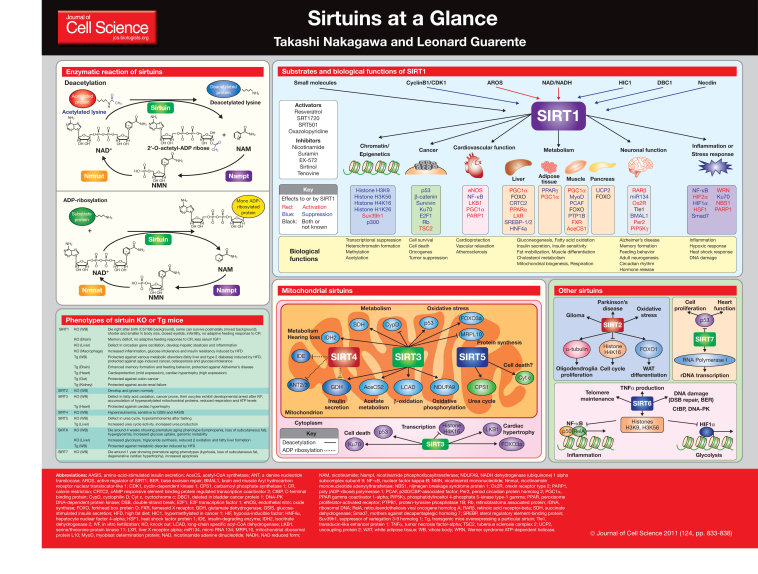
Sirtuins are NAD-dependent deacetylases that are highly conserved from bacteria to human and SIR2 was originally shown to extend lifespan in budding yeast (Imai et al., 2000; Kaeberlein et al., 1999). Since then, sirtuins have been shown to also regulate longevity in other lower organisms, such as flies and worms (Tissenbaum and Guarente, 2001; Rogina and Helfand, 2004). In mammals, there are seven sirtuins (SIRT1-7). All mammalian sirtuins contain a conserved NAD-binding and catalytic domain, termed the sirtuin core domain, but differ in their N- and C-terminal domains (Frye, 2000). They have different specific substrates and biological functions, and are found in various cell compartments. The fact that sirtuins require NAD for their enzymatic activity connects metabolism to aging and aging-related diseases. In this Cell Science at a Glance article, we summarize the recent data related to the role of sirtuins in aging and aging-related diseases, and describe the underlying molecular mechanisms.
Enzymatic activity of sirtuins and NAD biosynthesis
Sirtuins belong to the class III protein deacetylase family, which are the only histone deactyalses (HDACs) that require NAD for their enzymatic activity (Imai et al., 2000). NAD is an important co-factor for the electron transport chain and is also involved in many enzymatic reactions (Houtkooper et al., 2010). Owing to the characteristic NAD requirement for their enzymatic reaction, the activity of sirtuins is directly linked to the metabolic state in the cell. Initially, yeast SIR2 was discovered as a histone deacetylase, but mammalian sirtuins have also various non-histone protein substrates. The deacetylation reaction of sirtuins consists of two steps. In the first, sirtuins cleave NAD and produce nicotinamide (NAM), and in the second step the acetyl group is transferred from the substrate to the ADP-ribose moiety of NAD to generate O-acetyl-ADP ribose and the deacetylated substrate (Tanner et al., 2000). Although most of sirtuins have deacetylase activity, SIRT4 has been shown to have only ADP-ribosyltransferase activity, whereas SIRT1 and SIRT6 have both deacetylation and a relatively weak ADP-ribosyltransferase activity
(Haigis et al., 2006; Imai et al., 2000; Liszt et al., 2005; Michishita et al., 2008). Interestingly, the generated NAM is an endogenous inhibitor of sirtuins and is usually converted to nicotinamide mononucleotide (NMN) by the NAD salvage pathway enzyme, nicotinamide phosphoribosyl-transferase (Nampt). Subsequently, nicotinamide mononucleotide adenyltransferase (Nmnat) regenerates NAD from NMN (Houtkooper et al., 2010) (see poster). Recent studies suggest that NAD biosynthesis critically regulates the activity of sirtuins. For example, Nampt-mediated NAD biosynthesis controls SIRT1 activity in several tissues; and SIRT3, SIRT4 and SIRT5 are also regulated by NAD biosynthesis in response to nutrient stress (Nakagawa et al., 2009; Revollo et al., 2007; Yang et al., 2007). Nampt also mediates the production of tumor necrosis factor α (TNFα) in a SIRT6 dependent manner (Van Gool et al., 2009). Additionally, NAD biosynthesis is linked to the circadian clock cycle, because Nampt is regulated by a complex consisting of the circadian locomotor output cycles kaput (CLOCK), brain and muscle aryl hydrocarbon receptor nuclear translocator-like 1 (BMAL1) and SIRT1, and in turn regulates SIRT1 activity through NAD (Nakahata et al., 2009; Ramsey et al., 2009).
SIRT1
Roles in chromatin and epigenetics
As well as yeast SIR2, its mammalian homolog SIRT1 was shown to be able to deacetylate histones and preferentially deacetylates lysine (K) residue 9 of histone 3 (H3K9) and lysine 16 of histone 4 (H4K16) in vitro (Imai et al., 2000; Vaquero et al., 2004). SIRT1-deficient mouse embryonic fibroblasts (MEFs) also accumulate hyperacetylated histones, thereby promoting heterochromatin formation and transcription repression (Vaquero et al., 2007; Wang et al., 2008). SIRT1 deletion in MEFs also leads to histone H3K9 hypomethylation (Vaquero et al., 2007). SIRT1 directly binds to the histone methyltransferase Suv39h1 and upregulates its methyltransferase activity by deacetylating lysine residue 266, which resides in the SET domain of Suv39h1. SIRT1 also interacts with linker histone H1b and deacetylates its lysine 26 (H1K26) (Vaquero et al., 2004). In response to oxidative stress, SIRT1 is redistributed and so binds to acetylated histone H1K26, which leads to the repression of various set of genes. Interestingly, the transcriptional repression pattern is similar to that observed in the mouse brain with aging (Oberdoerffer et al., 2008). Thus, it can be speculated that SIRT1 is responsible for aging-dependent global transcriptional changes through chromatin modification.
Roles in metabolism pathways and metabolic diseases
SIRT1 regulates various metabolic processes that allow the cell to adapt to nutrient stress and has a pivotal role in aging-related metabolic diseases. In response to fasting, SIRT1 modulates gluconeogenesis in the liver through deacetylation of important factors, such as peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α), forkhead box protein O1 (FOXO1) and cAMP response element-binding (CREB)-regulated transcription coactivator 2 (CRTC2) (Brunet et al., 2004; Liu et al., 2008; Motta et al., 2004; Rodgers et al., 2005). In the early phase of fasting, CRTC2 is activated through its acetylation by the co-activator CBP/p300, which then promotes transcription of gluconeogenic genes. If fasting is prolonged, SIRT1 deacetylates both CRTC2 and FOXO1, which leads to a switch from activation of early gluconeogenic genes through CRTC2 to the activation of genes involved in the late phase of gluconeogenesis through FOXO1 (Liu et al., 2008). Deacetylation of PGC-1α by SIRT1 not only controls gluconeogenesis, but also fatty acid oxidation in coordination with peroxisome proliferator-activated receptor alpha (PPARα) (Purushotham et al., 2009). PGC1α also regulates mitochondria biogenesis and oxidative phosphorylation, and it has been proposed that these effects are mediated through the AMP-activated protein kinase (AMPK)–SIRT1–PGC1α pathway (Canto et al., 2009; Iwabu et al., 2010). SIRT1 also targets other nuclear receptors, such as the liver X receptor LXRα and the farnesoid X receptor (FXR), which regulate hepatic metabolic processes (Kemper et al., 2009; Li et al., 2007b). SIRT1 deacetylates LXRα and promotes its ubiquitylation, which results in its activation and induction of cholesterol efflux (Li et al., 2007b). In white adipose tissue, SIRT1 regulates fat mobilization through the repression of peroxisome proliferator-activated receptor gamma (PPARγ) by binding to its cofactors, the nuclear receptor co-repressor (NCoR) and silencing mediator of retinoid and thyroid hormone receptors (SMRT) (Picard et al., 2004). In pancreatic β cells, SIRT1 regulates glucose-stimulated insulin secretion through the synthesis of uncoupling protein 2 (UCP2) (Bordone et al., 2006; Moynihan et al., 2005). Transgenic mice that specifically overexpress SIRT1 in pancreatic β cells (the so-called BESTO mice) have improved glucose tolerance when fed with a high fat diet (HFD) (Moynihan et al., 2005). In addition, mice that overexpress SIRT1 from a bacterial artificial chromosome (BAC) construct (the so-called SIRT1 BAC mice) are also protected against HFD-induced type 2 diabetes (Banks et al., 2008; Pfluger et al., 2008). Consistent with these results, compounds that are able to activate SIRT1, such as resveratrol and SRT1720, protect mice from HFD-induced metabolic disorders (Baur et al., 2006; Feige et al., 2008; Lagouge et al., 2006; Milne et al., 2007). In humans, genetic variation in SIRT1 was shown to be correlated with obesity and type 2 diabetes in studies of Dutch populations (Zillikens et al., 2009b; Zillikens et al., 2009a). Taken together, these findings provide strong evidence that SIRT1 has significant roles in metabolic diseases, such as type 2 diabetes and obesity, and controlling SIRT1 activity with nutrients or small molecules could be a valuable treatment strategy.
Roles in inflammation and stress response
A number of studies have revealed that SIRT1 mediates different stress responses, including inflammation, hypoxic stress, heat shock and genotoxic stress, and inflammation in particular is a highly important cause of aging and aging related diseases. SIRT1 can suppress inflammation through its effect on nuclear factor-κB (NF-κB) (Yeung et al., 2004); it physically interacts with its RelA subunit and deacetylates lysine 310, which inactivates NF-κB, thereby inhibiting the expression of its target genes. By contrast, during hypoxic condition, SIRT1 activates hypoxia inducible factor 2 alpha (HIF2α) through its deacetylation and thus initiates hypoxic stress responses (Dioum et al., 2009). However, SIRT1 also deacetylates HIF1α at lysine 647 – which, in this case, inhibits its activity – to control glycolysis in response to hypoxic conditions (Lim et al., 2010). During hypoxia, NAD level gradually decrease and, subsequently, SIRT1 is deactivated. Therefore, it has been speculated that SIRT1 triggers a switch from HIF2α to HIF1α activation to coordinate metabolism, vascular formation and hypoxic stress responses (Lim et al., 2010).
SIRT1 is also involved in the transmission of the heat shock response through the heat shock factor protein1 (HSF1) (Westerheide et al., 2009). Upon protein-damage stress that is associated with the accumulation of misfolded proteins, SIRT1 deacetylates and, thereby, positively regulates HSF1 activity, which promotes the transcription of heat shock response genes (Westerheide et al., 2009). Thus, SIRT1 acts as a sensor of various stresses and organizes the survival signals in response to these stresses.
Roles in cardiovascular disease
Cardiovascular diseases increase with aging and are also closely influenced by the metabolism. Several lines of evidence show that SIRT1 has pivotal roles in cardiovascular functions. For example, transgenic mice that overexpress SIRT1 in the heart, are protected against age-related cardiac hypertrophy as well as ischemia or reperfusion injury (Alcendor et al., 2007; Hsu et al., 2010). SIRT1 also regulates vascular endothelial cell functions through deacetylation of endothelial nitric oxide synthase (eNOS) (Mattagajasingh et al., 2007). In addition, activation of SIRT1 with resveratrol can ameliorate heart ischemia or reperfusion injury and also improve vascular functions (Orallo et al., 2002). SIRT1 also functions in reactive oxygen species (ROS)-mediated cell death through deacetylation of poly (ADP-ribose) polymerase 1 (PARP1) and, furthermore, deletion of SIRT1 results in the promotion of cardiomyocyes cell death during heart failure (Pillai et al., 2005; Kolthur-Seetharam et al., 2006).
Roles in cancer
The role of SIRT1 in tumor progression is controversial, as SIRT1 might have dual functions as an oncogene and tumor suppressor. The initial evidence that SIRT1 might function as an oncogene was the observation that it deacetylates lysine residue 383 of p53, thereby repressing its transcriptional activity (Luo et al., 2001; Vaziri et al., 2001). As p53 is a key tumor suppressor, its downregulation by SIRT1 could drive cells to tumorgenesis. In addition, two other tumor suppressors, deleted in bladder cancer 1 (DBC1) and hypermethylated in cancer 1 (HIC1) have been found to affect SIRT1. DBC1 binds to the N-terminus of SIRT1 and inhibits its enzymatic activity (Kim et al., 2008; Zhao et al., 2008), whereas HIC1 binds to the promoter region of the SIRT1 gene and represses its expression (Chen et al., 2005). As DBC1 and HIC1 are silenced in certain types of cancer cell, it has been speculated that they indirectly promote tumorgenesis – at least in part – through the resulting activation of SIRT1.
By contrast, several studies have shown that SIRT1 has also potential tumor suppressor effects. As discussed above, SIRT1 inactivates HIF1α through deacetylation and so protects against the growth of tumors and vascular formation – prerequisites for tumor progression (Lim et al., 2010). In addition, overexpression of SIRT1 in murine intestines reduces the incidence of cancer and growth in a mouse colon cancer model: SIRT1 was shown to deacetylate β-catenin and inhibit its accumulation in the nucleus, which lead to the hyperactivation of β-catenin and the formation of tumors (Firestein et al., 2008). Another study showed that the SIRT1 activator resveratrol also protects mice that are heterozygous for p53 from cancer (Boily et al., 2009; Oberdoerffer et al., 2008). Consistent with these results, it was shown that SIRT1 haploinsufficiency facilitates tumorigenesis in these mice by accelerating tumorigenesis (Wang et al., 2008). These data imply that SIRT1 functions as a tumor suppressor in vivo. However, the exact molecular effects of SIRT1 might vary in different tissues and/or cancer types.
Roles in neuronal functions and neurodegenerative diseases
Recent accumulating evidence shows that SIRT1 has also crucial roles in neuronal physiology and pathology. One of the most important neuronal functions of SIRT1 is its role in promoting feeding behavior during dietary limited conditions, including calorie restriction (CR) and fasting. Several studies suggest that SIRT1 expression is induced in hypothalamic pro-opiomelanocortin (POMC) expressing neurons, where it regulates various feeding behaviors (Cakir et al., 2009; Ramadori et al., 2010; Satoh et al., 2010). Correspondingly, mice in which SIRT1 was specifically knocked out in the brain lack the increase of adaptive physical activity in response to CR – a characteristic that is enhanced in mice overexpressing SIRT1 in their brains (Cohen et al., 2009; Satoh et al., 2010). SIRT1 also controls the central endocrine axes in the hypothalamus and pituitary gland. Mice in which SIRT1 was specifically knocked out in the brain have lower serum levels of growth hormones (Cohen et al., 2009). SIRT1 also positively regulates the secretion of thyroid-stimulating hormone (TSH) from the pituitary gland by deacetylating phosphatidylinositol 4-phosphate 5-kinase type-1 gamma (PIP5Kγ), which has a crucial role in exoctytosis of TSH from pituitary cells (Akieda-Asai et al., 2010).
More recently, SIRT1 was found to modulate memory formation and synaptic plasticity (Michan et al., 2010; Gao et al., 2010). Here, SIRT1 promotes CREB expression through repression of the micro RNA 134 (miR134) and induces transcription of brain-derived neurotrophic factor (BDNF), which has crucial roles in normal cognitive function. Indeed, activation of SIRT1 in brain enhances, and its deletion impairs, memory formation and synaptic plasticity in mice (Gao et al., 2010).
Another recent study in mice indicates that SIRT1 is also implicated in Alzheimer's disease (AD) (Donmez et al., 2010). Using an AD mouse model, we have shown that overexpression of SIRT1 in the brain could prevent the formation of amyloid β (Aβ) plaques and rescue behavioral deficits. Here, SIRT1 directly activates retinoid acid receptor β (RARβ) through its deacetylation. RARβ activation, in turn, promotes transcription of a disintegrin and metallopeptidase 10 (ADAM10), which encodes for a component of α-secretase. The resulting increase in α-secretase activity leads to a reduced processing of toxic amyloid precursor protein (APP) (Donmez et al., 2010). In another AD mice model (mice that overexpress CDK5 p25), activation of SIRT1 by genetic or pharmacological means ameliorated neurodegeneration and memory decline (Kim et al., 2007). This study also uncovered a protective role for SIRT1 in amyotrophic lateral sclerosis (ALS) (Kim et al., 2007).
SIRT2
SIRT2 resides mainly in the cytoplasm but can also shuttle to the nucleus. SIRT2 can function as a α-tubulin deacetylase and was suggested to have a role in oligodendroglial differentiation (Li et al., 2007a; North et al., 2003). In the nucleus, SIRT2 acts as a H4K16 deacetylase and controls the cell cycle. MEFs, in which SIRT2 has been knocked out, accumulate H4K16 acetylation during mitosis and exhibit a delay in S-phase entry (Vaquero et al., 2006). SIRT2 also regulates adipocyte differentiation through FOXO1 deacetylation (Jing et al., 2007). Of particular note, a SIRT2-specific inhibitor can ameliorate α-synuclein-mediated toxicity in a Parkinson's disease model (Outeiro et al., 2007), but the mechanism underlying this effect is remains unknown. Further investigations are required to determine in greater depth the physiological and pathological functions of SIRT2.
Mitochondrial sirtuins SIRT3, SIRT4 and SIRT5
SIRT3, SIRT4 and SIRT5 are found in the mitochondrial matrix because they contain mitochondrion-targeting sequences in their N-termini (Haigis et al., 2006; Lombard et al., 2007; Nakagawa et al., 2009). SIRT3 is the best-characterized mitochondrial sirtuin. It interacts with acetyl-CoA synthetase 2 (AceCS2) and deacetylates lysine 642 in vitro and in vivo (Hallows et al., 2006; Schwer et al., 2006), thereby increasing its acetyl-CoA synthesis activity. Interestingly, the bacterial sirtuin CobB also regulates acetyl-CoA synthetase through its deacetylation, and cytoplasmic AceCS1 is regulated by SIRT1 in mammals (Hallows et al., 2006; Starai et al., 2002). Therefore, this mechanism of AceCS regulation appears to be evolutionary conserved. SIRT3 also deacetylates NDUFA9, a component of the OXPHOS complex I, which is involved in regulating cellular ATP levels (Ahn et al., 2008).
A recent study also revealed a new function of SIRT3 in fatty acid oxidation. Here, SIRT3 deacetylates long-chain acyl CoA dehydrogenase (LCAD) and upregulates its enzymatic activity during fasting (Hirschey et al., 2010). Moreover, SIRT3-knockout mice accumulate higher levels of fatty acid oxidation intermediates and have lower ATP levels during fasting (Hirschey et al., 2010).
In contrast to SIRT3, SIRT4 has only ADP-ribosylation but no deacetylation activity. It has been shown that SIRT4 can ADP-ribosylate glutamate dehydrogenase (GDH) and control insulin secretion in pancreatic β-cells in response to CR (Haigis et al., 2006). GDH is also deacetylated by SIRT3, but the physiological significance of this reaction is unknown (Lombard et al., 2007). SIRT4 also negatively regulates fatty acid oxidation and this effect is dependent on SIRT1 (Nasrin et al., 2010).
SIRT5 is also known to have a deacetylase activity. We have recently reported that SIRT5 has a role in the regulation of the urea cycle (Nakagawa et al., 2009). In this study, we uncovered that SIRT5 deacetylates and activates carbamoyl phosphate synthetase 1 (CPS1), an enzyme, which catalyzes the first and rate-limiting step of the urea cycle. SIRT5-knockout mice fail to upregulate CPS1 activity and are hyperammonemic during fasting, and similar effects were also observed with mice on a high protein diet or undergoing CR (Nakagawa et al., 2009). Conversely, SIRT5-overexpressing mice show increased CPS1 activity and urea production (Ogura et al., 2010). Thus, taken together, these findings support the idea that mitochondrial sirtuins directly control the activity of metabolic enzymes and have crucial roles in the metabolic adaptation to dietary conditions, such as calorie restriction and fasting.
SIRT6
SIRT6 is a nuclear sirtuin and has roles that differ from those of SIRT1. SIRT6-knockout mice exhibit aging-like phenotypes, such as lymphopenia, loss of subcutaneous fat and severe hypoglycemia, which lead to their death by 4-5 weeks (Mostoslavsky et al., 2006; Xiao et al., 2010). MEFs and embryonic stem cells that are deficient in SIRT6 show genomic instability and impaired base-excision repair (Mostoslavsky et al., 2006). SIRT6 is also involved in DNA double-strand break repair by regulating C-terminal binding protein (CtBP) and DNA protein kinase (McCord et al., 2009; Kaidi et al., 2010). Biochemically, SIRT6 was shown to have ADP-ribosyltransferase activity, but recent studies suggest that SIRT6 has also a deacetylase activity, preferentially acting on histone H3K9 and H3K56 (Liszt et al., 2005; Michishita et al., 2008; Michishita et al., 2009).
SIRT6 also associates with telomeric chromatin and depletion of SIRT6 results in premature cellular senescence and telomere dysfunction (Michishita et al., 2008). Similarly to SIRT1, SIRT6 also interacts with the RelA subunit of NF-κB and deacetylates histone H3K9 at NF-κB target gene promoters, which leads to their repression (Kawahara et al., 2009). Disruption of the SIRT6 gene leads to hyperactivation of NF-κB signaling and is, thus, the likely cause for the premature aging phenotype observed in SIRT6-knockout mice, as haploinsufficiency of RelA attenuates the lethality and aging-like phenotypes of SIRT6-knockout mice. Another recent paper has revealed that SIRT6 also selectively binds to the promoter regions of HIF1α target genes, at which it deacetylates histone H3K9 (Zhong et al., 2010). Here, SIRT6 functions as a co-repressor of HIF1α and inhibits the transcription of glycolytic genes. Consistent with this notion, MEFs and mice that are deficient in SIRT6 exhibit amplified HIF1α activity and increased glycolysis together with diminished mitochondrial respiration, thus providing a possible explanation for the observed hypoglycemia in SIRT6-knockout mice. In addition, a recent report shows that liver-specific deletion of SIRT6 in mice leads to increased glycolysis, triglyceride synthesis, reduced β oxidation, and fatty liver formation (Kim et al., 2010). Taken together, these data suggest that SIRT6, similar to SIRT1, also has pivotal roles in metabolism.
SIRT7
Among the sirtuins, SIRT7 is the least studied. SIRT7 has been shown to be a positive regulator of RNA polymerase I transcription (Ford et al., 2006), but the enzymatic activity of SIRT7 remains undetermined thus far. Recently, SIRT7-knockout mice were reported to exhibit various signs of aging-related changes, such as kyphosis, loss of subcutaneous fat and premature death (Vakhrusheva et al., 2008), and they also suffer from degenerative heart hypertrophy. However, SIRT7-specific substrates have not been reported yet and additional studies are necessary to reveal the biological roles of SIRT7.
Perspectives
Over the last decade, the study of sirtuins has made remarkable progress and expanded our knowledge regarding aging and aging-related diseases. However, our understanding of sirtuin biology is still far from complete and many important questions remain to be answered. For example, it has been shown that activation of SIRT1 can slow down phenotypes of aging, such as diabetes and bone loss, but an extension of life span has only been demonstrated in mice that are fed a high-fat diet. It is possible that this crucially important sirtuin turns out to have opposing roles in aging, which might make it a good target for specific aging diseases, but not for life span extension. Moreover, one or several of the other sirtuins (SIRT2 to SIRT7) might need to be modulated in concert with SIRT1 to be able to extend life span. What has become clear is that sirtuins are required to exert many of the beneficial aspects of calorie restriction. Most recently, SIRT3 has been shown essential in CR-mediated protection against aging-related hearing loss (Someya et al., 2010).
Finally, it will be extremely important to determine what happens to the activity of sirtuins during aging. There are hints that their activity declines, which provide further rationales to develop drugs that activate sirtuins to combat aging. Thus, to achieve the goal of a therapeutic intervention of aging, it will be important to fully elucidate the functions of all seven sirtuins and in many different tissues.
Acknowledgments
We apologize to those authors whose work has not been cited because of space limitations. T.N. is supported by JSPS Postdoctoral Fellowships for Research Abroad and L.G. is funded by grants from the NIH and the Paul F. Glenn Foundation. Deposited in PMC for release after 12 months.
References
- Ahn B. H., Kim H. S., Song S., Lee I. H., Liu J., Vassilopoulos A., Deng C. X., Finkel T. (2008). A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc. Natl. Acad. Sci. USA 105, 14447-14452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akieda-Asai S., Zaima N., Ikegami K., Kahyo T., Yao I., Hatanaka T., Iemura S., Sugiyama R., Yokozeki T., Eishi Y., et al. (2010). SIRT1 regulates thyroid-stimulating hormone release by enhancing PIP5Kgamma activity through deacetylation of specific lysine residues in mammals. PLoS One 5, e11755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcendor R. R., Gao S., Zhai P., Zablocki D., Holle E., Yu X., Tian B., Wagner T., Vatner S. F., Sadoshima J. (2007). Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ. Res. 100, 1512-1521 [DOI] [PubMed] [Google Scholar]
- Banks A. S., Kon N., Knight C., Matsumoto M., Gutierrez-Juarez R., Rossetti L., Gu W., Accili D. (2008). SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 8, 333-341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur J. A., Pearson K. J., Price N. L., Jamieson H. A., Lerin C., Kalra A., Prabhu V. V., Allard J. S., Lopez-Lluch G., Lewis K., et al. (2006). Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444, 337-342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boily G., He X. H., Pearce B., Jardine K., McBurney M. W. (2009). SirT1-null mice develop tumors at normal rates but are poorly protected by resveratrol. Oncogene 28, 2882-2893 [DOI] [PubMed] [Google Scholar]
- Bordone L., Motta M. C., Picard F., Robinson A., Jhala U. S., Apfeld J., McDonagh T., Lemieux M., McBurney M., Szilvasi A., et al. (2006). Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 4, e31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., et al. (2004). Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303, 2011-2015 [DOI] [PubMed] [Google Scholar]
- Cakir I., Perello M., Lansari O., Messier N. J., Vaslet C. A., Nillni E. A. (2009). Hypothalamic Sirt1 regulates food intake in a rodent model system. PLoS One 4, e8322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C., Gerhart-Hines Z., Feige J. N., Lagouge M., Noriega L., Milne J. C., Elliott P. J., Puigserver P., Auwerx J. (2009). AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458, 1056-1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. Y., Wang D. H., Yen R. C., Luo J., Gu W., Baylin S. B. (2005). Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell 123, 437-448 [DOI] [PubMed] [Google Scholar]
- Cohen D. E., Supinski A. M., Bonkowski M. S., Donmez G., Guarente L. P. (2009). Neuronal SIRT1 regulates endocrine and behavioral responses to calorie restriction. Genes Dev. 23, 2812-2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dioum E. M., Chen R., Alexander M. S., Zhang Q., Hogg R. T., Gerard R. D., Garcia J. A. (2009). Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science 324, 1289-1293 [DOI] [PubMed] [Google Scholar]
- Donmez G., Wang D., Cohen D. E., Guarente L. (2010). SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell 142, 320-332 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Feige J. N., Lagouge M., Canto C., Strehle A., Houten S. M., Milne J. C., Lambert P. D., Mataki C., Elliott P. J., Auwerx J. (2008). Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 8, 347-358 [DOI] [PubMed] [Google Scholar]
- Firestein R., Blander G., Michan S., Oberdoerffer P., Ogino S., Campbell J., Bhimavarapu A., Luikenhuis S., de Cabo R., Fuchs C., et al. (2008). The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS One 3, e2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford E., Voit R., Liszt G., Magin C., Grummt I., Guarente L. (2006). Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 20, 1075-1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye R. A. (2000). Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273, 793-798 [DOI] [PubMed] [Google Scholar]
- Gao J., Wang W. Y., Mao Y. W., Graff J., Guan J. S., Pan L., Mak G., Kim D., Su S. C., Tsai L. H. (2010). A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 466, 1105-1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis M. C., Mostoslavsky R., Haigis K. M., Fahie K., Christodoulou D. C., Murphy A. J., Valenzuela D. M., Yancopoulos G. D., Karow M., Blander G., et al. (2006). SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell 126, 941-954 [DOI] [PubMed] [Google Scholar]
- Hallows W. C., Lee S., Denu J. M. (2006). Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. USA 103, 10230-10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey M. D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D. B., Grueter C. A., Harris C., Biddinger S., Ilkayeva O. R., et al. (2010). SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464, 121-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper R. H., Canto C., Wanders R. J., Auwerx J. (2010). The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr. Rev. 31, 194-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C. P., Zhai P., Yamamoto T., Maejima Y., Matsushima S., Hariharan N., Shao D., Takagi H., Oka S., Sadoshima J. (2010). Silent information regulator 1 protects the heart from ischemia/reperfusion. Circulation 122, 2170-2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Armstrong C. M., Kaeberlein M., Guarente L. (2000). Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795-800 [DOI] [PubMed] [Google Scholar]
- Iwabu M., Yamauchi T., Okada-Iwabu M., Sato K., Nakagawa T., Funata M., Yamaguchi M., Namiki S., Nakayama R., Tabata M., et al. (2010). Adiponectin and AdipoR1 regulate PGC-1alpha and mitochondria by Ca(2+) and AMPK/SIRT1. Nature 464, 1313-1319 [DOI] [PubMed] [Google Scholar]
- Jing E., Gesta S., Kahn C. R. (2007). SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab. 6, 105-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., McVey M., Guarente L. (1999). The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13, 2570-2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaidi A., Weinert B. T., Choudhary C., Jackson S. P. (2010). Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science 329, 1348-1353 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kawahara T. L., Michishita E., Adler A. S., Damian M., Berber E., Lin M., McCord R. A., Ongaigui K. C., Boxer L. D., Chang H. Y., et al. (2009). SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell 136, 62-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper J. K., Xiao Z., Ponugoti B., Miao J., Fang S., Kanamaluru D., Tsang S., Wu S. Y., Chiang C. M., Veenstra T. D. (2009). FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab. 10, 392-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Nguyen M. D., Dobbin M. M., Fischer A., Sananbenesi F., Rodgers J. T., Delalle I., Baur J. A., Sui G., Armour S. M., et al. (2007). SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer's disease and amyotrophic lateral sclerosis. EMBO J. 26, 3169-3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. S., Xiao C., Wang R. H., Lahusen T., Xu X., Vassilopoulos A., Vazquez-Ortiz G., Jeong W. I., Park O., Ki S. H., et al. (2010). Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 12, 224-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. E., Chen J., Lou Z. (2008). DBC1 is a negative regulator of SIRT1. Nature 451, 583-586 [DOI] [PubMed] [Google Scholar]
- Kolthur-Seetharam U., Dantzer F., McBurney M. W., de Murcia G., Sassone-Corsi P. (2006). Control of AIF-mediated cell death by the functional interplay of SIRT1 and PARP-1 in response to DNA damage. Cell Cycle 5, 873-877 [DOI] [PubMed] [Google Scholar]
- Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., et al. (2006). Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell 127, 1109-1122 [DOI] [PubMed] [Google Scholar]
- Li W., Zhang B., Tang J., Cao Q., Wu Y., Wu C., Guo J., Ling E. A., Liang F. (2007a). Sirtuin 2, a mammalian homolog of yeast silent information regulator-2 longevity regulator, is an oligodendroglial protein that decelerates cell differentiation through deacetylating alpha-tubulin. J. Neurosci. 27, 2606-2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang S., Blander G., Tse J. G., Krieger M., Guarente L. (2007b). SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol. Cell 28, 91-106 [DOI] [PubMed] [Google Scholar]
- Lim J. H., Lee Y. M., Chun Y. S., Chen J., Kim J. E., Park J. W. (2010). Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol. Cell 38, 864-878 [DOI] [PubMed] [Google Scholar]
- Liszt G., Ford E., Kurtev M., Guarente L. (2005). Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 280, 21313-21320 [DOI] [PubMed] [Google Scholar]
- Liu Y., Dentin R., Chen D., Hedrick S., Ravnskjaer K., Schenk S., Milne J., Meyers D. J., Cole P., Yates J., 3rd, et al. (2008). A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature 456, 269-273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard D. B., Alt F. W., Cheng H. L., Bunkenborg J., Streeper R. S., Mostoslavsky R., Kim J., Yancopoulos G., Valenzuela D., Murphy A., et al. (2007). Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell. Biol. 27, 8807-8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Nikolaev A. Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. (2001). Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107, 137-148 [DOI] [PubMed] [Google Scholar]
- Mattagajasingh I., Kim C. S., Naqvi A., Yamamori T., Hoffman T. A., Jung S. B., DeRicco J., Kasuno K., Irani K. (2007). SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc. Natl. Acad. Sci. USA 104, 14855-14860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord R. A., Michishita E., Hong T., Berber E., Boxer L. D., Kusumoto R., Guan S., Shi X., Gozani O., Burlingame A. L., et al. (2009). SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging 1, 109-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michan S., Li Y., Chou M. M., Parrella E., Ge H., Long J. M., Allard J. S., Lewis K., Miller M., Xu W., et al. (2010). SIRT1 is essential for normal cognitive function and synaptic plasticity. J. Neurosci. 30, 9695-9707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E., McCord R. A., Berber E., Kioi M., Padilla-Nash H., Damian M., Cheung P., Kusumoto R., Kawahara T. L., Barrett J. C., et al. (2008). SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452, 492-496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E., McCord R. A., Boxer L. D., Barber M. F., Hong T., Gozani O., Chua K. F. (2009). Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 8, 2664-2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne J. C., Lambert P. D., Schenk S., Carney D. P., Smith J. J., Gagne D. J., Jin L., Boss O., Perni R. B., Vu C. B., et al. (2007). Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 450, 712-716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R., Chua K. F., Lombard D. B., Pang W. W., Fischer M. R., Gellon L., Liu P., Mostoslavsky G., Franco S., Murphy M. M., et al. (2006). Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124, 315-329 [DOI] [PubMed] [Google Scholar]
- Motta M. C., Divecha N., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L. (2004). Mammalian SIRT1 represses forkhead transcription factors. Cell 116, 551-563 [DOI] [PubMed] [Google Scholar]
- Moynihan K. A., Grimm A. A., Plueger M. M., Bernal-Mizrachi E., Ford E., Cras-Meneur C., Permutt M. A., Imai S. (2005). Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2, 105-117 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Lomb D. J., Haigis M. C., Guarente L. (2009). SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell 137, 560-570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y., Sahar S., Astarita G., Kaluzova M., Sassone-Corsi P. (2009). Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324, 654-657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrin N., Wu X., Fortier E., Feng Y., Bare O. C., Chen S., Ren X., Wu Z., Streeper R. S., Bordone L. (2010). SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J. Biol. Chem. 285, 31995-32002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North B. J., Marshall B. L., Borra M. T., Denu J. M., Verdin E. (2003). The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11, 437-444 [DOI] [PubMed] [Google Scholar]
- Oberdoerffer P., Michan S., McVay M., Mostoslavsky R., Vann J., Park S. K., Hartlerode A., Stegmuller J., Hafner A., Loerch P., et al. (2008). SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell 135, 907-918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura M., Nakamura Y., Tanaka D., Zhuang X., Fujita Y., Obara A., Hamasaki A., Hosokawa M., Inagaki N. (2010). Overexpression of SIRT5 confirms its involvement in deacetylation and activation of carbamoyl phosphate synthetase 1. Biochem. Biophys. Res. Commun. 393, 73-78 [DOI] [PubMed] [Google Scholar]
- Orallo F., Alvarez E., Camina M., Leiro J. M., Gomez E., Fernandez P. (2002). The possible implication of trans-Resveratrol in the cardioprotective effects of long-term moderate wine consumption. Mol. Pharmacol. 61, 294-302 [DOI] [PubMed] [Google Scholar]
- Outeiro T. F., Kontopoulos E., Altmann S. M., Kufareva I., Strathearn K. E., Amore A. M., Volk C. B., Maxwell M. M., Rochet J. C., McLean P. J., et al. (2007). Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science 317, 516-519 [DOI] [PubMed] [Google Scholar]
- Pfluger P. T., Herranz D., Velasco-Miguel S., Serrano M., Tschop M. H. (2008). Sirt1 protects against high-fat diet-induced metabolic damage. Proc. Natl. Acad. Sci. USA 105, 9793-9798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., Machado De Oliveira R., Leid M., McBurney M. W., Guarente L. (2004). Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature 429, 771-776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai J. B., Isbatan A., Imai S., Gupta M. P. (2005). Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2alpha deacetylase activity. J. Biol. Chem. 280, 43121-43130 [DOI] [PubMed] [Google Scholar]
- Purushotham A., Schug T. T., Xu Q., Surapureddi S., Guo X., Li X. (2009). Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 9, 327-338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadori G., Fujikawa T., Fukuda M., Anderson J., Morgan D. A., Mostoslavsky R., Stuart R. C., Perello M., Vianna C. R., Nillni E. A., et al. (2010). SIRT1 deacetylase in POMC neurons is required for homeostatic defenses against diet-induced obesity. Cell Metab. 12, 78-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey K. M., Yoshino J., Brace C. S., Abrassart D., Kobayashi Y., Marcheva B., Hong H. K., Chong J. L., Buhr E. D., Lee C., et al. (2009). Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324, 651-654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revollo J. R., Korner A., Mills K. F., Satoh A., Wang T., Garten A., Dasgupta B., Sasaki Y., Wolberger C., Townsend R. R., et al. (2007). Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab. 6, 363-375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. (2005). Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 434, 113-118 [DOI] [PubMed] [Google Scholar]
- Rogina B., Helfand S. L. (2004). Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. USA 101, 15998-16003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh A., Brace C. S., Ben-Josef G., West T., Wozniak D. F., Holtzman D. M., Herzog E. D., Imai S. (2010). SIRT1 promotes the central adaptive response to diet restriction through activation of the dorsomedial and lateral nuclei of the hypothalamus. J. Neurosci. 30, 10220-10232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B., Bunkenborg J., Verdin R. O., Andersen J. S., Verdin E. (2006). Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. USA 103, 10224-10229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya S., Yu W., Hallows W. C., Xu J., Vann J. M., Leeuwenburgh C., Tanokura M., Denu J. M., Prolla T. A. (2010). Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell 143, 802-812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starai V. J., Celic I., Cole R. N., Boeke J. D., Escalante-Semerena J. C. (2002). Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298, 2390-2392 [DOI] [PubMed] [Google Scholar]
- Tanner K. G., Landry J., Sternglanz R., Denu J. M. (2000). Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. USA 97, 14178-14182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum H. A., Guarente L. (2001). Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410, 227-230 [DOI] [PubMed] [Google Scholar]
- Vakhrusheva O., Smolka C., Gajawada P., Kostin S., Boettger T., Kubin T., Braun T., Bober E. (2008). Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ. Res. 102, 703-710 [DOI] [PubMed] [Google Scholar]
- Van Gool F., Galli M., Gueydan C., Kruys V., Prevot P. P., Bedalov A., Mostoslavsky R., Alt F. W., De Smedt T., Leo O. (2009). Intracellular NAD levels regulate tumor necrosis factor protein synthesis in a sirtuin-dependent manner. Nat. Med. 15, 206-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A., Scher M., Lee D., Erdjument-Bromage H., Tempst P., Reinberg D. (2004). Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell 16, 93-105 [DOI] [PubMed] [Google Scholar]
- Vaquero A., Scher M. B., Lee D. H., Sutton A., Cheng H. L., Alt F. W., Serrano L., Sternglanz R., Reinberg D. (2006). SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 20, 1256-1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A., Scher M., Erdjument-Bromage H., Tempst P., Serrano L., Reinberg D. (2007). SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature 450, 440-444 [DOI] [PubMed] [Google Scholar]
- Vaziri H., Dessain S. K., Ng Eaton E., Imai S. I., Frye R. A., Pandita T. K., Guarente L., Weinberg R. A. (2001). hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107, 149-159 [DOI] [PubMed] [Google Scholar]
- Wang R. H., Sengupta K., Li C., Kim H. S., Cao L., Xiao C., Kim S., Xu X., Zheng Y., Chilton B., et al. (2008). Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell 14, 312-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerheide S. D., Anckar J., Stevens S. M., Jr, Sistonen L., Morimoto R. I. (2009). Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science 323, 1063-1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Kim H. S., Lahusen T., Wang R. H., Xu X., Gavrilova O., Jou W., Gius D., Deng C. X. (2010). SIRT6 deficiency results in severe hypoglycemia by enhancing both basal and insulin-stimulated glucose uptake in mice. J. Biol. Chem. 285, 36776-36784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yang T., Baur J. A., Perez E., Matsui T., Carmona J. J., Lamming D. W., Souza-Pinto N. C., Bohr V. A., Rosenzweig A., et al. (2007). Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell 130, 1095-1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung F., Hoberg J. E., Ramsey C. S., Keller M. D., Jones D. R., Frye R. A., Mayo M. W. (2004). Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23, 2369-2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Kruse J. P., Tang Y., Jung S. Y., Qin J., Gu W. (2008). Negative regulation of the deacetylase SIRT1 by DBC1. Nature 451, 587-590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L., D'Urso A., Toiber D., Sebastian C., Henry R. E., Vadysirisack D. D., Guimaraes A., Marinelli B., Wikstrom J. D., Nir T., et al. (2010). The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell 140, 280-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillikens M. C., van Meurs J. B., Rivadeneira F., Amin N., Hofman A., Oostra B. A., Sijbrands E. J., Witteman J. C., Pols H. A., van Duijn C. M., et al. (2009a). SIRT1 genetic variation is related to BMI and risk of obesity. Diabetes 58, 2828-2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillikens M. C., van Meurs J. B., Sijbrands E. J., Rivadeneira F., Dehghan A., van Leeuwen J. P., Hofman A., van Duijn C. M., Witteman J. C., Uitterlinden A. G., et al. (2009b). SIRT1 genetic variation and mortality in type 2 diabetes: interaction with smoking and dietary niacin. Free Radic. Biol. Med. 46, 836-841 [DOI] [PubMed] [Google Scholar]