Abstract
Crescentic glomerulonephritis (CRGN) is a major cause of rapidly progressive renal failure for which the underlying genetic basis is unknown. WKY rats show marked susceptibility to CRGN, while Lewis rats are resistant. Glomerular injury and crescent formation are macrophage-dependent and mainly explained by seven quantitative trait loci (Crgn1-7). Here, we used microarray analysis in basal and lipopolysaccharide (LPS)-stimulated macrophages to identify genes that reside on pathways predisposing WKY rats to CRGN. We detected 97 novel positional candidates for the uncharacterised Crgn3-7. We identified 10 additional secondary effector genes with profound differences in expression between the two strains (>5-fold change, <1% False Discovery Rate) for basal and LPS-stimulated macrophages. Moreover, we identified 8 genes with differentially expressed alternatively spliced isoforms, by using an in depth analysis at probe-level that allowed us to discard false positives due to polymorphisms between the two rat strains. Pathway analysis identified several common linked pathways, enriched for differentially expressed genes, which affect macrophage activation. In summary, our results identify distinct macrophage transcriptome profiles between two rat strains that differ in susceptibility to glomerulonephritis, provide novel positional candidates for Crgn3-7, and define groups of genes that play a significant role in differential regulation of macrophage activity.
Keywords: Crescentic glomerulonephritis, macrophage, microarray, rat
Introduction
Crescentic glomerulonephritis (CRGN) is a major cause of rapidly progressive renal failure for which the underlying genetic basis is largely unknown 1. The Wistar-Kyoto (WKY) rat shows marked susceptibility to CRGN, as administration of a small dose of nephrotoxic serum (NTS), that is subnephritogenic in other strains, leads to rapid onset of nephrotoxic nephritis (NTN). In WKY rats, NTN is characterised by the development of albuminuria by day 4, crescent formation in the majority of glomeruli by day 11 and progression to severe scarring, that is characteristic of CRGN, with renal failure by 6 weeks 2. Monocytes/macrophages and CD8+ cells are the major cell types in the glomerular infiltrate, detected as early as 2.5 hours after injection of NTS, and reaching a maximal number between days 4 and 8 2. The model is highly reproducible and the histology closely resembles that seen in human glomerulonephritis. In contrast Lewis (LEW) rats, which share the same MHC haplotype (RT1-l), develop only mild glomerular hypercellularity with no crescents after administration of the same dose of NTS 3.
In WKY rats, macrophage depletion studies 4 have shown that glomerular injury in CRGN is macrophage dependent. This suggests at least in part, shared pathophysiology with human CRGN, where macrophage accumulation correlates with the degree of histological and functional injury 5-7. Furthermore, bone marrow-derived macrophages (BMDMs) from WKY rats show a number of phenotypic differences compared with those from LEW rats including enhanced antibody-dependent cytotoxicity, Fc receptor-mediated phagocytosis and Fc-receptor-dependent oxidative burst, as well as increased inducible nitric oxide synthase gene (Nos2) expression upon lipopolysaccharide (LPS) stimulation 3,8. However, bone marrow and kidney transplantation studies between WKY and LEW rats show that, in addition to circulating cells, genetic susceptibility to CRGN is also partly dependent on intrinsic factors within the kidney itself 9,10.
In previous studies, we carried out a genome-wide linkage analysis of an F2 population derived from WKY and LEW rats and mapped seven CRGN quantitative trait loci (QTL) (Crgn1-7) linked to crescent formation, proteinuria or macrophage infiltration 3. The two most significant linkage peaks, with lod score over 8, were on chromosomes 13 (Crgn1) and 16 (Crgn2). By a combination of expression studies, fine mapping and functional assays, we identified deletion of an Fc receptor gene, Fcgr3-rs, as a cause of macrophage over-activity and CRGN susceptibility at Crgn1. Subsequently, we identified over-activity of the AP-1 transcription factor JunD as a cause of enhanced macrophage oxygen burst activity as well as iNOS synthesis and WKY susceptibility to CRGN at Crgn2 8. Recently we generated a double congenic rat strain where both Crgn1 and Crgn2 from NTN-resistant LEW rats were introgressed into the genetic background of the WKY rat 10. Our results show that Crgn1 and Crgn2 have an additive protective effect against glomerular crescent formation 10. However, this study also highlighted the effects of Crgn3-7, as bone marrow transplantation experiments showed that the double congenic line shows residual susceptibility to CRGN, which is mainly attributable to macrophage activation rather than macrophage number 10.
The aim of this study was to compare macrophage gene expression between the CRGN-susceptible WKY and CRGN-resistant LEW strains to identify genes that reside on effector pathways for macrophage-mediated damage to glomeruli in WKY rats. To achieve this aim we used rat exon arrays, in basal (unstimulated) and LPS-stimulated macrophages. Our results revealed extensive strain differences in macrophage gene expression, identified several differentially expressed genes that positionally map to Crgn3-7 (primary effector candidates), as well as a subset of genes, that although they map outside the known CRGN QTL regions, showed highly significant differential expression between WKY and LEW macrophages, before and after LPS stimulation. We also identified a small number of alternative splicing events that cause transcript isoform differences between macrophages from the two strains.
Results
Differential gene expression between WKY and LEW macrophages
Exon array analysis identified 800 transcripts that were differentially expressed between WKY and LEW bone marrow-derived macrophages, in the basal state, with a 5% false discovery rate (FDR) threshold (Supplementary Table S1). 52.9% of these transcripts showed higher expression in WKY compared to LEW. Notably, a subset of 19 genes showed very strong differential expression in basal conditions (>5 fold change, <1% FDR) (Table 1). Following LPS stimulation, 887 transcripts were differentially expressed (Supplementary Table S2), and 60.0% of these showed higher expression in WKY compared to LEW. There was an overlap of exactly 400 genes between the basal and LPS conditions, while 487 genes were differentially expressed between the two strains only after LPS stimulation. In addition, 15 genes showed very strong macrophage gene differential expression (>5 fold change, <1% FDR) between the two strains following LPS stimulation (Table 2). Eleven of these genes (Arg1, Fcgr3-rs, Grit, Igf2bp1, IMAGE:5598800, Lilrb3l, Ly49si1, Ly49si2, Mmp7, Orl1684 and Pirb) showed similarly strong differential expression in both the basal and LPS-stimulated state.
Table 1.
Microarray results of transcripts showing greater than 5-fold difference in expression between WKY and LEW, in basal macrophages, with <1% false discovery rate (FDR). Data are sorted according to fold change
| Affymetrix transcript identity |
Gene name | Gene Symbol | QTL position |
p-value (LEW vs. WKY basal) |
Fold- Change (LEW vs. WKY basal) |
|---|---|---|---|---|---|
| 7113955 | Fc receptor, IgG, low affinity III, related sequence | Fcgr3-rs | Crgn1 | 2.30E-07 | 52.4 |
| 7028278 | Leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3-like |
Lilrb3l | 6.91E-09 | 36.7 | |
| 7332712 | Matrix metallopeptidase 7 | Mmp7 | 4.34E-06 | 12.1 | |
| 7216236 | Olfactory receptor 1684 | Olr1684 | 1.64E-07 | 12.0 | |
| 7047485 | Paired-Ig-like receptor B | Pirb | 3.49E-09 | 11.6 | |
| 7332774 | Transient receptor potential cation channel, subfamily C, member 6 | Trpc6 | 4.35E-05 | 11.2 | |
| 7317031 | Nephroblastoma overexpressed gene | Nov | 4.46E-09 | 10.2 | |
| 7367852 | Rattus norvegicus cDNA clone IMAGE:5598800 5-, mRNA sequence | IMAGE:5598800 | 1.97E-05 | 7.9 | |
| 7147913 | Ligase IV, DNA, ATP-dependent | Lig4 | 2.31E-07 | 6.0 | |
| 7080736 | Chemokine (C-C motif) ligand 9 | Ccl9 | 1.04E-05 | 5.4 | |
| 7294643 | Pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 | Plekhh2 | 3.56E-06 | −5.0 | |
| 7037483 | HtrA serine peptidase 1 | Htra1 | 3.28E-06 | −5.7 | |
| 7270146 | Immunoreceptor Ly49si2 | Ly49si2 | 3.14E-06 | −5.8 | |
| 7334400 | Rho GTPase-activating protein | Grit/Rics | 1.39E-07 | −6.8 | |
| 7025757 | Arginase 1, liver | Arg1 | 1.54E-05 | −6.9 | |
| 7080663 | Schlafen 8 | Slfn8 | 4.64E-06 | −8.3 | |
| 7374731 | Carbonic anhydrase VB, mitochondrial | Ca5b | 1.30E-07 | −10.0 | |
| 7081895 | Insulin-like growth factor 2 mRNA binding protein 1 | Igf2bp1 | 1.81E-08 | −11.3 | |
| 7270138 | Immunoreceptor Ly49si1 | Ly49si1 | 5.04E-08 | −12.8 |
p-value - uncorrected ANOVA p-value; Fold-change - positive values show genes down-regulated in WKY, while negative values show genes up-regulated in WKY compared to LEW macrophages
Table 2.
Microarray results of transcripts showing greater than 5-fold difference in expression between WKY and LEW, in LPS-stimulated macrophages, with <1% false discovery rate (FDR). Data are sorted according to fold change
| Affymetrix transcript identity |
Gene name | Gene Symbol | QTL position |
p-value (LEW vs. WKY LPS) |
Fold- Change (LEW vs. WKY LPS) |
|---|---|---|---|---|---|
| 7113955 | Fc receptor, IgG, low affinity III, related sequence | Fcgr3-rs | Crgn1 | 1.35E-07 | 40.58 |
| 7028278 | Leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3-like | Lilrb3l | 2.44E-09 | 37.47 | |
| 7332712 | Matrix metallopeptidase 7 | Mmp7 | 2.04E-07 | 29.46 | |
| 7216236 | Olfactory receptor 1684 | Olr1684 | 2.92E-07 | 7.22 | |
| 7047485 | Paired-Ig-like receptor B | Pirb | 9.15E-09 | 6.33 | |
| 7367852 | Rattus norvegicus cDNA clone IMAGE:5598800 5-, mRNA sequence | IMAGE:5598800 | 2.01E-05 | 5.97 | |
| 7080742 | Chemokine (C-C motif) ligand 6 | Ccl6 | 1.88E-07 | 5.52 | |
| 7361223 | Similar to putative protein (5S487) | RGD1310819 | 2.34E-07 | 5.31 | |
| 7334400 | Rho GTPase-activating protein | Grit/Rics | 1.23E-07 | −5.39 | |
| 7270146 | Immunoreceptor Ly49si2 | Ly49si2 | 1.60E-06 | −5.40 | |
| 7238766 | Rho family GTPase 3 | Rnd3 | 8.74E-07 | −5.84 | |
| 7132879 | Stathmin-like 4 | Stmn4 | Crgn7 | 1.58E-07 | −7.85 |
| 7270138 | Immunoreceptor Ly49si1 | Ly49si1 | 5.03E-08 | −9.07 | |
| 7025757 | Arginase 1, liver | Arg1 | 7.09E-07 | −14.09 | |
| 7081895 | Insulin-like growth factor 2 mRNA binding protein 1 | Igf2bp1 | 5.65E-10 | −31.52 |
p-value - uncorrected ANOVA p-value; LPS - lipopolysaccharide; Fold-change - positive values show genes down-regulated in WKY, while negative values show genes up-regulated in WKY compared to LEW macrophages
Validation of differentially expressed genes
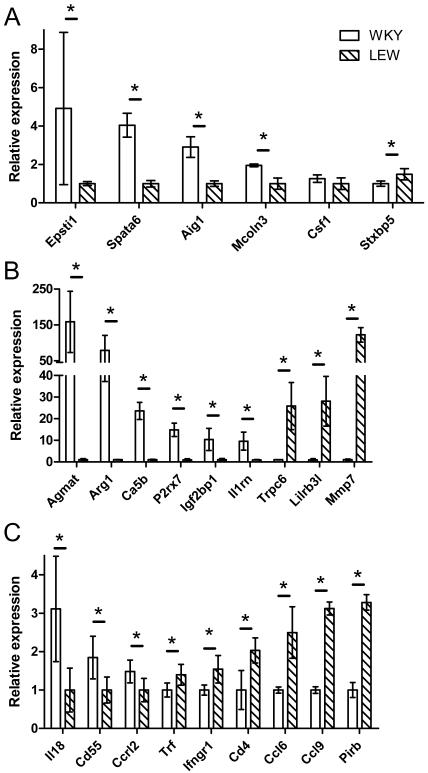
Sixty-one of the genes showing differential expression (<5% FDR) between WKY and LEW in basal macrophages and 68 genes showing differential expression in LPS-stimulated macrophages mapped within the 1.5-LOD support interval of our previously localised crescentic glomerulonephritis susceptibility QTLs (Crgn3-7) 3 (Table 3). Because these genes are differentially expressed and map to a known QTL, they can be considered as candidate genes for Crgn3-7 that may confer primary susceptibility to CRGN. Thirty-two of these genes showed similarly strong differential expression in both the basal and LPS-stimulated state. To test the accuracy of the microarray results, we randomly selected six genes (Aig1, Csf1, Epsti1, Mcoln3, Spata6 and Stxbp5) and carried out quantitative real-time PCR (qPCR) on these six differentially expressed transcripts (Figure 1A). With the exception of Csf1, the differences in gene expression shown by the microarray were confirmed for all of these genes.
Table 3.
Candidate genes for crescentic glomerulonephritis (CRGN) quantitative trait loci
| QTL | Chr | Markers | Position (Mb) | Phenotypes | Candidate Genes Basal macrophages |
Candidate Genes LPS stimulated macrophages |
|---|---|---|---|---|---|---|
| Crgn3 | 1 | D1Rat246 - D1Rat4 | 2.4 - 11.86 | Proteinuria | RGD1309759, Map3k7ip2, Stxbp5, Pex3, Aig1, Gpr126 |
Map3k7ip2, Stxbp5, RGD1564259, Pex3, Aig1, Gpr126 |
|
| ||||||
| Crgn4 | 2 | D2Rat234-D2Rat70 | 201.2 - 254.9 | Crescents, Proteinuria |
Tmem77, Csf1, Gstm1, Slc25a24, Ptbp2, Alpk1, Manba, Gbp2, Clca4, Mcoln3 |
Tmem77, Kcna3, Csf1, Sars, Slc25a24, Slc44a3, Bcar3, Camk2d, LOC691931, Manba, Ppp3ca, Eif4e, Mcoln3, Mcoln2 |
|
| ||||||
| Crgn5 | 5 | D5Rat22 - D5Rat33 | 102 - 143.5 | Crescents |
Adfp, Cdkn2b, Cyp2j4, Atg4c, Jak1, Zcchc11, Kti12, Eps15, Spata6, Cmpk1, Mmachc, Atp6v0b, Ipo13, Med8, RGD1309802, Zmpste24, Mycl1 |
RGD1305797, Cdkn2b, Cyp2j4, Atg4c, Jak1, Ak3l1, Zfyve9, Btf3l4, Kti12, Osbpl9, Spata6, Toe1, Plk3, Atp6v0b, Ipo13, RGD1309802, Cap1 |
|
| ||||||
| Crgn6 | 10 | D10Rat186-D10Rat45 | 0 - 20.2 | Proteinuria | RGD1563547, Mgrn1, Zfp263, Thoc6, LOC100158225, Tsc2, Clcn7, RGD1307381, Tmem8, Dusp1 |
Abcc1, Rrn3, RGD1306568, Emp2, Crebbp, Tmem204, RGD1307381, Tmem8, RGD1311343, Dusp1 |
|
| ||||||
| Crgn7 | 15 | D15Rat54 - D15Rat96 | 14.1 - 71.5 | Proteinuria |
Flnb, Lrp10, Slc7a8, Zfhx2, Irf9, Kpna3, Mtmr9, Ephx2, Ptk2b, Trim35, Stmn4, Hr, Htr2a, Cog3, Slc25a30, Epsti1, Elf1, Pcdh8 |
Flnb, Wdhd1, Slc7a7, Acin1, Slc7a8, Ripk3, Spata13, Kpna3, Mtmr9, Ccdc25, Ephx2, Trim35, Stmn4, Dpysl2, Hr, Dok2, Lcp1, Slc25a30, Nufip1, Epsti1, Diap3 |
Genes highlighted in bold were tested by quantitative PCR. Chr - Chromosome
Figure 1. Validation of microarray data with quantitative PCR of selected differentially expressed genes.
A) positional differentially expressed QTL candidate genes; B and C) secondary effector genes. Relative gene expression was compared to Hprt for WKY and LEW basal macrophages. The secondary effector genes are presented in two separate graphs (B, C) according to scale of relative gene expression. All samples were amplified using an independent set of biological duplicates with three technical replicates per sample. * = p < 0.05 using a Mann-Whitney non-parametric test (one-tailed). Error bars represent standard deviation.
In addition to the sixty-one genes that map to CRGN QTL regions, a large number of genes (739 transcripts) that were differentially expressed (<5% FDR) between WKY and LEW basal macrophages, do not map to previously localised CRGN QTLs. Although these genes are not primary positional candidates for the Crgn3-7 QTLs, they may contribute indirectly to the effector pathways that lead to WKY susceptibility to CRGN. To separate these genes from the Crgn3-7 positional candidates, we refer to these 739 transcripts as “secondary effector” genes. To validate these results, we selected a set of 18 of these genes for analysis by qPCR. Eight of the 18 genes (Arg1, Ca5b, Ccl9, Igf2bp1, Lilrb3l, Mmp7, Pirb and Trpc6) were selected because they were amongst the 19 genes that show profound differences in expression (>5 fold change, <1% FDR) between WKY and LEW basal macrophages, while the remaining ten (Agmat, Ccl6, Ccrl2, Cd4, Cd55, Ifngr1, Il18, Il1rn, P2rx7, and Trf) were selected based on literature searches, as having known or suspected links to glomerulonephritis. Consistent with the microarray results, qPCR showed that all eighteen genes were significantly differentially expressed between WKY and LEW macrophages and matched the directional change of the microarray data (Figure 1 B and C). Two of the genes Agmat and Arg1 showed more than 50 fold greater expression by qPCR analysis in WKY compared to LEW. Conversely, three genes Mmp7, Lilrb3l and Trpc6 were expressed at over 20-fold higher levels in LEW compared to WKY macrophages. To ensure that the strong differential expression detected by qPCR for Agmat, Arg1 and Mmp7 (the three most strongly differentially expressed genes) is not an artefact due to genomic sequence variation, we sequenced the qPCR primer binding regions by capillary sequencing and found no SNPs in the WKY or LEW rat strains that were able to affect the binding properties of the primers used for qPCR amplification. Overall, therefore, we successfully validated 23 out of the 24 transcripts selected on the basis that they either i) map to Crgn3-7, ii) showed marked inter-strain differential expression, or iii) have previously reported links to glomerular inflammation.
Testing for influence of Crgn1 and Crgn2 on selected gene expression
To establish whether differential expression of these genes was under the genetic control of our previously identified QTL genes at Crgn1 and Crgn2, we measured the expression of the 23 validated selected genes in a panel of congenic strains. The panel of congenic strains consisted of: congenic rats WKY.LCrgn1 and WKY.LCrgn2, in which we previously introgressed the Crgn1 or Crgn2 QTL region from the resistant LEW strain into the WKY genetic background 8,10; a reciprocal congenic strain on the Lewis background for Crgn1 (LEW.WCrgn1); and a double congenic strain (WKY.LCrgn1,2) where both Crgn loci from the resistant LEW strain were introgressed into the WKY genetic background. The expression levels in the macrophages of the congenic animals matched those of the genetic background parental strain for all genes, indicating that they are not influenced in trans by genes encoded at Crgn1 and/or Crgn2 (Figure 2). Most significantly our data shows that none of the five validated positional candidate genes at Crgn3-7 are influenced by Crgn1 or Crgn2.
Figure 2. Selected gene expression in a panel of congenic strains.
Relative gene expression compared to Hprt for WKY, LEW, WKY.LCrgn1, LEW.WCrgn1, WKY.LCrgn2, and WKY.LCrgn1,2 in basal macrophages. The sample order shown in the top row of graphs is maintained for all subsequent graphs. All samples were amplified using an independent set of biological duplicates with three technical replicates per sample. * = p < 0.05 **P < 0.01, ***P < 0.001 statistically significantly different to WKY, using a one-way ANOVA with Bonferonni’s correction. Error bars represent standard deviation.
Pathway and functional annotation analysis
To characterise functionally the sets of genes that were differentially expressed between WKY and LEW macrophages, we performed pathway and functional annotation analysis using the DAVID Bioinformatics Resources 11,12. We used the lists of 800 and 887 genes that were differentially expressed between WKY and LEW macrophages before and after LPS stimulation respectively (<5% FDR) and found five KEGG (Kyoto Encyclopedia of Genes and Genomes) or PANTHER (Protein ANalysis THrough Evolutionary Relationships) pathways that were enriched (EASE score < 0.01) in basal macrophages and four pathways that were enriched in LPS-stimulated macrophages (Table 4). These results suggest that differential gene expression between the two strains may be functionally related to altered endocytosis, regulation of the actin cytoskeleton, leukocyte transendothelial migration and Fc gamma receptor mediated phagocytosis. Furthermore, we identified 33 Biological Process annotation terms, that were significantly enriched (Benjamini correction < 0.05) in basal macrophages and five Biological Process annotation terms, that were significantly enriched in LPS-stimulated macrophages (Table 5). These results provide strong evidence that the differences in gene expression between WKY and LEW (before and after LPS stimulation) are likely to impact on regulation of macrophage activation, as the top functional annotation terms, in both basal and LPS-stimulated macrophages, are cell activation and leukocyte or lymphocyte activation.
Table 4.
Canonical pathways enriched for genes differentially expressed between Lewis and WKY macrophages, with EASE score values less than 0.01
| Pathway database |
Pathway | Genes (n) |
p-value | % | Differentially expressed genes |
|---|---|---|---|---|---|
| Basal macrophages | |||||
| KEGG | rno04144:Endocytosis | 22 | 6.89E-04 | 2.9 |
Igf1r, Vps37c, Pip5k1b, Tfrc, Ap2m1, Git2, Cxcr4, Kit,Fam125a,Psd3, Grk6, Chmp1b, Pld1, RT1-CE10, Itch, Met, Agap3, Ret, Eps15, Erbb3, Ldlr, Rab31 |
| KEGG | rno04640:Hematopoietic cell lineage |
12 | 1.25E-03 | 1.6 |
Anpep, Tfrc, Cd55, Kit, Cd14, Itga1, Csf1, H2-Ea, Cd24, Cd44, Cd8b, Cd4 |
| KEGG | rno04810:Regulation of actin cytoskeleton |
20 | 5.99E-03 | 2.6 |
Itgal, Pip5k1b, Myh10, Baiap2, Ssh1, Arpc1b, Arpc3, Pxn, Enah, Cd14, Pdgfc, Itga1, Pak6, Slc9a1, Pik3cd, Pik3cg, Scin, Itgb8, Rac2, Arhgef12 |
| KEGG | rno04670:Leukocyte transendothelial migration |
13 | 9.65E-03 | 1.7 |
Itgal, Sipa1, Pxn, Cxcr4, Rassf5, Ptk2b, Ocln, Pik3cd, Pik3cg, Rac2, Icam1, Esam, Cybb |
| KEGG | rno04666:Fc gamma R-mediated phagocytosis |
11 | 9.90E-03 | 1.4 |
Pip5k1b, Arpc1b, Arpc3, Pla2g4a, Fcgr2b III Fcgr3 III Fcgr3-rs, Prkcd, Pld1, Pik3cd, Pik3cg, Scin, Rac2 |
|
| |||||
| LPS-stimulated macrophages | |||||
| KEGG | rno04666:Fc gamma R-mediated phagocytosis |
15 | 1.98E-04 | 1.8 |
Pak1, Pip5k1b, Arpc1b, Pla2g4a, Fcgr3-rs, Prkcd, Syk, Ppap2a, Myo10, Pld1, Gsn, Pik3cg, Asap1, Pla2g6, Was |
| KEGG | rno04144:Endocytosis | 22 | 2.20E-03 | 2.6 |
Igf1r, Vps37c, Adrbk1, Pip5k1b, Arrb2, Cblb, Tfrc, Git2, Grk4, Fam125a, Chmp1b, Dab2, Pld1, RT1-N2, Psd4, Ehd4, Ret, Erbb3, Mdm2, Asap1, Rab31, Sh3kbp1 |
| PANTHER |
P00016:Cytoskeletal regulation by Rho GTPase |
15 | 5.57E-03 | 1.8 |
Pak1, Myh10, Cdgap, Ssh1, Arpc1b, Stmn4, Diaph3, Tubb6, Diaph1, Tubb5, Pak6, Rics, Arhgef12, Was, Diaph2 |
| KEGG | rno04810:Regulation of actin cytoskeleton |
21 | 7.50E-03 | 2.5 |
Pak1, Actn4, Pip5k1b, Myh10, LOC685269, Ssh1, Arpc1b, Pxn, Vcl, Diaph3, Fgd3, Diaph1, Gsn, Pak6, Gng12, Slc9a1, Pik3cg, Golga4, Arhgef12, Was, Diaph2 |
p value - modified Fisher’s exact test (EASE score); % - percentage (differentially expressed genes/total genes in pathway) involved genes
Table 5.
Enrichment of functional annotation terms using DAVID for genes differentially expressed between Lewis and WKY macrophages, with values less than 0.05 after Benjamini multiple testing correction
| BP_FAT GO Term | Genes (n) | p-value | Benjamini |
|---|---|---|---|
| Basal macrophages | |||
| GO:0001775~cell activation | 29 | 6.13E-07 | 1.80E-03 |
| GO:0045321~leukocyte activation | 26 | 1.89E-06 | 2.78E-03 |
| GO:0002250~adaptive immune response | 14 | 2.07E-06 | 2.03E-03 |
| GO:0002460~adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains |
14 | 2.07E-06 | 2.03E-03 |
| GO:0046649~lymphocyte activation | 22 | 4.66E-06 | 3.43E-03 |
| GO:0010033~response to organic substance | 67 | 5.20E-06 | 3.06E-03 |
| GO:0006955~immune response | 38 | 6.04E-06 | 2.96E-03 |
| GO:0043067~regulation of programmed cell death | 52 | 2.13E-05 | 8.93E-03 |
| GO:0010941~regulation of cell death | 52 | 2.40E-05 | 8.78E-03 |
| GO:0002684~positive regulation of immune system process | 25 | 2.63E-05 | 8.56E-03 |
| GO:0002443~leukocyte mediated immunity | 13 | 4.64E-05 | 1.36E-02 |
| GO:0051338~regulation of transferase activity | 28 | 9.21E-05 | 2.44E-02 |
| GO:0001666~response to hypoxia | 21 | 1.03E-04 | 2.50E-02 |
| GO:0043549~regulation of kinase activity | 27 | 1.05E-04 | 2.35E-02 |
| GO:0030155~regulation of cell adhesion | 16 | 1.06E-04 | 2.20E-02 |
| GO:0045859~regulation of protein kinase activity | 26 | 1.19E-04 | 2.31E-02 |
| GO:0042981~regulation of apoptosis | 49 | 1.22E-04 | 2.21E-02 |
| GO:0001816~cytokine production | 10 | 1.39E-04 | 2.37E-02 |
| GO:0048871~multicellular organismal homeostasis | 13 | 1.44E-04 | 2.32E-02 |
| GO:0042127~regulation of cell proliferation | 48 | 1.55E-04 | 2.38E-02 |
| GO:0002449~lymphocyte mediated immunity | 11 | 1.61E-04 | 2.34E-02 |
| GO:0042110~T cell activation | 15 | 1.79E-04 | 2.48E-02 |
| GO:0051174~regulation of phosphorus metabolic process | 34 | 1.85E-04 | 2.44E-02 |
| GO:0019220~regulation of phosphate metabolic process | 34 | 1.85E-04 | 2.44E-02 |
| GO:0050778~positive regulation of immune response | 17 | 2.00E-04 | 2.52E-02 |
| GO:0070482~response to oxygen levels | 21 | 2.37E-04 | 2.86E-02 |
| GO:0009611~response to wounding | 34 | 2.78E-04 | 3.22E-02 |
| GO:0051347~positive regulation of transferase activity | 21 | 3.27E-04 | 3.64E-02 |
| GO:0002252~immune effector process | 15 | 4.03E-04 | 4.30E-02 |
| GO:0042592~homeostatic process | 49 | 4.25E-04 | 4.37E-02 |
| GO:0042325~regulation of phosphorylation | 32 | 4.42E-04 | 4.39E-02 |
| GO:0033674~positive regulation of kinase activity | 20 | 5.00E-04 | 4.79E-02 |
| GO:0048584~positive regulation of response to stimulus | 22 | 5.27E-04 | 4.89E-02 |
| LPS stimulated macrophages | |||
| GO:0001775~cell activation | 30 | 2.01E-06 | 5.87E-03 |
| GO:0046649~lymphocyte activation | 23 | 8.36E-06 | 1.21E-02 |
| GO:0045321~leukocyte activation | 26 | 1.46E-05 | 1.41E-02 |
| GO:0048872~homeostasis of number of cells | 17 | 2.15E-05 | 1.56E-02 |
| GO:0006955~immune response | 38 | 7.69E-05 | 4.40E-02 |
BP_FAT - subset of Biological Process Gene Ontology (GO) terms, generated by the DAVID team. This subset is missing the broadest GO terms for ease of data interpretation; p value - modified Fisher’s exact test (EASE score); Benjamini - Benjamini multiple testing correction value.
Alternative splicing analysis between WKY and LEW macrophages
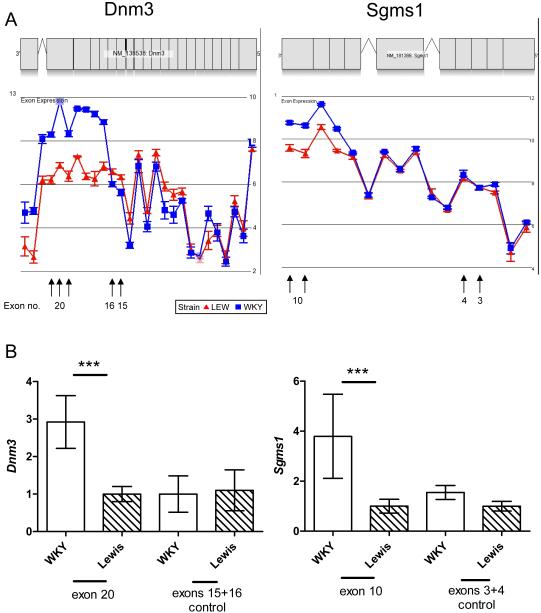
Affymetrix exon arrays allow the detection of splicing differences in two groups of data. We initially used the intersect of three commonly used alternative splicing detection algorithms (MiDAS, Rank Product of Slice Index and Alternative Splice ANOVA) and found 154 and 94 transcripts with isoform differences between WKY and LEW for basal or LPS-stimulated macrophages, respectively. However, capillary sequencing of the probe-set regions of 7 selected transcripts that showed evidence of alternative exon use (Akr7a2, Coq10b, Cxcl10, Pa2g4, Pttg1ip, Reps2 and Slfn3), revealed that all 7 selected transcripts contained SNPs within the probe-binding sequence of the exon that was predicted to be alternatively spliced (Supplementary Table S3). This suggested that the set of transcripts showing apparent alternative splicing contained a high number of false positives, due to SNPs affecting the hybridisation properties of the corresponding probe-sets. We therefore carried out a more in depth analysis of the data at probe level, permitting us to remove SNP containing probe-sets, by using a filter that marks probe-sets as false positives when not all four probes within a probe-set are differentially expressed between the two rat strains. Our revised analysis identified a much smaller number of potentially alternatively spliced genes. Four transcripts (Dnm3, Hes1, Ndrg4 and Rtel1) showed isoform differences between WKY and LEW macrophages in the basal state, and four transcripts (Mical2, Polr1a, Sgms1 and Ube3a) showed isoform differences following LPS stimulation (Table 6). None of these transcripts show a single internal exon that is included or excluded in a strain specific pattern. Rather, the isoform differences that we detect concern differential expression of probe-sets mapping to the 5′ or 3′ UTR of a gene, or multiple consecutive exons of a transcript. To validate these data, we selected two transcripts (Dmn3 and Sgms1), sequenced their differentially expressed exons and confirmed that they did not contain any SNPs. Next we used qPCR to test whether specific isoforms of the two selected transcripts are differentially expressed between WKY and LEW macrophages. We used pairs of primers to amplify the exons showing differential expression between strains (Dnm3 exon 20, and Sgms1 exon 10) and compared the results to a second control pair of primers designed to amplify exons showing similar expression levels between the two strains (Dnm3 exons 15-16 and Sgms1 exons 3-4). Our data confirms the microarray data predictions for i) Dnm3 where differential expression between strains was found for exon 20, while no differential expression was detected for exons 15 and 16; and ii) Sgms1 where differential expression between strains was found for exon 10, while no differential expression was detected for exons 3 and 4 (Figure 3). In summary, although our analysis has revealed a high number of polymorphisms between the two strains that account for the majority of transcripts showing apparent splicing differences, we also identified a small number of alternatively spliced transcripts that are differentially expressed between WKY and LEW macrophages.
Table 6.
Microarray results of transcripts showing showing isoform differences between WKY and LEW, in basal or LPS-stimulated macrophages
| Gene Symbol | Ensembl ID | Transcript ID |
Probe-set ID |
p-value | Fold-Change (LEW vs. WKY) |
Differential exon |
|---|---|---|---|---|---|---|
| Basal macrophages | ||||||
| Hes1 | ENSRNOT00000002346 | 7094386 | 6372610 | 1.52E-03 | 1.61 | 1 |
| Dnm3 | ENSRNOT00000067653 | 7113169 | 6694062 | 1.69E-04 | −7.83 | 18 |
| Dnm3 | ENSRNOT00000067653 | 7113169 | 6589493 | 1.98E-08 | −4.57 | 19 |
| Dnm3 | ENSRNOT00000067653 | 7113169 | 5968420 | 1.52E-06 | −8.40 | 19 |
| Dnm3 | ENSRNOT00000067653 | 7113169 | 5697034 | 4.25E-04 | −3.87 | 20 |
| Dnm3 | ENSRNOT00000067653 | 7113169 | 5799664 | 4.59E-06 | −7.75 | 20 |
| Ndrg4 | ENSRNOT00000017413 | 7182656 | 6239394 | 6.97E-04 | 2.94 | 11 |
| Ndrg4 | ENSRNOT00000017413 | 7182656 | 6202670 | 1.03E-03 | 4.75 | 13 |
| Rtel1 | ENSRNOT00000055030 | 7236395 | 5939980 | 2.49E-05 | 2.16 | novel 5′utr |
| Rtel1 | ENSRNOT00000055030 | 7236395 | 5780635 | 7.71E-04 | 2.96 | 1 |
| Rtel1 | ENSRNOT00000055030 | 7236395 | 6399147 | 7.43E-06 | 4.78 | 1 |
| Rtel1 | ENSRNOT00000055030 | 7236395 | 6522856 | 1.64E-04 | 2.33 | 2 |
| LPS macrophages | ||||||
| Ube3a | ENSRNOT00000021366 | 7032220 | 5976529 | 2.17E-04 | 2.30 | 11 |
| Ube3a | ENSRNOT00000021366 | 7032220 | 6269903 | 5.78E-05 | 12.24 | novel extention of 11 |
| Mical2 | ENSRNOT00000021858 | 7035525 | 6277897 | 2.99E-03 | −1.93 | 8 |
| Mical2 | ENSRNOT00000021858 | 7035525 | 5940132 | 5.82E-04 | −3.07 | 9 |
| Mical2 | ENSRNOT00000021858 | 7035525 | 6161609 | 2.91E-04 | −3.08 | novel 3′ UTR |
| Sgms1 | ENSRNOT00000054761 | 7061894 | 6030014 | 9.26E-06 | −2.03 | 8 |
| Sgms1 | ENSRNOT00000054761 | 7061894 | 5676967 | 1.03E-04 | −2.10 | 9 |
| Sgms1 | ENSRNOT00000054761 | 7061894 | 6269809 | 4.10E-04 | −2.25 | 10 |
| Sgms1 | ENSRNOT00000054761 | 7061894 | 6528432 | 7.29E-04 | −2.47 | 10 |
| Polr1a | ENSRNOT00000013587 | 7254811 | 6282280 | 4.42E-04 | −1.62 | 31 |
| Polr1a | ENSRNOT00000013587 | 7254811 | 6206810 | 3.05E-05 | −1.70 | 32 |
| Polr1a | ENSRNOT00000013587 | 7254811 | 6591259 | 3.45E-05 | −1.85 | 33 |
ID - identity; p-value - ANOVA p-value; Fold-change - positive values show exons down-regulated in WKY, while negative values show exons up-regulated in WKY compared to LEW macrophages; Differential Exon - Exon differentially expressed between strains. No other exons in each gene showed statistically significant differences between WKY and LEW.
Figure 3. Comparison of alternative splicing microarray data predictions with quantitative PCR of selected transcript isoforms.
A) Microarray data showing increased expression of probe-sets that correspond to Dnm3 exons 17-21, in WKY basal macrophages and increased expression of probe-sets that correspond to Sgms1 exons 8-10, for WKY LPS-stimulated macrophages. Data is illustrated as a gene view plot showing mean log2 signal probe-set intensities by strain. Error bars represent standard errors. B) qPCR data confirming the microarray predictions. Relative expression, compared to Hprt, for WKY and LEW basal macrophages of Dnm3 exon 20 and control exons 15-16, together with WKY and LEW LPS-stimulated macrophages of Sgms1 exon 10 and control exons 3-4. All samples were amplified using an independent set of biological duplicates with three technical replicates per sample. ***P < 0.001, using a one-way ANOVA with Bonferonni’s correction. Error bars represent standard deviation.
Discussion
The aim of this study was to identify differentially expressed macrophage genes that reside on effector pathways for macrophage-mediated damage to glomeruli following CRGN induction in WKY rats. To achieve this aim, we compared gene expression in basal and LPS-stimulated bone marrow-derived macrophages between CRGN-susceptible WKY and CRGN-resistant LEW inbred rat strains using rat exon arrays that provide extensive coverage of the rat genome. We used LPS stimulation because LPS, an outer membrane component of Gram-negative bacteria, is widely used as an in vitro stimulus that mirrors pro-inflammatory macrophage activation. Our results reveal extensive strain differences in macrophage gene expression, with several genes that reside within Crgn3-7 showing major expression differences, as well as a subset of genes that, although they map outside the known CRGN QTL regions, also show highly significant differential expression between WKY vs. LEW macrophages, before and after LPS stimulation.
We found 61 genes, differentially expressed between WKY and LEW in basal macrophages and 68 genes differentially expressed in LPS-stimulated macrophages, which map within our previously localised CRGN QTLs Crgn3-7. Most of these genes are novel candidates as they have not been previously identified to have a role in renal disease. We validated the microarray results by an independent assay, for five out of six selected genes (Aig1, Epsti1, Mcoln3, Spata6 and Stxbp5). Analysis in congenic strains showed that none of these five genes appear to be influenced by Crgn1 or Crgn2. Therefore, as these five genes both are differentially expressed and map to a CRGN susceptibility locus, they can be considered as attractive differentially expressed positional candidate genes for glomerulonephritis susceptibility encoded at Crgn3-7. The potential role of these genes, Aig1 (an androgen inducible gene), Epsti1 (an epithelial-stromal interaction protein), Mcoln3 (an inward rectifying cation channel), Spata6 (a spermatogenesis associated gene) and Stxbp5 (a syntaxin-binding protein implicated in the formation of complexes involved in neurotransmitter release), in macrophage function is currently unknown.
This study has identified a number of differentially expressed genes for Crgn3-7. However, it should be noted that one or more of these QTLs may account for genetic susceptibility mediated by intrinsic rather than circulating factors. In our recently published research aiming to dissect the contribution of Crgn3-7 to susceptibility to CRGN in the WKY rat, we performed bone marrow transplantation experiments between congenic and parental rats 10. Specifically, when Lewis bone marrow is transplanted to a WKY rat, this leads to an almost 90% protection in the glomerular crescent formation following glomerulonephritis induction. These results suggest that a residual 10 % of the CRGN susceptibility may be due to intrinsic cell activation in the glomerulus. As this current work focused on circulating cells, candidates for intrinsic factor CRGN susceptibility are not detectable via this screen.
We found 11 genes (Arg1, Fcgr3-rs, Grit, Igf2bp1, IMAGE:5598800, Lilrb3l, Ly49si1, Ly49si2, Mmp7, Orl1684 and Pirb) that showed profound differences in expression between the two strains (>5-fold change, <1% FDR), for both basal and LPS-stimulated macrophages. Aside from Fcgr3-rs, the remaining 10 genes do not map to previously identified CRGN QTL regions, although they show significant differential expression both between WKY and LEW macrophages in the basal state and after LPS stimulation. These 10 genes may therefore play a significant role in differential regulation of macrophage activation between WKY and LEW rats and may have a secondary effector role in mediating CRGN susceptibility. Of these 10 genes, Arg1, Mmp7 and Pirb have been identified in other studies as having potential roles in macrophage function or glomerulonephritis. Particularly, Arg1 competes for substrate with inducible nitric oxide synthase (iNOS) (encoded by Nos2) an enzyme that controls the production of nitric oxide (NO), and this can prevent NO-mediated apoptosis in activated macrophages 13,14. In addition to a primary role in macrophage function, Arg1 has been implicated in having a role in glomerulonephritis, as up-regulation of this gene has previously been shown to occur in macrophages and glomeruli isolated from rats following induction of acute immune complex glomerulonephritis 15-18. We found Arg1 to be markedly over-expressed in WKY compared to LEW basal macrophages. Mmp7 (also referred to as matrilysin) is a secreted protease, which is important for mediating proteolytic release of tumor nercrosis factor from macrophages 19 as well as the activation of anti-pathogenic and chemotactic proteins such as defensins (reviewed by Burke 20). Macrophage-produced Mmp-7 has also been shown to be necessary for macrophage infiltration 21. We found this gene to be highly under-expressed in WKY compared to LEW basal macrophages. Pirb (also known as Lilrb3) is a receptor expressed on immune cells where it binds to MHC class I molecules on antigen-presenting cells and transduces a negative signal that inhibits stimulation of an immune response (reviewed by Takai 22). We found this gene to be down-regulated in WKY compared to LEW, and studies from Pirb-deficient mice have shown that its absence in macrophages causes increased macrophage infiltration and affects integrin signaling 23.
Both of our previously identified Crgn1 and Crgn2 genes 3;8 were detected as significantly differentially expressed (<5% FDR) by this study. The molecular basis for Crgn1 is a deletion within Fcgr3-rs which abolishes the expression of this gene in WKY macrophages. Such disruptions in gene expression are easily detected by microarrays and Fcgr3-rs was found to be amongst the 11 genes that showed profound differences in expression between WKY and LEW (>5-fold change, <1% FDR), for both basal and LPS-stimulated macrophages. The molecular basis for Crgn2 is over-expression of JunD in WKY rats caused at least in part by a promoter polymorphism. This gene was detected by the exon microarray as having modest (1.42 fold) yet highly significant (p = 3.5e−4; < 5% FDR) differences in macrophages gene expression between the two strains. We find that the differences in expression for this gene are detected more easily when qPCR is used, where over 4 fold relative gene expression changes are detected between WKY and LEW basal macrophages 8. Compared to quantitative PCR based methods microarrays may show a compression of measured fold changes 24, which could have led to missing subtle differences between transcripts if the data was filtered according to fold change. However, our data analysis is not based on fold change filtering, but based on use of statistical tests (ANOVA) together with false-discovery rate correction.
Our previous work has showed that WKY macrophages are primed for an inflammatory response and show a number of phenotypic differences relating to macrophage activation compared with those from LEW rats, including enhanced antibody-dependent cytotoxicity, Fc receptor-mediated phagocytosis and Fc-receptor-dependent oxidative burst, as well as increased inducible nitric oxide synthase gene (Nos2) expression upon lipopolysaccharide (LPS) stimulation 3,8. Our functional annotation and pathway analysis supports these results, because the canonical pathways which associate with the inter-strain differential expression in basal and LPS-stimulated macrophages also relate to macrophage activation. For example, Fc receptor-mediated phagocytosis is widely used as one of the measures of macrophage activation. The canonical pathways which associate with the inter-strain differential expression in basal and LPS-stimulated macrophages are also connected. Specifically, phagocytosis is a specific form of endocytosis, while remodelling of the actin cytoskeleton is both an integral part of Fc gamma receptor-mediated phagocytosis 25, as well as being important for macrophage migration 26. Dysregulation of Fc gamma receptors is known to associate with glomerulonephritis susceptibility both in WKY rats and in humans (3,27,28). Therefore, some of the differentially expressed genes that associate with the above connected pathways may represent novel targets for therapeutic intervention in nephritis.
RNA splicing is an important mechanism for generating transcriptional diversity and regulating gene expression. We used alternative splicing analysis to identify a total of 8 genes with alternatively spliced isoforms differentially expressed between WKY and LEW, in basal (Dnm3, Hes1, Ndrg4 and Rtel1) or LPS-stimulated macrophages (Mical2, Polr1a, Sgms1 and Ube3a). None of these genes map to our previously localised CRGN QTLs and their role in macrophage function is currently unknown. However, Dnm3, a dynamin GTPase, has been implicated in phagocytosis 29, while Sgms1, a key component of lipid rafts, is involved in T cell receptor signalling and T cell activation 30. These genes may merit further investigation as regulators of activity in macrophages from the WKY or other rat strains. It is interesting that the inter-strain alternative splicing differences detected by this study were dependant on the activation state of the macrophages. Following our stringent alternative splicing detection analysis, the four differentially alternatively spliced transcripts in basal state did not show any inter-strain difference following the LPS treatment. This was also the case for the four differentially alternatively spliced transcripts in LPS-treated macrophages as the differences were no longer observed in basal conditions. Our results therefore suggest treatment-specific alternative splicing differences in rat BMDMs.
Our alternative splicing analysis also revealed that there are a high number of polymorphisms between the WKY and LEW strains, which affect the hybridisation properties of probe-sets and lead to the appearance in the microarray data of alternatively spliced transcripts. We resolved this difficulty by analysing the data at the probe, rather than the conventionally used probe-set level, together with a filter that marks probe-sets as false positives when not all four probes within a probe-set are differentially expressed between the two rat strains. The problem of SNPs resulting in the appearance of false positive alternative splicing events has been noticed before 31,32. In humans, data from the 1000 Genomes Project has allowed production of a list of all the SNP-containing probe-sets in the Affymetrix Gene Chip Human Exon 1.0 ST array, which can now be masked 33,33. Similarly, full sequencing of the WKY and LEW genomes will be needed to identify definitively and exclude all SNP-containing probe-sets from the rat exon array.
In summary our findings reveal distinct macrophage transcriptome profiles between two rat strains that differ in susceptibility to glomerulonephritis. We have detected novel positional candidate genes for the CRGN susceptibility loci Crgn3-7 and identified highly differentially expressed genes that may play a part in regulation of macrophage activity. Many of these genes have not previously been implicated in CRGN and represent novel candidate disease susceptibility genes and potential targets for therapeutic intervention in immunologically-mediated glomerulonephritis.
Materials and Methods
Animals
Wistar-Kyoto (WKY/NCrl, designated here as WKY) and Lewis (LEW/Crl, designated here as LEW) rats were purchased from Charles River (Margate, UK). All procedures were performed in accordance with the United Kingdom Animals (Scientific Procedures) Act.
Bone marrow collection and macrophage cell culture
Femurs from adult (8-10 weeks) WKY and Lewis rats were isolated and flushed with Hanks buffer (Invitrogen, Paisley, UK). Total BM-derived cells were plated and cultured for 6 days in DMEM (Invitrogen) containing 25 mM HEPES (Sigma, Dorset, UK), 25% L929 conditioned medium, 25% decomplemented FBS (Biosera, Ringmer, UK), penicillin (100 U/ml; Invitrogen), streptomycin (100 μg/ml; Invitrogen), and L-glutamine (2 mM; Invitrogen). These cells were characterized as macrophages by immunohistochemistry for CD68 (Supplementary Figure S1). BMDMs were cultured in serum/antibiotic-free HBSS (Invitrogen) for 16 hours. Next, cells were split into two groups of 1 × 106 cells each and cultured for a further 24 hours in the presence/absence of 10ng/ml lipopolysachharide (LPS) (Sigma). Cells were finally homogenized in TRIzol (Invitrogen) and stored at −70°C.
RNA Extraction, array hybridization and data analysis
Total RNA was extracted, labeled and hybridized to Gene Chip Rat Exon 1.0 ST Arrays (Affymetrix, Santa Clara, CA, USA) as previously described 34. Four BMDM preparations, each generated from a single animal (biological replicates), were used for each condition. The microarray data is available in MIAME-compliant (minimum information about a microarray experiment) format at the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) 35 under accession code E-MEXP-2624. CEL intensity files were produced using GeneChip Operating Software version 1.4 (Affymetrix) and quality tested using the Affymetrix Expression Console. All 16 files were deemed suitable for further study. Probe-level data were normalized using Robust Multi-array Average (RMA) 36. Detection of differential expression at the gene level was performed in Partek Genomics Suite 6.4 (Partek Incorporated, St.Louis, MO, USA) using transcripts with “extended” annotations. Data was summarized at the gene level using a One-Step Tukey’s Biweight Algorithm, which reduces the effect of outlier probe-sets (Statistical Algorithms Description Document; Affymetrix-White-Paper). An ANOVA model, using strain, condition and batch as co-factors, was used to generate raw p-values, while step-up FDR 37 was used for multiple test corrections.
Interval estimates of QTL location
To calculate 1.5-LOD support interval estimates, data from the original genome screen for NTN susceptibility loci in F2 rats 3 was re-analysed using the CRAN package R/qtlbim v1.9.4 38.
Functional Association Analysis
Lists of differentially expressed genes were uploaded into The Database for Annotation, Visualization and Integrated Discovery v6.7 (DAVID; http://david.abcc.ncifcrf.gov/) 11,12 to determine differentially regulated pathways using the full rat genome as reference background. Data were analyzed with the “Functional Annotation Chart” tool using KEGG and PANTHER pathways or the Biological Process (BP) Gene Ontology (GO) FAT set term. Cut off criteria used were either a Benjamini multiple testing correction p-value less than 0.05, or an EASE score p-value of less than 0.01. EASE score is a modified Fisher’s exact test 39, and it is the default test used by DAVID to identify statistical significance for functional association analysis.
Quantitative PCR
Primers used are listed in Supplementary Table S4. PCRs were performed with an ABI 7900 Sequence Detection System (Applied Biosystems, Warrington, UK). Macrophage quantitative real-time PCRs (qPCRs) were performed using a two-step protocol starting with cDNA synthesis using Cells-to-cDNA II (Applied Biosystems/Ambion, Austin, TX, USA), followed by PCR using SYBR Green Jumpstart Taq Ready mix (Sigma). A total of 10ng of cDNA per sample was used. After an initial denaturation step (94°C for 2min), samples were cycled 40 times at 94°C for 15 s, 60°C for 1 min and 72°C for 1min. All samples were amplified using an independent set of biological duplicates with three technical replicates per sample. The 7900 Fast system SDS software (Applied Biosystems) was used to obtain CT values. Results were analysed using the comparative CT method 40. Each sample was normalized to the reference gene Hprt, to account for cDNA loading differences.
Alternative Splicing Analysis
The Robust Multi-array Analysis (RMA) 36 algorithm was used for probe-set (exon-level) intensity analysis. Detection of alternative splicing events was performed using three parallel approaches: 1) Microarray Detection of Alternative Splicing (MiDAS) (Alternative Transcript Analysis Methods for Exon Arrays; Affymetrix-White-Paper) was calculated using the Bioconductor v2.4 41 package OneChannelGUI v1.4.5 42; 2) Rank Product of Splice Index 43 (100 permutations) was calculated also using OneChannelGUI v1.4.5 42; 3) An Alternative Splice ANOVA model was applied using Partek Genomics Suite v6.4 (Partek Incorporated) together with a filter to select for probe-sets showing significant alternative splicing score determined at a 5% False Discovery Rate (FDR) 37. Next, transcripts predicted to be alternatively spliced using an intersect of the above three approaches, were filtered to remove data in which apparent alternative use of exons was most likely due to single nucleotide polymorphisms (SNPs) within the probe-binding regions. For this filter, data was analysed at the probe level, as follows: Affymetrix perfect match (pm) probe intensities were measured using Affymetrix Power Tools version apt-1.12.0 (http://www.affymetrix.com/partners_programs/programs/developer/tools/powertools.affx#1_1). The data obtained were imported on to R statistical tools version 2.11.0 and subjected to quantiles normalization using the Bioconductor package preprocessCore version 1.8 44. The normalized probe intensities were mapped to the Affymetrix transcript annotation (NetAffx release 28). Probes were median summarized for the four biological replicates and the respective medians from both strains were subtracted and squared. The values obtained were subjected to the empirical fluctuation test based on moving sum estimate (MOSUM) implemented in R package strucchange version 1.4-0 45. P-values less than 0.1 suggested that a transcript contained SNPs and was a false positive for alternative splicing. Such transcripts were filtered out. Finally, all results were subjected to manual analysis using the Partek gene view plots as well as probe-level expression signal plots to remove false positives that had escaped the different analytical filtering steps.
Sequencing
WKY and LEW genomic DNA was isolated using a standard protocol. PCR was performed using KOD DNA polymerse (Merk Chemicals Ltd, Nottingham, UK) with primers listed in Supplementary Table S5. The amplified products were run on a 4% agarose gel. Bands were extracted and purified using a QIAquick Gel Extraction Kit (Qiagen, Crawley, UK). Samples were Cycle Sequenced using BigDye v3.1 (Applied Biosystems) chemistry with an ABI 3730 capillary- sequencer (Applied Biosystems). Alignment was performed in Sequencher v4.9 (Gene Codes Corporation, Ann Arbor, MI, USA).
Supplementary Material
Acknowledgements
We thank Kylie McDonald and Hetal Patel for technical support, and Dr Richard Hull for useful discussions. This work was supported by the 6th Framework Program of the European Union, EURATools (LSHG-CT-2005-019015), by an MRC intramural grant to Tim Aitman and by an Imperial College Junior Research Fellowship to Jacques Behmoaras.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on Genes and Immunity website
Reference List
- 1.Segelmark M, Hellmark T. Autoimmune kidney diseases. Autoimmun Rev. 2010;9:A366–A371. doi: 10.1016/j.autrev.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Tam FW, Smith J, Morel D, Karkar AM, Thompson EM, Cook HT, et al. Development of scarring and renal failure in a rat model of crescentic glomerulonephritis. Nephrol Dial Transplant. 1999;14:1658–66. doi: 10.1093/ndt/14.7.1658. [DOI] [PubMed] [Google Scholar]
- 3.Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, Smith J, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–5. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- 4.Isome M, Fujinaka H, Adhikary LP, Kovalenko P, El-Shemi AG, Yoshida Y, et al. Important role for macrophages in induction of crescentic anti-GBM glomerulonephritis in WKY rats. Nephrol Dial Transplant. 2004;19:2997–3004. doi: 10.1093/ndt/gfh558. [DOI] [PubMed] [Google Scholar]
- 5.Hooke DH, Hancock WW, Gee DC, Kraft N, Atkins RC. Monoclonal antibody analysis of glomerular hypercellularity in human glomerulonephritis. Clin Nephrol. 1984;22:163–8. [PubMed] [Google Scholar]
- 6.Hooke DH, Gee DC, Atkins RC. Leukocyte analysis using monoclonal antibodies in human glomerulonephritis. Kidney Int. 1987;31:964–72. doi: 10.1038/ki.1987.93. [DOI] [PubMed] [Google Scholar]
- 7.Markovic-Lipkovski J, Muller CA, Risler T, Bohle A, Muller GA. Association of glomerular and interstitial mononuclear leukocytes with different forms of glomerulonephritis. Nephrol Dial Transplant. 1990;5:10–7. doi: 10.1093/ndt/5.1.10. [DOI] [PubMed] [Google Scholar]
- 8.Behmoaras J, Bhangal G, Smith J, McDonald K, Mutch B, Lai PC, et al. Jund is a determinant of macrophage activation and is associated with glomerulonephritis susceptibility. Nat Genet. 2008;40:553–9. doi: 10.1038/ng.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith J, Lai PC, Behmoaras J, Roufosse C, Bhangal G, McDaid JP, et al. Genes expressed by both mesangial cells and bone marrow-derived cells underlie genetic susceptibility to crescentic glomerulonephritis in the rat. J Am Soc Nephrol. 2007;18:1816–23. doi: 10.1681/ASN.2006070733. [DOI] [PubMed] [Google Scholar]
- 10.Behmoaras J, Smith J, D’Souza Z, Bhangal G, Chawanasuntoropoj R, Tam FW, et al. Genetic Loci modulate macrophage activity and glomerular damage in experimental glomerulonephritis. J Am Soc Nephrol. 2010;21:1136–44. doi: 10.1681/ASN.2009090968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 12.Huang dW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 13.Mori M. Regulation of nitric oxide synthesis and apoptosis by arginase and arginine recycling. J Nutr. 2007;137:1616S–20S. doi: 10.1093/jn/137.6.1616S. [DOI] [PubMed] [Google Scholar]
- 14.Sonoki T, Nagasaki A, Gotoh T, Takiguchi M, Takeya M, Matsuzaki H, et al. Coinduction of nitric-oxide synthase and arginase I in cultured rat peritoneal macrophages and rat tissues in vivo by lipopolysaccharide. J Biol Chem. 1997;272:3689–93. doi: 10.1074/jbc.272.6.3689. [DOI] [PubMed] [Google Scholar]
- 15.Jansen A, Lewis S, Cattell V, Cook HT. Arginase is a major pathway of L-arginine metabolism in nephritic glomeruli. Kidney Int. 1992;42:1107–12. doi: 10.1038/ki.1992.394. [DOI] [PubMed] [Google Scholar]
- 16.Cook HT, Jansen A, Lewis S, Largen P, O’Donnell M, Reaveley D, et al. Arginine metabolism in experimental glomerulonephritis: interaction between nitric oxide synthase and arginase. Am J Physiol. 1994;267:F646–F653. doi: 10.1152/ajprenal.1994.267.4.F646. [DOI] [PubMed] [Google Scholar]
- 17.Waddington SN, Mosley K, Cook HT, Tam FW, Cattell V. Arginase AI is upregulated in acute immune complex-induced inflammation. Biochem Biophys Res Commun. 1998;247:84–7. doi: 10.1006/bbrc.1998.8755. [DOI] [PubMed] [Google Scholar]
- 18.Waddington SN, Tam FW, Cook HT, Cattell V. Arginase activity is modulated by IL-4 and HOArg in nephritic glomeruli and mesangial cells. Am J Physiol. 1998;274:F473–F480. doi: 10.1152/ajprenal.1998.274.3.F473. [DOI] [PubMed] [Google Scholar]
- 19.Gearing AJ, Beckett P, Christodoulou M, Churchill M, Clements JM, Crimmin M, et al. Matrix metalloproteinases and processing of pro-TNF-alpha. J Leukoc Biol. 1995;57:774–7. doi: 10.1002/jlb.57.5.774. [DOI] [PubMed] [Google Scholar]
- 20.Burke B. The role of matrix metalloproteinase 7 in innate immunity. Immunobiology. 2004;209:51–6. doi: 10.1016/j.imbio.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Haro H, Crawford HC, Fingleton B, Shinomiya K, Spengler DM, Matrisian LM. Matrix metalloproteinase-7-dependent release of tumor necrosis factor-alpha in a model of herniated disc resorption. J Clin Invest. 2000;105:143–50. doi: 10.1172/JCI7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takai T. Paired immunoglobulin-like receptors and their MHC class I recognition. Immunology. 2005;115:433–40. doi: 10.1111/j.1365-2567.2005.02177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pereira S, Zhang H, Takai T, Lowell CA. The inhibitory receptor PIR-B negatively regulates neutrophil and macrophage integrin signaling. J Immunol. 2004;173:5757–65. doi: 10.4049/jimmunol.173.9.5757. [DOI] [PubMed] [Google Scholar]
- 24.Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol. 2006;24:1115–22. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 25.Coppolino MG, Krause M, Hagendorff P, Monner DA, Trimble W, Grinstein S, et al. Evidence for a molecular complex consisting of Fyb/SLAP, SLP-76, Nck, VASP and WASP that links the actin cytoskeleton to Fcgamma receptor signalling during phagocytosis. J Cell Sci. 2001;114:4307–18. doi: 10.1242/jcs.114.23.4307. [DOI] [PubMed] [Google Scholar]
- 26.Jones GE. Cellular signaling in macrophage migration and chemotaxis. J Leukoc Biol. 2000;68:593–602. [PubMed] [Google Scholar]
- 27.Tarzi RM, Cook HT. Role of Fcgamma receptors in glomerulonephritis. Nephron Exp Nephrol. 2003;95:e7–12. doi: 10.1159/000073018. [DOI] [PubMed] [Google Scholar]
- 28.Kovalenko P, Fujinaka H, Yoshida Y, Kawamura H, Qu Z, El-Shemi AG, et al. Fc receptor-mediated accumulation of macrophages in crescentic glomerulonephritis induced by anti-glomerular basement membrane antibody administration in WKY rats. Int Immunol. 2004;16:625–34. doi: 10.1093/intimm/dxh058. [DOI] [PubMed] [Google Scholar]
- 29.Otsuka A, Abe T, Watanabe M, Yagisawa H, Takei K, Yamada H. Dynamin 2 is required for actin assembly in phagocytosis in Sertoli cells. Biochem Biophys Res Commun. 2009;378:478–82. doi: 10.1016/j.bbrc.2008.11.066. [DOI] [PubMed] [Google Scholar]
- 30.Jin ZX, Huang CR, Dong L, Goda S, Kawanami T, Sawaki T, et al. Impaired TCR signaling through dysfunction of lipid rafts in sphingomyelin synthase 1 (SMS1)-knockdown T cells. Int Immunol. 2008;20:1427–37. doi: 10.1093/intimm/dxn100. [DOI] [PubMed] [Google Scholar]
- 31.Benovoy D, Kwan T, Majewski J. Effect of polymorphisms within probe-target sequences on olignonucleotide microarray experiments. Nucleic Acids Res. 2008;36:4417–23. doi: 10.1093/nar/gkn409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nurtdinov RN, Vasiliev MO, Ershova AS, Lossev IS, Karyagina AS. PLANdbAffy: probe-level annotation database for Affymetrix expression microarrays. Nucleic Acids Res. 2010;38:D726–D730. doi: 10.1093/nar/gkp969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gamazon ER, Zhang W, Dolan ME, Cox NJ. Comprehensive survey of SNPs in the Affymetrix exon array using the 1000 Genomes dataset. PLoS One. 2010;5:e9366. doi: 10.1371/journal.pone.0009366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gillett A, Maratou K, Fewings C, Harris RA, Jagodic M, Aitman T, et al. Alternative splicing and transcriptome profiling of experimental autoimmune encephalomyelitis using genome-wide exon arrays. PLoS One. 2009;4:e7773. doi: 10.1371/journal.pone.0007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parkinson H, Kapushesky M, Kolesnikov N, Rustici G, Shojatalab M, Abeygunawardena N, et al. ArrayExpress update--from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37:D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 37.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful apprach to multiple testing. J Roy Statist Soc Ser B (Methodological) 1995;57:289–300. [Google Scholar]
- 38.Yandell BS, Mehta T, Banerjee S, Shriner D, Venkataraman R, Moon JY, et al. R/qtlbim: QTL with Bayesian Interval Mapping in experimental crosses. Bioinformatics. 2007;23:641–3. doi: 10.1093/bioinformatics/btm011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hosack DA, Dennis G, Jr., Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 41.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanges R, Cordero F, Calogero RA. oneChannelGUI: a graphical interface to Bioconductor tools, designed for life scientists who are not familiar with R language. Bioinformatics. 2007;23:3406–8. doi: 10.1093/bioinformatics/btm469. [DOI] [PubMed] [Google Scholar]
- 43.Della BC, Cordero F, Calogero RA. Dissecting an alternative splicing analysis workflow for GeneChip Exon 1.0 ST Affymetrix arrays. BMC Genomics. 2008;9:571. doi: 10.1186/1471-2164-9-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 45.Zeileis A, Leisch F, Hornik K, Kleiber C. strucchange: An R Package for Testing for Structural Chang in Linear Regression Models. Journal of Statistical Software. 2002;7:1–38. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.