Abstract
Cartilage has a poor intrinsic repair capacity, requiring surgical intervention to effect biological repair. Tissue engineering technologies or regenerative medicine strategies are currently being employed to address cartilage repair. Mesenchymal stem cells (MSCs) are considered to be an excellent cell source for this application. However, the different gene expression profiles between the MSCs and differentiated cartilage remain unclear. In this report, we first examined the gene expression profiles between the MSCs, hyaline and elastic chondrocytes, and then identify candidate genes, which may be important in the process of MSC differentiation into hyaline and elastic cartilage. Several hundred differentially expressed genes were screened initially by microarray, including 417 simultaneously up-regulated genes in both hyaline and elastic chondrocytes, with 313 down-regulated genes. Several genes were identified that were up-regulated in hyaline chondrocytes while down-regulated in elastic chondrocytes. Both RT-PCR and western blot analysis were consistent with those results obtained by microarray analysis. Chondromodulinl (Chm1) was found to be highly expressed in MSCs differentiating to hyaline and elastic cartilage. Both collagen type II, alpha 1 (Col2a1) and cartilage homeo protein 1 (Cart1) were also highly upregulated and may be important early differentiation of MSCs to hyaline cartilage.
Keywords: Gene chip, mesenchymal stem cells, hyaline cartilage, elastic cartilage
Introduction
Defects of the cartilages of the nose and ear are commonly encountered in the fields of head and neck surgery and plastic surgery. Excellent quality of repair is required at these anatomical sites as it can influence not only the function of nose and ear but also the esthetic appearance. The poor regenerative capacity of cartilage, likely due to its avascular nature, requiresthe need for surgical intervention [1,2]. The field of tissue engineering is one approach to address this problem. Recent experimental evidence suggests that mesenchymal stem cells (MSCs) are a candidate cell source with the capacity to differentiate into multiple mesenchymal tissue lineages including bone, adipose, and cartilage tissue [3-7]. MSCs are easily obtained from various tissue sources, such as bone marrow, fat, and muscle, and can be expanded more than one million-fold in culture while retaining their capacity to differentiate [3,8-13]. These characteristics make them ideally suitable for a wide spectrum of clinical applications for repair of damaged or defective tissue.
However, it remains unclear which genes are critical in the process of differentiation toward the cartilage phenotype. To address this question, we utilized microarray analysis [gene chip] to identify the differential expression of genes between MSCs, elastic, and hyaline chondrocytes. In a further set of experiments, human MSCs were cultured under conditions known to promote chondrogenesis differentiation in vitro. Genes known to be important in cartilage were analyzed by reverse transcription PCR (RT-PCR) and western blot analysis.
Materials and methods
Isolation, cultivation and identification of MSCs
Male Sprague-Dawley (SD) rats, two months old and 120-160gms were used (Experimental Animal Center of Dalian Medicine University, Dalian, China). Rats were fed a standard rodent diet and water ad libitum according to the guidelines approved by the China association of laboratory animal care and the institutional animal care committee. Femurs were aseptically harvested, washed in a mixture of phosphate buffered saline (PBS) and antibiotics for 5 minutes, dissected of all soft tissue, transected at their epiphysis, and their marrow cavity was rinsed repeatedly with a mixture of heparin and Dulbecco's minimum essential medium (DMEM, Gibco, MD, USA). The harvested cells were collected, centrifuged at 1000 rpm for 10 minutes. Cell pellets were resuspended with DMEM, and equal-volume percoll separator liquid (Boster, Wuhan, China) with a density of 1.082 g/mL was added to the tube. After centrifugation at 2000rpm for 30 minutes, the single nucleated cells layer were separated, the MSC layer was resuspended in DMEM, and centrifuged at 2000rpm for 10 minutes. Following washes, the cells were placed in DMEM culture medium supplemented with 10% Fetal Bovine Serum (FBS, GIBCO), and cultured to the third passage. Cells were identified as MSCs by flow cytometry (FACSCaliburBD, USA).
Isolation and culture of elastic chondrocytes
Sprague-Dawley rat ear cartilage was harvested under aseptic conditions. Cartilage was minced and digested at 37°C for 11 hours in media containing 0.1% type II collagenase (Sigma, USA); the digest was centrifuged at 1500rpm for 3 minutes. The cell pellet was washed three times in culture medium, and placed into DMEM/F12 (GIBCO) containing 10% FBS and cultured. Chondrocyte cultures were split 1:2 and used in passage three for all experiments.
Isolation and culture of hyaline chondrocytes
The adventitia of Sprague-Dawley rat's costal cartilage was isolated, incised, and the underlying cartilage tissue was collected and used for RNA isolation. The cartilage was placed in chilled sterile 0.9% sodium chloride. Cells were isolated within five hours. Cartilage specimens were minced and washed three times in culture medium containing DMEM/F12 medium supplemented with HEPES buffer (10 mmol/l), gen-tamicin sulfate (70 μmol/l), amphotericin B (2.2 μmol/l), and l-ascorbic acid (300 μmol/l). The minced cartilage was digested for 12 hours in a spinner bottle in 10 ml of culture medium containing clostridial collagenase (150U/I) and de-oxyribonuclease I (25,000 U/l). The cells were then filtered through nylon mesh with a pore diameter of 25 micrometers, washed three times, resuspended in culture medium supplemented, and seeded.
Preparation of gene chips and hybridization: microarray analysis
Total RNA was extracted from the cells and minced cartilage tissue by the 1-step method using TRIzol reagent (Invitrogen, MD, USA), column purified by the NucleoSpin RNA clean-up kit (Macherey-nagel, Germany), quantified spec-trophotometric measurement, and electrophore-sed with 1% agarose. Total RNA was extracted with T7-Oligo (dT) 15 as the primer, compounded into the double stranded cDNA with the cDNA Synthesis Kit (Promega, USA), and purified by the PCR NucleoSpin Extract II Kit (Promega, USA). The double stranded cDNA was transcribed in vitro, compounded into cRNA and purified, then retrotranscribed and the purified production was then marked by KLEN0W enzyme (Takara, Japan). The chondrocytes and MSCs were marked individually by the Cy3-dCTP and Cy5-dCTP (Amersham Pharmacia Biotech, Inc., NJ, USA). Hybridization was performed at 42°C by 27K Rat Genome Array gene chip (Capital Bio Corporation, Beijing, China) containing 26962 probes. Three groups and each group containing three arrays for microarray analysis (n=9).
Chip scan and data analysis
The chip images were scanned using a double-channel laser scanner LuxScan 10KA (Capital Bio). The signals refer to unified data of the light intensity detected by the scanner and were analyzed with the image analysis software GenePix Pro 4.0 (Axon Instruments, USA). The image signals were transmitted into the digital ones, and then the data on the chips were normalized by the Lowess method [14]. Finally, the difference of expressing genes was determined by the standard of two-fold difference.
Reverse transcription PCR
To verify the microarray results, the most significantly up-regulated gene, chondromodulin (Chm1) was assayed by RT-PCR. Total RNAfrom three specimens was reverse transcribed in 20μL of reaction mixture containing 1μL of reverse transcriptase at 42°C for 50 minutes. To this was added complementary DNA, the forward (5'-GGCATGATCTTGCCTTCCAG-3') and reverse (5'-GGAAGGCAAGATCATGCCAGT-3') primers. All primers were used at a final concentration each of 1 mmol/Land PCR was performed. Denaturation was performed at 95°C for 5 minutes before 30 amplification cycles. Each cycle included 94°C degeneration for 45 seconds, 55°C annealing for 45 seconds and extension for 45 seconds, followed by a final extension of 72°C for 7 minutes. The RT-PCR products were analyzed by electrophoresis on a 2% agarose gel and were analyzed by ethidium bromide staining. The identity of each product was confirmed by molecular weight profile from the agarose gels.
Human MSCs were obtained from bone marrow as previously described and placed into tissue culture with medium containing transforming growth factor (TGF)beta [10ng/ml]. Control cultures were fed media minus TGFb. Cultures were harvested for RNA 1, 3, 7 and 14 days in vitro and prepared for RT-PCR. Primers for collagen type II, aggrecan, sox-9, and COMP were used to screen the cultures for expression of these cartilage specific proteins. Data was normalized by expression of the housekeeping gene GAPDH. Data was analyzed using a 2×2×4 factorial ANOVA.
Western blot analysis
To further verify the microarray results, we validated the differential expression of Chm1, Col2a 1 and Cart 1 protein levels in MSCs, hyaline and elastic chondrocytes by Western blot analysis. Protein was extracted from the cells. Briefly, cells were suspended in ice-cold hypo-tonic buffer [100 mmol/L HEPES (pH 7.6), 10 mmol/L KCI, 3 mmol/L NaCI, 3 mmol/L MgCI2, 1 mmol/L EDTA, 1 mmol/L EGTA, 2 mmol/L DTT, and 10% (v/v) glycerol] plus the complete protease inhibitor cocktail (Roche, Indianapolis, IN) and phosphatase inhibitor (20 mmol/L p-glycerophosphate, 1 mmol/L sodium orthova-nadate, and 30 mmol/L sodium fluoride). Protein concentration was measured using a BCA protein kit (Beyotime Institute of Biotechnology, Jiangsu, China). Proteins were separated using SDS-PAGE, then transferred to a PVDF membrane, blocked with 5% dried milk, incubated with anti-Chm1 antibody solution (1:300), anti-Cart 1 antibody (1:500), anti-Col2a 1 antibody (1:500) and anti-β-actin at 1:1,000 (Zhongshan golden bridge, Beijing, China) overnight at 4°C, washed with TBST, and incubated with secondary antibody goat anti-rabbit-lgG-ALP (Zhongshan golden bridge) conjugated at a concentration of 1:3000 for one hour at room temperature. After detection, blots were stripped and reprobed with antibodies to β-actin to confirm equivalence in loading and transfer. For color development, the Lumi-light plus western blotting substrate (Roche, USA) was added and the membrane kept at 37°C until the development of color. The reaction was stopped by washing the membrane in ddhbO and photographed using the Biolmaging system (UVP, USA). Signal intensity was quantified by UN-SCAN-IT Program (Silk Scientific, Inc., Orem, UT).
Statistical analysis
Western blot was analyzed by SPSS, version 13.0. A P<0.05 was considered statistically significant.
Results
Gene expression profiles
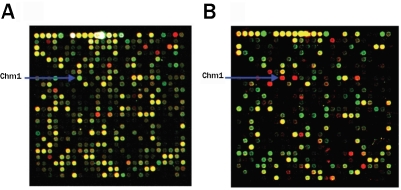
Overlay image 1 (Figure 1A) shows the dot (cy5/ cy3) of Chm1 in the group comprised of MSCs and elastic chondrocytes. Overlay image 2 (Figure IB) shows the dot (cy5/cy3) of Chm1 in the group of MSCs and hyaline chondrocytes. The up-regulated and down-regulated genes in both hyaline and elastic chondrocytes are shown in Table 1.
Figure 1.
Chm1 located on gene chips. The Overlay image (A) shows the dot (cy5/cy3) of Chm1 in the group of MSCs and elastic chondrocytes . Overlay image (B) shows the dot (cy5/cy3) representing Chm1 in the group of MSCs and hyaline chondrocytes.
Table 1.
Part of genes in hyaline and elastic chondrocytes
| Gene Name | NCBI Accession | Gene Definition | Fold Change |
|---|---|---|---|
| Part of up-regulated genes | |||
| Chm1 | NM_030854 | Chondromodulinl | 9.3 |
| Fmod | NM_080698 | Fibromodulin | 4.5 |
| Col11a1 | XM_342326 | procollagen, type XI, alpha 1 Co1l2a1 | 7.7 |
| Tnmd | NM_022290 | Tenomodulin | 3.1 |
| Cspg4 | NM_031022 | chondroitin sulfate proteoglycan 4 | 3.2 |
| S100b | NM_013191 | S100 protein, beta polypeptide | 3.7 |
| Omd | NM_031817 | Osteomodulin | 5.8 |
| Col10a1 | XM_345113 | procollagen, type X, alpha 1 Col10a1 | 4.1 |
| Fxyd1 | NM_031648 | FXYD domain-containing ion transport regulator 1 | 2.5 |
| Mia | NM_030852 | cartilage derived retinoic acid sensitive protein | 2.8 |
| Phyh | NM_053674 | phytanoyl-CoA hydroxylase | 2.9 |
| Part of down-regulated genes | |||
| Mmp8 | NM_022221 | matrix metalloproteinase-8 | -5.8 |
| Hmox1 | NM_012580 | heme oxygnase-1 | -4.2 |
| Hck | BC078890 | hemopoietic cell kinase | -3.7 |
| Lyz | NM_012771 | lysozyme c | -4.1 |
| Ela1 | NM_012552 | elastase 1 | -5.6 |
| Alcam | NM 031753 | activated leukocyte cell adhesion molecule | -4.8 |
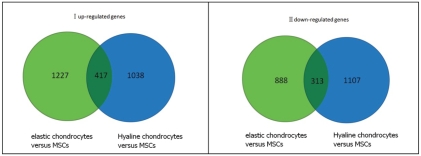
With the two-fold level of difference as the standard for significance in gene expression, the number of up-regulated genes in elastic chondrocytes versus MSCs group was 1227. In hyaline chondrocytes versus MSCs group the number was 1038, and the number of the genes which were up-regulated simultaneously in both of the two groups was 417. The number of down-regulated genes in elastic chondrocytes versus MSCs group was 888, in the hyaline chondrocytes versus MSCs group the number was 1107, and the number of the genes which were down-regulated simultaneously in both of the two groups was 313 (Figure 2).
Figure 2.
Venn diagram I (the number of up-regulated genes in elastic chondrocytes versus MSCs group was 1227, in hyaline chondrocytes versus MSCs Group the number was 1038. The number of the genes which were up-regulated simultaneously in both of the two groups was 417). Venn diagram II (the number of down-regulated genes in elastic chondrocytes versus MSCs group was 888, in hyaline chondrocytes versus MSCs Group the number was 1107. The number of the genes which were down-regulated simultaneously in both of the two groups was 313).
Figure 3A shows scatter plots in gene chip hybridization of MSCs and elastic chondrocytes (x-axis represents the signal value of elastic chondrocytes, and y-axis represents the signal value of MSCs). Figure 3B shows scatter plots in gene chip hybridization of MSCs and hyaline chondrocytes (x-axis represents the signal value of hyaline chondrocytes, and y-axis represents the signal value of MSCs). In both scatter plots, the green dots show the up-regulated genes, the red dots show the down-regulated genes and the black dots show genes that were not differentially expressed.
Figure 3.
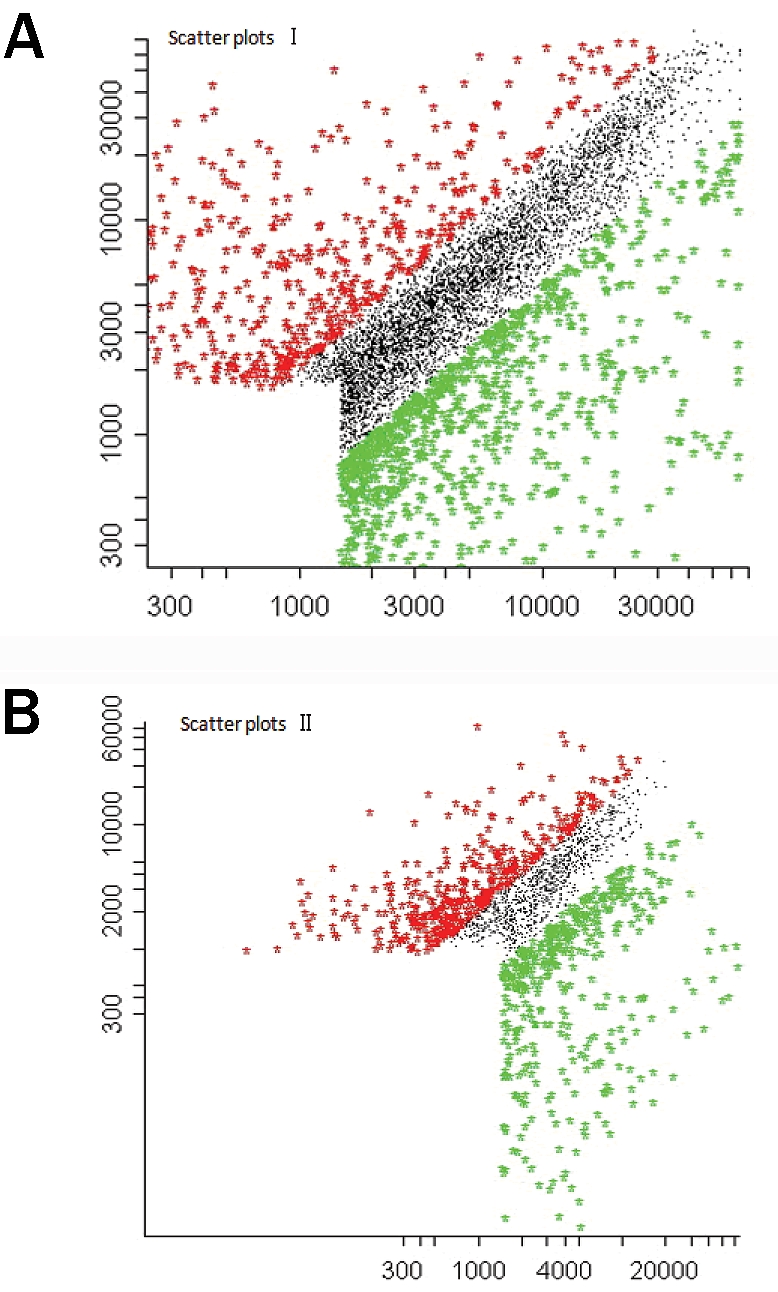
Scatter plots I: gene chip hybridization of MSCs and elastic chondrocytes (x-axis represents the signal value of elastic chondrocytes, and y-axis represents the signal value of MSCs). B. Scatter plots II: gene chip hybridization of MSCs and hyaline chondrocytes (x-axis represents the signal value of hyaline chondrocytes, and y-axis represents the signal value of MSCs). In both scatter plots, the green dots show the up-regulated genes, the red dots show the down-regulated genes and the black dots show the genes were not differentially expressed.
Identification of MSCs by morphology and flow cytometry
Third passage MSCs were uniform in shape, elongated and spindle shaped, arranged in a cobble stone appearance. Through identification by the flow cytometry, the CD13 antigen of the cells was positive (the expression rate was 98.74%), the CD45 antigen was negative (the expression rate was 1.62%). These findings were consistent with classical features of MSCs.
RNA extraction and denaturing electrophoresis
The total RNA (0D260/280) in each group was more than 1.8 and less than 2.0. Formaldehyde Denaturing Gel Electrophoresis showed two master bands, and their brightness of 28s/18s is about 2 to 1. These criteria met the requirement for continuation with the microarray analysis.
Results of RT-PCR
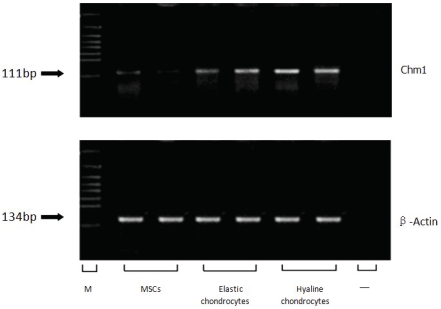
The results of the RT-PCR showed that the expression level of the gene Chm1, chondromodulinl, in hyaline and elastic chondrocytes were significantly higher than in MSCs (Figure 4).
Figure 4.
Expression of Chondromodulinl (Chm1) messenger RNA in mesenchymal stem cells (MSCs), elastic chondrocytes and hyaline chondrocytes by reverse transcription-polymerase chain reaction (RT-PCR). The PCR products are 111 base pair (bp) for Chm1. As an internal control, 134 bp bands for (β-Actin are shown. “M” indicates marker, “-"indicates negative control.
The addition of transforming growth factor (TGF) beta to MSC cultures resulted in significant upregulation of collagen type II, aggrecan, sox-9, and COMP gene expression compared to MSCs cultured under standard conditions. The increase of expression of these genes was over the 14 day time period, but was not observed in cultures without the TGFb present in the media. The results of this data set are shown in Table 2.
Table 2.
Chondrogenic gene expression of MSCs in culture
| Fold Change in Culture time | ||||
|---|---|---|---|---|
| Gene Name | Day 1 | Day 3 | Day 7 | Day 14 |
| Aggrecan | -4.8 | 2.3 | 3.3 | 4.1 |
| Collagen type II | -1.2 | 1.9 | -3.1 | -3.4 |
| COMP | 6.8 | 5.7 | 6 | 6.7 |
| Sox-9 | 7.8 | 5.2 | 7.1 | 7.5 |
Western blot analysis
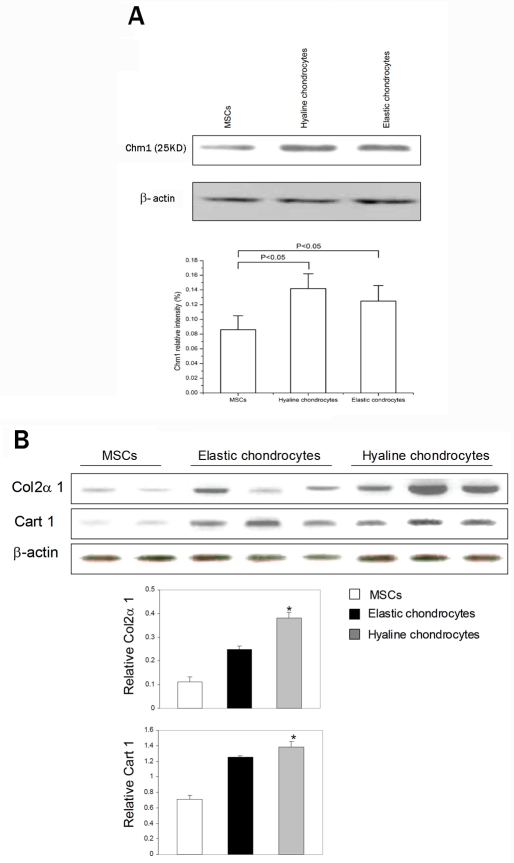
Western blot analysis showed that the relative Chm1 levels (normalized with β-actin) in hyaline chondrocytes were significantly higher than that in MSCs (P<0.05). The relative Chm1 levels in elastic chondrocytes were also higher than that in MSCs (P<0.05, n=5) (Figure 5A). However, the average expression level of Chm1 in hyaline cartilage cells compared with that in elastic cartilage cells was not significant different (P> 0.05). In addition, Col2a1 and Cart1 (normalized with β-actin) were highly expressed in hyaline chondrocytes comparing to MSCs (P<0.05, n=8) (Figure 5B).
Figure 5.
Protein levels in MSCs, hyaline and elastic chondrocytes were detected by Western blot analysis. A. Chm1 Expression was significantly increased in the hyaline and elastic chondrocyte groups compared to MSCs group (P<0.05, n=5). B. High levels of Col2a 1 and Cart 1 protein were in hyaline comparing to MSCs. Bar graphs show mean ± SD of multiple samples of each type, normalized to b-actin (P< 0.05, n=8).
Discussion
Established theories show that different cells of the same individual have a set of the same genome, and that different gene expression profiles determine the direction of cell differentiation and ultimately phenotype of the cell. Based on this theory, we evaluated the differential expression of genes of MSCs and chondrocytes, both from elastic and hyaline cartilage, by microarray analysis. The results of our study showed that there exist several hundred differentially expressed genes between the three groups. In the differential gene expression profile, Chm1 was, compared with MSCs, significantly up-regulated simultaneously in both of the two kinds of cartilages, and the Col2a1 gene and Cart1 gene were up-regulated in the hyaline cartilage, and down-regulated in elastic cartilage. To verify the results of the gene chips, the expression of the Chm1 was demonstrated at the mRNA level by RT-PCR and protein level by western blot. These genes may be critical in the differentiation pathway from the MSCs to the hyaline chondrocytes.
In theory, growth factors or morphogens able to regulate and control the expression of these genes could be used as a strategy to induce the MSCs differentiation toward cartilaginous phe-notype able to repair cartilage lesions. With the utilization of the high throughout gene chips, our studies detected the expressed genes of MSCs, hyaline chondrocytes and elastic chondrocytes and acquired differentially expressed gene patterns.
Chondromodulin, the protein product of the Chm1 gene, is a growth factor with a cartilaginous origin, and its molecular structure is similar to that of transforming growth factor. This molecule can promote the proliferation of chondrocytes, and the synthesis of the cartilage specific extracellular protein aggrecan. Chm1 was first discovered and isolated from epiphyseal cartilage, and could strongly promote the synthesis of proteoglycans such as aggrecan in the epiphyseal growth plate and in chondrocytes cultured three dimensionally in agarose. Wata-hiki et al [15] reported that Chm1 could inhibit in vitro proliferation of vascular endothelium cells and inhibit formation of tubular morphology. Thus Chm1 functions not only as a growth factor for chondrocytes, but also as a growth inhibitor of the vascular endothelium, thus imparting cartilage with its anti angiogenic capacity. Hayami et al [2] observed Chm1 in the process of joint cartilage degeneration. The expression of the Chm1 of the healthy rat joint cartilage with normal health was expressed adequately. In the early stages of osteoarthritis, Chm1 of the superficial cartilage zone was expressed in a decreasing manner. In the development of osteoarthritis, Chm1 of all layers of cartilages was shown to be decreasingly expressed, and vascular endothelial growth factor was expressed in an increasing manner. The studies mentioned above showed the normal development of Chm1 in chondrocytes of the cartilage tissues and the maintenance of the characteristic of anti angiogenic properties of cartilage tissues.
Col2a1 genes-coded proteins are assembled to form type II collagen fibers, which are main components of the extra-cellular matrix of articular cartilage. The extra-cellular matrix has an impact on the differentiation of the chondrocytes and the maintenance of their phenotypic characteristics. Type II collagen fibers participate in regulating and controlling formation of the collagen fibers [16] and play an important role in the process of cartilage development [17]. Our results showed that Col2a1 expression was up-regulated in the hyaline cartilage, while down-regulated in elastic cartilage. These results suggest that Col2a1 gene plays an important role in the differentiation of hyaline cartilage, and to increase the type II collagen fiber content in the extra-cellular matrix probably is one of the key factors determining the structural characteristics of the hyaline cartilage. Currently, the various methods for engineering cartilage are not able to form hyaline cartilage with native tissue characteristics. Combining our experimental results, the analytic reasons likely were: the oriented differentiation of type II collagen fibers was related to not only whether they were expressed or not, but also how many were expressed, and that it is necessary to promote type II collagen fibers to be expressed in large numbers in the process of the formation of tissue engineering hyaline cartilage.
Located in the No. 7 rat chromosome, the genetic sequence of Cart1 gene consists of three parts, and the middle section is the coded protein sequence. The proteins coded by the Cart1 genes are homeotic proteins, located in the nucleus. Through combining the particular locus of the target genes, the expression of target genes are activated and play the role of transcription factors [18], but the specific target genes are unknown yet. Cart 1 mRNA expression in the cartilaginous precursor aggregate was coincident to the time of the expression of the type II collagen, and this demonstrates that early cartilage formation in vitro was at the time when chondrocytes were aggregated and formed a connection between cells, providing the conditions for differentiation of cartilage [19]. A certain relationship exists between the Cart1 and Col2a1, inferring that Col2a1 was probably the target gene of the Cart1. These studies also showed that mRNA transcribed from Cart1 was expressed and up-regulated in differentiation of the rat chondrosarcoma, the cartilaginous precursor mesenchymal cells and the early stage chondrocytes, while expressed lower in the mature chondrocytes. The expression of this mRNA was not found in the various fibers of original cells, the myeloma cells and the embryonic stem cells. In addition, the Cart1 was also up-regulated in tissues with cartilaginous mesenchymal cells which would form the cartilage tissues in the future [20]. The gene chip results showed that the Cart1 gene was up-regulated in hyaline cartilage, while down-regulated in elastic cartilage. Based on this, we inferred that the Cart1 gene was able to regulate and control the expression of the Col2a1, and had an impact on the differentiation of the cartilage. The conclusions are: Cart1 in the cell nucleus of MSCs is able to be expressed and up-regulated, the product of its expression acts on its downstream gene Col2a1, the Col2a1 is able to be expressed highly until a certain quantity is present to formation of the type II collagen fiber. The interaction between the extra-cellular matrix and the cells may allow the MSCs to aggregate and differentiate toward the direction of the hyaline cartilage, and finally produce the tissue engineering hyaline cartilage with the appropriate tissue characteristics. The additional set of experiments with MSC differentiation toward cartilage demonstrate that several genes that are critical to the cartilage pheno-type are not present in undifferentiated MSCs but begin to be expressed in the transition toward a hyaline cartilage phenotype.
Acknowledgments
We acknowledge the support of Liaoning Province Education Committee Grants 2004D125, 2009A210.
References
- 1.Yu J, Yan YH, Wan T. Progress of Technology Cartilage Tissue-Engineering. Orthopaedic Bio-mechanics Materials and Clinical Study. 2005;2:35–40. [Google Scholar]
- 2.Hayami T, Funaki H, Yaoeda K, Mitui K, Yamagiwa H, Tokunaga K, Hatano H, Kondo J, Kiraki Y, Yamamoto T, Duong LT, Endo N. Expression of the cartilage derived anti— angiogenic factor chondromodulin-1 decreases in the early stage of experimental osteoarthritis. J Rheumatol. 2003;30:2207–2217. [PubMed] [Google Scholar]
- 3.Gregory CA, Prockop DJ, Spees JL. Nonhematopoietic bone marrow stem cells: molecular control of expansion and differentiation. Exp Cell Res. 2005;23:306–330. doi: 10.1016/j.yexcr.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 4.Friedenstein AJ, Chailakhyan RK, Latsinik NV, Panasyuk AF, Keiliss-Borok IV. Stromal cells responsible for transferring the microenviron-ment of the hemopoietic tissues. Cloning in vitro and retransplantation in vivo. Transplantation. 1974;17:331–340. doi: 10.1097/00007890-197404000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Pereira RF, Halford KW, O'Hara MD, Leeper DB, Sokolov BP, Pollard MD, Bagasra O, Prockop DJ. Cultures of adherent cells from marrow can serve as long-lasting precursor cells for bone, cartilage and lung in irradiated mice. Proc Natl Acad Sci USA. 1995;92:4857–4861. doi: 10.1073/pnas.92.11.4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–148. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 7.Sekiya I, Vuoristo JT, Larson BL, Prockop DJ. In vitro cartilage formation by human adult stem cells from bone marrow stroma defines the sequence of cellular and molecular events during chondrogenesis. Proc Natl Acad Sci USA. 2002;99:4397–4402. doi: 10.1073/pnas.052716199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakaguchi Y, Sekiya I, Yagishita K, Ichinose S, Shinomiya K, Muneta T. Suspended cells from trabecular bone by collagenase digestion become virtually identical to mesenchymal stem cells obtained from marrow aspirates. Blood. 2004;104:2728–2735. doi: 10.1182/blood-2003-12-4452. [DOI] [PubMed] [Google Scholar]
- 9.Yoshimura H, Muneta T, Nimura A, Yokoyama A, Koga H, Sekiya I. Comparison of rat mesenchymal stem cells derived from bone marrow, synovium, periosteum, adipose tissue, and muscle. Cell Tissue Res. 2007;327:449–462. doi: 10.1007/s00441-006-0308-z. [DOI] [PubMed] [Google Scholar]
- 10.Sakaguchi Y, Sekiyal I, Yagishita K, Muneta T. Comparison of human stem cells derived from various mesenchymal tissues: superiority of synovium as a cell source. Arthritis Rheum. 2005;52:2521–2529. doi: 10.1002/art.21212. [DOI] [PubMed] [Google Scholar]
- 11.De Bari C, Dell'Accio F, Tylzanowski P, Luyten FP. Multipotent mesenchymal stem cells from adult human synovial membrane. Arthritis Rheum. 2001;44:1928–1942. doi: 10.1002/1529-0131(200108)44:8<1928::AID-ART331>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 12.Zuk PA, Zhu M, Ashjian P, De Ugarte DA, Huang JI, Mizuno H, Alfonso ZC, Fraser JK, Benhaim P, Hedrick MH. Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell. 2002;13:4279–4285. doi: 10.1091/mbc.E02-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miura M, Gronthos S, Zhao M, Lu B, Fisher LW, Robey PG, Shi S. SHED: stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci USA. 2003;100:5807–5812. doi: 10.1073/pnas.0937635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucl Acids Res. 2002;30:el5. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watahiki J, Yamaguchi T, Enomoto A, Irie T, Yoshie K, Tachikawa T, Maki K. Identification of differentially expressed genes in mandibular condylar and tibial growth cartilages using laser microdissection and fluorescent differential display: chondromodulin-l (ChM-1) and tenomodulin (TeM) are differentially expressed in mandibular condylar and other growth cartilages. Bone. 2008;42:1053–1060. doi: 10.1016/j.bone.2007.09.048. [DOI] [PubMed] [Google Scholar]
- 16.Bosnakovski D, Mizuno M, Kim G, Takagi S, Okumura M, Fujinaga T. Chondrogenic differentiation of bovine bone marrow mesenchymal stem cells (MSCs) in different hydrogels: influence of collagen type II extracellular matrix on MSC chondrogenesis. Biotechnol Bioeng. 2006;93:1152–1163. doi: 10.1002/bit.20828. [DOI] [PubMed] [Google Scholar]
- 17.Imai K, Dalai SS, Hambor J, Mitchell P, Okada Y, William C, Horton WC, D'Armiento J. Bone growth retardation in mouse embryos expressing human collagenase 1. Am J Physiol Cell Physiol. 2007;293:1209–1215. doi: 10.1152/ajpcell.00213.2007. [DOI] [PubMed] [Google Scholar]
- 18.Furukawa K, Iioka T, Morishita M, Yamaguchi A, Shindo H, Namba H, Yamashita S, Tsukazaki T. Functional domains of paired-like homeoprotein Cart1 and the relationship between dimerization and transcription activity. Genes Cells. 2002;7:1135–1147. doi: 10.1046/j.1365-2443.2002.00587.x. [DOI] [PubMed] [Google Scholar]
- 19.Loty S, Foil C, Forest N, Sautier JM. Association of enhanced expression of gap junctions with in vitro chondrogenic differentiation of rat nasal septal cartilage-released cells following their dedifferentiation and redifferentiation. Arch Oral Biol. 2000;45:843–856. doi: 10.1016/s0003-9969(00)00062-5. [DOI] [PubMed] [Google Scholar]
- 20.Zhao GQ, Eberspaecher H, Seldin MF, de Crombrugghe B. The gene for the homeodomain-containing protein Cart-1 is expressed in cells that have a chondrogenic potential during embryonic development. Mech Dev. 1994;48:245–254. doi: 10.1016/0925-4773(94)90063-9. [DOI] [PubMed] [Google Scholar]