Abstract
The cellular factor TRIM5α inhibits infection by numerous retroviruses in a species-specific manner. The TRIM5α protein from rhesus macaques (rhTRIM5α) restricts infection by HIV-1 while human TRIM5α (huTRIM5α) restricts infection by murine leukemia virus (MLV). In owl monkeys a related protein TRIM-Cyp restricts HIV-1 infection. Several models have been proposed for retroviral restriction by TRIM5 proteins (TRIM5α and TRIM-Cyp). These models collectively suggest that TRIM5 proteins mediate restriction by directly binding to specific determinants in the viral capsid. Through their ability to self-associate TRIM5 proteins compartmentalize the viral capsid core and mediate its abortive disassembly via a poorly understood mechanism that is sensitive to proteasome inhibitors. In this review, we discuss TRIM5-mediated restriction in detail. We also discuss how polymorphisms within human and rhesus macaque populations have been demonstrated to affect disease progression of immunodeficiency viruses in these species.
Introduction
HIV-1, a major causative agent of AIDS in humans, was introduced into the human population from chimpanzees. Although HIV-1 is highly pathogenic in humans and chimpanzees, it cannot replicate in most Old World monkey species. Similar cross-species barriers to infection are present in most primates and can dictate the ability of many strains of simian immunodeficiency viruses (SIV) to replicate in individual primate species. This intrinsic immunity to retroviruses is attributed to certain restriction factors constitutively expressed in host cells that inhibit retroviruses at different stages of infection.1,2 Evidence that host cells express inhibitors of retroviral replication first originated in the 1960s when the Friend virus susceptibility (Fv) factor-1 that dictates the susceptibility of mice to two different strains of murine leukemia virus (MLV) was discovered.1,2 Recent advances in understanding this innate, intracellular immunity against retroviruses have led to the discovery of the Tripartite motif 5 (TRIM5) proteins involved in antiviral responses. In 2004, using a genetic screen, the Sodroski laboratory identified TRIM5α as the protein responsible for preventing HIV-1 infection in rhesus macaques (rhTRIM5α).3 A similar protein, TRIM-Cyp, identified in owl monkeys, also restricts HIV-1 infection.4 Since then, TRIM5 proteins (TRIM5α and TRIM-Cyp) have been identified as being responsible for previously identified restrictions to retroviral infection naturally present in humans,5,6 other primate species,5,7–14 cattle,15,16 and nonprimates.17,18 It has been shown that strong positive selective pressure has been exerted on the regions of TRIM5α proteins that confer species specificity in recognition of viral capsid determinants.19,20 This suggests that TRIM5α-mediated restriction of retroviral infection is a critical, evolutionarily conserved antiviral mechanism. Here we review the recent advances in our understanding of how TRIM5 proteins restrict retroviral infection. We also discuss the consequences of this restriction on viral replication and disease progression in host species infected with primate immunodeficiency viruses.
TRIM5 Proteins (TRIM5α and TRIM-Cyp)
The tripartite motif (TRIM) family of proteins is defined by the three domains (RING, B-Box2, Coiled-Coil) present throughout this family.21,22 The RING domain is the N-terminal domain of all TRIM proteins,22 and possesses E3 ubiquitin ligase activity.23 The B-Box2 and Coiled-Coil (CC) domains are thought to contribute to the higher and low-order multimerization of TRIM5α, respectively. TRIM5 proteins also possess a C-terminal capsid binding domain that mediates specific recognition and restriction of certain retroviruses.24
Self-Association of TRIM5α and Cytoplasmic Body Formation
TRIM family members are characterized by the ability to form protein assemblies or bodies of numerous shapes and sizes in both the nucleus and the cytoplasm.22 TRIM5α is known to localize to cytoplasmic bodies, having been described as the “cytoplasmic body component TRIM5α” in the article in which its antiviral activity was first reported.3 However, the relevance of the cytoplasmic bodies to which TRIM5α localizes has been somewhat controversial. Two studies have found that preexisting cytoplasmic bodies are not required for the ability of TRIM5 proteins to restrict retroviral infection.25,26 One study found that treating cells with the heat shock protein 90 inhibitor geldanamycin prevented the cytoplasmic body localization of rhTRIM5α without significantly perturbing the ability of rhTRIM5α to restrict HIV-1 infection.26 Another study observed that a cell line expressing relatively low amounts of owl monkey TRIM-Cyp did not localize to cytoplasmic bodies and was still able to restrict HIV-1 infection.25 Alternatively, our studies have observed fluorescently labeled HIV-1 virions associating with rhTRIM5α cytoplasmic bodies, and live cell imaging has observed the de novo formation of cytoplasmic bodies around individual virions.27 We have also found that two discrete regions of the Linker 2 (L2) region, located between the CC domain and C-terminal capsid binding domain, are required for the localization of TRIM5α to cytoplasmic bodies. rhTRIM5α variants that lose the ability to localize to cytoplasmic bodies are completely unable to restrict HIV-1 infection, though they still form low-order and higher-order multimers to the same degree as the wild-type (WT) protein.28
Although these works appear contradictory on the surface, they collectively suggest that although preexisting cytoplasmic bodies are not required for restriction, the ability to form cytoplasmic bodies around a restriction-sensitive virion is a critical aspect of TRIM5α restriction. In point of this fact, the studies that found cytoplasmic bodies are not required for TRIM5α-mediated restriction visualized the localization of TRIM5α only prior to infection, not following the addition of restriction-sensitive virus.25,26 Our study, which observed that rhTRIM5α cytoplasmic body localization correlates with the ability to restrict infection, examined rhTRIM5α localization in the absence of virus.28 However, we believe it is likely that the localization of TRIM5α to cytoplasmic bodies in the absence of virus simply reflects the ability of rhTRIM5α to assemble into cytoplasmic bodies during restriction. It is this ability to self-associate and ultimately assemble into a cytoplasmic body during restriction that is required for TRIM5α function. This self-association occurs at a number of levels, all of which are required for the biological activity of TRIM5α.
Low-order multimerization: dimerization
Dimerization appears to be the lowest order of TRIM5α self-association.29–32 Previous studies have demonstrated that the CC domain plays a critical role in the multimerization of TRIM family proteins,22 including TRIM5α and TRIM-Cyp.30,33 Early work suggested that the multimeric form of TRIM5α was a trimer.29,30 However, these studies were hampered by the tendency of TRIM5α to self-associate by numerous mechanisms, as well as migrate at electrophoretic mobility inconsistent with its actual molecular weight. A more rigorous analysis, which required the replacement of the TRIM5α RING domain with the RING domain of TRIM21 (TRIM5α-21R), was recently performed by the Sodroski and Sundquist laboratories. These studies reveal that the dimeric form is the predominant multimeric form assumed by the protein.31,32 The functionality of this multimerization was demonstrated by the fact that dimeric TRIM5α − 21R could directly bind restriction-sensitive viral cores, whereas monomeric forms could not.32 Other studies also show that CC-mediated multimerization is clearly critical for TRIM5-mediated restriction, as deletion of this domain abrogates the ability of rhTRIM5α to restrict infection.6,29 Moreover, expression of CC domains in the absence of other TRIM5 domains has been shown by numerous studies to exert a dominant negative effect on restriction mediated by full length TRIM5 proteins.6,29,30,34
Higher-order multimerization
In addition to lower-order dimerization, TRIM5α also exhibits a higher-order multimerization that is visible by protein electrophoresis when cellular proteins are cross-linked biochemically with an agent such as ethylene glycol-bis(succinimidyl succinate) (EGS) or glutaraldehyde.28–30,35 Work from the Sodroski laboratory has identified the B-Box2 and the L2 region of TRIM5α and TRIM-Cyp as contributing to the higher-order multimerization of TRIM5 proteins.11,24,34,36,37 A cysteine point mutation within the B-Box2 region (C96A) greatly reduces the level of restriction of HIV-1 by TRIM-Cyp,11 and a similar mutant (C97A/H100A) abrogates restriction of HIV-1 by rhTRIM5α.34 More recent studies have demonstrated that these residues mediate the higher-order multimerization of TRIM5 proteins.36,37 The relevance of this activity to TRIM5α function is underscored by the observation that this ability to form higher-order multimers is required for efficient binding of TRIM5α to the retroviral capsid.36 The nature of these higher-order multimers is typically difficult to resolve in most biochemical cross-linking experiments.28–30 However, one recent study has observed apparent hexameric forms of TRIM5α in MDTF cells, irrespective of the presence or absence of restriction-sensitive viral cores.35 They concluded that TRIM5α multimerization possibly involves multiple protein:protein interfaces. As discussed below, this observation of apparent hexamers may be in accordance with emerging models of TRIM5α self-association.
Assembly
In addition to its role in facilitating higher-order multimers, the L2 region also contains determinants that mediate the assembly of higher-order multimers into even higher-order assemblies that are manifested as cytoplasmic bodies microscopically. This L2-mediated assembly activity appears to be distinct from the higher-order assemblies mediated by the B-Box2 region or other parts of the L2 region because mutations that disrupt TRIM5α cytoplasmic body localization do not affect the formation of higher-order multimers, as measured by biochemical cross-linking.28 However, just as was the case with mutations that disrupt lower-order or higher-order TRIM5α multimerization, mutations that disrupt cytoplasmic body assembly are no longer able to restrict infection.28
A recent and exciting study by the Sundquist and Yeager laboratories provides significant insight into and support for the numerous studies mentioned above. Using negative-stain electron microscopy of recombinant purified TRIM5α − 21R, these authors observe spontaneous assembly of this protein into two-dimensional hexagonal arrays.38 They observe that these assemblies require protein dimerization and higher-order multimerization mediated by the CC and B-Box2 domains, respectively. Assemblies of HIV-1 capsid protein enhanced the formation of TRIM5α-21R hexameric assemblies, though they were not absolutely required.38 By considering the size of the viral and TRIM5α hexameric assemblies, these authors provide a model of TRIM5α self-assembly around a viral core that is mediated by the CC and B-Box2 domains and L2 region. In this model, B-Box2-dependent self-association mediates the formation of a trimeric interface of TRIM5α dimers, which facilitates the tripodial protein extensions required in a hexameric lattice. Assuming they are mediated by the B-Box2 domain, this may explain the apparent hexameric forms of TRIM5α observed by the Berthoux group.35 In this model, the L2 region mediates the self-association of opposing dimers,38 allowing the projection of B-Box2 domains on both ends of each segment of the hexameric lattice, facilitating the formation of hexameric assemblies.
Capsid Recognition by TRIM5 Proteins
Work by numerous groups has demonstrated that the recognition of viral capsid determinants depends on regions in the C-terminal B30.2/PRYSPRY (hereafter SPRY) domain of TRIM5α. This domain has been shown to possess three variable regions that have evolved to recognize the capsids of restricted retroviruses.19,20,39,40 This is best illustrated by the fact that, while human TRIM5α (huTRIM5α) can only weakly interfere with HIV-1 infection,3 replacing a single amino acid in the SPRY domain of huTRIM5α can confer the ability to restrict HIV-1 to a level similar to rhTRIM5α.39,41 Notably, the same regions and amino acids that have been shown to contribute to restriction specificity have been shown to have undergone strong selective pressure during primate evolution.19,20 In support of this notion, the SPRY domain has been functionally replaced, on more than one occasion, by the cellular cyclophillin A (CypA) gene in a number of monkey species,4,42,43 which is known to bind the HIV-1 capsid alone or in the context of the TRIM-Cyp fusion protein.21,44,45 The fact that different species of monkey have convergently evolved similar mechanisms to tether TRIM5 proteins to cytoplasmic retroviral capsids underscores the critical role this family of proteins plays in the defense against retroviral infection.
Although all available evidence suggests that TRIM5 proteins interact directly with determinants present in retroviral capsids, measuring this interaction using existing scientific methods has proven difficult. This is due to the fact that TRIM5 proteins appear to recognize and bind the viral capsid protein only in the context of a mature retroviral core. The most direct evidence for this has come from the Aiken laboratory, which found that saturation of TRIM5α restriction is achievable with wild-type or hyperstable viral cores. Cores that are unstable or not fully processed are not able to saturate TRIM5-mediated restriction.46 Therefore, because of the relative instability of viral cores isolated from an active infection, classical molecular biology techniques have been unable to measure this interaction in cells. However, an interaction between huTRIM5α and N-MLV virions has been detected in vitro using detergent-stripped MLV virions47 and between rhTRIM5α or TRIM-Cyp and HIV-1 capsid complexes assembled from purified recombinant capsid-nucleocapsid (CA–NC) proteins.11,48 Taken together, these data suggest that TRIM5-mediated retroviral restriction involves the specific recognition of capsid determinants in the context of an intact, mature viral core. This interaction occurs via the C-terminal SPRY domain in the case of TRIM5α and via the cyclophilin A domain in the case of TRIM-Cyp proteins.
Although C-terminal capsid-binding domains appear to be the primary determinants conferring retroviral specificity, some studies have additionally implicated the CC domain as being capable of modulating specificity of restriction. Comparison of the primate TRIM5α gene sequences suggests that residues not only in the SPRY domain but also in the CC domain have been subjected to strong positive selection during evolution.19,49,50 Studies by the Trono and Stoye groups, using interspecies TRIM5α chimeric derivatives, demonstrate a possible cooperativity between the SPRY and CC domains to determine the specificity of TRIM5α-mediated retroviral restriction.41,51 How the CC domain affects specificity is unclear. Some residues not involved in mediating TRIM5α self-association may be mediating a low-affinity interaction with the viral capsid lattice. Alternatively, changes in the CC domain may alter the geometry of the TRIM5α dimer, which could affect the geometry of the hexameric assemblies formed by TRIM5α. For example, both rhesus and human isoforms of TRIM5α localize to cytoplasmic assemblies. However, rhTRIM5α tends to form spherical assemblies and is very efficient at restricting a conical retrovirus such as HIV-1. HuTRIM5α forms elongated or filamentous assemblies,52 which may make it slightly better equipped to disrupt the structure of spherical retroviral cores such as MLV.
Proposed Models of TRIM5α-Mediated Restriction
Although the regions responsible for mediating TRIM5α binding to retroviral capsids, including HIV-1, are well defined by concordant data from numerous groups,19,39–41 the mechanism by which TRIM5α prevents viral infection remains unclear. Numerous models have been proposed to explain this restriction, which are described below.
Accelerated uncoating
The accelerated uncoating model, supported by the work of the Sodroski laboratory, suggests that TRIM5α accelerates uncoating of the viral core in a manner that prevents subsequent steps in infection. The term “uncoating” is used to describe the loss of the p24 capsid protein from the viral ribonucleoprotein complex during infection.53 The accelerated uncoating model is based primarily on a “fate of capsid assay” that attempts to measure the amount of intact viral capsids in cells following infection. This is accomplished by centrifugation of cellular extracts through a sucrose cushion, which separates cytosolic capsid protein into pelletable (intact cores) and supernatant (dissociated or uncoated cores) fractions. These studies indicate that TRIM5α induces a loss of the pelletable capsid population while not affecting the total amount of capsid in the sample.11,48,54–56 This suggests a model in which rhTRIM5α promotes dissociation of capsid from the viral ribonucleoprotein complex, but not via a mechanism that involves the TRIM5α-induced degradation of cytosolic capsid protein. As rhTRIM5α mediates the more rapid, observable loss of pelletable complexes containing capsid protein, this model has been described as the accelerated uncoating model. However, it remains to be seen if the loss of p24 from the viral complex during restriction shares mechanistic similarities with the events occurring during the normal uncoating process that facilitates viral infection.
Proteasome independent capsid degradation
Work from the Gallay laboratory has found a specific loss of cytosolic capsid in cells expressing rhTRIM5α.57 This work separated cytosolic and vesicular fractions from cells infected with HIV-1 and found that cells expressing rhTRIM5α induced the degradation of viral capsid without affecting other components of the viral nucleoprotein complex. This work also found that the rhTRIM5α-mediated degradation of capsid was independent of proteasome function, as proteasome inhibitors did not prevent capsid degradation measured in these experiments.57 These data form a model in which the stripping of capsid protein from the viral nucleoprotein complexes prevents the complex from completing subsequent steps in the infectious pathway.
Two-step mechanism of restriction
This model proposes that TRIM5-mediated restriction occurs in two distinct phases. In the first step, which is sufficient to inhibit retroviral infection, the TRIM5 proteins bind the retroviral capsid via the SPRY domain, or CypA in the case of owl monkey TRIM-Cyp. In the second step, TRIM5 proteins induce the proteasome-dependent, abortive disassembly of the bound virion. This model is primarily based on the observation that proteasome inhibitors relieve the TRIM5α-mediated restriction to reverse transcription (RT) products while not affecting the ability of TRIM5 proteins to restrict infection.58,59 In collaboration with the Engelman laboratory, we have demonstrated that proteasome inhibition allows reverse transcription to complete in restricted cells, and that these viral ribonucleoprotein complexes exhibit unrestricted integration activity in vitro.58 This suggests that viral preintegration complexes are unable to achieve their nuclear translocation under these conditions due to an interaction with TRIM5 proteins, but otherwise remain intact. This notion is supported by our observation that proteasome inhibition leads to the accumulation of fluorescently labeled HIV-1 virions in the cytoplasm of restricted cells, where they accumulate in enlarged cytoplasmic bodies.27 The role of a proteasome-dependent step in the restriction process is also supported by data from the Aiken laboratory, which has observed that the addition of restriction-sensitive virus results in the proteasome-mediated degradation of the TRIM5α protein.60 However, the mechanism by which TRIM5α engages the proteasomal machinery has not been elucidated.
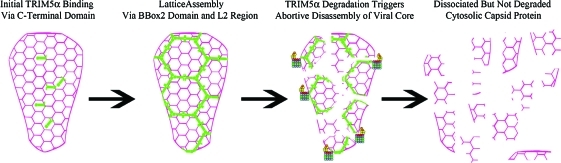
Although these previously proposed models suggest different mechanisms of TRIM function, they are obviously not mutually exclusive. Proteasome inhibitors have been observed to prevent the TRIM5α-mediated acceleration of uncoating measured by the Sodroski laboratory55 and also result in the accumulation of fluorescently labeled virions in the cytoplasm of restricted cells.27 The most obvious explanation of these data is that TRIM5α induces the proteasomal degradation of components of the viral ribonucleoprotein complex in a fashion that abrogates the infectivity of the complex. However, no studies have observed a TRIM5α-mediated proteasome-dependent loss of any viral proteins during infection, although certainly many researchers have looked for such an occurrence. The only proteasome-dependent loss of protein that has been observed during restriction has been the loss of TRIM5α itself.60 Studies using the fate of capsid assay observe that, during restriction, TRIM5α disrupts the amount of pelletable capsid cores in cells without affecting the total amount of cytoplasmic capsid protein.11,48,54–56 Taken together, these data suggest a model whereby TRIM5α degradation by the proteasome is functionally tethered to the disruption of the viral capsid core. This would explain why there is no observable loss of total capsid protein in the cytoplasm of restricted cells, despite the numerous studies that suggest that the viral capsid protein is the target of TRIM5α restriction. Once bound to the viral capsid core, TRIM5α autoubiquitination23 may facilitate its recognition by the proteasome. Translocation of TRIM5α through the proteasomal complex would therefore generate the mechanical force responsible for inducing the abortive disassembly of the viral capsid core (Fig 1).
FIG. 1.
Model for TRIM5α-mediated retroviral restriction. Following entry of the retroviral core (pink) in the host cell cytoplasm TRIM5 proteins directly bind the retroviral capsid by recognizing specific determinants in the viral capsid. Following binding, the B-box2 and L2 regions mediate high-order multimerization and assembly of individual TRIM5 dimers, respectively, resulting in the formation of a hexameric lattice surrounding the viral core. Given the specificity of the SPRY domain for the viral capsid proteins it is possible that multiple TRIM5 lattices assemble around a single virion. A single TRIM5 lattice (green) is shown for clarity. Proteasomal degradation of TRIM5 proteins then triggers “abortive disassembly” of the viral core. This results in dissociation but not degradation of the capsid proteins keeping the cytosolic capsid levels unchanged. The viral capsid is shown in pink and the TRIM5 lattice is shown in green. Color images available online at www.liebertonline.com/aid.
Cellular Degradation of TRIM5α
The fact that the TRIM5α RING domain possesses E3 ubiquitin ligase activity made proteasome-dependent virion degradation a popular model early in the characterization of this process. However, two groups independently found that proteasome function was not required for TRIM5-mediated restriction.25,48 However, other studies have demonstrated that although restriction to infection is not relieved, proteasome inhibition relieves the TRIM5α-mediated block to RT product formation.58,59 TRIM5α has been observed to possess functional ubiquitin ligase activity, capable of ubiquitinating itself in vitro and in 293 cells.23 This suggests the ubiquitin ligase activity of TRIM5α may play a role in the ability of TRIM5α to restrict retroviral infection. However, this hypothesis has been difficult to test because mutations to the critical zinc coordinating cysteines in the RING domain (C15/C18) result in a nonfunctional protein that localizes to large, nonfunctional aggregates when expressed in cells. Studies monitoring the degradative fate of TRIM5α have revealed an interesting and potentially important aspect of TRIM5α cell biology. Namely, TRIM5α is degraded in a proteasome-independent fashion in the absence of viral infection23,56,59 but in a proteasome-dependent fashion following exposure to restriction sensitive virus.60 In either case, TRIM5α is turned over rapidly, suggesting that the degradation of the protein is not simply activated following exposure to virus. Rather, the pathway mediating the degradation of TRIM5α is conditionally altered in the presence of virus. We have recently demonstrated, using FRET and coimmunoprecipitation, that TRIM5α associates with the protein p62/sequestosome 1.61 p62 is an interferon-inducible protein62 involved in cellular degradative and signaling pathways.63,64 p62 binds ubiquitinated proteins via its UBA domain and targets them for proteasomal or autophagic degradation. Interestingly, p62 also stabilizes TRIM5α, as demonstrated by reduced levels of exogenously and endogenously expressed TRIM5α upon siRNA knockdown of p62. Inhibition of autophagy results in increased TRIM5α levels (E.M. Campbell, unpublished data). Another study has observed TRIM5α associating with components of the autophagic degradation pathway.65 How and why restriction-sensitive virus alters the degradative fate of TRIM5α remains unclear. However, as p62 mediates both proteasomal64,66 and autophagic degradation,67–69 it may play a role in dictating the degradative fate of TRIM5α.
The Emerging Relationship Between Capsid Recognition Domains and Viral Replication, Viral Loads, and Pathogenesis
Most studies examining the antiviral activity of TRIM5α proteins describe them as cross-species barriers to infection. This is not surprising, as the context in which TRIM5 proteins were first hypothesized and screened for involved observations stemming from the inability of certain retroviruses to infect cells from different species of primates. The inability of HIV-1 to infect cells of rhesus macaques and owl monkeys led to the identification of rhTRIM5α3 and TRIM-Cyp, respectively.4 As described above, the C-terminal capsid-binding domain appears to be the primary determinant affecting the spectrum of viruses that are susceptible or resistant to TRIM5 proteins from individual primate species. However, overexpression of huTRIM5α in human cells or the expression of huTRIM5α in cell lines that lack a TRIM5α gene consistently shows a modest but reproducible degree of HIV-1 restriction. Given this modest level of restriction exhibited by huTRIM5α against HIV-1, a number of studies examined the possibility that intraspecies genetic variation may modulate or even control replication of a virus endemic to that species.
Studies of human populations have yet to identify polymorphisms that exhibit increased antiviral activity, although some polymorphisms have been shown to result in a TRIM5α protein with reduced antiviral activity. For example, a single nucleotide polymorphism (SNP), H43Y, is present in a significant percentage of indigenous Central and South Americans and results in a TRIM5α protein with impaired ability to restrict N-MLV and HIV-1 in single-cycle infectivity assays.70,71 However, this polymorphism did not affect replication in primary T cell cultures.72 One report found that individuals homozygous for this allele exhibited accelerated disease progression.73 However, another study did not observe a statistically significant difference in viral set point in patients homozygous for this allele, although the trend seems visible despite a small number of individuals within the H43Y patient population analyzed.74 Similarly neutral observations have been made for other common SNPs present within the human population, including V112F, R136Q, E238W, G249D, and H419Y. Although some of these SNPs have, in individual studies, shown some small alteration of susceptibility to HIV-1 infection, these appear to be spurious, as other studies examining the same SNP have failed to confirm the small differences observed in some individual studies.70–74
Although numerous studies of TRIM5α polymorphisms in humans do not support the idea that alleles with increased ability to prevent HIV-1 infection or alter the course of disease exist, recent studies in rhesus macaques have found precisely the opposite phenomenon. A number of independent studies have examined polymorphisms in rhesus macaques and identified alleles that exhibit significant differences in their ability to restrict diverse retroviruses.75–78 One of these studies, by Lim and colleagues, observed significant differences in the ability of different TRIM5α alleles present in these macaques to inhibit infection by SIVmac239, a viral strain commonly used to infect rhesus macaques to induce an AIDS-like disease in studies of AIDS pathogenesis and vaccine strategies.79–81 Critically, not only did intraspecies allelic differences affect restriction of SIVmac239 in single cycle infectivity assays, these differences also conferred altered susceptibility to critical aspects of AIDS-like disease progression, including peak and set-point plasma RNA levels, CD4 T cell depletion, and survival.76 Another similar study by Kirmaier et al. found that many of the same rhTRIM5α alleles examined in the Lim study also differentially affected the replication of another commonly used laboratory strain, SIVsmE543-3.75 This SIV strain is derived from a strain of SIV isolated from sooty mangabeys and experimentally passaged through two rhesus macaques.82 It is also used to induce AIDS-like disease in rhesus macaques, although the degree of pathogenicity and viremia induced by this strain is highly variable.75,82 Similar to the Lim study, this study also observed that SIVsmE543-3 replication in rhesus macaques is modulated by individual TRIM5α alleles.75 By following the replication of virus in these animals over an extended period, the emergence of a TRIM5α-resistant strain of this virus in infected macaques that were originally able to control viral replication quite well was also noted.75 Taken together, these two studies demonstrate that allelic diversity of the TRIM5α gene has the potential to differentially modulate the replication of viruses that individuals within a species may encounter.
The very different results obtained from the examination of allelic diversity and the relative ability of these alleles to differentially regulate viral replication in humans and rhesus macaques is likely due to the nature of the polymorphisms present in each species. In humans, the majority of polymorphisms occur within the RBCC motif in regions of the protein that are not thought to be responsible for modulating TRIM5α specificity to individual retroviruses.70–72,74 Conversely, almost all of the nonsynonymous SNPs identified in rhesus macaques are located in the SPRY and CC domains,75–78 which are the regions of the protein reported to dictate the specificity of TRIM5α restriction (Fig. 2). It appears that these two very different outcomes are mediated by different evolutionary pressures applied to each species. Rhesus macaques appear to have maintained alleles with functional variability as a consequence of balancing selection during their evolution.77 Alternatively, humans appear to have undergone purifying selection of the TRIM5α locus, at least in the regions that govern retroviral specificity.70 A review examining evolution of the TRIM5α gene has been published for readers interested in a more thorough coverage of this topic.49
FIG. 2.
TRIM5α polymorphisms present in humans and rhesus macaques. A summary of the studies examining single nucleotide polymorphisms (SNPs) present in human and rhesus macaque TRIM5a genes. The majority of human SNPs are located within the RBCC motif of TRIM5a in regions not thought to govern retroviral specificity (SNPs above the TRIM5a domain structure). Conversely, rhesus macaque SNPs occur predominantly in those regions of the protein that dictate restriction specificity (SNPs below the TRIM5α domain structure).
If the polymorphisms present in the human population are not responsible for variation of AIDS progression in humans, what is the relevance of this phenomenon in rhesus macaques? Although it is true that functional polymorphisms within rhesus macaques are unlikely to have a direct impact on the generation of treatment options in humans, the functional variability of TRIM5α activity in rhesus macaques, or perhaps other primate species, must now be considered when designing and interpreting vaccine or pathogenesis studies in primates. Such studies are frequently hampered by a high degree of variability that confounds data interpretation or prevents results from obtaining statistical significance. Future studies may benefit greatly from careful selection of animals with consideration of the TRIM5α alleles they possess. When feasible, it may even be worthwhile to reexamine data from previous studies to understand the TRIM5α genotype of animals in the study, especially where a high degree of variability between animals in treatment groups was observed.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Bieniasz PD. Intrinsic immunity: A front-line defense against viral attack. Nat Immunol. 2004;5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- 2.Goff SP. Retrovirus restriction factors. Mol Cell. 2004;16:849–859. doi: 10.1016/j.molcel.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 4.Sayah DM, et al. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 5.Keckesova Z, et al. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc Natl Acad Sci USA. 2004;101:10780–10785. doi: 10.1073/pnas.0402474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perez-Caballero D, et al. Human tripartite motif 5alpha domains responsible for retrovirus restriction activity and specificity. J Virol. 2005;79:8969–8978. doi: 10.1128/JVI.79.14.8969-8978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song B, et al. Retrovirus restriction by TRIM5alpha variants from Old World and New World primates. J Virol. 2005;79:3930–3937. doi: 10.1128/JVI.79.7.3930-3937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pacheco B, et al. Species-specific inhibition of foamy viruses from South American monkeys by New World Monkey TRIM5{alpha} proteins. J Virol. 2010;84:4095–4099. doi: 10.1128/JVI.02631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennan G, et al. TRIMCyp expression in Old World primates Macaca nemestrina and Macaca fascicularis. Proc Natl Acad Sci USA. 2008;105:3569–3574. doi: 10.1073/pnas.0709511105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carthagena L, et al. Implication of TRIM alpha and TRIMCyp in interferon-induced anti-retroviral restriction activities. Retrovirology. 2008;5:59. doi: 10.1186/1742-4690-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz-Griffero F, et al. Requirements for capsid-binding and an effector function in TRIMCyp-mediated restriction of HIV-1. Virology. 2006;351:404–419. doi: 10.1016/j.virol.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 12.Liao CH, et al. A novel fusion gene, TRIM5-cyclophilin A in the pig-tailed macaque determines its susceptibility to HIV-1 infection. AIDS. 2007;21(Suppl 8):S19–S26. doi: 10.1097/01.aids.0000304692.09143.1b. [DOI] [PubMed] [Google Scholar]
- 13.Newman RM, et al. Evolution of a TRIM5-CypA splice isoform in Old World monkeys. PLoS Pathog. 2008;4:e1000003. doi: 10.1371/journal.ppat.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nisole S, et al. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc Natl Acad Sci USA. 2004;101:13324–13328. doi: 10.1073/pnas.0404640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Si Z, et al. Evolution of a cytoplasmic tripartite motif (TRIM) protein in cows that restricts retroviral infection. Proc Natl Acad Sci USA. 2006;103:7454–7459. doi: 10.1073/pnas.0600771103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ylinen LM, et al. Isolation of an active Lv1 gene from cattle indicates that tripartite motif protein-mediated innate immunity to retroviral infection is widespread among mammals. J Virol. 2006;80:7332–7338. doi: 10.1128/JVI.00516-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fletcher AJ, et al. Hare TRIM5alpha restricts divergent retroviruses and exhibits significant sequence variation from closely related lagomorpha TRIM5 genes. J Virol. 2010;84:12463–12468. doi: 10.1128/JVI.01514-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaller T, et al. An active TRIM5 protein in rabbits indicates a common antiviral ancestor for mammalian TRIM5 proteins. J Virol. 2007;81:11713–11721. doi: 10.1128/JVI.01468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawyer SL, et al. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song B, et al. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5alpha exhibits lineage-specific length and sequence variation in primates. J Virol. 2005;79:6111–6121. doi: 10.1128/JVI.79.10.6111-6121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nisole S, et al. TRIM family proteins: Retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 22.Reymond A, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamauchi K, et al. Ubiquitination of E3 ubiquitin ligase TRIM5 alpha and its potential role. FEBS J. 2008;275:1540–1555. doi: 10.1111/j.1742-4658.2008.06313.x. [DOI] [PubMed] [Google Scholar]
- 24.Diaz-Griffero F, et al. A B-box 2 surface patch important for TRIM5alpha self-association, capsid binding avidity, and retrovirus restriction. J Virol. 2009;83:10737–10751. doi: 10.1128/JVI.01307-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez-Caballero D, et al. Restriction of human immunodeficiency virus type 1 by TRIM-CypA occurs with rapid kinetics and independently of cytoplasmic bodies, ubiquitin, and proteasome activity. J Virol. 2005;79:15567–15572. doi: 10.1128/JVI.79.24.15567-15572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song B, et al. TRIM5alpha association with cytoplasmic bodies is not required for antiretroviral activity. Virology. 2005;343:201–211. doi: 10.1016/j.virol.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 27.Campbell EM, et al. Visualization of a proteasome-independent intermediate during restriction of HIV-1 by rhesus TRIM5alpha. J Cell Biol. 2008;180:549–561. doi: 10.1083/jcb.200706154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sastri J, et al. Identification of residues within the L2 region of TRIM5α that are required for retroviral restriction and cytoplasmic body localization. Virology. 2010;405:259–266. doi: 10.1016/j.virol.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Javanbakht H, et al. Characterization of TRIM5alpha trimerization and its contribution to human immunodeficiency virus capsid binding. Virology. 2006;353:234–246. doi: 10.1016/j.virol.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 30.Mische CC, et al. Retroviral restriction factor TRIM5alpha is a trimer. J Virol. 2005;79:14446–14450. doi: 10.1128/JVI.79.22.14446-14450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kar AK, et al. Biochemical and biophysical characterization of a chimeric TRIM21-TRIM5alpha protein. J Virol. 2008;82:11669–11681. doi: 10.1128/JVI.01559-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langelier CR, et al. Biochemical characterization of a recombinant TRIM5alpha protein that restricts human immunodeficiency virus type 1 replication. J Virol. 2008;82:11682–11694. doi: 10.1128/JVI.01562-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Javanbakht H, et al. The ability of multimerized cyclophilin A to restrict retrovirus infection. Virology. 2007;367:19–29. doi: 10.1016/j.virol.2007.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Javanbakht H, et al. The contribution of RING and B-box 2 domains to retroviral restriction mediated by monkey TRIM5alpha. J Biol Chem. 2005;280:26933–26940. doi: 10.1074/jbc.M502145200. [DOI] [PubMed] [Google Scholar]
- 35.Nepveu-Traversy ME, et al. TRIM5alpha and TRIMCyp form apparent hexamers and their multimeric state is not affected by exposure to restriction-sensitive viruses or by treatment with pharmacological inhibitors. Retrovirology. 2009;6:100. doi: 10.1186/1742-4690-6-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X. Sodroski J. The TRIM5alpha B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. J Virol. 2008;82:11495–11502. doi: 10.1128/JVI.01548-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X, et al. Functional interplay between the B-box 2 and the B30.2(SPRY) domains of TRIM5alpha. Virology. 2007;366:234–244. doi: 10.1016/j.virol.2007.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ganser-Pornillos BK, et al. Hexagonal assembly of a restricting TRIM5{alpha} protein. Proc Natl Acad Sci USA. 2011;108:534–539. doi: 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stremlau M, et al. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohkura S, et al. All three variable regions of the TRIM5alpha B30.2 domain can contribute to the specificity of retrovirus restriction. J Virol. 2006;80:8554–8565. doi: 10.1128/JVI.00688-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yap MW, et al. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr Biol. 2005;5:3–8. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 42.Virgen CA, et al. Independent genesis of chimeric TRIM5-cyclophilin proteins in two primate species. Proc Natl Acad Sci USA. 2008;105:3563–3568. doi: 10.1073/pnas.0709258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson SJ, et al. Independent evolution of an antiviral TRIMCyp in rhesus macaques. Proc Natl Acad Sci USA. 2008;105:3557–3562. doi: 10.1073/pnas.0709003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colgan J, et al. Binding of the human immunodeficiency virus type 1 Gag polyprotein to cyclophilin A is mediated by the central region of capsid and requires Gag dimerization. J Virol. 1996;70:4299–4310. doi: 10.1128/jvi.70.7.4299-4310.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Franke EK, et al. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 46.Forshey BM, et al. Structural requirements for recognition of the human immunodeficiency virus type 1 core during host restriction in owl monkey cells. J Virol. 2005;79:869–875. doi: 10.1128/JVI.79.2.869-875.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sebastian S. Luban J. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stremlau M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5{alpha} restriction factor. Proc Natl Acad Sci USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson WE. Sawyer SL. Molecular evolution of the antiretroviral TRIM5 gene. Immunogenetics. 2009;61:163–176. doi: 10.1007/s00251-009-0358-y. [DOI] [PubMed] [Google Scholar]
- 50.Ortiz M, et al. Evolutionary trajectories of primate genes involved in HIV pathogenesis. Mol Biol Evol. 2009;26:2865–2875. doi: 10.1093/molbev/msp197. [DOI] [PubMed] [Google Scholar]
- 51.Maillard PV, et al. The specificity of TRIM5 alpha-mediated restriction is influenced by its coiled-coil domain. J Virol. 2010;84:5790–5801. doi: 10.1128/JVI.02413-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Campbell EM, et al. TRIM5 alpha cytoplasmic bodies are highly dynamic structures. Mol Biol Cell. 2007;18:2102–2111. doi: 10.1091/mbc.E06-12-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dvorin JD. Malim MH. Intracellular trafficking of HIV-1 cores: Journey to the center of the cell. Curr Top Microbiol Immunol. 2003;281:179–208. doi: 10.1007/978-3-642-19012-4_5. [DOI] [PubMed] [Google Scholar]
- 54.Perron MJ, et al. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc Natl Acad Sci USA. 2004;101:11827–11832. doi: 10.1073/pnas.0403364101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diaz-Griffero F, et al. Comparative requirements for the restriction of retrovirus infection by TRIM5alpha and TRIMCyp. J Virol. 2007;8:32. doi: 10.1016/j.virol.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diaz-Griffero F, et al. Modulation of retroviral restriction and proteasome inhibitor-resistant turnover by changes in the TRIM5{alpha} B-box 2 Domain. J Virol. 2007;81:10362–10378. doi: 10.1128/JVI.00703-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chatterji U, et al. Trim5alpha accelerates degradation of cytosolic capsid associated with productive HIV-1 entry. J Biol Chem. 2006;281:37025–37033. doi: 10.1074/jbc.M606066200. [DOI] [PubMed] [Google Scholar]
- 58.Anderson JL, et al. Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J Virol. 2006;80:9754–9760. doi: 10.1128/JVI.01052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu X, et al. Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc Natl Acad Sci USA. 2006;103:7465–7470. doi: 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rold CJ. Aiken C. Proteasomal degradation of TRIM5alpha during retrovirus restriction. PLoS Pathog. 2008;4:e1000074. doi: 10.1371/journal.ppat.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Connor C, et al. p62/sequestosome1 associates with and sustains the expression of the retroviral restriction factor TRIM5{alpha} J Virol. 2010;84:5997–6006. doi: 10.1128/JVI.02412-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim JY. Ozato K. The sequestosome 1/p62 attenuates cytokine gene expression in activated macrophages by inhibiting IFN regulatory factor 8 and TNF receptor-associated factor 6/NF-kappaB activity. J Immunol. 2009;182:2131–2140. doi: 10.4049/jimmunol.0802755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moscat J, et al. Signal integration and diversification through the p62 scaffold protein. Trends Biochem Sci. 2007;32:95–100. doi: 10.1016/j.tibs.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 64.Seibenhener ML, et al. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hwang CY, et al. Hsp70 interacts with the retroviral restriction factor TRIM5alpha and assists the folding of TRIM5alpha. J Biol Chem. 2010;285:7827–7837. doi: 10.1074/jbc.M109.040618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wooten MW, et al. Essential role of sequestosome 1/p62 in regulating accumulation of Lys63-ubiquitinated proteins. J Biol Chem. 2008;283:6783–6789. doi: 10.1074/jbc.M709496200. [DOI] [PubMed] [Google Scholar]
- 67.Kim PK, et al. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci USA. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Komatsu M, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 69.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 70.Sawyer SL, et al. High-frequency persistence of an impaired allele of the retroviral defense gene TRIM5alpha in humans. Curr Biol. 2006;16:95–100. doi: 10.1016/j.cub.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 71.Javanbakht H, et al. Effects of human TRIM5alpha polymorphisms on antiretroviral function and susceptibility to human immunodeficiency virus infection. Virology. 2006;354:15–27. doi: 10.1016/j.virol.2006.06.031. [DOI] [PubMed] [Google Scholar]
- 72.Goldschmidt V, et al. Role of common human TRIM5alpha variants in HIV-1 disease progression. Retrovirology. 2006;3:54. doi: 10.1186/1742-4690-3-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Manen D, et al. The effect of Trim5 polymorphisms on the clinical course of HIV-1 infection. PLoS Pathog. 2008;4:e18. doi: 10.1371/journal.ppat.0040018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Speelmon EC, et al. Genetic association of the antiviral restriction factor TRIM5alpha with human immunodeficiency virus type 1 infection. J Virol. 2006;80:2463–2471. doi: 10.1128/JVI.80.5.2463-2471.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kirmaier A, et al. TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lim SY, et al. TRIM5alpha modulates immunodeficiency virus control in rhesus monkeys. PLoS Pathog. 2010;6:e1000738. doi: 10.1371/journal.ppat.1000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Newman RM, et al. Balancing selection and the evolution of functional polymorphism in Old World monkey TRIM5alpha. Proc Natl Acad Sci USA. 2006;103:19134–19139. doi: 10.1073/pnas.0605838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wilson SJ, et al. Rhesus macaque TRIM5 alleles have divergent antiretroviral specificities. J Virol. 2008;82:7243–7247. doi: 10.1128/JVI.00307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ourmanov I, et al. Recombinant modified vaccinia virus ankara expressing the surface gp120 of simian immunodeficiency virus (SIV) primes for a rapid neutralizing antibody response to SIV infection in macaques. J Virol. 2000;74:2960–2965. doi: 10.1128/jvi.74.6.2960-2965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Polacino P, et al. Limited breadth of the protective immunity elicited by simian immunodeficiency virus SIVmne gp160 vaccines in a combination immunization regimen. J Virol. 1999;73:618–630. doi: 10.1128/jvi.73.1.618-630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yasutomi Y, et al. Simian immunodeficiency virus-specific CD8 + lymphocyte response in acutely infected rhesus monkeys. J Virol. 1993;67:1707–1711. doi: 10.1128/jvi.67.3.1707-1711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hirsch V, et al. A molecularly cloned, pathogenic, neutralization-resistant simian immunodeficiency virus, SIVsmE543-3. J Virol. 1997;71:1608–1620. doi: 10.1128/jvi.71.2.1608-1620.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]