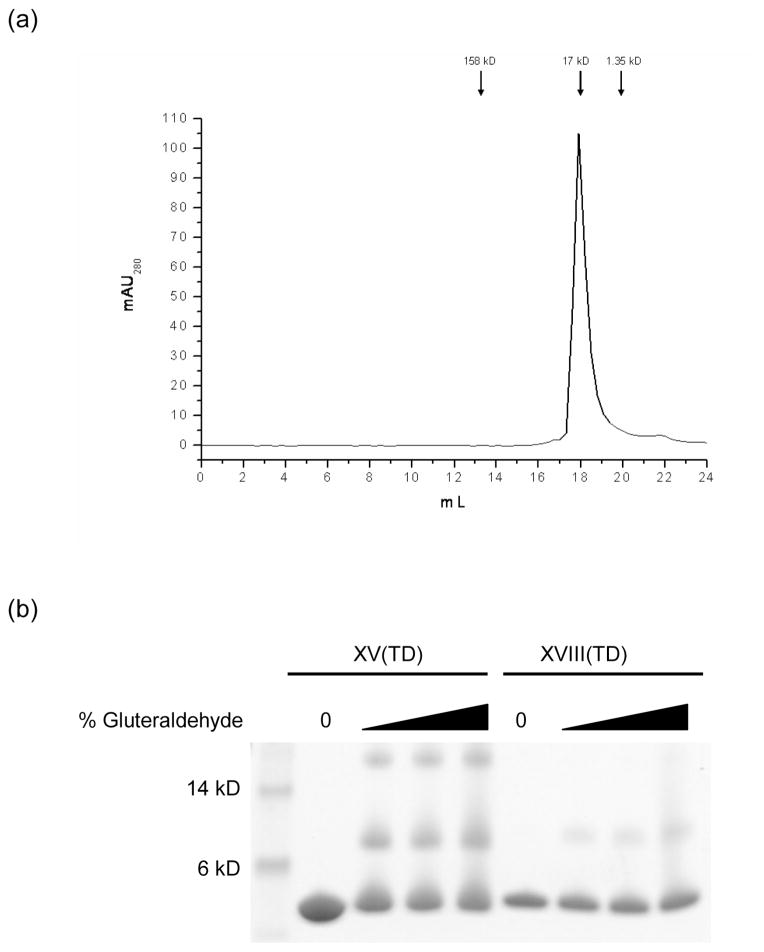
Gel filtration chromatography was performed to assess the oligomeric state of the XV(TD) protein. The protein eluted as a single peak with a molecular weight of approximately 18 kDa (Fig. 5), which corresponds to a trimer. Chemical crosslinking was performed with glutaraldehyde and analyzed by SDS-PAGE (Fig. 5). The XV(TD) readily crosslinks to form trimers. The XVIII(TD) protein has only two potential sites for crosslinking which results in little formation of crosslinked trimer under similar conditions when compared to the XV(TD) protein. Analytical ultracentrifugation was performed in phosphate-buffered saline, and results independently confirm the trimeric state of purified XV(TD) in solution. The trimerization domain of type XVIII collagen was also analyzed by ultracentrifugation, and the results are also consistent with a trimer in solution (Table 2).
Fig 5.
Gel filtration chromatography and chemical crosslinking. (a) XV(TD) protein were run on a Superose 6 column in PBS. Arrows represent elution peaks for the protein standards BSA, myoglobin and vitamin B12. (b) Chemical crosslinking experiments were performed with zero, 0.625, 0.125 and 0.25 % glutaraldehyde for 5 minute with either XV(TD) or XVIII(TD) protein at a final concentration of 0.75 or 0.5 mg/mL. Samples were run on a 12% Bis-Tris poly acrylamide gel.
Table 2.
Mass Spectrometry and Sedimentation Equilibrium Data
| Calculated MW (Da) | Mass Spectrometry (Da) | < v> (cm3g−1) | Analytical Ultracentrifuge (kDa) | Oligomeric State | |
|---|---|---|---|---|---|
| XV(TD) | 6552 | 6552 | 0.74823 | 18.7 ± 0.1 | 2.85 |
| XVIII(TD) | 6434 | 6434 | 0.73874 | 19.2 ± 0.1 | 2.98 |
Samples were run in phosphate-buffered saline at 20 °C
XV(TD) and XVIII(TD) samples were run at a concentration of 0.375 and 0.235 mg/mL, respectively.
<v> were calculated using UltrascanII (Demeler, 2010).
2.3 Denaturant Induced Equilibrium Transitions
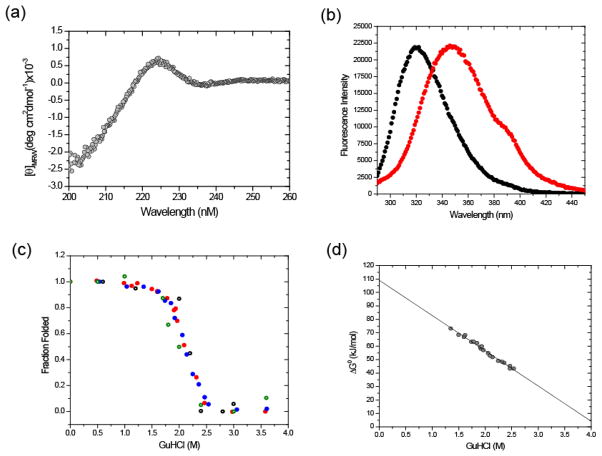
Biophysical properties of the XV(TD) protein were tested using CD and fluorescence. Figure 6a shows the CD spectrum of the XV(TD) protein in 50 mM sodium phosphate buffer pH 8.0. A positive peak is located at 224 nm and a slight negative peak is located at 236 nm. The maximum positive CD signal at 224 nm was observed for initial monitoring of guanidinium chloride (GuHCl) induced transitions. Complete unfolding as assessed by loss of signal at 224 nm was observed at 4 M GuHCl (data not shown).
Fig 6.
CD and fluorescence spectra and GuHCl-induced equilibrium transitions of the XV(TD). XV(TD was measured at 23.5 μM chain concentration in 50 mM sodium phosphate buffer pH 8.0. (a) CD spectrum of the native protein. (b) Fluorescence spectra of protein in 50 mM sodium phosphate buffer pH 8.0 with 0M GuHCl (●) or 4M GuHCl ( ). (c) Fraction of folded protein as a function of GuHCl concentration. Unfolding by CD(○), refolding by CD (
). (c) Fraction of folded protein as a function of GuHCl concentration. Unfolding by CD(○), refolding by CD ( ), unfolding by fluorescence (
), unfolding by fluorescence ( ), refolding by fluorescence (
), refolding by fluorescence ( ). (d) ΔG° dependence on GuHCl concentration with a linear fit.
). (d) ΔG° dependence on GuHCl concentration with a linear fit.
Fluorescence spectra of the XV(TD) protein in 0 and 4 M GuHCl buffer show a shift in the emission peak from 320 nm to 347 nm (Fig. 6b). Denaturant induced equilibrium transitions were monitored as a function of peak shift at a final protein concentration of 26 μM. Initial refolding samples were prepared at a concentration of 260 μM in 4 M GuHCl, 50 mM phosphate pH 8.0 buffer. Samples were subsequently refolded by dilution to a final concentration of 26 μM into buffers with a final GuHCl concentration of 0.4 to 4 M GuHCl. Samples used for unfolding were prepared in a buffer with no GuHCl and diluted to a final protein concentration of 26 μM into buffers with a final concentration of GuHCl of 0 to 3.6 M. The agreement between folding and unfolding curves indicates that refolding is reversible.
The standard free energy of trimerization (ΔG°) in water was calculated to be 109.0 (±1.0) kJ/mol. The calculated concentration at which half of the total chain concentration is incorporated into a trimer is equal to 119 pM for the collagen XV(TD). This is comparable to the calculated value of 56.6 pM for the midpoint concentration of the trimerization domain of type XVIII collagen.
3. Discussion
Collagen molecules are characterized by the presence of one or more triple helical regions formed by a repeating Gly-X-Y sequence. Much research has been performed to better understand the structure and function of the classic collagen triple helix (Shoulders and Raines, 2009). However, in recent years more attention has been paid to non-collagenous (NC) domains found in all collagen molecules. The NC domains for many collagen families have been shown to mediate trimerization. Here we present the crystal structure and biophysical characterization of the trimerization domain for human type XV collagen. The trimerization domain is structurally and biophysically very similar to the trimerization domain of type XVIII collagen.
Four crystal structures of collagen trimerization domains are known to date: the NC1 domain of network forming collagens IV, VIII and X (Bogin et al., 2002; Kvansakul et al., 2003; Sundaramoorthy et al., 2002; Than et al., 2002) as well as the only other known multiplexin, collagen XVIII (Boudko et al., 2009). The trimerization domains of the multiplexin family are ~50 residues in length, which are significantly smaller than the trimerization domains for the network forming collagens that are between 160 and 230 residues in length. The multiplexin trimerization domains appear to be stabilized primarily by their hydrophobic core. In contrast, multiple methods of trimer stabilization are found in the network forming collagens. The crystal structures of type IV collagen are stabilized by intersubunit exchange of β-sheets in addition to extensive hydrophobic and hydrophilic interactions. Solvent molecules and polar residues are found in the trimeric hydrophobic core of collagens VIII and X. Furthermore, the type X trimerization core is further strengthened by a buried cluster of calcium ions.
More significantly, the NC1 domains of network forming collagens form supramolecular assemblies via trimer oligomerization. Collagens VIII and X NC1 structures display strips of solvent exposed aromatic residues on the trimer surface that are believed to facilitate supramolecular assembly. The type IV NC1 domain derived from human placenta forms a stable hexamer with a novel covalent crosslink between a Met and Lys residue on opposite trimers (Than et al., 2002; Vanacore et al., 2009). The multiplexin trimerization domains are, in contrast, discrete entities that have no known additional function outside of chain association.
Multiplexins are present in all human tissues examined to date, but are present in very low concentrations (Myers et al., 2007). Collagen XV and XVIII trimers are capable of forming at picomolar concentrations, and are thus able to successfully associate even at low physiological concentrations. Additionally, the small size of the trimerization domain and the lack of disulphide bonds make trimerization highly efficient.
The multiplexin trimerization domains share relatively low sequence identity of 32 %. In contrast, the trimerization domains of collagens VIII and X which have similar structures but different mechanisms of trimer stabilization share over 60% sequence identity (Kvansakul et al., 2003). The endostatin and endostatin-like domains of type XV and XVIII collagens also share over 60 % identity (Sasaki et al., 2000), significantly more than the trimerization domains themselves. Analysis of the trimerization domain of multiplexins from a variety of organisms reveals little sequence homology, with the highest levels of homology involving key residues now known to be in the monomer or trimer hydrophobic core(Boudko et al., 2009). Sequence analysis of multiplexins predicted that the trimerization domain would contain a coiled-coil sequence (McAlinden et al., 2002). The crystal structures of both type XV(TD) and XVIII(TD) reveal no such structures, indicating that more accurate details of the multiplexin trimerization domain fold can be identified by structural rather than sequence analysis.
The structures of type XV and XVIII trimerization domains are highly similar (Fig. 4), with 1.5 Å RMSD between Cα atoms along the entire backbone of the trimerization domain. The overall topology and organization of both the monomer and trimeric hydrophobic cores are comparable. Key residues in hydrophobic core formation are located in identical positions, although the sequence identity among these selected residues is only 33 %. Thus, the relative location of hydrophobic residues plays a more important role in formation of a multiplexin trimerization domain than the absolute sequence identity. This further demonstrates the importance of empirically derived structural analysis, as traditional sequence analysis did not predict the structural similarity observed among these multiplexin trimerization domains.
A homologue to multiplexin collagen has been identified in both C. elegans and Drosophila (Ackley et al., 2001; Momota et al., 2008). Types XV/XVIII and IV collagens are the only known collagens that are conserved from nematode to human, which suggests that multiplexins may play an evolutionarily important role. The NC1 domain of the C. elegans multiplexin homologue was shown to trimerize in vitro (Ackley et al., 2001); the multiplexin fold may therefore represent an ancient and well conserved folding motif.
It would be of interest to see how accommodating the multiplexin fold is, specifically through mutational analysis of residues key to either hydrophobic core. It is of note that the multiplexin trimerization domain fold can form trimers at extremely low concentrations, but is unfolded by relatively low concentrations of GuHCl (~2 M can unfold half of the total population). The hydrophobic core may be optimized for fast kinetics, but modifications to the core may also be able to enhance its chemical stability.
The first three residues of the XV(TD) protein were not visible, indicating that they are flexible and thus not resolvable by X-ray crystallography. In contrast, the N-terminus of the XVIII(TD) protein has ordered structure through the analogous residues. These residues were laid out across and stabilized by interactions with the trimer surface such that their ends are more than 20 Å apart. It is unlikely that this conformation would be biologically relevant, since a triple helical domain is immediately adjacent and must place the three chains in a close orientation. The positions of these residues may result from an artifact in the crystallization of type XVIII. The XV(TD) structure here, in contrast, indicates that the first four residues are able to move freely and can thus come in close enough contact to align triple helix. Since the multiplexins are homotrimers, chain selection for staggering is not a vital issue. However, the N-terminus may still play a role in accomodating staggering. The exact nature of the triple helix and trimerization domain interface remains to be determined.
In conclusion, the type XV trimerization domain is remarkably similar to the type XVIII trimerization domain, despite low sequence identity. Its regulatory role in multiplexin NC1 domain processing remains to be determined, but as a structural fold it adds to a new class of small, efficient trimerization domains capable of folding at very low concentrations. The multiplexin trimerization fold may be used in future studies as a tool to create or increase the amount of correctly oligomerized recombinant trimeric proteins.
4. Experimental Procedures
4.1 Cloning, Expression and Purification
The trimerization domain of human α1(XV) sequence was synthetically constructed by Integrated DNA Technologies. The pIDTSMART-KAN vector contained the coding sequence with an embedded BamHI site and the 5′ end and a flanking Sal1 end after the stop codon at the 3′ end. Plasmids were expressed and purified by Midi Prep (Qiagen), the insert was excised with BamHI + Sal1 and cloned into the pET23-HisTag-Trx vector (Boudko et al., 2010). The DNA inserts were verified by Sanger dideoxy DNA sequencing.
The recombinant plasmids were expressed at 30 °C in the E. coli BL21(DE3) strain (Novagen). Once the cells reached an OD600 ~ 0.6, cells were induced with IPTG to a final concentration of 1 mM. The temperature was lowered to 30 °C and growth proceeded for an additional 6 hours prior to harvest. Purification of His and thioredoxin tagged protein from soluble cell lysate was performed using immobilized metal affinity chromatography on a Chelating HP Column (GE Healthcare) charged with NiCl2 following the manufacturer’s instructions. Tagged protein was cleaved with 1U/mg thrombin (ICN) or 0.1 mg/mL trypsin in 50 mM Tris/HCl (pH 8.0) buffer with 150 mM NaCl. The resulting fragments were separated with a Chelating HP Column as above. The non-tagged flow-through was further purified with a HiTrap Q Column (GE Healthcare) in 50 mM Tris pH 8.0 using a salt gradient from 0 to 0.5 M NaCl. The resulting purified proteins were analyzed for sequence by amino acid analysis and correct masses were confirmed by mass spectroscopy.
The trimerization domain (TD) of human collagen XVIII was cloned into the pET23-HisTag-Trx vector and purified as above.
4.2 Gel Filtration Chromatography and Analytical Ultracentrifugation
Gel filtration chromatography was performed using a 10/300 Superose 6 column (GE Healthcare) at 0.5 mL/min in phosphate buffered saline. Gel filtration standards (BioRad) were run to calibrate the column prior to injection of the XV(TD). Absorbance was monitored at 280 nm. Sedimentation equilibrium experiments were performed with a Beckman model XLA analytical centrifuge. Runs were carried out using an An60-Ti rotor with 12 mm cells and Epon two-channel centerpieces at 20 °C in phosphate buffered saline. Absorbance was monitored at 280 nm. V-bar values were calculated using UltrascanII (Demeler, 2010).
4.3 Crystallization and Data Collection
Crystallization was performed using hanging-drop vapor diffusion by mixing 1.5 μl of reservoir solution with 1.5 μl of protein solution at 5 mg/mL. The best crystals were grown at 25 °C using a reservoir solution of 0.1 to 0.2 M MgCl2, 0.1 M Tris (pH 7.0 – 8.0) and 28 – 32% (w/v) polyethylene glycol 3350. The crystals grew to a final size of roughly 0.25 × 0.25 × 0.25 mm after 7–14 days. Immediately prior X-ray exposure, the crystals were dipped into a cryoprotectant solution containing the reservoir solution supplemented with PEG 400 to a final concentration of 20 % (v/v) PEG 400. Data collection was performed using a RIGAKU RU200-H rotating anode generator. Data statistics are summarized in Table 1.
4.4 Crystal Structure Determination
The images collected were indexed, integrated and scaled using MOSFLM and SCALA from the CCP4 suite (CCP4, 1994). The AMORE (Navaza, 2001; Trapani and Navaza, 2008) program was used within the CCP4 suite (CCP4, 1994) to find initial molecular replacement solutions. A monomer of the collagen XVIII trimerization domain (PDB:3HSH, chain A) was used as a search model. Multiple solutions generated by AMORE were analyzed using two criteria: crystal packing without severe clashes and the generation of a trimer, applying crystal symmetry operators. Two chains were found as a solution, neither of which was able to form the crystal packing by itself. Iterative cycles of model correction and refinement were performed using COOT (Emsley et al., 2010) and CNS (Brunger et al., 1998), respectively.
4.5 Circular Dichroism and Fluorescence Analysis
CD Spectra were recorded on an AVIV model 202 spectropolarimeter (AVIV Instruments, Inc.). Quartz cells of 0.05- to 0.1-mm path length were used (Hellma), depending on protein concentration. Spectra were buffer subtracted and normalized for concentration and path length to obtain the mean molar residue ellipticity. Protein concentrations were determined by amino acid analysis. Fluorescence spectra were recorded on a LM8000C instrument (SLM Instruments) with modified electronics (ISS Corp., Champaign, IL) using 104F-QS cells (Hellma). Emission spectra were obtained by excitation of protein samples at 280 nm at 25 °C.
4.5 GuHCl Induced CD & Fluorescence Transitions
Folded protein samples were in 50 mM sodium phosphate buffer (pH 8.0) at a stock concentration of 260 μM. Unfolded protein samples were at the same concentration in 50 mM sodium phosphate buffer (pH 8.0) supplemented with 4 M GuHCl. Proteins were diluted to a final concentration of 26 μM into buffers of varying concentrations of GuHCl. Final GuHCl concentrations were determined using refractive indices. Change in secondary structure with varying concentrations of GuHCl was monitored by CD at 224 nm. Change in tertiary structure with varying concentrations of GuHCl was monitored by a peak shift in fluorescence emission spectra from 320 nm to 347 nm.
To calculate the standard free energy (ΔG°), unfolding and refolding curves were interpreted using a two state system where unfolded monomers (U) combine into a native trimer (N) in a cooperative mechanism (Boudko et al., 2009):
Each GuHCl concentration can be used to calculate an equilibrium constant (Keq):
where [M]0 is the total monomer concentration ([M]0=[U] + 3[N]) and fu is the fraction of unfolded monomer (fu=[U]/[M]0). The standard free energy ΔG° is related to Keq by:
The denaturant dependence of ΔG° is linear and can be described by:
where ΔG°(H2O) is the free energy for unfolding in water.
The calculated concentration at which half of the total chain concentration is incorporated into a trimer (3[N]1/2=[U]1/2) can be derived from Keq:
4.6 Protein Data Bank Accession Number
The refined atomic model and the observed structure factors were deposited into the RCSB Protein Data Bank with the accession number 3N3F.
Acknowledgments
We would like to thank Keith Zientek for MS analysis and Jesse Vance for DNA sequencing and amino acid analysis. This work was supported by a grant from Shriners Hospital for Children.
Abbreviations
- COL
collagenous
- NC
non-collagenous
- NC1
carboxyl-terminal non-collagenous domain
- GuHCl
Guanidine Hydrochloride
References
- Ackley BD, Crew JR, Elamaa H, Pihlajaniemi T, Kuo CJ, Kramer JM. The NC1/endostatin domain of Caenorhabditis elegans type XVIII collagen affects cell migration and axon guidance. J Cell Biol. 2001;152:1219–1232. doi: 10.1083/jcb.152.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amenta PS, Scivoletti NA, Newman MD, Sciancalepore JP, Li D, Myers JC. Proteoglycan-collagen XV in human tissues is seen linking banded collagen fibers subjacent to the basement membrane. J Histochem Cytochem. 2005;53:165–176. doi: 10.1369/jhc.4A6376.2005. [DOI] [PubMed] [Google Scholar]
- Bogin O, Kvansakul M, Rom E, Singer J, Yayon A, Hohenester E. Insight into Schmid metaphyseal chondrodysplasia from the crystal structure of the collagen X NC1 domain trimer. Structure. 2002;10:165–173. doi: 10.1016/s0969-2126(02)00697-4. [DOI] [PubMed] [Google Scholar]
- Boudko SP, Engel J, Bachinger HP. Trimerization and Triple Helix Stabilization of the Collagen XIX NC2 Domain. J Biol Chem. 2008;283:34345–34351. doi: 10.1074/jbc.M806352200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudko SP, Sasaki T, Engel J, Lerch TF, Nix J, Chapman MS, Bachinger HP. Crystal structure of human collagen XVIII trimerization domain: A novel collagen trimerization Fold. J Mol Biol. 2009;392:787–802. doi: 10.1016/j.jmb.2009.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudko SP, Zientek KD, Vance JM, Hacker JL, Engel J, Bachinger HP. The NC2 domain of collagen IX provides chain selection and heterotrimerization. J Biol Chem. 2010 doi: 10.1074/jbc.M110.128405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Demeler B. Methods for the design and analysis of sedimentation velocity and sedimentation equilibrium experiments with proteins. Curr Protoc Protein Sci. 2010;Chapter 7(Unit 7):13. doi: 10.1002/0471140864.ps0713s60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doege KJ, Fessler JH. Folding of carboxyl domain and assembly of procollagen I. J Biol Chem. 1986;261:8924–8935. [PubMed] [Google Scholar]
- Eklund L, Piuhola J, Komulainen J, Sormunen R, Ongvarrasopone C, Fassler R, Muona A, Ilves M, Ruskoaho H, Takala TE, Pihlajaniemi T. Lack of type XV collagen causes a skeletal myopathy and cardiovascular defects in mice. Proc Natl Acad Sci U S A. 2001;98:1194–1199. doi: 10.1073/pnas.031444798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukai N, Eklund L, Marneros AG, Oh SP, Keene DR, Tamarkin L, Niemela M, Ilves M, Li E, Pihlajaniemi T, Olsen BR. Lack of collagen XVIII/endostatin results in eye abnormalities. Embo J. 2002;21:1535–1544. doi: 10.1093/emboj/21.7.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon MK, Hahn RA. Collagens. Cell Tissue Res. 2010;339:247–257. doi: 10.1007/s00441-009-0844-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter W, Dong S, Schurer B, Cole GJ. Collagen XVIII is a basement membrane heparan sulfate proteoglycan. J Biol Chem. 1998;273:25404–25412. doi: 10.1074/jbc.273.39.25404. [DOI] [PubMed] [Google Scholar]
- Iozzo RV. The biology of the small leucine-rich proteoglycans. Functional network of interactive proteins. J Biol Chem. 1999;274:18843–18846. doi: 10.1074/jbc.274.27.18843. [DOI] [PubMed] [Google Scholar]
- Khoshnoodi J, Cartailler JP, Alvares K, Veis A, Hudson BG. Molecular recognition in the assembly of collagens: terminal noncollagenous domains are key recognition modules in the formation of triple helical protomers. J Biol Chem. 2006;281:38117–38121. doi: 10.1074/jbc.R600025200. [DOI] [PubMed] [Google Scholar]
- Kvansakul M, Bogin O, Hohenester E, Yayon A. Crystal structure of the collagen alpha1(VIII) NC1 trimer. Matrix Biol. 2003;22:145–152. doi: 10.1016/s0945-053x(02)00119-1. [DOI] [PubMed] [Google Scholar]
- Marneros AG, Olsen BR. Physiological role of collagen XVIII and endostatin. Faseb J. 2005;19:716–728. doi: 10.1096/fj.04-2134rev. [DOI] [PubMed] [Google Scholar]
- McAlinden A, Crouch EC, Bann JG, Zhang P, Sandell LJ. Trimerization of the amino propeptide of type IIA procollagen using a 14-amino acid sequence derived from the coiled-coil neck domain of surfactant protein D. J Biol Chem. 2002;277:41274–41281. doi: 10.1074/jbc.M202257200. [DOI] [PubMed] [Google Scholar]
- McLaughlin SH, Bulleid NJ. Molecular recognition in procollagen chain assembly. Matrix Biol. 1998;16:369–377. doi: 10.1016/s0945-053x(98)90010-5. [DOI] [PubMed] [Google Scholar]
- Momota R, Naito I, Ninomiya Y, Ohtsuka A. Characterization of drole, drosophila type XV/XVIII collagen. Matrix Biology. 2008;27:23–24. [Google Scholar]
- Myers JC, Amenta PS, Dion AS, Sciancalepore JP, Nagaswami C, Weisel JW, Yurchenco PD. The molecular structure of human tissue type XV presents a unique conformation among the collagens. Biochem J. 2007;404:535–544. doi: 10.1042/BJ20070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers JC, Kivirikko S, Gordon MK, Dion AS, Pihlajaniemi T. Identification of a previously unknown human collagen chain, alpha 1(XV), characterized by extensive interruptions in the triple-helical region. Proc Natl Acad Sci U S A. 1992;89:10144–10148. doi: 10.1073/pnas.89.21.10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 2004;20:33–43. doi: 10.1016/j.tig.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr D Biol Crystallogr. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- Pihlajaniemi T, Rehn M. Two new collagen subgroups: membrane-associated collagens and types XV and XVII. Prog Nucleic Acid Res Mol Biol. 1995;50:225–262. doi: 10.1016/s0079-6603(08)60816-8. [DOI] [PubMed] [Google Scholar]
- Pushpakom SP, Herrick AL, Kumar S, Worthington J. Polymorphisms in COL15 gene are not associated with systemic sclerosis. J Rheumatol. 2008;35:251–253. [PubMed] [Google Scholar]
- Rehn M, Hintikka E, Pihlajaniemi T. Primary structure of the alpha 1 chain of mouse type XVIII collagen, partial structure of the corresponding gene, and comparison of the alpha 1(XVIII) chain with its homologue, the alpha 1(XV) collagen chain. J Biol Chem. 1994;269:13929–13935. [PubMed] [Google Scholar]
- Rehn M, Pihlajaniemi T. Alpha 1(XVIII), a collagen chain with frequent interruptions in the collagenous sequence, a distinct tissue distribution, and homology with type XV collagen. Proc Natl Acad Sci U S A. 1994;91:4234–4238. doi: 10.1073/pnas.91.10.4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarela J, Rehn M, Oikarinen A, Autio-Harmainen H, Pihlajaniemi T. The short and long forms of type XVIII collagen show clear tissue specificities in their expression and location in basement membrane zones in humans. Am J Pathol. 1998;153:611–626. doi: 10.1016/S0002-9440(10)65603-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Fukai N, Mann K, Gohring W, Olsen BR, Timpl R. Structure, function and tissue forms of the C-terminal globular domain of collagen XVIII containing the angiogenesis inhibitor endostatin. Embo J. 1998;17:4249–4256. doi: 10.1093/emboj/17.15.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Larsson H, Tisi D, Claesson-Welsh L, Hohenester E, Timpl R. Endostatins derived from collagens XV and XVIII differ in structural and binding properties, tissue distribution and anti-angiogenic activity. J Mol Biol. 2000;301:1179–1190. doi: 10.1006/jmbi.2000.3996. [DOI] [PubMed] [Google Scholar]
- Shoulders MD, Raines RT. Collagen structure and stability. Annu Rev Biochem. 2009;78:929–958. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snellman A, Tu H, Vaisanen T, Kvist AP, Huhtala P, Pihlajaniemi T. A short sequence in the N-terminal region is required for the trimerization of type XIII collagen and is conserved in other collagenous transmembrane proteins. Embo J. 2000;19:5051–5059. doi: 10.1093/emboj/19.19.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy M, Meiyappan M, Todd P, Hudson BG. Crystal structure of NC1 domains. Structural basis for type IV collagen assembly in basement membranes. J Biol Chem. 2002;277:31142–31153. doi: 10.1074/jbc.M201740200. [DOI] [PubMed] [Google Scholar]
- Than ME, Henrich S, Huber R, Ries A, Mann K, Kuhn K, Timpl R, Bourenkov GP, Bartunik HD, Bode W. The 1.9-A crystal structure of the noncollagenous (NC1) domain of human placenta collagen IV shows stabilization via a novel type of covalent Met-Lys cross-link. Proc Natl Acad Sci U S A. 2002;99:6607–6612. doi: 10.1073/pnas.062183499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapani S, Navaza J. AMoRe: classical and modern. Acta Crystallogr D Biol Crystallogr. 2008;64:11–16. doi: 10.1107/S0907444907044460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utriainen A, Sormunen R, Kettunen M, Carvalhaes LS, Sajanti E, Eklund L, Kauppinen R, Kitten GT, Pihlajaniemi T. Structurally altered basement membranes and hydrocephalus in a type XVIII collagen deficient mouse line. Hum Mol Genet. 2004;13:2089–2099. doi: 10.1093/hmg/ddh213. [DOI] [PubMed] [Google Scholar]
- Vanacore R, Ham AJ, Voehler M, Sanders CR, Conrads TP, Veenstra TD, Sharpless KB, Dawson PE, Hudson BG. A sulfilimine bond identified in collagen IV. Science. 2009;325:1230–1234. doi: 10.1126/science.1176811. [DOI] [PMC free article] [PubMed] [Google Scholar]