Abstract
Whales in the suborder Mysticeti are filter feeders that use baleen to sift zooplankton and small fish from ocean waters. Adult mysticetes lack teeth, although tooth buds are present in foetal stages. Cladistic analyses suggest that functional teeth were lost in the common ancestor of crown-group Mysticeti. DNA sequences for the tooth-specific genes, ameloblastin (AMBN), enamelin (ENAM) and amelogenin (AMEL), have frameshift mutations and/or stop codons in this taxon, but none of these molecular cavities are shared by all extant mysticetes. Here, we provide the first evidence for pseudogenization of a tooth gene, enamelysin (MMP20), in the common ancestor of living baleen whales. Specifically, pseudogenization resulted from the insertion of a CHR-2 SINE retroposon in exon 2 of MMP20. Genomic and palaeontological data now provide congruent support for the loss of enamel-capped teeth on the common ancestral branch of crown-group mysticetes. The new data for MMP20 also document a polymorphic stop codon in exon 2 of the pygmy sperm whale (Kogia breviceps), which has enamel-less teeth. These results, in conjunction with the evidence for pseudogenization of MMP20 in Hoffmann's two-toed sloth (Choloepus hoffmanni), another enamel-less species, support the hypothesis that the only unique, non-overlapping function of the MMP20 gene is in enamel formation.
Keywords: baleen whale, enamel, enamelysin, macroevolution, pseudogene, teeth
1. Introduction
The evolution of Cetacea from terrestrial ancestors is one of the best-documented macroevolutionary transitions in the fossil record [1]. Stem cetaceans (pakicetids, ambulocetids, remingtonocetids, protocetids and basilosaurids) document progressive limb reduction, posterior migration of the external nares and other specializations for a fully aquatic lifestyle. Archaeocetes retained teeth, as do living cetaceans in the suborder Odontoceti. Cetaceans in the suborder Mysticeti, by contrast, have lost their adult teeth and instead use racks of baleen to filter zooplankton and small fish from ocean waters. Baleen is a key innovation that facilitated the exploitation of an unexploited ecological niche, bulk filter-feeding and laid the foundation for the evolution of the largest animals on the Earth [2]. In addition to including the largest extant mammal (blue whale), Mysticeti also includes the putatively longest living extant mammal (bowhead whale) [3–5]. The fossil record of stem mysticetes includes primitive forms that had teeth but not baleen (e.g. Janjucetus, Mammalodon), intermediate forms that had teeth, and by inference baleen, based on the presence of lateral nutrient foramina and sulci on the palate (e.g. Aetiocetus) and more derived forms with baleen but not teeth (e.g. Eomysticetus, Micromysticetus) [2,6–10]. Ontogenetic observations provide additional evidence for the occurrence of teeth in ancestral baleen whales; tooth buds develop in mysticete foetuses, but are subsequently aborted and resorbed prior to enamel formation [11–14]. The presence of teeth in foetal whales was even known to Darwin [15], who discussed the significance of this rudiment in his long argument for evolution.
Cladistic analyses of living and extinct mysticetes support the hypothesis that mineralized teeth were lost in the common ancestor of crown Mysticeti [2,8,9]. Molecular sequences of three enamel-specific genes, ameloblastin (AMBN), enamelin (ENAM) and amelogenin (AMEL) contain stop codons and/or frameshift mutations in various mysticete species ([2,16]; J. Gatesy 2010, unpublished data), but none of the inactivating mutations are common to all extant mysticetes even though a total of approximately 3650 bp have been sequenced from exonic regions of AMBN, ENAM and AMEL. Thus, the current body of molecular evidence agrees with the phylogenetic studies of fossils in documenting the pseudogenization of enamel-specific genes in living mysticetes, but falls short of supporting the hypothesis that the genetic toolkit for manufacturing enamel was knocked out on the same branch on which functional, mineralized teeth were apparently lost, i.e. the common ancestor of crown Mysticeti.
Several explanations can account for this inconsistency between fossil and molecular evidence. First, given that only partial protein-coding sequences were generated for AMBN, ENAM and AMEL, it is possible that frameshift mutations and/or stop codons will be discovered in the unsequenced protein-coding regions of one or more of these extracellular matrix protein (EMP) genes. A second possibility is that one or more of these genes were initially silenced by mutations in a regulatory gene region on the ancestral mysticete branch, and that mutations in protein-coding regions accumulated subsequently on descendant branches within crown-group Mysticeti. A third possibility is that a different enamel- or tooth-specific gene was knocked out in the common ancestor of mysticetes, and that AMBN, ENAM and AMEL acquired molecular cavities on descendant branches within crown-group Mysticeti. Indeed, mysticetes have the slowest rates of nuclear gene evolution among mammals [16–18], which reduces the likelihood that all enamel-specific genes acquired their first inactivating mutation on the common mysticete branch. Finally, enamel may have been lost independently in several mysticete lineages, rather than once in the common ancestor of crown mysticetes. Edentulous stem mysticetes (e.g. Eomysticetus) and early crown mysticetes (‘cetotheres’) may have retained rudimentary, enamel-capped teeth that were embedded in soft tissue, rather than set in bony alveoli, as is the case for maxillary teeth in Ziphiidae (beaked whales) and Physeteroidea (sperm whales) [19–21]. In summary, the occurrence of stem mysticete fossils that lack teeth suggests that we may find evidence of molecular cavities in one or more tooth-specific genes that are shared by all living mysticetes, unless enamel was lost independently in several extant mysticete lineages.
In an attempt to discriminate among competing hypotheses, we amplified and sequenced three of the longer exons (2, 3, 4) of the enamelysin gene of representative mysticetes, and searched for shared frameshift mutations and/or stop codons. Enamelysin belongs to the matrix metalloproteinase gene family and is otherwise known as matrix metalloproteinase 20 (MMP20). The MMP20 gene is located in a cluster of matrix metalloproteinase genes at human chromosome 11q22 [22,23]. MMP20 diverged from other matrix metalloproteinase loci prior to the common ancestry of tetrapods [24], and plays a key role in processing structural proteins (amelogenin, ameloblastin, enamelin) that are secreted into the extracellular matrix by ameloblasts during the secretory stage of enamel formation [23,25–28]. MMP20 may also be necessary to activate kallikrein-related peptidase 4 (KLK4; [29,30]), which cleaves and degrades remnants of enamel matrix proteins during the maturation stage of amelogenesis. MMP20-deficient mice have an amelogenesis imperfecta phenotype that is characterized by thin, hypomineralized enamel that easily chips away from the underlying dentin [31,32]. There are also mutations in the human MMP20 gene that cause non-syndromic amelogenesis imperfecta [30]. Other evidence suggests a role for MMP20, along with matrix metalloproteinase 2 (MMP2), in cleaving dentin sialophosphoprotein (DSPP), which consists of three parts: dentin sialoprotein (DSP), dentin glycoprotein (DGP) and dentin phosphoprotein (DPP) [33]. Specifically, MMP20 cleaves DSP–DGP to generate DSP and DGP, and also cleaves DSP at multiple sites to yield smaller DSP products [33]. However, the absence of conspicuous dentin phenotypes in humans and mice that lack a functional copy of MMP20 suggests that this enzyme and MMP2 are functionally redundant [33]. Given that the only unique, non-overlapping functions of MMP20 are enamel- or tooth-specific, we hypothesized that the MMP20 gene should show evidence of pseudogenization in crown mysticetes as it occurs for AMBN, ENAM and AMEL ([2,16]; J. Gatesy 2010, unpublished data).
2. Material and methods
(a). Laboratory procedures
PCR amplifications for three different exons of MMP20 (2, 3, 4) were performed with primers from flanking introns to negate the possibility of amplifying processed pseudogenes. PCR primers were designed based on aligned sequences for Bos taurus, Sus scrofa, Tursiops truncatus and Vicugna pacos that were obtained from Ensembl 56. Exons 3 and 4 were each amplified with a single set of primers. Exon 2 was amplified with a nested set of primers after initial amplification with an outer set of primers. We used 1 µl of the first PCR reaction product as the template DNA in nested reactions. Primer sequences are provided in electronic supplementary material, table S1. Amplifications were performed with Denville Scientific Inc. Ramp-Taq DNA polymerase in 50 µl reactions with the following thermal cycling profile: pre-activation step at 95°C for 7 min; initial denaturation at 95°C for 2 min; 45 cycles of 1 min at 95°C (denaturation), 1 min at 50°C (annealing) and 2 min at 72°C (extension); and a final extension at 72°C for 10 min. In our initial screen, we attempted to amplify exons 2–4 from Eubalaena australis (southern right whale), Caperea marginata (pygmy right whale), Eschrichtius robustus (grey whale) and Megaptera novaeangliae (humpback whale), which are representative of the four extant mysticete families. After discovering a SINE insertion in exon 2 of MMP20 in each of these mysticetes, we performed additional amplifications with the mysticete taxa Balaena mysticetus (bowhead whale), Balaenoptera acutorostrata (common minke whale), Balaenoptera physalus (fin whale) and Balaenoptera musculus (blue whale). We also amplified exon 2 of MMP20 from representatives of most odontocete families, and additional cetartiodactyl outgroups, as follows: Monodontidae (Monodon monoceros (narwhal), Delphinapterus leucas (beluga)); Phocoenidae (Phocoena phocoena (harbour porpoise)); Iniidae (Inia geoffrensis (Amazon River dolphin)); Pontoporiidae (Pontoporia blainvillei (La Plata dolphin)); Platanistidae (Platanista minor (Indus River dolphin)); Physeteridae (Physeter macrocephalus (giant sperm whale)); Kogiidae (Kogia sima (dwarf sperm whale), Kogia breviceps (pygmy sperm whale)); Ziphiidae (Mesoplodon bidens (Sowerby's beaked whale)); Hippopotamidae (Hippopotamus amphibius (hippopotamus)); Cervidae (Cervus nippon (sika deer)); Giraffidae (Okapia johnstoni (okapi)); Moschidae (Moschus sp. (musk deer)); Antilocapridae (Antilocapra americana (pronghorn)); Tayassuidae (Pecari tajacu (collared peccary)); and Camelidae (Lama glama (llama)). Specimen numbers for genomic DNA samples are given in electronic supplementary material, table S2. Successfully amplified PCR products were electrophoresed on 1 per cent agarose gels, excised and cleaned with the Bioneer AccuPrep Gel Purification Kit. Cleaned PCR products were sequenced at the UCR Core Instrumentation Facility with an ABI 3730xl automated DNA sequencer. Sequencher 4.8 was used to assemble contigs. GenBank accession numbers for the new MMP20 sequences are HQ171778–HQ171814. Sequences for B. taurus, S. scrofa and T. truncatus were obtained from Ensembl 56. The identification of the insertion in mysticete MMP20 exon 2 as a CHR-2 short interspersed nuclear element (SINE) retroposon resulted from BLASTing the Megaptera novaeangliae insertion against nucleotide sequences in GenBank. Accession numbers for additional CHR-2 SINEs that were employed in alignments and/or phylogenetic analyses are as follows: AB054403, AB054436, AB054471, AB054480, AB071537, AB071542, AB071567, AB071586, AB195475, AB195478, AB195479, AB195481–AB195486, AB195488, AB195492, AB195495.
(b). Alignments and phylogenetic analyses
Sequences were aligned manually with Se-Al [34]. The MMP20 exon 2 alignment (578 base pairs (bp)) consisted of complete exonic sequences, including the CHR-2 SINE retroposon in mysticetes, and the 5′ end of intron 2 for 28 species (electronic supplementary material, alignment 1). We also constructed a CHR-2 SINE alignment (263 bp) that consisted of sequences from the MMP20 locus, additional sequences from the CD (cetacean deletion) subfamily of CHR-2 SINEs and an outgroup sequence (Megaptera novaeangliae) from the Hump14 locus of the CHR-2 SINE DT (deletion type) subfamily (electronic supplementary material, alignment 2). Separate phylogenetic analyses were performed on: (i) the exonic region of the MMP20 exon 2 alignment, which comprised 248 bp after excluding frameshift insertions and intronic sequences, and (ii) the CHR-2 SINE alignment. The Akaike Information Criterion (AIC) of jModeltest [35] was used to select the model of molecular evolution that was implemented in maximum-likelihood (ML) analyses (MMP20 exon 2 = K80 + Γ; CHR-2 SINE = HKY + Γ). ML searches were performed with PAUP* 4.0b10 [36] and employed stepwise addition with 100 randomized input orders and tree bisection and reconnection branch swapping. ML bootstrap analyses were performed with neighbour-joining starting trees and 500 pseudoreplicate datasets.
(c). dN/dS analyses
The codeml program of PAML 4.2 [37] was used to estimate the ratio (ω) of the non-synonymous substitution rate (dN) to the synonymous substitution rate (dS) for functional and pseudogenic branches of exon 2 of MMP20 after removing frameshift insertions and recoding the stop codon in B. acutorostrata as missing data. Given that none of the highest supported nodes (>70%) on the MMP20 tree conflicted with cetartiodactyl species trees (see below), and that differences pertain to nodes that are weakly supported by MMP20 alone and typically require larger datasets to achieve improved resolution, we employed a composite species tree based on McGowen et al. [38] for relationships within Mysticeti, and Gatesy [39] for relationships among other cetartiodactyl taxa. The branch model of PAML [37] was used to estimate ω values for functional and pseudogenic branches following Meredith et al.'s [16] branch-coding method. Functional branches lead to external nodes (i.e. extant taxa) having enamel or to internal nodes having enamel based on parsimony reconstructions, and are expected to evolve under purifying selection with ω < 1. Pseudogenic branches post-date the first detected occurrence of a frameshift mutation or stop codon on an earlier branch and are expected to evolve at the neutral rate with ω = 0. We used χ2-tests to compare the observed numbers of non-synonymous and synonymous substitutions, which were estimated using PAML [37], with the expected numbers of non-synonymous and synonymous substitutions according to a neutral model of evolution with ω = 1. PAML estimates for the number of non-synonymous and synonymous sites in the MMP20 alignment were 170.8 and 75.2, respectively. Estimated numbers of non-synonymous and synonymous substitutions on functional branches were 30.4 and 70.7, respectively, whereas expected numbers of non-synonymous and synonymous changes were 70.2 and 30.9 for ω = 1. Estimated numbers of non-synonymous and synonymous substitutions on pseudogenic branches were 17.3 and 4.2, respectively, whereas expected numbers of non-synonymous and synonymous changes were 14.9 and 6.6 for ω = 1. dN/dS analyses were run with the CodonFreq = 3 option in PAML.
(d). Ancestral sequence reconstructions
The baseml program of PAML 4.2 [37] was used to estimate ancestral DNA and amino acid sequences.
3. Results and discussion
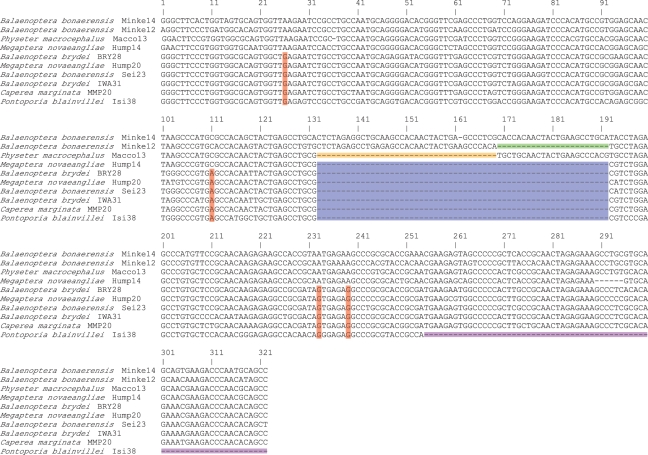
Representative mysticete sequences from exons 3 and 4 of MMP20 lack frameshift mutations and stop codons, but we discovered a CHR-2 SINE insertion in exon 2 that is shared by eight mysticete species that are representative of all extant mysticete genera. CHR SINEs were originally described by Shimamura et al. [40] and given the name CHR based on their exclusive occurrence in the genomes of cetaceans (C), hippopotamuses (H) and ruminants (R). CHR SINEs are divided into two families, CHR-1 and CHR-2, with the latter derived from the former [41]. In the CHR-2 group, Nikaido et al. [42] defined FL (full-length), MDI (middle deletion I), MDII (middle deletion II), DT (deletion type), CD (cetacean deletion) and CDO (cetacean deletion odontocete) subfamilies. The CHR-2 SINE in MMP20 belongs to the CD subfamily (figure 1). MMP20 SINE sequences share diagnostic features with members of the CD and CDO subfamilies [42], including a centrally located deletion in the tRNA unrelated region and several diagnostic nucleotide substitutions (figure 1), but lack the terminal deletion that defines the CDO subfamily. The length of the MMP20 SINE ranges from 302 bp (B. musculus) to 318 bp (B. physalus) and includes a tRNA-related region, tRNA-unrelated region, poly-AT region and 14-nucleotide target site duplication at the 3′ end of the SINE (electronic supplementary material, alignment 1). All of the length variation occurs in the poly-AT region.
Figure 1.
Alignment of CHR-2 SINE sequences for representatives of the FL subfamily (Balaenoptera bonaerensis Minke14), MDI subfamily (B. bonaerensis Minke12), MDII subfamily (Physeter macrocephalus Macco13), DT subfamily (Megaptera novaeangliae Hump14), CD subfamily (B. brydei BRY28, M. novaeangliae Hump20, B. bonaerensis Sei23, B. brydei IWA31, Caperea marginata MMP20) and CDO subfamily (Pontoporia blainvillei Isi38). Diagnostic features of different subfamilies are highlighted with coloured boxes as follows: green, MDI subfamily deletion; yellow, MDII subfamily deletion; blue, central deletion of DT, CD and CDO subfamilies; red, examples of diagnostic substitutions that are shared by members of the CD and CDO subfamilies; purple, CDO subfamily deletion. Poly-AT regions of SINEs are not shown.
The preferential insertion of SINEs into introns, rather than exons, reflects selection against the deleterious effects of SINE insertions in protein-coding regions [43]. There are numerous examples of disease-causing SINE insertions in exons [44–46]. The CHR-2 SINE in the MMP20 gene is located in the propeptide-coding region of exon 2, and would result in premature truncation of the MMP20 protein owing to stop codons in all possible reading frames of the CHR-2 SINE. Three mutations in the human MMP20 gene that cause amelogenesis imperfecta have been characterized, and in every case the inheritance pattern is autosomal recessive. One of these mutations encodes a stop signal in the propeptide-coding region of exon 1 [30]. Enamel in the afflicted individual is thin, hypomineralized and chips away from the underlying dentin [30]. Disease-causing SINE insertions in exons are sometimes associated with multiple transcripts, including mRNAs in which the SINE-afflicted exon has been spliced out [46]. However, exon 2 of MMP20 encodes the carboxyl-terminal region of the propeptide, as well as 18 amino-terminal residues of the catalytic (Zn2+, Ca2+) subdomain, and removal of this exon from the mRNA would presumably render MMP20 non-functional. Indeed, the homologous 18-amino acid region of the catalytic (Zn2+, Ca2+) subdomain encoded by exon 2 is completely conserved in human, cow, pig and spectacled caiman, and shows only one amino acid difference in mouse [24]. This pattern of evolutionary conservation validates the critical importance of the catalytic subdomain that is partially encoded by exon 2.
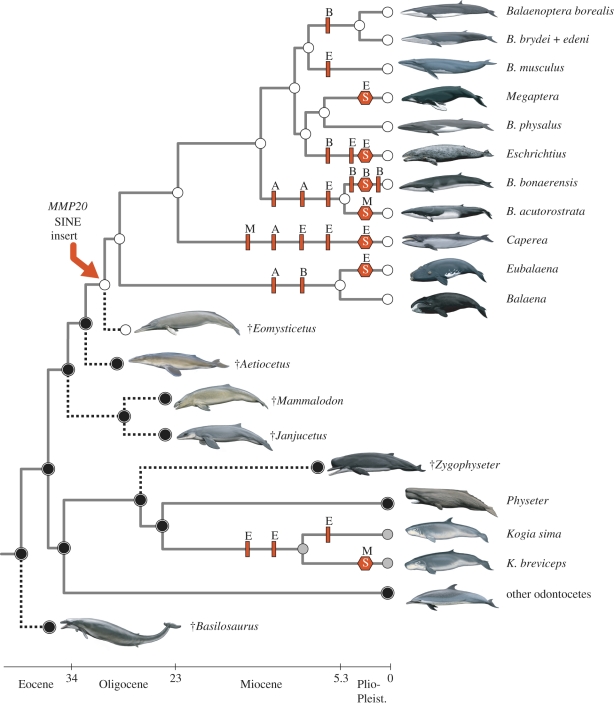
The SINE insertion in exon 2 of MMP20 provides the first molecular evidence for pseudogenization of the genetic toolkit for enamel production in the common ancestor of extant mysticetes (figure 2). Given that MMP20 is required for proper processing of amelogenin, ameloblastin and enamelin, and may also be required for KLK4 activation; this insert provides compelling evidence that normal enamel formation was abrogated no later than the date of this SINE insertion. There is, therefore, congruent genomic and fossil evidence for the loss of enamel-capped teeth prior to the last common ancestor of crown-group mysticetes (figure 2). Approximately 3.9 kb from among the exons of four tooth-specific genes (AMBN, AMEL, ENAM and MMP20) have been sequenced for representative mysticetes, but the SINE insertion in MMP20 is the only example of a shared frameshift mutation (figure 2). However, mysticete pseudogenes have low rates of frameshift accumulation. Previously, Meredith et al. [16] calculated a rate of 0.0081 frameshifts kb−1 myr−1 for neutrally evolving mysticete DNA. Assuming this rate, and a stem mysticete branch that comprises 7.6 myr of evolutionary history [38], then we should expect only 0.24 shared frameshifts per 3.9 kb for exonic segments that have evolved under neutral evolution.
Figure 2.
A phylogenetic hypothesis for living and extinct taxa that summarizes the evolution of teeth, enamel and enamel-specific genes within Cetacea. Cetaceans in the tree are toothless as adults (white circles), have enamel-less teeth (grey circles) or have enamel-capped teeth (black circles). Circles at internal nodes of the tree show parsimony reconstructions of these three states and indicate the loss of teeth within Mysticeti (baleen whales) and the loss of enamel in Kogia (pygmy and dwarf sperm whales). Frameshift mutations (red bars) and nonsense substitutions (red hexagons) in four enamel genes (AMEL, A; AMBN, B; ENAM, E; MMP20, M) are mapped onto the tree (deltran parsimony optimization). The MMP20 SINE insertion in the common ancestor of extant baleen whales is indicated with a red arrow, and may have occurred on the branch before or after the indicated node. Phylogenetic relationships and divergence times among extant lineages (grey branches) are according to McGowen et al. [38]; the placements of extinct lineages (dotted lines) are as in Bianucci & Landini [47] for the physeteroid, Zygophyseter and as in Fitzgerald [9] for stem mysticetes (Eomysticetus, Aetiocetus, Mammalodon, Janjucetus) and the archaeocete outgroup, Basilosaurus.
In addition to the CHR-2 SINE insertion that is shared by all mysticete genera, there is additional evidence for the inactivation of MMP20 in toothless and enamel-less cetaceans (figure 2). Within Mysticeti, a 1 bp insertion at position 552 is present in the pygmy right whale, Caperea marginata, and a G to T point mutation at position 532 in the common minke whale, Balaenoptera acutorostrata, results in an ochre (‘TAA’) stop codon (electronic supplementary material, alignment 1). We also discovered an opal stop codon (‘TGA’) in the propeptide-coding region of MMP20 exon 2 in a single individual of the pygmy sperm whale, Kogia breviceps (electronic supplementary material, alignment 1). This species and its congener, Kogia sima (dwarf sperm whale), have enamel-less teeth [47]. Previously, three frameshift mutations were reported in the enamelin (ENAM) genes of Kogia, two in the common ancestor of the two extant species, and the other in K. sima [16]. Although available data suggest that the MMP20 gene was incapacitated before ENAM in baleen whales, current evidence suggests that ENAM was pseudogenized before MMP20 in Kogia (figure 2). Indeed, the stop codon in K. breviceps was only present in one of three individuals surveyed here, and also was absent in five individuals of K. sima (electronic supplementary material, alignment 1).
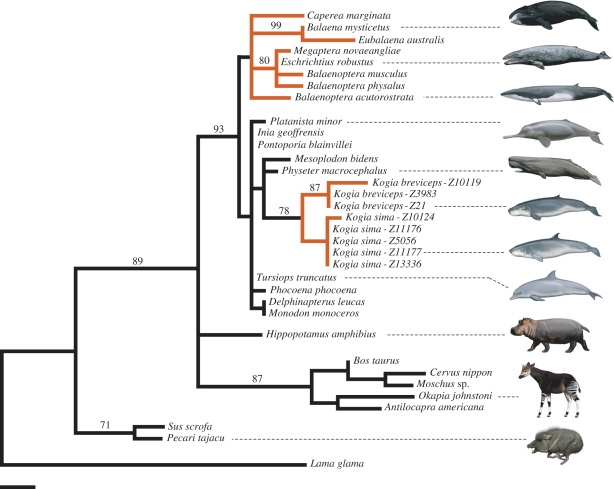
An ML phylogram based on exon 2 sequences from MMP20 (figure 3) is broadly congruent with cetartiodactyl trees based on large supermatrices [38,48,49] even though the exon 2 alignment is only 248 bp. Only eight clades were supported above the 70 per cent bootstrap level (figure 3), but in every case these clades were concordant with the analyses based on supermatrices. Within Cetacea, visual inspection of the MMP20 tree confirms that branches leading exclusively to taxa without enamel (mysticetes, Kogia) are longer than branches leading to taxa that retain enamel. dN/dS values were calculated for functional versus pseudogenic branches of Cetartiodactyla following Meredith et al. [16] and in each case tested against the null hypothesis of no selection (ω = 1) using a χ2-test. Functional branches have a low dN/dS, consistent with strong purifying selection (ω = 0.191, χ2 = 73.82, p < 0.001), whereas pseudogenic branches had dN/dS nearly an order of magnitude higher and are compatible with the absence of selective constraints (ω = 1.84, χ2 = 1.26, 0.25 < p < 0.50).
Figure 3.
One of the two ML phylograms for MMP20 exon 2 protein-coding sequences (−ln L = 1027.63005). The second tree (not shown) includes a short branch (1.94 × 10−7 substitutions per site) that groups Hippopotamus with Cetacea to the exclusion of other cetartiodactyls. Branches coloured red indicate evolutionary lineages that lack enamel-capped teeth according to parsimony reconstructions. Bootstrap scores ≥70% are shown. Branch lengths are proportional to the amount of change in nucleotide substitutions per site. Scale bar, 0.01 substitutions per site.
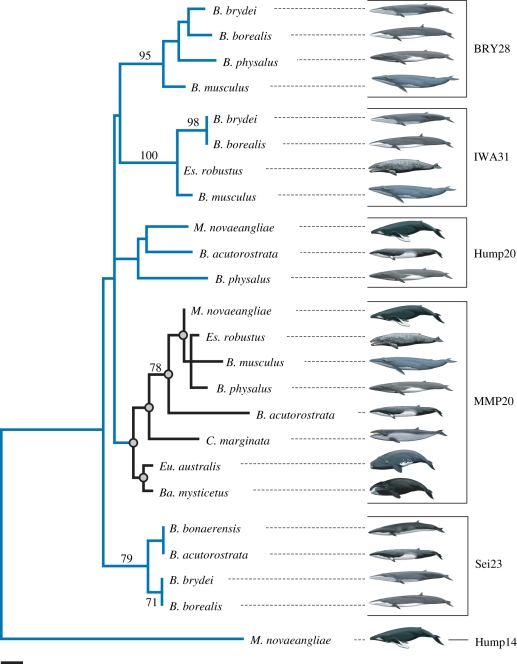
ML analysis of CD subfamily CHR-2 SINE sequences (figure 4) groups the mysticete MMP20 SINEs to the exclusion of several other loci (BRY28, Hump20, IWA31, Sei23) that have been sequenced for multiple mysticetes [50], albeit with weak bootstrap support. Relationships among mysticete MMP20 SINE sequences are generally congruent with trees supported by large concatenations of molecular data (e.g. [38]).
Figure 4.
Maximum-likelihood phylogram for mysticete CHR-2 SINE sequences (−ln L = 1068.74802). The clade of SINE sequences from the MMP20 locus is coloured black, and groupings that are congruent with the supermatrix topology of McGowen et al. [38] are marked by grey circles at nodes. The CHR-2 SINE tree was rooted with Megaptera novaeangliae Hump14, which belongs to the DT subfamily. Bootstrap scores greater than 70% are shown. Branch lengths are proportional to the amount of change in nucleotide substitutions per site. Mysticete genera are abbreviated as follows: Balaenoptera, B.; Balaena, Ba.; Caperea, C.; Eschrichtius, Es.; Eubalaena, Eu.; Megaptera, M. Scale bar, 0.01 substitutions per site.
MMP20 is primarily expressed in developing teeth, but expression has been reported in human lung [51,52] and in mouse large intestine [53]. There are also SNPs in MMP20 that are significantly associated with kidney ageing [54]. Nevertheless, MMP20 generally has been considered a tooth-specific gene owing to its primary expression pattern, and the occurrence of non-syndromic amelogenesis imperfecta in mice and humans that lack a functional copy of this gene [53]. Pseudogenization of MMP20 in two enamel-less cetacean lineages, Mysticeti and Kogia breviceps, provides additional evidence for the tooth-specific function of MMP20. Likewise, the genomic assembly for Hoffmann's two-toed sloth (Choloepus hoffmanni), another species with enamel-less teeth, revealed frameshift mutations in exon 1 (8 bp deletion), exon 4 (1 bp deletion), exon 5 (7 bp insertion), exon 6 (1 bp deletion) and exon 9 (2 bp deletion) of MMP20 (Ensembl 57; electronic supplementary material, figure S1). A polymorphic stop codon in K. breviceps and multiple frameshift indels in C. hoffmanni further suggest that MMP20 is not only tooth-specific, but also enamel-specific; K. breviceps and C. hoffmanni both retain dentin in their mineralized, enamel-less teeth. These findings do not invalidate reports of MMP20 expression in other tissues, but imply that the only critical, unique and non-overlapping role of this gene is in enamel formation. In other instances, MMP20 expression may be incidental and entirely overlapping with other components of the transcriptome. Along these lines, the molecular redundancy of MMP2 and MMP20 in cleaving DSPP [33] may have permitted pseudogenization of MMP20 once it was released from performing its unique function in enamel formation.
Mammalian diversity provides a natural laboratory, complete with replicated experiments, for testing hypotheses of tooth-specific gene function. Multiple lineages of enamel-less and edentulous mammals have descended from ancestors with enamel-capped teeth, and we expect to find degraded remnants of enamel-specific genes in these taxa owing to their evolutionary history. Previous work has documented pseudogenization of three genes that code for structural EMPs (enamelin, ameloblastin, amelogenin) in one or more lineages that lack enamel ([2,16]; J. Gatesy 2010, unpublished data). Molecular cavities in the MMP20 gene in three different lineages of enamel-less mammals (Mysticeti, K. breviceps, C. hoffmanni) provide the first evidence for pseudogenization of an enzymatic EMP gene. Further, the insertion of a CHR-2 SINE retroposon in MMP20 shows that the genetic toolkit for enamel production was knocked out in the common ancestor of living mysticetes. The combination of palaeontological and molecular data now provide support for the gain and loss of two complex adaptations, baleen and enamel-capped teeth, respectively, on the stem mysticete branch. Pseudogenization may occur through neutral evolution when changes in the genetic background or environment render a formerly useful gene worthless, or through positive selection when a previously useful gene becomes harmful to an organism [55,56]. It remains unclear if the SINE insertion in MMP20 was favoured by natural selection because it was advantageous to stop enamel production or was simply a consequence of neutral evolution owing to relaxed functional constraints on tooth-specific genes subsequent to the origin of baleen. Beyond the SINE insertion, the only other reconstructed change in exon 2 of MMP20 on the stem mysticete branch is a non-synonymous transition from A to G at nucleotide position 38 that replaced histidine with arginine. This change has a smaller Grantham matrix distance (29) than the reconstructed change from histidine to proline (77) at the same amino acid position in ruminants, and is an unlikely candidate for adaptive loss of protein function. Whether adaptive or neutral, the SINE insertion in MMP20 fills an important gap in our understanding of the macroevolutionary transition leading from the last common ancestor of crown Cetacea to the last common ancestor of crown Mysticeti.
Acknowledgements
This work was supported by NSF (EF0629860 to M.S.S. and J.G.; DEB0743724 to J.G.). For providing DNA samples, we thank Southwest Fisheries Science Center, South Australian Museum, North Slope Borough (Barrow, Alaska), The Marine Mammal Center (Sausalito), Smithsonian Institution, New York Zoological Society, World Wildlife Fund, Greenland Institute of Natural Resources, Alaska Department of Fish and Game, P. Morin, K. Robertson, S. Chivers, A. Dizon, M. Milinkovitch, G. Amato, G. Schaller, M. Cronin, W. Murphy, M. P. Heide-Jørgensen, Ú. Árnason, H. Rosenbaum and G. Braulik. C. Buell painted living and extinct mammals. Two anonymous reviewers provided helpful comments on an earlier version of this manuscript.
References
- 1.Uhen M. D. 2010. The origin(s) of whales. Annu. Rev. Earth Planet. Sci. 38, 189–219 10.1146/annurev-earth-040809-152453 (doi:10.1146/annurev-earth-040809-152453) [DOI] [Google Scholar]
- 2.Deméré T. A., McGowen M. R., Berta A., Gatesy J. 2008. Morphological and molecular evidence for a stepwise evolutionary transition from teeth to baleen in mysticete whales. Syst. Biol. 57, 15–37 10.1080/10635150701884632 (doi:10.1080/10635150701884632) [DOI] [PubMed] [Google Scholar]
- 3.George J. C., Bada J., Zeh J., Scott L., Brown S. E., O'Hara T., Suydam R. 1999. Age and growth estimates of bowhead whales (Balaena mysticetus) via aspartic acid racemization. Can. J. Zool. 77, 571–580 10.1139/cjz-77-4-571 (doi:10.1139/cjz-77-4-571) [DOI] [Google Scholar]
- 4.Deméré T. A., Berta A., McGowen M. R. 2005. The taxonomic and evolutionary history of fossil and modern balaenopteroid mysticetes. J. Mammal. Evol. 12, 99–143 10.1007/s10914-005-6944-3 (doi:10.1007/s10914-005-6944-3) [DOI] [Google Scholar]
- 5.de Magalhães J. P., Sedivy J. M., Finch C. E., Austad S. N., Church G. M. 2007. A proposal to sequence genomes of unique interest for research on aging. J. Gerontol. 62A, 583–584 [DOI] [PubMed] [Google Scholar]
- 6.Sanders A. E., Barnes L. G. 2002. Paleontology of the Late Oligocene Ashley and Chandler Bridge formations of South Carolina, 2: Micromysticetus rothauseni, a primitive cetotheriid mysticete (Mammalia: Cetacea). Smithsonian Contrib. Paleobiol. 93, 271–293 [Google Scholar]
- 7.Sanders A. E., Barnes L. G. 2002. Paleontology of the Late Oligocene Ashley and Chandler Bridge formations of South Carolina, 3: Eomysticetidae, a new family of primitive mysticetes (Mammalia: Cetacea). Smithsonian Contrib. Paleobiol. 93, 313–356 [Google Scholar]
- 8.Fitzgerald E. M. G. 2006. A bizarre new toothed mysticete (Cetacea) from Australia and the early evolution of baleen whales. Proc. R. Soc. B 273, 2955–2963 10.1098/rspb.2006.3664 (doi:10.1098/rspb.2006.3664) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzgerald E. M. G. 2010. The morphology and systematics of Mammalodon colliveri (Cetacea: Mysticeti), a toothed mysticete from the Oligocene of Australia. Zool. J. Linn. Soc. 158, 367–476 10.1111/j.1096-3642.2009.00572.x (doi:10.1111/j.1096-3642.2009.00572.x) [DOI] [Google Scholar]
- 10.Deméré T. A., Berta A. 2008. Skull anatomy of the Oligocene toothed mysticete Aetioceus weltoni (Mammalia: Cetacea): implications for mysticete evolution and functional anatomy. Zool. J. Linn. Soc. 154, 308–352 10.1111/j.1096-3642.2008.00414.x (doi:10.1111/j.1096-3642.2008.00414.x) [DOI] [Google Scholar]
- 11.Ridewood W. G. 1923. Observations on the skull in foetal specimens of whales of the genera Megaptera and Balaenoptera. Phil. Trans. R. Soc. Lond. B 211, 209–292 10.1098/rstb.1923.0005 (doi:10.1098/rstb.1923.0005) [DOI] [Google Scholar]
- 12.Dissel-Scherft M. C. V., Vervoort W. 1954. Development of the teeth in fetal Balaenoptera physalus (L.) (Cetacea, Mystacoceti). Proc. K. Ned. Akad. Wet. Ser. C 57, 196–210 [Google Scholar]
- 13.Karlsen K. 1962. Development of tooth germs and adjacent structures in the whalebone whale (Balaenoptera physalus (L.)). Hvalrådets Skrifter 45, 5–56 [Google Scholar]
- 14.Ishikawa H. H., Amasaki H., Dohguchi A., Furuya A., Suzuki K. 1999. Immunohistological distributions of fibronectin, tenascin, type I, III and IV collagens, and laminin during tooth development and degeneration in fetuses of minke whale, Balaenoptera acutorostrata. J. Vet. Med. Sci. 61, 227–232 10.1292/jvms.61.227 (doi:10.1292/jvms.61.227) [DOI] [PubMed] [Google Scholar]
- 15.Darwin C. 1859. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London, UK: John Murray; [PMC free article] [PubMed] [Google Scholar]
- 16.Meredith R. W., Gatesy J., Murphy W. J., Ryder O. A., Springer M. S. 2009. Molecular decay of the tooth gene enamelin (ENAM) mirrors the loss of enamel in the fossil record of placental mammals. PLoS Genet. 5, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alter S. E., Rynes E., Palumbi S. R. 2007. DNA evidence for historic population size and past ecosystem impacts of gray whales. Proc. Natl Acad. Sci. USA 104, 15 162–15 167 10.1073/pnas.0706056104 (doi:10.1073/pnas.0706056104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson J. A., Baker C. S., Vant M., Steel D. J., Medrano-Gonzalez L., Palumbi S. R. 2009. Big and slow: phylogenetic estimates of molecular evolution in baleen whales (Suborder Mysticeti). Mol. Biol. Evol. 26, 2427–2440 10.1093/molbev/msp169 (doi:10.1093/molbev/msp169) [DOI] [PubMed] [Google Scholar]
- 19.Boschma H. 1938. On the teeth and other particulars of the sperm whale (Physeter macrocephalus L.). Temminckia 3, 151–278 [Google Scholar]
- 20.Boschma H. 1950. Maxillary teeth in specimens of Hyperoodon rostratus (Müller) and Mesoplodon grayi von Haast stranded on the Dutch coast. Proc. K. Ned. Akad. Wet. Ser. C 53, 3–14 [Google Scholar]
- 21.Rice D. W. 1989. Sperm whale, Physeter macrocephalus Linnaeus 1758. Handbook of marine mammals (eds Ridgway S. H., Harrison R. J.), vol. 4, pp. 177–233 London, UK: Academic Press [Google Scholar]
- 22.Llano E., et al. 1997. Identification and structural and functional characterization of human enamelysin (MMP-20). Biochemistry 36, 15 101–15 108 10.1021/bi972120y (doi:10.1021/bi972120y) [DOI] [PubMed] [Google Scholar]
- 23.Bartlett J. D., Ganss B., Goldberg M., Moradian-Oldak J., Paine M. L., Snead M. L., Wen X., White S. N., Zhou Y. L. 2006. Protein-protein interactions of the developing enamel matrix. Curr. Topics Dev. Biol. 74, 57–115 10.1016/S0070-2153(06)74003-0 (doi:10.1016/S0070-2153(06)74003-0) [DOI] [PubMed] [Google Scholar]
- 24.Shintani S., Kobata M., Kamakura N., Toyosawa S., Ooshima T. 2007. Identification and characterization of matrix metalloproteinase-20 (MMP20: enamelysin) genes in reptile and amphibian. Gene 392, 89–97 10.1016/j.gene.2006.11.014 (doi:10.1016/j.gene.2006.11.014) [DOI] [PubMed] [Google Scholar]
- 25.Ryu O. H., Fincham A. G., Hu J. C.-C., Zhang C., Qian Q., Bartlett J. D., Simmer J. P. 1999. Characterization of recombinant pig enamelysin activity and cleavage of recombinant pig and mouse amelogenins. J. Dent. Res. 78, 743–750 [DOI] [PubMed] [Google Scholar]
- 26.Iwata T., Yamakoshi Y., Hu J. C.-C., Ishikawa K., Bartlett J. D., Krebsbach P. H., Simmer J. P. 2007. Processing of ameloblastin by MMP-20. J. Dent. Res. 86, 153–157 10.1177/154405910708600209 (doi:10.1177/154405910708600209) [DOI] [PubMed] [Google Scholar]
- 27.Lu Y., Papagerakis P., Yamakoshi Y., Hu J. C.-C., Bartlett J. D., Simmer J. P. 2008. Functions of KLK4 and MMP-20 in dental enamel formation. Biol. Chem. 389, 695–700 10.1515/BC.2008.080 (doi:10.1515/BC.2008.080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Hashimi N., Sire J.-Y., Delgado S. 2009. Evolutionary analysis of mammalian enamelin, the largest enamel protein, supports a crucial role for the 332-kDa peptide and reveals selective adaptation in rodents and primates. J. Mol. Evol. 69, 635–656 10.1007/s00239-009-9302-x (doi:10.1007/s00239-009-9302-x) [DOI] [PubMed] [Google Scholar]
- 29.Ryu O., Hu J. C.-C., Yamakoshi Y., Villemain J. L., Cao X., Zhang C., Bartlett J. D., Simmer J. P. 2002. Porcine kallikrein-4 activation, glycosylation, activity, and expression in prokaryotic and eukaryotic hosts. Eur. J. Oral Sci. 110, 358–365 10.1034/j.1600-0722.2002.21349.x (doi:10.1034/j.1600-0722.2002.21349.x) [DOI] [PubMed] [Google Scholar]
- 30.Papagerakis P., Lin H.-K., Lee K. Y., Hu Y., Simmer J. P., Bartlett J. D., Hu J. C.-C. 2008. Premature stop codon in MMP20 causing amelogenesis imperfecta. J. Dent. Res. 87, 56–59 10.1177/154405910808700109 (doi:10.1177/154405910808700109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caterina J. J., Skobe Z., Shi J., Ding Y., Simmer J. P., Birkedal-Hansen H., Bartlett J. D. 2002. Enamelysin (matrix metalloproteinase 20)-deficient mice display an amelogenesis imperfecta phenotype. J. Biol. Chem. 277, 49598–59604 10.1074/jbc.M209100200 (doi:10.1074/jbc.M209100200) [DOI] [PubMed] [Google Scholar]
- 32.Fanjul-Fernandez M., Folgueras A. R., Cabrera S., Lopez-Otin C. 2009. Matrix metalloproteinases: evolution, gene regulation and functional analysis in mouse models. Biochim. Biophys. Acta 1803, 3–19 [DOI] [PubMed] [Google Scholar]
- 33.Yamakoshi Y., Hu J. C.-C., Iwata T., Kobayashi K., Fukae M., Simmer J. P. 2006. Dentin sialophosphoprotein is processed by MMP-2 and MMP-20 in vitro and in vivo. J. Biol. Chem. 281, 38235–38243 10.1074/jbc.M607767200 (doi:10.1074/jbc.M607767200) [DOI] [PubMed] [Google Scholar]
- 34.Rambaut A.2002. Se-Al: sequence alignment editor, v2.0a1.1. Oxford, UK: University of Oxford. Available at http://evolve.zoo.ox.ac.uk/
- 35.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256 10.1093/molbev/msn083 (doi:10.1093/molbev/msn083) [DOI] [PubMed] [Google Scholar]
- 36.Swofford D. L. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods, version 4.0b10). Sunderland, MA: Sinauer Associates [Google Scholar]
- 37.Yang Z. 2007. PAML 4: a program package for phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 10.1093/molbev/msm088 (doi:10.1093/molbev/msm088) [DOI] [PubMed] [Google Scholar]
- 38.McGowen M. R., Spaulding M., Gatesy J. 2009. Divergence date estimation and a comprehensive molecular tree of extant cetaceans. Mol. Phylogenet. Evol. 53, 891–906 10.1016/j.ympev.2009.08.018 (doi:10.1016/j.ympev.2009.08.018) [DOI] [PubMed] [Google Scholar]
- 39.Gatesy J. 2009. Whales and even-toed ungulates (Cetartiodactyla). In The timetree of life (eds Hedges S. B., Kumar S.), pp. 511–515 Oxford, UK: Oxford University Press [Google Scholar]
- 40.Shimamura M., Yasue H., Ohshima K., Abe H., Kato H., Kishiro T., Goto M., Munechika I., Okada N. 1997. Molecular evidence from retroposons that whales form a clade within even-toed ungulates. Nature 388, 666–670 10.1038/41759 (doi:10.1038/41759) [DOI] [PubMed] [Google Scholar]
- 41.Shimamura M., Abe H., Nikaido N., Ohshima K., Okada N. 1999. Genealogy of families of SINEs in cetaceans and artiodactyls: the presence of a huge superfamily of tRNAGlu-derived families of SINEs. Mol. Biol. Evol. 16, 1046–1060 [DOI] [PubMed] [Google Scholar]
- 42.Nikaido M., Matsuno F., Abe H., Shimamura M., Hamilton H., Matsubayashi H., Okada N. 2001. Evolution of CHR-2 SINEs in cetartiodactyl genomes: possible evidence for the monophyletic origin of tooth whales. Mamm. Genome 12, 909–915 10.1007/s0033501-1015-4 (doi:10.1007/s0033501-1015-4) [DOI] [PubMed] [Google Scholar]
- 43.Sela N., Mersch B., Gal-Mark N., Lev-Maor G., Hotz-Wagenblatt A., Ast G. 2007. Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu's unique role in shaping the human transcriptome. Genome Biol. 8, R127. 10.1186/gb-2007-8-6-r127 (doi:10.1186/gb-2007-8-6-r127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mustajoki S., Ahola H., Mustajoki P., Kauppinen R. 1999. Insertion of Alu element responsible for acute intermittent porphyria. Hum. Mutat. 13, 431–438 (doi:10.1002/(SICI)1098-1004(1999)13:6<431::AID-HUMU2>3.0.CO;2-Y) [DOI] [PubMed] [Google Scholar]
- 45.Ricci V., Regis S., Di Duca M., Filocamo M. 2003. An Alu-mediated rearrangement as cause of exon skipping in Hunter disease. Hum. Genet. 112, 419–425 [DOI] [PubMed] [Google Scholar]
- 46.Pelé M., Tiret L., Kessler J.-L., Blot S., Panthier J.-J. 2005. SINE exonic insertion in the PTPLA gene leads to multiple splicing defects and segregates with the autosomal recessive centronuclear myopathy in dogs. Hum. Mol. Genet. 14, 1417–1427 10.1093/hmg/ddi151 (doi:10.1093/hmg/ddi151) [DOI] [PubMed] [Google Scholar]
- 47.Bianucci G., Landini W. 2006. Killer sperm whale: a new basal physeteroid (Mammalia, Cetacea) from the Late Miocene of Italy. Zool. J. Linn. Soc. 148, 103–131 10.1111/j.1096-3642.2006.00228.x (doi:10.1111/j.1096-3642.2006.00228.x) [DOI] [Google Scholar]
- 48.O'Leary M. A., Gatesy J. 2008. Impact of increased character sampling on the phylogeny of Cetartiodactyla (Mammalia): combined analysis including fossils. Cladistics 24, 397–442 10.1111/j.1096-0031.2007.00187.x (doi:10.1111/j.1096-0031.2007.00187.x) [DOI] [PubMed] [Google Scholar]
- 49.Steeman M. E., et al. 2009. Radiation of extant cetaceans driven by restructuring of the oceans. Syst. Biol. 58, 573–585 10.1093/sysbio/syp060 (doi:10.1093/sysbio/syp060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nikaido M., Piskurek O., Okada N. 2007. Toothed whale monophyly reassessed by SINE insertion analysis: the absence of lineage sorting effects suggests a small population of a common ancestral species. Mol. Phylogenet. Evol. 40, 216–224 [DOI] [PubMed] [Google Scholar]
- 51.Ryu J., Vicencio A. G., Yeager M. E., Kashgarian M., Haddad G. G., Eickelberg O. 2005. Differential expression of matrix metalloproteinases and their inhibitors in human and mouse lung development. Thromb. Haem. 94, 175–183 [DOI] [PubMed] [Google Scholar]
- 52.Greenlee K. J., Werb Z., Kheradmand F. 2007. Matrix metalloproteinases in lung: multiple, multifarious, and multifaceted. Physiol. Rev. 87, 69–98 10.1152/physrev.00022.2006 (doi:10.1152/physrev.00022.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turk B. E., et al. 2006. MMP-20 is predominately a tooth-specific enzyme with a deep catalytic pocket that hydrolyzes type V collagen. Biochemistry 45, 3863–3874 10.1021/bi052252o (doi:10.1021/bi052252o) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wheeler H. E., et al. 2009. Sequential use of transcriptional profiling, expression quantitative trait mapping, and gene association implicates MMP20 in human kidney aging. PLoS Genet. 5, e1000685. 10.1371/journal.pgen.1000685 (doi:10.1371/journal.pgen.1000685) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jeffrey W. R., Strickler A. G., Yamamoto Y. 2003. To see or not to see: evolution of eye degeneration in Mexican blind cavefish. Integr. Comp. Biol. 43, 531–541 [DOI] [PubMed] [Google Scholar]
- 56.Zhang J. 2008. Positive selection, not negative selection, in the pseudogenization of rcsA in Yersinia pestis. Proc. Natl Acad. Sci. USA 105, E69. 10.1073/pnas.0806419105 (doi:10.1073/pnas.0806419105) [DOI] [PMC free article] [PubMed] [Google Scholar]