Abstract
The heat shock (HS) response, a phylogenetically conserved ubiquitous response to stress, is generally characterized by the induced expression of heat shock protein (HSP) genes. Our earlier studies showed that the stress-activated transcription factor, heat shock factor-1 (HSF1), activated at febrile range or HS temperatures also modified expression of non-HSP genes including cytokine and chemokine genes. We also showed by in silico analysis that 28 among 29 human and mouse CXC chemokine genes had multiple putative heat shock response elements (HSEs) present in their gene promoters. To further determine whether these potential HSEs were functional and bound HSF1, we analyzed the recruitment of HSF1 to promoters of 5 human CXC chemokine genes (CXCL-1, 2, 3, 5 and 8) by chromatin immunoprecipitation (ChIP) assay and analyzed the effect of HS exposure on tumor necrosis factor-α (TNFα)-induced expression of these genes in human lung epithelial-like A549 cells. HSF1 ChIP analysis showed that HSF1 was recruited to all but one of these CXC chemokine genes (CXCL-3) and HS caused a significant increase in recruitment of HSF1 to one or multiple HSEs present in the promoters of CXCL-1, 2, 5 and 8 genes. However, the effect of HS exposure on expression of these genes showed a variable gene-specific effect. For example, CXCL8 expression was markedly enhanced (p<0.05) whereas CXCL5 expression was significantly repressed (p<0.05) in cells exposed to HS coincident with TNFα stimulation. In contrast, expression of CXCL1 and CXCL2, despite HSF1 recruitment to their promoters, was not affected by HS exposure. Our results indicate that some, if not all, putative HSEs present in the CXC chemokine gene promoters are functional and recruit HSF1 in vivo but the effects on gene expression are variable and gene specific. We speculate, the physical proximity and interactions of other transcription factors and co-regulators with HSF1 could be critical to determining the effects of HS on the expression of these genes.
Keywords: Hyperthermia, stress, TNF, A549, IL-8, transcription
1. Introduction
The heat shock (HS) response is an ubiquitous and phylogenetically conserved response that protects cells against a myriad of physical and chemical stresses [1-5]. It is generally characterized by a shift in the transcriptional program resulting in the induced expression of a set of evolutionarily conserved heat shock protein (HSP) genes that preserve cell viability by sequestering denatured cellular proteins and facilitating their refolding or degradation. Expression of HSP genes is regulated by the stress-activated transcription factor, heat shock factor-1 (HSF1), which binds to cis-acting heat shock response elements (HSEs) comprising inverted triad/dyad nGAAn repeats [6-8]. Activation of HSF1 following stress is a complex multi-step process comprising trimerization, nuclear translocation, acquisition of DNA binding activity, and transactivation of responsive genes [8, 9]. Trimerization of HSF1 directly confers DNA-binding capacity but transactivation requires subsequent modifications that include phosphorylation and sumoylation [10-12]. Although identified as an activator of HSP genes, recent studies have indicated an expanding role of HSF1, including its role as a transcriptional regulator of several non-HSP genes, several of which participate in the regulation of inflammatory responses and immune defenses [13-24]. For example, HSF1 mediates HS-induced transcriptional repression of human pro-interleukin (IL)-1β, c-fos, urokinase-type plasminogen activator [13, 19, 20], and TNFα genes [14, 15, 17, 18, 22]. We found that exposure to febrile-range hyperthermia (FRH) markedly enhanced intrapulmonary expression of the CXC chemokines, KC, LIX and MIP-2 in mouse models of pneumonia and pulmonary oxygen toxicity [23-25] and demonstrated that this phenomenon requires HSF1 [24]. We showed that HSF1 binds to at least two 5′-flanking regions in the human CXC chemokine, IL-8/CXCL8, gene and that interaction of HSF1 with these CXCL8 promoter sequences augments tumor necrosis factor-α (TNFα)-induced activation of IL-8/CXCL8 transcription [24]. Similar effects of HS were also demonstrated for the iNOS gene in which HSF1 binds to the iNos gene promoter and enhances LPS-induced iNos gene expression in heat shocked RAW macrophage cell line [26].
We previously performed a computational analysis of potential HSEs in CXC chemokine 5′-flanking sequences that revealed the presence of putative HSE sequences in 28 of 29 human and mouse CXC chemokines [21]. However, the capacity of these promoter sequences to bind HSF1 and the consequences for gene activation have not yet been analyzed. In the present study we extend our previous observations of CXCL8 expression by analyzing the interaction of HSF1 with the human CXC chemokines, CXCL1, CXCL2, CXCL3, and CXCL5 and analyzed the effects of HS on expression of each of these genes.
2. Materials and methods
2.1. Cell culture
Human pulmonary epithelial-like A549 cells were maintained in RPMI 1640 supplemented with 50 U/ml penicillin, 50 μg/ml streptomycin, 2 mM L-glutamine, 1 mM sodium pyruvate, 10 mM N-2-hydroxyethylpiperazine-N’-2-ethanesulfonic acid (HEPES) buffer (GIBCO/Invitrogen, Carlsbad, CA), pH 7.3 (Complete RPMI; CRPMI) and 5% defined fetal bovine serum (FBS; Hyclone, Logan, UT) as described previously [24]. For analysis of CXC chemokine gene expression, A549 cells were seeded at 0.5×105/ml in 35mm culture dishes and maintained at 37°C. After 2 days, the cells were stimulated with 1 ng/ml of human TNFα and incubated either at 37°C or immediately subjected to heat shock at 42°C for 2h followed by incubation at 37°C. Cells were harvested either immediately after HS (0h) or 1, 2, or 4h after HS for isolation of RNA. Similarly treated non-heat shocked cells at 37°C were also harvested at the corresponding time points.
2.2. Immunoblotting
A549 cells were lysed in Cell Culture Lysis buffer, resolved on 10% SDS-polyacrylamide gels, transferred to polyvinylidene fluoride membranes and immunoblotted for HSP72 (rat monoclonal, Santa Cruz) and β-tubulin (mouse monoclonal, Chemicon) essentially as described earlier [24, 27, 28].
2.3. RNA Extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated using RNeasy Kit (QIAGEN) according to the manufacturer’s instructions. Isolated RNA was reverse-transcribed using oligo-dT primers and a cDNA synthesis kit according to the manufacturer’s protocol (Promega). Duplicate 25 μl real-time PCR reactions were performed in 96 well plates using a SYBR-Green reaction mix (BioRad) in BioRad iCycler (Thermocycler) according to the supplier’s protocol. Primer pairs for CXCL-1, 2, 3 and 5 were obtained from SuperArray while primer pairs for CXCL8, HSPA1A and GAPDH were obtained from Realtimeprimers.com. Data were quantified using the gene expression Ct difference method as described by Schefe et al. [29] and standardized to levels of the housekeeping gene, GAPDH using Ct values automatically determined by the thermocycler. Efficiency of amplification for each primer pair was calculated and data analyzed essentially as descried earlier [17, 24, 27].
2.4. Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were performed using a kit from Millipore according to the manufacturer’s protocol as we have previously described [15, 18, 24]. Briefly, A549 cells were heat shocked at 42°C for 1h and fixed by adding formaldehyde (Sigma, St. Louis, MO) to the medium to a final concentration of 1%. After 15 min the cells were washed, resuspended in SDS lysis buffer and sonicated. Immunoprecipitation was carried out at 4°C overnight using 2 μg anti-HSF-1 rabbit polyclonal antibody or non-immune rabbit IgG as a control (both from SantaCruz) and immune complexes were washed, eluted, and protein-DNA cross linking was reversed according to the manufacturer’s protocol. Immunoprecipitated DNA was quantified by qRT-PCR using primer pairs spanning the HRE consensus sequence for the respective genes (Table 1) as described earlier [18]. qRT-PCR data was analyzed using a template from SA Biosciences, and fold enrichment with specific anti-HSF1 antibody against non-specific rabbit polyclonal antibody was determined [18].
Table 1.
CXCL genes, number of HSE sites and primer pairs for amplification of the flanking sequences following HSF1 ChIP assays
| CXCL | Name/ (Accession No.) |
Amplified Region* |
Primer Pairs (F/R) | HSE sites |
|---|---|---|---|---|
| 1 | Gro1, Groα, MGSA, NAP3 (NM 001511) |
−873/−723 | gagtgacaaccagtgccgta/ gccagggctgtataacatgaa |
1 |
| −676/−551 | gttgccaccaaatggaaaac/ tgctgtctgtgaaagtacagtcc |
3 | ||
| −300/−185 | cctctcaggtggtatcttcagc/ gcctcgcccttcagagtaac |
1 | ||
| −177/−19 | atcagtggacccccacac/ accccttttatgcatggttg |
1 | ||
| 2 | MIP2α, Groβ (NM 002089) |
−1511/1362 | tacttctaacaaccccgtgagg/ gagatcacttgataaggatgtcagg |
2 |
| −652/−502 | gctccactacgttgaaacaca/ gctttaacagtacatgtgtcatctca |
2 | ||
| −371/−212 | cccccggtaaggatgtagc/ tctttctgccccgaatcc |
1 | ||
| −88/41 | ctggagctccgggaattt/ gaggagagctggcaaggag |
1 | ||
| 3 | MIP2β, Groγ (NM 002090) |
−1109/−989 | ccagagaaagatcccccagt/ ttcccactgaaggagcaaac |
1 |
| −892/−736 | agcttcagagtgacagccagt/ cagggctgtataacatgacctct |
1 | ||
| −674/−525 | ggaaaacgtaaacaaggtattctaa/ ttttcggtttagcgttatttca |
2 | ||
| −154/−11 | ctacccgtatccgactccac/ gatcggcgaaccctttttat |
1 | ||
| 5 | ENA78 (NM 002994) |
−1172/−1019 | ttcaatttcaggcagcagtg/ cccagtctgttttccctcaa |
1 |
| −495/−345 | ctgcaaggaagacaggaagg/ gagagaaaaggtggaaccaaa |
1 | ||
| 8 | IL8, NAF, GCP1 (NM 000584) |
−1230/−1081 | tactatcataagaacccttccttgg/ ctagcaaaagggatggagtga |
4 |
| −820/−717 | tgaagccctcctattcctca/ gcagaatagacaagtggtactaagaca |
1 |
numbers denote positions relative to transcription start site.
2.5. ELISA for analysis of CXCL8 and CXCL5 production
IL-8 (CXCL8) and ENA 78 (CXCL5) production was measured by ELISA essentially as described earlier [24]. Briefly, A549 cells were plated in 24-well culture plates at 5.0×104 cells/ml/well 24 h before stimulation with TNFα. Cells were stimulated with recombinant human TNFα (R & D Systems; Minneapolis, MN) for 15 min at 37°C and then either were incubated at 37°C for an additional 6h (control) or were heat-shocked by incubating for 2h at 42°C then returned to 37°C for an additional 4h. IL-8 and ENA 78 concentration in culture supernatants was measured in the University of Maryland Cytokine Core Lab by ELISA using paired antibodies from R & D Systems (Minneapolis, MN). The lower detection limit for these assays were 3.9 and 15 pg/ml, respectively.
2.6. Statistical analysis
Data are presented as mean ± standard error (SE). Differences between two groups were tested using an unpaired Student t-test. Differences among more than two groups were analyzed by applying a Tukey Honestly Significant Difference test to a one-way analysis of variance (ANOVA). Differences in chemokine gene expression levels over time between heat-shocked and control cells were analyzed by Multivariate analysis of variance (MANOVA). Differences with p<0.05 were considered significant.
3. Results
3.1. HS activates HSF1 and induces HSP72 expression in A549 cells
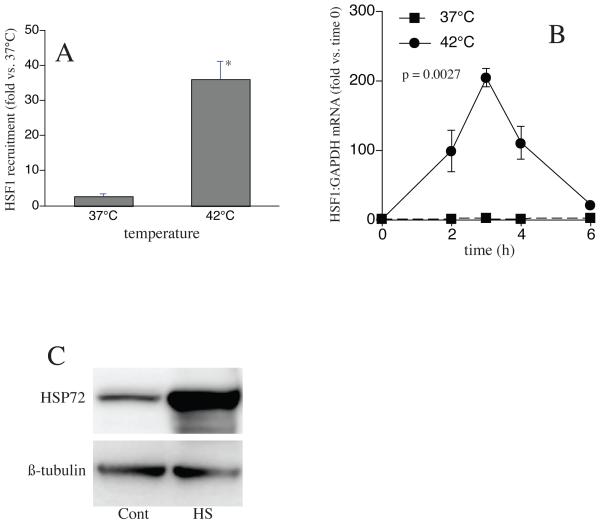
We analyzed the effects of HS on CXC chemokine gene expression in human pulmonary epithelial-like A549 cells, in which we previously demonstrated augmented CXCL8 expression following HS. To confirm that the HS protocol used in this study stimulated a significant HS response, we exposed the cells to HS and analyzed activation of HSF1 and induction of HSP72 gene expression. HSF1-ChIP assays showed that exposing A549 cells at 42°C for 60 min increased the recruitment of HSF1 to the HSP72 chromatin by approximately 14-fold compared with non-heat shock control cells (Fig 1a). To analyze the induction of HSP72 expression, A549 cells were heat shocked at 42°C for 2h, then switched to 37°C and total RNA was prepared either immediately after completion of the HS exposure (0h) or after a 1, 2 or 4h recovery. To analyze HSP72 protein expression, A549 cells were heat shocked for 2h, recovered at 37°C for 4h and cell extracts analyzed for HSP72 protein by immunoblotting. As expected, HS caused an 80-100-fold increase in HSP72 mRNA (Fig 1b) and a 5-fold increase in HSP72 protein (Fig 1c) compared with non-heat shocked control cells.
1. Exposing A549 cells to HS recruits HSF1 to the HSP72 gene and induces HSP72 mRNA and protein expression.
A. A549 cells were heat-shocked at 42°C for 1h, DNA and protein were cross-linked with formaldehyde, and HSF1 recruitment to the HSP72 gene analyzed by ChIP assay. B. For analysis of HSP72 mRNA levels, A549 cells were heat-shocked at 42°C for 2h then switched to 37°C. Total RNA was prepared either immediately after HS or after 1, 2 or 4h recovery at 37°C and analyzed by qRT-PCR for HSP72 and standardized for GAPDH. Data are presented as mean±SE (n=4), * denotes p<0.05, in comparison to non-heat shocked controls. C. For analysis of HSP72 protein levels, cells were heat shocked at 42°C for 2h then switched to 37°C for 4h. Cells were lysed and HSP72 protein was analyzed by immunoblot and standardized to β-tubulin. Figure representative of two independent experiments.
3.2. HS recruits HSF1 to CXC chemokine gene promoters in A549 cells
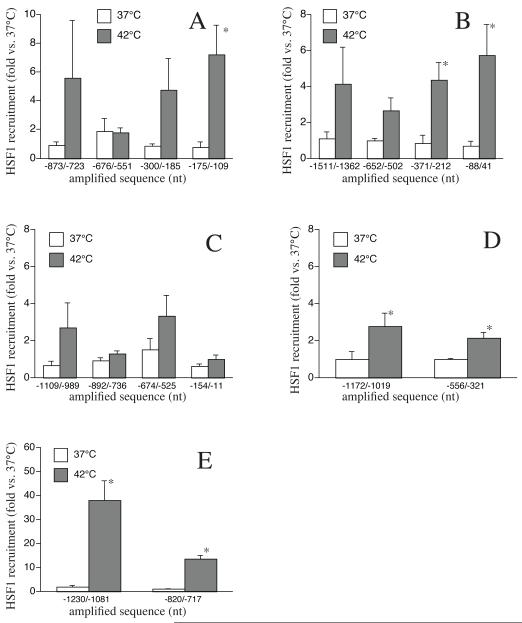
We have previously shown that the genes of several CXC chemokines including CXCL1, CXCL2, CXCL3, CXCL5, and CXCL8 contain multiple putative HSE sequences in their 5′-flanking sequences [21]. To test whether these HSE sequences were functional, we analyzed HS-induced recruitment of HSF1 to the HSE-containing regions of each of these genes using ChIP assays. This analysis showed that some of the HSEs were nonfunctional, but that HSF1 was recruited to at least one of the HSE-containing promoter regions in all but one of these CXC chemokine genes. The CXCL1 gene contains six putative HSE sequences distributed within four 5′-flanking regions, −873/−723 (1 HSE), −676/−551 (3 HSEs), −300/−185 (1 HSE) and −177/−19 (1 HSE) (see Table 1). Of the four regions analyzed by ChIP, only the −175/−19 region exhibited significant HS-induced HSF1 recruitment (Fig 2a). In the case of CXCL2 gene promoter, which also contained six HSEs distributed within four regions of the 5′-flanking sequence, only regions −371/−212 and −88/+41 (each containing 1 HSE) exhibited significant HS-induced HSF1 recruitment (Fig 2b). In contrast, exposure to HS did not induce HSF1 recruitment to any of the four 5′-flanking regions of CXCL3, which contain five putative HSE sequences. However, both of the two putative HSE-containing 5′-flanking regions in CXCL5 exhibited significant HS-induced HSF1 (Fig 2d). Finally, as we have previously reported, CXCL8 gene, which has five putative HSEs distributed over two 5′flanking regions, exhibited strong HS-induced recruitment of HSF1 to both regions, −1230/−1081 (4 HSEs) and −820/−717 (1 HSE) (Fig 2e).
2. HSF1 is recruited to CXC chemokine gene promoters in heat shocked A549 cells.
A549 cells were heat shocked at 42°C for 1h, and cross-linked chromatin was immunoprecipitated with either anti-HSF1 monoclonal antibody or isotype-specific control IgG (SantaCruz). Primer pairs for the indicated 5′-flanking regions containing HSEs (Table 1) were used to quantify enrichment of HSF1 to the respective sequences. A. CXCL1, B. CXCL2, C. CXCL3, D. CXCL5, and E. CXCL8. Data presented as mean±SE (n=4), * denotes p<0.05, in comparison to non-heat shocked controls.
3.3. HS exposure differentially affects TNFα-induced CXC chemokine gene expression in A549 cells
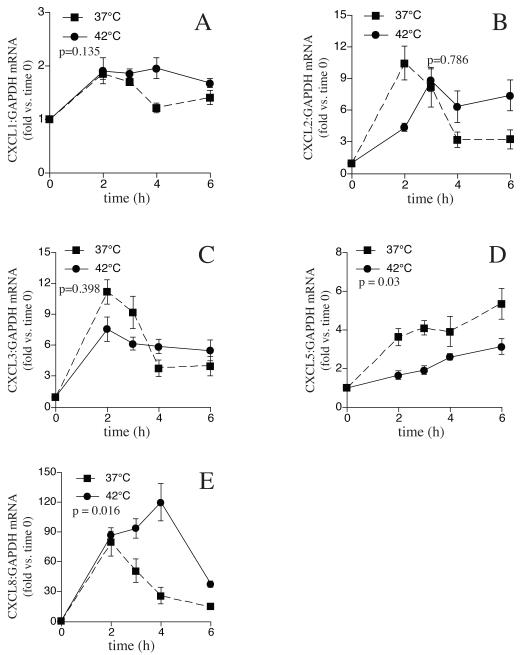
Having determined that at least some of the putative HSEs contained in the 5′-flanking regions of four of the five CXC chemokine genes studied exhibited HS-induced recruitment of HSF1, we analyzed the consequences of these interactions with respect to CXC chemokine gene expression. CXC chemokines are inducible genes and have little or no basal level expression in the absence of pro-inflammatory stimuli in A549 cells. As we previously demonstrated for CXCL8 [24], HS exposure in the absence of pro-inflammatory stimuli failed to activate detectable expression of any of the five CXC chemokine genes (data not shown). Following stimulation with TNFα at 37°C, mRNA levels of all five CXC chemokines increased several fold (Fig 3). Coincident exposure to HS exerted distinct effects on the TNFα-induced expression of the five CXC chemokines studied and the magnitude and direction of the effect did not correlate with HSF1 recruitment to the putative HSEs present on their gene promoters. For example, as we have shown earlier [24] treating A549 cells with TNFα for 2, 3, 4, and 6h stimulated 80-, 51-, 26, and 15-fold increases in CXCL8 mRNA levels compared with untreated control cells (Fig 3e). Co-treatment with TNFα and HS did not increase the initial increase in CXCL8 mRNA above levels seen with TNFα stimulation at 37°C, but sustained the increased expression so that the levels at the 3, 4, and 6h time points were 1.8-, 4.6-, and 2.5-fold higher compared with cells exposed to TNFα alone without HS. (Fig 3e). In contrast to the effects of HS on CXCL8 expression, levels of CXCL5 mRNA, which increased ~4-6 fold following stimulation with TNFα at 37°C, were reduced by about half at all time points analyzed when TNFα treatment was accompanied by HS exposure (Fig 3d). For CXCL1 and CXCL2, mRNA levels increased 2-10-fold after treatment with TNFα at 37°C, but coincident exposure to HS had no effect on the TNFα-induced expression level of either gene (Fig 3a, b). As expected, the expression of CXCL3, which did not exhibit HS-induced HSF1 recruitment to its 5′-flanking region, was not affected by HS exposure (Fig 3c).
3. TNFα-induced CXC chemokine gene expression is differentially modified in heat shocked A549 cells.
A549 cells were treated with TNFα (1ng/ml) and incubated either at 37°C for up to 6h or heat-shocked at 42°C for 2h and then incubated at 37°C for up to 4h. Total RNA was prepared either immediately after HS or 1, 2 or 4h after HS from heat-shocked and corresponding non-heat-shocked controls. qRT-PCR for A. CXCL1, B. CXCL2, C. CXCL3, D. CXCL5, and E. CXCL8 and GAPDH was performed using primer pairs from SA Biosciences. Data presented as mean±SE (n=4). Comparison between heat-shocked and non-heat-shocked cells were analyzed by MANOVA and p-values less than 0.05 was considered statistically significant.
3.4. HS exposure exerts opposing affects on TNFα-induced secretion of ENA-78 and IL-8 in A549 cells
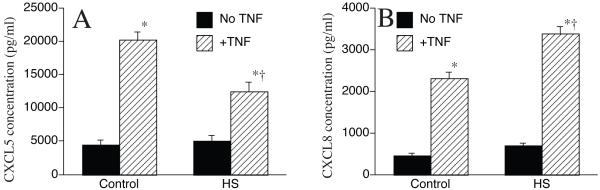
To further analyze the effect of heat shock on the CXC chemokines we determined the production and release of ENA-78 (CXCL-5) and IL-8 (CXCL-8). A549 cells were treated with TNFα and either incubated at 37°C for 6h or heat shocked for 2h and then incubated at 37°C for an additional 4h. Culture supernatants were collected and ENA-78 and IL-8 was measured by ELISA (Fig 4). As expected, both ENA-78 and IL-8 levels were markedly enhanced in culture supernatants of TNFα-stimulated A549 cells. However, in the heat shocked TNFα -stimulated cells, ENA-78 levels were significantly reduced (~40%) (Fig 4a) whereas IL-8 levels were significantly enhanced (~50%) (Fig 4b) in comparison to TNFα-stimulated cells incubated only at 37°C. Thus, similar to the mRNA levels, heat shock exposure had an opposing effect on TNFα-induced expression of CXCL-5 and CXCL-8 proteins.
4. TNFα-induced ENA-78 and IL-8 production is differentially modified in heat shocked A549 cells.
A549 cells were treated with TNFα (1ng/ml) and incubated either at 37°C for up to 6h or heat shocked at 42°C for 2h and then incubated at 37°C for up to 4h. Cell culture supernatant was collected and ENA-78/CXCL5 (A) and IL-8/CXCL8 (B) was assayed by ELISA. Data presented as mean±SE (n=8), * denotes p<0.05, in comparison to controls, † denotes p<0.05, in comparison to non-heat shocked TNFα-stimulated cells.
4. Discussion
Our results indicate that some but not all of the putative HSEs that were previously identified in five human CXC chemokine gene promoters [21] are capable of binding HSF1 in heat shocked cells. Recruitment of HSF1 to these putative sites varied markedly among the five CXC chemokine genes studied and among multiple putative HSE sequences in the same CXC chemokine gene. Moreover, interaction of HSF1 with each of the CXC chemokine genes analyzed elicits distinct effects, including both positive and negative effects on gene expression.
The first evidence that HSF1 may regulate expression of non-HSP genes was provided by Westwood et al [30] who showed that HS stimulated recruitment of Drosophila HSF to 150 chromosomal loci, which are far more than could be accounted for by the known HSP genes. Recently, Trinklein et al. [31] used ChIP and human promoter microarray analyses to demonstrate that of 182 promoters with putative HSE sequences, HSF1 was recruited to only 94 of these promoters and only 46 among them demonstrated HS-induced activation [31]. The pattern of these results is similar to those of the present study. Of the 24 predicted potential HSEs in the five CXC chemokine genes analyzed, only 10 exhibited HS-induced HSF1 recruitment, and among the four CXCL genes that did exhibit HS-inducible HSF1 recruitment, only two (CXCL8 and CXCL5) exhibited an effect of HS on TNFα-induced gene expression. One important difference between our study and the Trinklein [31] study is the treatment with the pro-inflammatory agonist, TNFα, to detect an effect of HS on CXCL gene expression and in its absence, HS alone failed to exert any significant effect on expression of the CXCL genes. These results confirm our previous studies of IL-8 [24] and TNFα [14, 15], indicating multiple distinct mechanisms through which HSF1 may modify gene activation.
Regarding the opposing effects of HS on CXCL8 and CXCL5 gene expression, HSF1 has been found to act both as an activator and a repressor of non-HSP genes. We previously demonstrated that activated HSF1 binds to the HSE sequence present in the TNFα 5′-UTR and represses TNFα transcription [14, 15]. Others have shown HSF1 to bind to the IL-1β gene promoter to inhibit IL-1β gene expression [13, 20]. In contrast, HSF1 binds to an HSE sequence in the iNos gene promoter and augments LPS-induced activation of iNos gene expression in heat-shocked murine macrophages [26]. Thus, binding of HSF1 to gene promoters can exert multiple patterns of gene regulation, including: (1) no effect; (2) transcriptional activation; (3) co-activation of transcription in the presence of a pro-inflammatory stimulus; and (4) transcriptional repression. That the interaction of HSF1 with gene promoters result in different, gene-specific effects underscores the complexity of the mechanisms through which HSF1 regulates transcription. For example, Xie et al. [13] showed that inhibition of IL-1β gene transcription was dependent upon direct interaction and mutual inhibition of HSF1 with the transcription factor, C/EBP-β that binds to adjacent sites in the IL-1β 5′-flanking sequence. Whereas in the case of iNOS, HSF1 binds to a site bordering an E-box in the iNOS gene promoter, which binds transcription factors belonging to the USF family [26]. Thus the spatial orientation of HSF1 binding relative to binding of other transcriptional regulators appear to be critical for HSF1 mediated gene expression. Additionally, post-translational modifications like acetylation, phosphorylation and sumoylation are also key regulators of HSF1 activity [8-12, 32-34]. For example, acetylation on certain lysine residues inhibit HSF1 DNA-binding activity [32], whereas phosphorylation of certain serines like S-303/307 inhibit transactivation but not HSF-1 DNA-binding activity [8, 33]. Conversely, phosphorylation at certain other serines like S-230 was shown to enhance HSF activity [34] indicating the crucial role of these processes in HSF1 regulated gene expression.
It is also important to note that increase in HSF1 transactivating capacity is dependent on both the degree of temperature elevation and the time of exposure [27]. Moreover, inflammatory mediators like arachidonic acid [35], type I interferons [36], or PGE2 [28], all modify activation of HSF1-dependent transcription. Mice deficient in HSF1 exhibit enhanced TNFα expression and increased mortality following experimental endotoxemia [37], providing further evidence for the role of HSF1 in modifying inflammatory responses and immune defenses in vivo. Our present studies also suggest that HSF1 may modify expression of CXC chemokines during stress and inflammation. Our own findings demonstrating enhanced intrapulmonary expression of murine KC (CXCL1), MIP-2 (CXCL2), and LIX (CXCL5/6) in experimental pneumonia, endotoxin aspiration, and pulmonary oxygen toxicity [23, 25] support this hypothesis and underscore the importance of understanding the complex biology of HSF1.
In conclusion, we have shown that in heat-shocked A549 cells, HSF1 is recruited to several of the predicted putative HSE-containing flanking sequences in CXC chemokine genes and that binding of HSF1 to each of these CXCL gene promoters has distinct consequences, including transcriptional activation and repression of CXCL gene expression.
Acknowledgements
Supported by NIH grants GM069431 (ISS) and GM066855, HL69057 and HL085256 (JDH), and by VA Merit Review grants to JDH and ISS.
Abbreviations
- HS
heat shock
- HSP
heat shock protein
- HSF
heat shock factor
- HSE
heat shock response element
- ChIP
chromatin immunoprecipitation
- TNF
tumor necrosis factor
- IL
interleukin
- FRH
febrile-range hyperthermia
- PCR
polymerase chain reaction
- qRT-PCR
quantitative real-time PCR
- ANOVA
analysis of variance
- MANOVA
multivariate analysis of variance
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Morimoto RI, Sarge KD, Abravaya K. Transcriptional regulation of heat shock genes. A paradigm for inducible genomic responses. J Biol Chem. 1992;267:21987–90. [PubMed] [Google Scholar]
- [2].Morimoto RI, Kline MP, Bimston DN, Cotto JJ. The heat-shock response: regulation and function of heat-shock proteins and molecular chaperones. Essays Biochem. 1997;32:17–29. [PubMed] [Google Scholar]
- [3].Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–96. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- [4].Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–11191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- [5].Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–82. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- [6].Sarge KD, Zimarino V, Holm K, Wu C, Morimoto RI. Cloning and characterization of two mouse heat shock factors with distinct inducible and constitutive DNA-binding ability. Genes Dev. 1991;5:1902–11. doi: 10.1101/gad.5.10.1902. [DOI] [PubMed] [Google Scholar]
- [7].Perisic O, Xiao H, Lis JT. Stable binding of Drosophila heat shock factor to head-to-head and tail-to-tail repeats of a conserved 5 bp recognition unit. Cell. 1989;59:797–806. doi: 10.1016/0092-8674(89)90603-x. [DOI] [PubMed] [Google Scholar]
- [8].Pirkkala L, Nykanen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. Faseb J. 2001;15:1118–31. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- [9].Sarge KD, Murphy SP, Morimoto RI. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol. 1993;13:1392–407. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, et al. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci U S A. 2006;103:45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hietakangas V, Ahlskog JK, Jakobsson AM, Hellesuo M, Sahlberg NM, Holmberg CI, et al. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol Cell Biol. 2003;23:2953–68. doi: 10.1128/MCB.23.8.2953-2968.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Guettouche T, Boellmann F, Lane WS, Voellmy R. Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem. 2005;6:4. doi: 10.1186/1471-2091-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Xie Y, Chen C, Stevenson MA, Auron PE, Calderwood SK. Heat shock factor 1 represses transcription of the IL-1beta gene through physical interaction with the nuclear factor of interleukin 6. J Biol Chem. 2002;277:11802–10. doi: 10.1074/jbc.M109296200. [DOI] [PubMed] [Google Scholar]
- [14].Singh IS, Viscardi RM, Kalvakolanu I, Calderwood S, Hasday JD. Inhibition of tumor necrosis factor-alpha transcription in macrophages exposed to febrile range temperature. A possible role for heat shock factor-1 as a negative transcriptional regulator. J Biol Chem. 2000;275:9841–8. doi: 10.1074/jbc.275.13.9841. [DOI] [PubMed] [Google Scholar]
- [15].Singh IS, He JR, Calderwood S, Hasday JD. A high affinity HSF-1 binding site in the 5′-untranslated region of the murine tumor necrosis factor-alpha gene is a transcriptional repressor. J Biol Chem. 2002;277:4981–8. doi: 10.1074/jbc.M108154200. [DOI] [PubMed] [Google Scholar]
- [16].Inouye S, Fujimoto M, Nakamura T, Takaki E, Hayashida N, Hai T, et al. Heat shock transcription factor 1 opens chromatin structure of interleukin-6 promoter to facilitate binding of an activator or a repressor. J Biol Chem. 2007;282:33210–7. doi: 10.1074/jbc.M704471200. [DOI] [PubMed] [Google Scholar]
- [17].Cooper ZA, Ghosh A, Gupta A, Maity T, Benjamin IJ, Vogel SN, et al. Febrile-range temperature modifies cytokine gene expression in LPS-stimulated macrophages by differentially modifying NF-{kappa}B recruitment to cytokine gene promoters. Am J Physiol Cell Physiol. 2010;298:C171–81. doi: 10.1152/ajpcell.00346.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cooper ZA, Singh IS, Hasday JD. Febrile range temperature represses TNF-alpha gene expression in LPS-stimulated macrophages by selectively blocking recruitment of Sp1 to the TNF-alpha promoter. Cell Stress Chaperones. 2010;15:665–73. doi: 10.1007/s12192-010-0179-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chen C, Xie Y, Stevenson MA, Auron PE, Calderwood SK. Heat shock factor 1 represses Ras-induced transcriptional activation of the c-fos gene. J Biol Chem. 1997;272:26803–6. doi: 10.1074/jbc.272.43.26803. [DOI] [PubMed] [Google Scholar]
- [20].Cahill CM, Waterman WR, Xie Y, Auron PE, Calderwood SK. Transcriptional repression of the prointerleukin 1beta gene by heat shock factor 1. J Biol Chem. 1996;271:24874–9. [PubMed] [Google Scholar]
- [21].Nagarsekar A, Hasday JD, Singh IS. CXC chemokines: a new family of heat-shock proteins? Immunol Invest. 2005;34:381–98. doi: 10.1081/imm-200067648. [DOI] [PubMed] [Google Scholar]
- [22].Singh IS, He JR, Hester L, Fenton MJ, Hasday JD. Bacterial endotoxin modifies heat shock factor-1 activity in RAW 264.7 cells: implications for TNF-alpha regulation during exposure to febrile range temperatures. J Endotoxin Res. 2004;10:175–84. doi: 10.1179/096805104225004851. [DOI] [PubMed] [Google Scholar]
- [23].Hasday JD, Garrison A, Singh IS, Standiford T, Ellis GS, Rao S, et al. Febrile-range hyperthermia augments pulmonary neutrophil recruitment and amplifies pulmonary oxygen toxicity. Am J Pathol. 2003;162:2005–17. doi: 10.1016/S0002-9440(10)64333-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Singh IS, Gupta A, Nagarsekar A, Cooper Z, Manka C, Hester L, et al. Heat shock co-activates interleukin-8 transcription. Am J Respir Cell Mol Biol. 2008;39:235–42. doi: 10.1165/rcmb.2007-0294OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rice P, Martin E, He JR, Frank M, DeTolla L, Hester L, et al. Febrile-range hyperthermia augments neutrophil accumulation and enhances lung injury in experimental gram-negative bacterial pneumonia. J Immunol. 2005;174:3676–85. doi: 10.4049/jimmunol.174.6.3676. [DOI] [PubMed] [Google Scholar]
- [26].Goldring CE, Reveneau S, Chantome A, Pance A, Fleury C, Hume DA, et al. Heat shock enhances transcriptional activation of the murine-inducible nitric oxide synthase gene. Faseb J. 2000;14:2393–5. doi: 10.1096/fj.98-0509fje. [DOI] [PubMed] [Google Scholar]
- [27].Tulapurkar ME, Asiegbu BE, Singh IS, Hasday JD. Hyperthermia in the febrile range induces HSP72 expression proportional to exposure temperature but not to HSF-1 DNA-binding activity in human lung epithelial A549 cells. Cell Stress Chaperones. 2009;14:499–508. doi: 10.1007/s12192-009-0103-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Shah NG, Tulapurkar ME, Singh IS, Shelhamer JH, Cowan MJ, Hasday JD. Prostaglandin E2 potentiates heat shock-induced heat shock protein 72 expression in A549 cells. Prostaglandins Other Lipid Mediat. 2010;93:1–7. doi: 10.1016/j.prostaglandins.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Schefe JH, Lehmann KE, Buschmann IR, Unger T, Funke-Kaiser H. Quantitative real-time RT-PCR data analysis: current concepts and the novel “gene expression’s CT difference” formula. J Mol Med. 2006;84:901–10. doi: 10.1007/s00109-006-0097-6. [DOI] [PubMed] [Google Scholar]
- [30].Westwood JT, Clos J, Wu C. Stress-induced oligomerization and chromosomal relocalization of heat-shock factor. Nature. 1991;353:822–7. doi: 10.1038/353822a0. [DOI] [PubMed] [Google Scholar]
- [31].Trinklein ND, Murray JI, Hartman SJ, Botstein D, Myers RM. The role of heat shock transcription factor 1 in the genome-wide regulation of the mammalian heat shock response. Mol Biol Cell. 2004;15:1254–61. doi: 10.1091/mbc.E03-10-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Westerheide SD, Anckar J, Stevens SM, Jr., Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–6. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chu B, Soncin F, Price BD, Stevenson MA, Calderwood SK. Sequential phosphorylation by mitogen-activated protein kinase and glycogen synthase kinase 3 represses transcriptional activation by heat shock factor-1. J Biol Chem. 1996;271:30847–57. doi: 10.1074/jbc.271.48.30847. [DOI] [PubMed] [Google Scholar]
- [34].Holmberg CI, Hietakangas V, Mikhailov A, Rantanen JO, Kallio M, Meinander A, et al. Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. Embo J. 2001;20:3800–10. doi: 10.1093/emboj/20.14.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Jurivich DA, Sistonen L, Sarge KD, Morimoto RI. Arachidonate is a potent modulator of human heat shock gene transcription. Proc Natl Acad Sci U S A. 1994;91:2280–4. doi: 10.1073/pnas.91.6.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pica F, Rossi A, Santirocco N, Palamara A, Garaci E, Santoro MG. Effect of combined alpha IFN and prostaglandin A1 treatment on vesicular stomatitis virus replication and heat shock protein synthesis in epithelial cells. Antiviral Res. 1996;29:187–98. doi: 10.1016/0166-3542(95)00834-9. [DOI] [PubMed] [Google Scholar]
- [37].Xiao X, Zuo X, Davis AA, McMillan DR, Curry BB, Richardson JA, et al. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. Embo J. 1999;18:5943–52. doi: 10.1093/emboj/18.21.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]