Abstract
The molecular signatures of the recent expansion of the western house mouse, Mus musculus domesticus, around the Mediterranean basin are investigated through the study of mitochondrial D-loop polymorphism on a 1313 individual dataset. When reducing the complexity of the matrilineal network to a series of haplogroups (HGs), our main results indicate that: (i) several HGs are recognized which seem to have almost simultaneously diverged from each other, confirming a recent expansion for the whole subspecies; (ii) some HGs are geographically delimited while others are widespread, indicative of multiple introductions or secondary exchanges; (iii) mice from the western and the eastern coasts of Africa harbour largely different sets of HGs; and (iv) HGs from the two shores of the Mediterranean are more similar in the west than in the east. This pattern is in keeping with the two-step westward expansion proposed by zooarchaeological data, an early one coincident with the Neolithic progression and limited to the eastern Mediterranean and a later one, particularly evident in the western Mediterranean, related to the generalization of maritime trade during the first millennium BC and onwards. The dispersal of mice along with humans, which continues until today, has for instance left complex footprints on the long ago colonized Cyprus or more simple ones on the much more recently populated Canary Islands.
Keywords: house mouse, mitochondrial D-loop, matrilineal phylogeography, zooarchaeology, Neolithic expansion
1. Introduction
In the present paper, we examine the expansion pattern of the western subspecies of the house mouse around the Mediterranean basin, which is thought to be the prominent theatre of its early expansion [1,2]. This subspecies, a well-known commensal of humans since Neolithic times [1–5], has been the focus of much attention during the last three decades, whether from a genetical [6–9] or a zooarchaeological standpoint [4,10]. The colonization of a new ecological niche (commensalism) and the subsequent association with humans has triggered a set of interesting questions as to the timing and pattern of its westward expansion. Most of this literature has been reviewed in a recent paper [8], which focused on the near-East and the Fertile Crescent as a possible cradle for the subspecies, and proposed an expansion onset around 10 000 years BP, in keeping with zooarchaeological data [10]. Recently, Searle and co-workers have addressed the more specific question of the origin of the mouse populations in the British Isles [11,12] as well as New Zealand [13] and Madeira [14]. The molecular data have provided support for a colonization pattern of Europe along two main routes, one termed the Mediterranean route and the other the Bosphorus/Black Sea route through Asia Minor [8]. Well-documented zooarchaeological records have suggested that the expansion through the northern Mediterranean shores took place in two waves, a first wave limited to the eastern Mediterranean, whereas western Europe was colonized only recently during the last millennium BC [10] with a possible separate colonization of its northern and southern part during the Iron Age [12]. This delay in the westward diffusion is thought to be related to both the large increase in maritime trade and the stability of the commensal niche in western Europe that occurred at this period [10]. Indeed, stowaway transport of the house mouse related to the Late Bronze Age Mediterranean trade has been directly documented by zooarchaeological data [15]. However, these studies mostly involved fossil remains as well as extant samples from the eastern and the northern Mediterranean regions, while few data were available concerning the African shore. By considerably extending our sampling around the Mediterranean basin particularly in the Levant and North Africa, including large island populations, we address the following questions.
Is it possible to identify an early southern Mediterranean route followed by house mice coinciding with the early Neolithic expansion? Is the timing and mode of colonization similar on both shores of the Mediterranean? Are the origins of the source populations the same in both cases?
The present study relies on the analysis of a fast-evolving region of the maternally transmitted mitochondrial genome (mtDNA) over a sample of 1313 individuals. Phylogeographic approaches based on mtDNA sequences have demonstrated their usefulness in reconstructing postglacial colonization pathways in many species. The house mouse, unlike less mobile species not likely to be transported by humans, is predicted to show complex historical patterns strongly influencing the distribution of its matrilines. As long as the sequences have diverged enough from each other, they will follow independent pathways and their distribution will bear the signature of the eventual complexity of the expansion of the subspecies. Namely, we expect to be able to tease apart recent versus old onsets of colonization by measuring the depth of the coalescent, and to separate multiple from single events by identifying the presence of independently derived matrilines in a given location. Two contrasting situations from large Mediterranean islands are compared to provide a referential signature of early- (Cyprus, first archaeological house mice dated from the middle of the ninth millennium BC [5]) versus late-colonization events (Canaries, first archaeological house mouse directly C14 dated around the fourth century AD [16]). Additionally, our sampling scheme also sheds light on the colonization of the western and eastern flanks of tropical Africa.
2. Material and methods
(a). Samples
The present work encompasses 321 sequences already analysed by Rajabi-Maham et al. [8], supplemented by new sequences obtained from 377 individuals from Algeria, Cyprus, Canary Islands, Tunisia, Kenya, Israel, Qatar, Senegal, Egypt, France, Germany, Lebanon, Syria, Morocco, Spain. Additionally, 615 sequences were retrieved from publications or GenBank. Altogether, 1313 sequences are included in the present study; a summary of their geographical origin is given in table 1, and detailed in electronic supplementary material, table S1. The geographical terms used to describe the origin of the samples necessarily entails some arbitrary choices. In most cases, it refers to political boundaries, but as the notion of country is not always meaningful, we instead defined a level called ‘Province’ to allow for instance the separation of European Turkey from its Asia Minor counterparts. For comparative purposes, we adopted the geographical delineations already used by Rajabi-Maham et al. [8], which themselves conserved those of Gündüz et al. [17]. Since we are aware of a possible imbalance in the geographical size of the subdivisions used, we also considered two higher order geographical levels, termed ‘Region’ and ‘Continent’; the hierarchical nesting of the three levels is easily seen in table 1.
Table 1.
Geographical location, sample abbreviation, origin and distribution across 40 population samples of 11 mitochondrial HGs as defined in figure 1b.
| region | province | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | N ind | N HG | HG div | references | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| west Africa | AFW | Cameroon | CMR | 15 | 2 | 3 | 7 | 27 | 4 | 0.61 | [41] | |||||||
| Canary Islands | CNR | Canary Island | CNR | 46 | 94 | 140 | 2 | 0.44 | this study | |||||||||
| north Africa (west) | AFN(W) | Algeria | DZA | 12 | 12 | 1 | 0.00 | this study | ||||||||||
| east Africa | AFE | Kenya | KEN | 8 | 1 | 1 | 1 | 11 | 4 | 0.45 | this study | |||||||
| Madeira | MAD | Madeira | MAD | 1 | 2 | 121 | 124 | 3 | 0.05 | [14,24] | ||||||||
| north Africa (west) | AFN(W) | Morocco | MAR | 2 | 1 | 16 | 1 | 12 | 32 | 5 | 0.60 | this study [28,42] | ||||||
| west Africa | AFW | Senegal | SEN | 1 | 1 | 10 | 12 | 3 | 0.29 | this study | ||||||||
| north Africa (east) | AFN(E) | Tunisia | TUN | 11 | 9 | 9 | 2 | 1 | 1 | 33 | 6 | 0.73 | this study | |||||
| west Europe | EUW | Austria | AUT | 1 | 1 | 1 | 0.00 | [28] | ||||||||||
| east Europe | EUE | Bulgaria | BGR | 2 | 17 | 1 | 3 | 1 | 24 | 5 | 0.47 | [8] | ||||||
| north Europe | EUN | British Isles | BRI | 1 | 58 | 7 | 1 | 29 | 96 | 5 | 0.54 | [7,11,27,28] | ||||||
| west Europe | EUW | Switzerland | CHE | 2 | 1 | 3 | 2 | 0.44 | [7] | |||||||||
| west Europe | EUW | Germany | DEU | 18 | 1 | 44 | 25 | 21 | 109 | 5 | 0.72 | this study [41] | ||||||
| north Europe | EUN | Denmark | DNK | 114 | 114 | 1 | 0.00 | [27,28] | ||||||||||
| west Europe | EUW | Spain | ESP | 3 | 1 | 1 | 2 | 4 | 11 | 5 | 0.74 | this study [7,26,27] | ||||||
| east Europe | EUE | European Turkey | ETU | 3 | 6 | 5 | 14 | 3 | 0.64 | [16] | ||||||||
| west Europe | EUW | France | FRA | 60 | 1 | 2 | 2 | 65 | 4 | 0.15 | this study [41] | |||||||
| east Europe | EUE | Greece | GRE | 1 | 2 | 1 | 1 | 3 | 8 | 5 | 0.75 | [7,27,28] | ||||||
| west Europe | EUW | Croatia | HRV | 1 | 2 | 3 | 2 | 0.44 | [27,28] | |||||||||
| west Europe | EUW | Italy | ITA | 18 | 5 | 10 | 2 | 1 | 2 | 1 | 23 | 62 | 8 | 0.74 | [7,8] | |||
| north Europe | EUN | Norway | NOR | 8 | 1 | 9 | 2 | 0.20 | [28] | |||||||||
| south America | SAM | Peru | PER | 1 | 1 | 1 | 0.00 | [27] | ||||||||||
| west Europe | EUW | Portugal | POR | 6 | 52 | 7 | 11 | 1 | 77 | 5 | 0.51 | [14,24,42] | ||||||
| north Europe | EUN | Sweden | SWE | 30 | 30 | 1 | 0.00 | [27] | ||||||||||
| north America | NAM | USA | USA | 2 | 2 | 1 | 0.00 | [27] | ||||||||||
| Asia minor | AMI | Black Sea Coast | BSC | 1 | 2 | 25 | 2 | 30 | 4 | 0.30 | [17] | |||||||
| Asia minor | AMI | centre East Anatolia | CEA | 11 | 2 | 13 | 2 | 0.26 | [17] | |||||||||
| Cyprus | CYP | Cyprus | CYP | 14 | 6 | 2 | 8 | 7 | 1 | 38 | 6 | 0.76 | this study [26] | |||||
| near-East (south) | NEA(S) | Egypt | EGY | 1 | 4 | 2 | 7 | 3 | 0.57 | this study [27] | ||||||||
| middle East | AMI | Georgia | GEO | 1 | 4 | 1 | 6 | 3 | 0.50 | this study [28] | ||||||||
| near-East (south) | NEA(S) | Israela | ISR | 5 | 2 | 1 | 7 | 1 | 8 | 24 | 5 | 0.75 | this study [27,28,43] | |||||
| near-East (north) | NEA(N) | Lebanon | LBN | 4 | 13 | 10 | 31 | 58 | 4 | 0.63 | this study | |||||||
| Asia minor | AMI | northwest Iran | NWI | 21 | 1 | 22 | 2 | 0.09 | [8] | |||||||||
| middle East | MEA | Qatar | QAT | 2 | 2 | 1 | 0.00 | this study | ||||||||||
| Fertile Crescent | FCR | south Anatolia | SAN | 3 | 2 | 5 | 10 | 3 | 0.62 | [17] | ||||||||
| middle East | MEA | southeast Iran | SEI | 1 | 3 | 14 | 18 | 3 | 0.36 | [8] | ||||||||
| Fertile Crescent | FCR | southwest Iran | SWI | 1 | 1 | 11 | 19 | 32 | 4 | 0.53 | [8] | |||||||
| near-East (north) | NEA(N) | Syria | SYR | 2 | 8 | 2 | 12 | 3 | 0.50 | this study | ||||||||
| Fertile Crescent | FCR | west Anatolia | WAN | 5 | 11 | 11 | 4 | 31 | 4 | 0.71 | [17] |
aSample encompassing 11 additional sequences from Geraldes et al. [43] not included in the main 1313 sequence alignment because of ambiguous indels.
(b). Sequencing and alignment
Sequences of the control region for the 377 samples new to this study were obtained in exactly the same way as in Rajabi-Maham et al. [8] between the positions 15 378 and 16 285 of the mouse mitochondrial genome and aligned with the Balb/c reference sequence. The complete alignment is available in electronic supplementary material, table S2. Additionally, 21 individuals were sequenced for the last 968 bp of the cytochrome b (Cytb) gene (positions 14 322–15 289 of the Balb/c mitochondrial genome; Genbank nos HQ270434-HQ270455).
(c). Data treatment
From the 1313 aligned sequences, we first reduced the dataset to the 479 different haplotypes it contained, and used these to construct a NeighbourNet network with the hypothesis-poor algorithm of Huson & Bryant [18] implemented in the Splitstree package (v. 4.10) with default settings (P distance). From this star-shaped network, we empirically defined haplogroups (HGs) as bundles of haplotypes connected through a series of potential reticulate pathways. This was performed by a search of the mutually compatible splits highlighted by the program upon edge-clicking. Additionally, the maximum-likelihood phylogenetic treatment as implemented by PhyML [19] was performed to evaluate the robustness of the so-defined HGs in the framework of a dichotomous tree.
We also computed the fixation index ΦST among geographical groups using Arlequin 3.11 [20] with default options and Kimura-2-Parameters distance model and visualized the corresponding Reynolds' coancestry distance matrix as a Neighbour-Joining tree. Mismatch distribution (MMD) analyses were performed with the same software. The between-sample comparisons were carried out using the uncorrected modal value of the observed MMD or its pairwise average, which is known to behave roughly as τ = 2μt after a demographic expansion [21], however the unevenness of the mutation rates in the D-loop region [22] renders this estimation dependent on the underlying mutation and expansion models and hence not necessarily very reliable. In addition, we applied the Spatial Analysis of Molecular Variance (SAMOVA) procedure [23] at the regional and provincial level to explore clustering of populations in our dataset.
3. Results
(a). NeighbourNet and HG differentiation
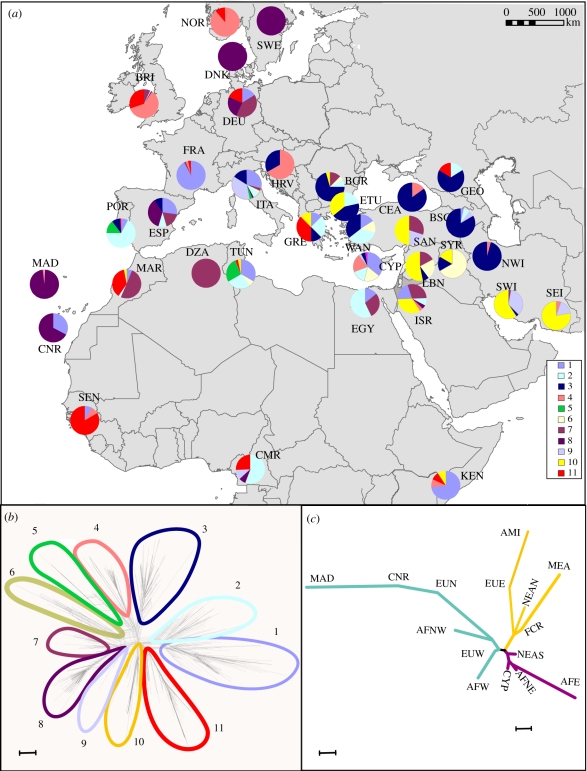
Figure 1b presents the 479 haplotype network showing 11 identifiable bundles that we term HGs rather than clades because with a few exceptions, the relative positions of these bundles are neither well-supported in standard phylogenetic analyses (electronic supplementary material, figure S3), nor it is possible to root the network precisely. It resembles more a multiple-arm sea star than a phylogenetic tree, as would be expected from a recent expansion phenomenon. Some of the HGs were the same as those visualized by Rajabi-Maham et al. [8], some were new to this study. For instance, Hg 3 encompasses exactly what was called the Turkish Main clade by Gündüz et al. [17] and possibly equates with clade B in the alphabetical nomenclature of Jones et al. [12], and Hg 4 contains the Orkney clade defined by Searle et al. [11], now called clade F by the same authors. Hg 1 and Hg 2 overlap largely with group 1 of Rajabi-Maham et al. [8], which itself contained the clade then called Ger3 and is mostly (but not only) constituted of European sequences. Former group Ger1 which was basal to group 1 in Rajabi-Maham et al. [8] is now identified as Hg 11 and would be similar to clade E of Jones et al. [12]. Former group 3 which was indeed not a well-delimited central group in the work of Rajabi-Maham et al. [8], is now unsurprisingly distributed among several HGs, the largest part being found in Hg 10, which probably overlaps largely with clade A [12]. The average within-HG nucleotidic divergence was 0.0040 for a between-HG value ranging between 0.0053 and 0.0142 (mean 0.0106). The per cent global diversity absorbed by the inter-HG differentiation reached a ΦST value of 0.71. Classical (dichotomous) phylogenetic analyses revealed that, while most of these SplitsTree-defined HGs appear as reasonably cohesive, only the terminal nodes are well supported (electronic supplementary material, figure S3). The high level of homoplasy present in the dataset prevented further exploration of the historical links between these HGs.
Figure 1.
(a) Geographical distribution of the matrilines attributed to one of the 11 haplogroups (HGs) according to their position in the network in (b). (b) NeighbourNet network for 1313 individual mitochondrial D-loop sequences. Elliptical envelopes delimitate the 11 HGs considered. (c) Neighbour-joining tree based on Reynolds' distance between regions. Colours help to visualize three main branches roughly reflecting geography. Scale bars, (a) 1–1000 km; (b) 0.0010; (c) 0.05.
(b). Geographical distribution
The distribution of HGs according to their frequency in each sample is given in table 1 and depicted graphically by pie charts in figure 1a. Several HGs are strongly associated with certain geographical regions. For instance, this is the case of Hgs 3, 4 and 7 that, respectively, corresponds for the major part (but not only) to samples from Asia Minor, the northern British Isles (together with Norway), and Germany. Similarly, Hg 6 is mostly composed of samples from Lebanon and Syria (near-East), while Hg 10 predominates in Southern Iran and Southern Anatolia (Fertile Crescent).
(c). Sample differentiation
(i). Global picture
HG diversity was quite variable from one sample to the other (range: 0–0.78; table 1), but on each continent some samples exhibited a high number of HGs and a high diversity. The AMOVA output yielded a highly significant global fixation index, ΦST of 0.28 (p < 10−5) and every pairwise comparison was highly significant. The SAMOVA procedure [23] applied to all possible partitions either at the Province or Region levels was unable to identify clusters of geographically close samples that would have been meaningful, since the between-cluster, ΦCT was a decreasing monotonic function of the number of partitions considered (not shown). When a hierarchical procedure was considered (i.e. when the provincial level was nested into the regional level), the standard AMOVA yielded a ΦCT of 0.20, while it was only 0.009 when the regional level was nested into the so-called continental level. This means that there is a significant heterogeneity of samples within the regional level while most if not all the differentiation is already present at the continental level. To further illustrate the information contained in table 1, the regional level was considered, with the insular samples treated separately; two regions were split into two subunits to account for their obvious heterogeneity: Tunisia (AFNE) was separated from the rest of the north-African sample (AFNW), and Syria plus Lebanon (NEAN) were considered separately from Israel plus Egypt (NEAS). Figure 1c shows the Neighbour-Joining tree obtained from the Reynolds' distance matrix. It is not strongly structured by long internal branches, in keeping with the fact that the SAMOVA analysis did not capture significant clusters. Basically, this tree is congruent with the one obtained on a smaller number of sequences by Rajabi-Maham et al. [8], since it opposes the same two branches: an Asia Minor offshoot through what we termed here eastern Europe (actually Greece, Bulgaria and European Turkey), another one encompassing the rest of Europe, and brings to light a third one including some of the new samples considered here. Several features concerning the samples new to this study are noteworthy. The Canary Islands plus Madeira appear at the tip of a very long branch sprouting from western Europe. Another noteworthy point is that both samples from North Africa are closely connected to the centre of the tree but on different offshoots. The Tunisian sample is more related to mice from the southern near-East, Cyprus and Kenya, than to those of Morocco and Algeria, which rather branch-off at the base of the western European shoot together with West Africa. Altogether, despite short internal branches and obvious discontinuities between geographically close samples, a rough geographical structure is present as underlined by the colour scheme in figure 1c.
(ii). Relative expansion time
The MMD of the global collection showed a clear signal of recent expansion (either under a demographic or a spatial model), with a peak at nine mismatches (average pairwise 7.4) over 802 informative nucleotidic sites (gaps not considered) corresponding to a global average pairwise divergence τ estimated to be in the order of 8.7 (8.53 and 8.88 according to the spatial and sudden expansion models implemented in Arlequin, respectively). Nevertheless, the shape of the MMD and the peak values varied considerably between samples (electronic supplementary material, figures S4 and S5). Several regional samples such as FCR (‘Fertile Crescent’) showed distributions that were almost identical to that of the overall collection of sequences suggesting an early expansion onset. Others such as MAD (Madeira) for instance, indicated a much more recent wave of expansion with a mode of zero differences and an average pairwise mismatch of 1.65, in agreement with the recent colonization of this island [14], as is the case also for the Canaries (see below). On the other hand, a sample like CYP (Cyprus) for instance displayed a clear multi-modal distribution owing to the simultaneous presence of divergent HGs that were also found elsewhere. Since it is very unlikely that the early Neolithic settlers of this island brought with them a large number of founders (see below), such a distribution is most probably the signature of secondary exchanges or multiple colonizations artificially inflating the MMD. How old is the expansion time for the whole subspecies? This estimation is strongly dependent on the calibration of the intra-subspecific substitution rate, a difficult question subject to controversy that we re-visit in §4.
(d). Detailed account on the samples new to this study
(i). Canaries
Table 1 clearly demonstrates that these islands have been populated by two very different HGs—Hg 1 and Hg 8. These two HGs are infrequent along the western African coast (Morocco, Senegal), but are predominant in Europe. In fact, one of them (Hg 8) is identical to the sole HG present in Madeira, strongly supporting the colonization of these islands following the European settlement. While a single HG founder event is evident on Madeira as already pointed out by Gündüz et al. [24], the presence of two divergent HGs in the Canary Islands indicates that at least two introductions occurred in the past. The 140 Canarian sequences analysed come from several sites on three islands; while Hg 1 is predominant on the island of Tenerife (frequency 96%), Hg 8 is found in high frequency on the island of El Hierro and La Palma (97% and 98%, respectively; data not shown). One clearly cannot consider that these two divergent HGs originated on the islands after their colonization, thus each HG was examined separately since our sample size was sufficient. MMD were even more skewed to the left for the Canaries when compared with Madeira (electronic supplementary material, figure S4), with modal values of 0 (Hg 1 on Tenerife, almost no polymorphism, average pairwise difference of 0.215) or 1 (Hg 8 on both La Palma-El Hierro, average difference of τ = 0.85). For this last HG, using 12 000 years BP as a reasonable onset of the domesticus global expansion following Rajabi-Maham et al. [8], a simple ratio of either the modal or the mean pairwise mismatch values would provide an age of about one-ninth of the total expansion time, i.e. 1300 years ago for these two latter islands, about twice as much for Madeira, and a more recent date that is quite difficult to estimate for Hg 1 on Tenerife. At face value, these rough estimates of the time of colonization do not agree well with the recent zooarchaeological studies. A re-appraisal of the chronology of the house mouse colonization of the Canary Islands by 14C dating of house mice collected from archaeological sites in La Palma, El Hierro, Fuerteventura and Lanzarote [16] indicates that the arrival of the house mouse could have happened between 756–414 BC and AD 128–313, discarding previous claims of an arrival of the house mouse in Canaries during Neolithic time [25]. The question we are left with is thus whether this dating of the house mouse arrival in the Canary Islands could correspond to the expansion age of one or the other HG in the archipelago? The available data limited to three islands favour neither of these dates: Hg 8, the most diversified HG in our Canary sample, is almost identical to the one on Madeira (net nucleotidic divergence between populations of only 3.6 × 10−4 between Hg 8 in Madeira and in the Canaries). This is consistent with the colonization of Madeira by the Portuguese (mid-fifteenth century) who established contact with the Canaries shortly thereafter. A possible earlier visit by Viking boats several centuries before [14,24] is neither contradicted nor supported by the similar net divergence values of 4.6 × 10−4 and 6.0 × 10−4 observed between Hg 8 in Madeira and Hg 8 in northern and western Europe, respectively. On the other hand, the other HG, Hg 1 on Tenerife is closest and very similar to the Hg 1s found in France (3.1 × 10−4 net nucleotidic divergence), and not to the Hg 1 found in Cyprus or the Levant for instance (3.6 × 10−3 and 1.8 × 10−3, respectively). These data on modern populations favour a recent colonization of the archipelago by house mice of European origin, which is in disagreement with the unquestionable much earlier documented occurrence of this species during the second to fourth century AD at the latest [14]. One of the main explanations would be that the first colonization wave left no footprints in the current mitochondrial lineages of the archipelago. Were the first house mouse founders extinct when the recent colonization happened, or were they outcompeted by a recent massive colonization? A molecular analysis of the earliest archaeological house mouse from Lanzarote, Fuerteventura and La Palma should provide some crucial insights on this matter.
(ii). Cyprus
Unlike the previous archipelago, this island is known as among the first Mediterranean islands colonized by both the house mouse and the first farmers at the very beginning of the Neolithic farming dispersal in the Mediterranean ca 8400 cal BC [5]. As such, we expected to find the presence of old endemic HGs on this island, which is not the case. To the contrary, an important haplotypic diversity (0.76) with seven differentiated HGs was observed including Hg 1, the one most represented in western Europe. This diversity is indicative of Cyprus being at a crossroads with many introductions of house mice from several origins all along its history.
The persistence through time of this diversity of haplotypes suggests that migrants found favourable ecological conditions to establish sufficiently large local populations and contribute significantly to the local gene pool. This is in agreement with the biology of a commensal species like the house mouse, which implies a metapopulation structure rather than a single large panmictic population. Had the latter been the case, precedence would have given advantage to the first haplotypes arriving, limiting the introduction of newcomer haplotypes. Such a process may be particularly true on Cyprus, where the non-commensal species Mus cypriacus probably restricts the installation of Mus m. domesticus to sparse human dwellings [26].
(iii). Western tropical Africa
The two samples from Senegal and Cameroon display primarily the presence of the two HGs—Hg 11 and Hg 2. Hg 11 is clearly largely European, although also present in Morocco, and Hg 2 may have been introduced from almost anywhere. The haplotypes present in Cameroon for instance are molecularly very close to several found in Portugal, Germany and France (see electronic supplementary material, figure S3), which is compatible with the political history of this region during the last five centuries. The colonization of the western coast of Africa by the house mouse thus appears as relatively recent and mostly influenced by European lineages. This is hardly surprising if one recalls that Mus musculus is probably entirely dependent on human habitats in this region and would likely not have reached these areas before the European colonization and the advent of modern transportation means.
(iv). Eastern tropical Africa
The Kenyan sample studied consisted of haplotypes from two subspecific origins. The data presented here correspond only to the domesticus HGs found in an otherwise predominantly Mus m. castaneus background (11/70 individuals, A. Orth & F. Bonhomme 2008, unpublished data). The predominant domesticus HG is Hg 1, which is well-represented in Europe. This predominance probably indicates that in this case also, the domesticus matrilines are probably recent newcomers.
4. General discussion
(a). When did the expansion occur?
As already pointed out by Rajabi-Maham et al. [8], there is a clear expansion signal in the MMD of the global sample, exemplified in the present study by the clear unimodal wave of expansion (electronic supplementary material, figures S4 and S5). The modal value of nine nucleotidic pairwise differences calculated over 802 informative sites (gaps removed) corresponds to an estimated average nucleotidic diversity of π = 0.98% and fits to a model with 2μt = 8.88 (sudden expansion) or 2μt = 8.56 (spatial expansion). This should roughly indicate the timing of the onset of expansion provided we know the mutation rate. Applying a value of 10 per cent per site Myr−1 for the mouse D-loop, as often set forth in the literature [27,28], the 1 per cent molecular divergence observed (0.5% per lineage) would correspond to a 54 000 year old expansion. This date is probably much too old as discussed by Rajabi-Maham et al. [8] who, with a somewhat higher modal value of 10 pairwise differences and a global sample one-third smaller than the present one, ascribed this expansion to the Holocene warming some 12 000 years ago on the basis of reasonable palaeobiological and archaeological arguments, and the observation that intraspecific and pedigree-based estimates of substitution rates are generally higher than interspecific phylogenetically calibrated rates [29]. Although this statement is still a matter of controversy, recent re-evaluations in the hominoid and human mitochondrial genome [30,31], all show that this nonlinearity of substitution rates is indeed the case (but see also [32]). Mutational hotspots represent one possible cause of this nonlinearity, since they may create an excess of undetected homoplasy in longer branches and an excess of detectable mutations in short branches. The mitochondrial control region with its two hypervariable regions may probably show this phenomenon, so that its long-term mutation rates estimated from the Rattus/Mus divergence are likely to be greatly underestimated. To evaluate this effect independently, we relied on the evolutionary substitution rate at the third codon position of cytochrome b, a non-coding and perhaps less homoplasy prone region of the mitochondrial genome. This rate has recently been re-evaluated in the Mus lineage at 22 per cent per third codon site Myr−1 using a multi-point calibration procedure of lineage-specific mutation rates across 1696 mammalian species [33]. We randomly chose 22 individuals among the 11 sub-HGs for which we sequenced the 968 bp of the Cyt b gene. Considering only the third codon position, a clear expansion signal was obtained as expected (electronic supplementary material, figure S6), with an average number of pairwise differences of τ = 4.29 and a mode of four mismatches. Using the previously mentioned μ, this would translate as 4.29/2/0.223/323 sites = 29 700 years since the beginning of the expansion. The first conclusion is that, if the substitution rate is correctly estimated by Nabholz et al. [33] and no time-dependency occurs, then that for D-loop which displays almost exactly twice as much variation over ca 802 informative sites should amount to 17.9 per cent per site Myr−1. This is already almost twice as much as the 10 per cent classically reported in the literature [17,27]. The second is that, if nonlinearity for recent evolution also applies to the Cyt b third position, then the expansion time is likely to be shorter and the intraspecific substitution rates for both molecules are likely to be about twice the value indicated above if the 12 000 years old expansion proposed by Rajabi-Maham et al. [8] holds. Whatever the controversy on this nonlinearity, the present discussion highlights the difficulty of using indirect methods with remote calibrations to date recent events, except in a rough comparative way.
(b). Where was the cradle of the M. m. domesticus subspecies?
Rajabi-Maham et al. [8] proposed that the Fertile Crescent (taken in a broad sense) could have been the region where the direct ancestors of the present day subspecies would have first become commensal. These authors based this scenario on the fact that the HGs present in this region stemmed directly from the central region of their haplotypic tree and were more diverse than the rest of their sample. In the present analysis, we extended the sampling to the near-East, and were thus able to have a more detailed picture of the haplotypic variation within several previously poorly sampled HGs as Hg 6 and Hg 10. From figure 1a, it appears that among the latter, Hg 10 is present mainly in southwest Iran and south Anatolia but exists as well in Syria, Lebanon and Israel, while Hg 6 is found predominantly in Lebanon and Syria but also on Cyprus. Thus, the presence of two independent HGs in this region supports the existence of two geographically close foci in which the initial association between mice and humans took place. Zooarchaeological data have evidenced the presence of M. m. domesticus in a southern Levant rock shelter in association with epipalaeolithic layers at a time when humans became sedentary [3]. This gave rise to the theory that sedentism was the driving force for house mouse commensalism [34,35]. Although the lack of evidence for the association between the house mouse remains and the human occupation in the cave has been raised, undermining the correlation between sedentism and commensalism [36,37], the house mouse presence in the natural habitat of southern Levant about 12 000 years ago remains irrefutable. Later studies have shown that the Neolithic niche construction in northern Levant with the rise of the farming practices provided the main factor triggering house mouse commensalism [4]. Recent discoveries of granaries in the earliest Pre-pottery Neolithic layers in Dhra' (Jordan) [38] indicate that the southern Levant became potentially highly attractive for commensal mice almost at the same time as in the northern Levant. So, two contemporaneous centres of commensalism could have occurred in the northern and southern Levant. Whether commensal or wild, M. m. domesticus populations were probably present throughout the Levant and probably all around Fertile Crescent during the warm episode of the Bølling/Allerød (12 700–10 700 BC). The following Younger Dryas colder episode that lasted about 1300 years [39] may have temporarily fragmented these populations, allowing the in situ differentiation of the two abovementioned HGs that seem to be predominant in this region. It is not clear where exactly the ancestors of M. m. domesticus spent the coldest Pleistocene episode, but since they were absent from the Jordan valley [3], the southern slopes of the Zagros or the palaeoshores of the Arabo-Persian gulf could have constituted this refugium, as suggested by Rajabi-Maham et al. [8]. Such a possibility is not contradicted by the present data since it is impossible to root correctly the matrilineal phylogeny nor to establish the order of precedence of the different HGs, which appear as having diverged almost simultaneously.
(c). Were both sides of the Mediterranean populated independently by house mice?
Despite a moderate sample size (no sample from Lybia, and a single geographical site in Algeria), the present dataset does not support a unique origin of the house mouse populating North Africa independently from that of eastern and western Europe, as would have been the case had the mice followed the Neolithic progression of agriculture. Rather, we have to invoke at least two events. The presence in Tunisia and in two individuals from Italy of an independent HG (Hg 5) related to sequences from Portugal and the near-East argues in favour of an early colonization. This happens pretty much symmetrically on the other side of the Mediterranean with clade Hg 9 predominantly found in Italy but also in the near- and middle East, indicative of an independent and probably ancient divergence. These HGs are far from alone in these countries, where traces of long-range exchanges are present (the well-represented Hg 7 exists in Italy but also in 12 other samples; in the same way, Hg 1 is present in Tunisia and in 16 other samples). Figure 1c (independent of HG definition) as well as table 1 indicate a proximity between the European and western North African samples, which share several HGs. Nevertheless, there seems to be no continuity between Tunisia and further west, the global picture depicted in figure 1c clearly showing instead a proximity between the Tunisian, Cyprus and southern near-East samples. Two alternative expansion scenarios can be proposed. The first one posits that an early and progressive colonization of North Africa occurred with populations differentiating from each other according to a geographical gradient starting in the near-East and ending on the Moroccan coast; this gradient would have since then been largely obliterated by subsequent immigration. The other possibility is that this gradient never existed and that the present day distribution of matrilines reflects a colonization of North Africa at different times: an early westward wave expanding into Tunisia and a second more recent one introducing migrants all along the coast.
How well does this match what is known from a zooarchaeological standpoint? It has been shown that along the northern shore of the Mediterranean, the initial progression of the house mouse from its near-Eastern cradle was not associated with the Neolithic farming dispersal. Indeed, in its westernmost part, it lagged several millennia behind until the commercial as well as demographic human expansions occurred during the last millennium BC [10]. The same phenomenon could very well have happened on both shores simultaneously. The case of the Canary Islands is highlighted in this respect since the arrival of the house mouse in the archipelago cannot be older than the second half of the last millennium BC at the earliest [16]. These data suggest that house mouse dispersal never reached western North Africa before the last millennium BC just as is postulated for the northern shores of the western Mediterranean. Mouse remains from Neolithic deposits of El Harhoura 2, Rabat-Temara in Morocco dated from 5800 BP have all been identified as Mus spretus [40] supporting the absence of a Neolithic-mediated house mouse dispersal at least in the western part of Northern Africa. Once again, a likely explanation of house mouse dispersal in the southern Mediterranean shore, at least on its western end, is linked with the commercial network of the Phoenicians which created colonies all along their main trade route situated in Northern Africa from their city–state ports on the southern Levantine shores. Indeed, Carthage controlled the maritime trade of western Mediterranean by the fifth century BC, which would have probably promoted admixture of house mouse metapopulations of this area. The subsequent take-over by Rome in the second century BC may explain the significant level of haplotype sharing between Italy, Tunisia and the Levant. Later, migrant flows of house mice carrying western European haplotypes would have contributed their mitochondrial footprint in most of the North African house mouse populations. Only the populations of Tunisia and Italy would have kept the signature of their Punic history.
5. Conclusion
The study of matrilineal variation in the western house mouse as revealed by nucleotidic variability of the D-loop provides evidence of the recent common ancestry of all extant populations, as well as a complex history of these populations owing to founder effects, genetic drift and secondary admixture, as expected for a species closely associated with human activity. Despite evidence of gene flow, there is nevertheless a rather high level of interpopulation differentiation providing the opportunity for local differentiation. The remaining geographical signal although somewhat difficult to interpret, points to the existence of two expansion processes in the Mediterranean: one group of populations shows traces of an early common history related to their geographical proximity during the early progression of Neolithic farming practices, while another shows a more pronounced influence of human activity related to a later onset of maritime trade across the Mediterranean. This last process has never ended, since the more recently populated areas, such as Atlantic islands for instance, show an unambiguous signal of recent dispersal, while anciently populated islands like Cyprus show the footprints of a succession of introduction events.
Acknowledgements
F.B. is the holder of a certificate authorizing experimentation on living vetebrates no. 34.72 and all work on mice was carried out under authorization from the veterinary services of the Hérault Préfecture no. C34-172-23.
Concerning the samples new to this study, authors wish to thank the collaboration of N. Khammes for sampling in Algeria, G. Ganem for the sample from Berlin, J. F. Agnese for trapping support in Kenya, M. Karam, R. Kabalan, R. Moussa as well as E. Davidian and J.-C. Davidian for their field support in Lebanon, M. Martinez in Qatar, K. Ba in Senegal, M. Hassan in Syria, and C. Montgelard and H. Croset in Italy. Sequences were generated on IFR 119 platform (Montpellier Environnement Biodiversité). This is contribution ISEM 2010-094.
References
- 1.Auffray J.-C., Vanlerberghe F., Britton-Davidian J. 1990. The house mouse progression in Eurasia: a palaeontological and archaeozoological approach. Biol. J. Linn. Soc. 41, 13–25 10.1111/j.1095-8312.1990.tb00818.x (doi:10.1111/j.1095-8312.1990.tb00818.x) [DOI] [Google Scholar]
- 2.Brothwell D. 1981. The Pleistocene and Holocene archaeology of the house mouse and related species. Symp. Zool. Soc. Lond. 47, 1–13 [Google Scholar]
- 3.Auffray J. C., Tchernov E., Nevo E. 1988. Origin of commensalism of the house mouse (Mus musculus domesticus) in relation to man. Comptes Rendus Acad. Sci. Ser. III-Sci. Vie-Life Sci. 307, 517–522 [Google Scholar]
- 4.Cucchi T., Vigne J.-D. 2006. Origin and diffusion of the house mouse in the Mediterranean. Hum. Evol. 21, 95–106 10.1007/s11598-006-9011-z (doi:10.1007/s11598-006-9011-z) [DOI] [Google Scholar]
- 5.Cucchi T., Vigne J. D., Auffray J. C., Croft P., Peltenburg E. 2002. Passive transport of the house mouse (Mus musculus domesticus) to Cyprus at the Early Preceramic Neolithic (late 9th and 8th millennia cal. BC). C. R. Palevol. 1, 235–241 10.1016/S1631-0683(02)00033-7 (doi:10.1016/S1631-0683(02)00033-7) [DOI] [Google Scholar]
- 6.Britton-Davidian J. 1990. Genic differentiation in M. m. domesticus populations from Europe the Middle East and North Africa: geographic patterns and colonization events. Biol. J. Linn. Soc. 41, 27–45 10.1111/j.1095-8312.1990.tb00819.x (doi:10.1111/j.1095-8312.1990.tb00819.x) [DOI] [Google Scholar]
- 7.Nachman M. W., Boyer S. N., Searle J. B., Aquadro C. F. 1994. Mitochondrial-DNA variation and the evolution of Robertsonian chromosomal races of house mice, Mus domesticus. Genetics 136, 1105–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajabi-Maham H., Orth A., Bonhomme F. 2008. Phylogeography and postglacial expansion of Mus musculus domesticus inferred from mitochondrial DNA coalescent, from Iran to Europe. Mol. Ecol. 17, 627–641 [DOI] [PubMed] [Google Scholar]
- 9.Sage R. D., Prager E. M., Tichy H., Wilson A. C. 1990. Mitochondrial-DNA variation in house mice, Mus domesticus (Rutty). Biol. J. Linn. Soc. 41, 105–123 10.1111/j.1095-8312.1990.tb00824.x (doi:10.1111/j.1095-8312.1990.tb00824.x) [DOI] [Google Scholar]
- 10.Cucchi T., Vigne J.-D., Auffray J.-C. 2005. First occurrence of the house mouse (Mus musculus domesticus Schwarz & Schwarz, 1943) in the Western Mediterranean: a zooarchaeological revision of subfossil occurrences. Biol. J. Linn. Soc. 84, 429–445 10.1111/j.1095-8312.2005.00445.x (doi:10.1111/j.1095-8312.2005.00445.x) [DOI] [Google Scholar]
- 11.Searle J. B., et al. 2009. Of mice and (Viking?) men: phylogeography of British and Irish house mice. Proc. R. Soc. B 276, 201–207 10.1098/rspb.2008.0958 (doi:10.1098/rspb.2008.0958) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones P. E., Johannesdottir F., Gündüz I., Richards M. B., Searle J. B. In press The expansion of the house mouse into North-western Europe. J. Zool. [Google Scholar]
- 13.Searle J. B., Jamieson P. M., Gündüz I., Stevens M. I., Jones E. P., Gemmill C. E. C., King C. M. 2009. The diverse origins of New Zealand house mice. Proc. R. Soc. B 276, 209–217 10.1098/rspb.2008.0959 (doi:10.1098/rspb.2008.0959) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Förster D. W., Gündüz I., Nunes A. C., Gabriel S., Ramalhinho M. G., Mathias M. L., Britton-Davidian J., Searle J. B. 2009. Molecular insights into the colonization and chromosomal diversification of Madeiran house mice. Mol. Ecol. 18, 4477–4494 10.1111/j.1365-294X.2009.04344.x (doi:10.1111/j.1365-294X.2009.04344.x) [DOI] [PubMed] [Google Scholar]
- 15.Cucchi T. 2008. Uluburun shipwreck stowaway house mouse: molar shape analysis and indirect clues about the vessel's last journey. J. Archaeol. Sci. 35, 2953–2959 10.1016/j.jas.2008.06.016 (doi:10.1016/j.jas.2008.06.016) [DOI] [Google Scholar]
- 16.Alcover J. A., Rando J. C., Garcia-Talavera F., Hutterer R., Michaux J., Trias M., Navarro J. F. 2009. A reappraisal of the stratigraphy of Cueva del Llano (Fuerteventura) and the chronology of the introduction of the house mouse (Mus musculus) into the Canary Islands. Paleogeogr. Paleoclimatol. Paleoecol. 277, 184–190 10.1016/j.palaeo.2009.03.016 (doi:10.1016/j.palaeo.2009.03.016) [DOI] [Google Scholar]
- 17.Gündüz I., Rambau R., Tez C., Searle J. 2005. Mitochondrial DNA variation in the western house mouse (Mus musculus domesticus) close to its site of origin: studies in Turkey. Biol. J. Linn. Soc. 84, 473–485 10.1111/j.1095-8312.2005.00448.x (doi:10.1111/j.1095-8312.2005.00448.x) [DOI] [Google Scholar]
- 18.Huson D., Bryant D. 2006. Applications of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 10.1093/molbev/msj030 (doi:10.1093/molbev/msj030) [DOI] [PubMed] [Google Scholar]
- 19.Guindon S., Gascuel O. 2003. A simple, fast accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 10.1080/10635150390235520 (doi:10.1080/10635150390235520) [DOI] [PubMed] [Google Scholar]
- 20.Excoffier L., Laval G., Schneider S. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. 1, 47–50 [PMC free article] [PubMed] [Google Scholar]
- 21.Rogers A. R., Harpending H. 1992. Population growth makes waves in the distribution of paiwise genetic differences. Mol. Biol. Evol. 9, 552–569 [DOI] [PubMed] [Google Scholar]
- 22.Schneider S., Excoffier L. 1999. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates very among sites: application to human mitochondrial DNA. Genetics 152, 1079–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dupanloup I., Schneider S., Excoffier L. 2002. A simulated annealing approach to define the genetic structure of populations. Mol. Ecol. 11, 2571–2581 10.1046/j.1365-294X.2002.01650.x (doi:10.1046/j.1365-294X.2002.01650.x) [DOI] [PubMed] [Google Scholar]
- 24.Gündüz I., Auffray J. C., Britton-Davidian J., Catalan J., Ganem G., Ramalhinho M. G., Mathias M. L., Searle J. B. 2001. Molecular studies on the colonization of the Madeiran archipelago by house mice. Mol. Ecol. 10, 2023–2029 10.1046/j.0962-1083.2001.01346.x (doi:10.1046/j.0962-1083.2001.01346.x) [DOI] [PubMed] [Google Scholar]
- 25.Coello J. J., Castillo C., Gonzalez E. M. 1999. Stratigraphy, chronology, and paleoenvironmental reconstruction of the quaternary sedimentary infilling of a volcanic tube in Fuerteventura, Canary Islands. Quatern. Res. 52, 360–368 10.1006/qres.1999.2074 (doi:10.1006/qres.1999.2074) [DOI] [Google Scholar]
- 26.Cucchi T., Orth A., Auffray J.-C., Renaud S., Fabre L., Catalan J., Hadjisterkotis E., Bonhomme F., Vigne J.-D. 2006. A new endemic species of the subgenus Mus (Rodentia, Mammalia) on the Island of Cyprus. Zootaxa 1241, 1–36 [Google Scholar]
- 27.Prager E. M., Sage R. D., Gyllensten U., Thomas W. K., Hübner R., Jones C. S., Noble L., Searle J. B., Wilson A. C. 1993. Mitochondrial DNA sequence diversity and the colonization of Scandinavia by house mice from East Holstein. Biol. J. Linn. Soc. 50, 85–122 10.1111/j.1095-8312.1993.tb00920.x (doi:10.1111/j.1095-8312.1993.tb00920.x) [DOI] [Google Scholar]
- 28.Prager E. M., Tichy H., Sage R. D. 1996. Mitochondrial DNA sequence variation in the eastern house mouse, Mus musculus: comparison with other house mice and report of a 75-bp tandem repeat. Genetics 143, 427–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho S., Shapiro B., Phillips M., Cooper A., Drummond A. 2007. Evidence for time dependency of molecular rate estimates. Syst. Biol. 56, 515–522 10.1080/10635150701435401 (doi:10.1080/10635150701435401) [DOI] [PubMed] [Google Scholar]
- 30.Henn B. M., Gignoux C. R., Feldman M. W., Mountain J. L. 2009. Characterizing the time dependency of human mitochondrial DNA mutation rate estimates. Mol. Biol. Evol. 26, 217–230 10.1093/molbev/msn244 (doi:10.1093/molbev/msn244) [DOI] [PubMed] [Google Scholar]
- 31.Loogväli E. L., Kivisild T., Margus T., Villems R. 2009. Explaining the imperfection of the molecular clock of hominid mitochondria. PLoS ONE 4, e8260. 10.1371/journal.pone.0008260 (doi:10.1371/journal.pone.0008260) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Debruyne R., Poinar H. N. 2009. Time dependency of molecular rates in ancient DNA data sets: a sampling artifact? Syst. Biol. 58, 348–359 10.1093/sysbio/syp028 (doi:10.1093/sysbio/syp028) [DOI] [PubMed] [Google Scholar]
- 33.Nabholz B., Glemin S., Galtier N. 2008. Strong variations of mitochondrial mutation rate across mammals: the longevity hypothesis. Mol. Biol. Evol. 25, 120–130 10.1093/molbev/msm248 (doi:10.1093/molbev/msm248) [DOI] [PubMed] [Google Scholar]
- 34.Tchernov E. 1984. Commensal animals and human sedentism in the Middle East. BAR Int. Ser. 202, 91–115 [Google Scholar]
- 35.Tchernov E. 1991. Of mice and men. Biological markers for long-term sedentism; a reply. Paléorient 17, 153–160 10.3406/paleo.1991.4548 (doi:10.3406/paleo.1991.4548) [DOI] [Google Scholar]
- 36.Tangri D., Wyncoll G. 1989. Of mice and men: is the presence of commensals animals in archaeological sites a positive correlate of sedentism? Paléorient 15, 85–94 10.3406/paleo.1989.4511 (doi:10.3406/paleo.1989.4511) [DOI] [Google Scholar]
- 37.Wyncoll G., Tangri D. 1991. The origins of commensalism and human sedentism. Paléoorient 17, 157. 10.3406/paleo.1991.5093 (doi:10.3406/paleo.1991.5093) [DOI] [Google Scholar]
- 38.Kuijt I., Finlayson B. 2009. Evidence for food storage and predomestication granaries 11,000 years ago in the Jordan Valley. Proc. Natl Acad. Sci. USA 106, 10 966–10 970 10.1073/pnas.0812764106 (doi:10.1073/pnas.0812764106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bottema S. 1995. The younger Dryas in the eastern Mediterranean. Quatern. Sci. Rev. 14, 883–891 10.1016/0277-3791(95)00069-0 (doi:10.1016/0277-3791(95)00069-0) [DOI] [Google Scholar]
- 40.Stoetzel E., Michel P., Nespoulet R., El Hajraout M. A. 2007. Holocene environments of the Moroccan Atlantic coastal zone: examples of small large vertebrates in archaeological context from El Harhoura 2 Cave, Temara. Quaternaire 18, 299–307 [Google Scholar]
- 41.Ihle S., Ravaoarimanana I., Thomas M., Tautz D. 2006. An analysis of signatures of selective sweeps in natural populations of the house mouse. Mol. Biol. Evol. 23, 790–797 10.1093/molbev/msj096 (doi:10.1093/molbev/msj096) [DOI] [PubMed] [Google Scholar]
- 42.Prager E. M., Orrego C., Sage R. D. 1998. Genetic variation and phylogeography of central Asian and other house mice, including a major new mitochondrial lineage in Yemen. Genetics 150, 835–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geraldes A., et al. 2008. Inferring the history of speciation in house mice from autosomal, X-linked, Y-linked and mitochondrial genes. Mol. Ecol. 17, 5349–5363 10.1111/j.1365-294X.2008.04005.x (doi:10.1111/j.1365-294X.2008.04005.x) [DOI] [PMC free article] [PubMed] [Google Scholar]