Abstract
The relationship between genotypic and phenotypic divergence over evolutionary time varies widely, and cases of rapid phenotypic differentiation despite genetic similarity have attracted much attention. Here, we report an extreme case of the reverse pattern—morphological stasis in a tropical fish despite massive genetic divergence. We studied the enigmatic African freshwater butterfly fish (Pantodon buchholzi), whose distinctive morphology earns it recognition as a monotypic family. We sequenced the mitochondrial genome of Pantodon from the Congo basin and nine other osteoglossomorph taxa for comparison with previous mitogenomic profiles of Pantodon from the Niger basin and other related taxa. Pantodon populations form a monophyletic group, yet their mitochondrial coding sequences differ by 15.2 per cent between the Niger and Congo basins. The mitogenomic divergence time between these populations is estimated to be greater than 50 Myr, and deep genetic divergence was confirmed by nuclear sequence data. Among six sister-group comparisons of osteoglossomorphs, Pantodon exhibits the slowest rate of morphological divergence despite a level of genetic differentiation comparable to both species-rich (e.g. Mormyridae) and species-poor (e.g. Osteoglossidae) families. Morphological stasis in these two allopatric lineages of Pantodon offers a living vertebrate model for investigating phenotypic stability over millions of generations in the face of profound fluctuations in environmental conditions.
Keywords: living fossil, morphological stasis, rate of evolution, cryptic diversity, Osteoglossomorpha
1. Introduction
Morphological disparities between contemporary species frequently capture the attention of evolutionary biologists when accompanied by little genetic divergence. Taxa exhibiting phenotypic divergence despite slight genetic differentiation serve as useful models for investigating the ultimate evolutionary mechanisms of rapid species radiation [1] and the proximate genetic underpinnings of morphological adaptation [2]. Yet, palaeontologists recognize that the opposite pattern, morphological conservatism, is also widespread and important for understanding the fossil record of macroevolutionary diversification [3–5]. Studying long-term morphological conservatism is difficult in contemporary systems because few extant animal lineages are known to have remained morphologically static over geological time scales. Horseshoe crabs, plethodontid salamanders and lampreys are thought to be among the best examples of such ‘living fossils’, although small to moderate levels of morphological differentiation are still measurable within each of these ancient lineages [6–8], leading to the recognition of different taxa. To better understand extrinsic and intrinsic constraints on phenotypic evolution in living populations, it would be ideal to study extant species that exhibit both deep genetic divergence and negligible morphological differentiation. Here, we show that allopatric populations of the bizarre butterfly fish of tropical African rivers represent exactly such a case, offering a unique vertebrate model for studying the mechanisms underlying long-term constraints on morphological stability in a fluctuating environment.
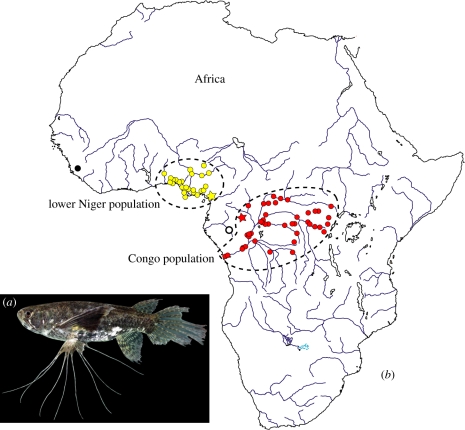
The African freshwater butterfly fish, Pantodon buchholzi (figure 1a), is the sole species in the family Pantodontidae. It is placed within the superorder Osteoglossomorpha, a Gondwanan clade with a rich fossil record beginning in the Early Cretaceous [9]. Pantodon's current distribution is restricted to the central Congo and lower Niger basins as well as a few adjacent coastal drainages (figure 1b), and no fossils of this lineage are known. Pantodon is a small (less than 12 cm), surface-oriented fish with an unusual morphology (figure 1a) including long oblique jaws, wing-like pectoral fins and pelvic fins bearing four elongated, filamentous rays. It locates prey by decoding information contained in surface ripples [10] and by using its specialized visual system to simultaneously view objects in the water, in the air and reflected by the air–water interface [11]. Pantodon's unique pectoral fins allow ballistic jumping to escape predators [12].
Figure 1.
Geographical distribution and morphology of P. buchholzi. (a) Photograph of a live specimen from Odzala National Park, Republic of Congo (CU 92896; standard length = 59.1 mm). (b) Collection localities for specimens housed in the Museum National d'Histoire Naturelle (Paris) and the Musée Royal de l'Afrique Centrale (Tervuren). Two main populations are evident (dashed lines): lower Niger basin and adjacent coastal rivers (yellow dots) and central Congo basin (red dots). Single records are known from the Upper Guinea province (black dot) and the Lower Guinea province (white dot). Yellow and red stars indicate the type locality and Odzala, respectively.
The combination of primitive and derived morphological characteristics exhibited by P. buchholzi has attracted the attention of morphologists since its original description [9]. Intraspecific variation has not been reported; therefore, Pantodon has been assumed to be a single species with a disjunct distribution. Here, we report, to our knowledge, the first assessment of genetic (mitochondrial and nuclear) and phenotypic differentiation between Pantodon populations, which reveals both remarkably long-standing divergence of populations and exceptional morphological stability.
2. Material and methods
(a). Sampling taxa and characters
We sequenced complete mitochondrial genomes (mitogenomes) of 10 osteoglossomorphs: P. buchholzi from the Congo basin at Odzala (GenBank: AP011564), Heterotis niloticus (AP009498), Arapaima gigas (AP009497), Hiodon tergisus (AP009499), Xenomystus nigri (AP009503), Gymnarchus niloticus (AP009610), Petrocephalus microphthalmus (AP009609), Petrocephalus soudanensis (AP009502), Gnathonemus petersii (AP009611) and Myomyrus macrops (AP009501). These new data were combined with five previously published mitogenomes: P. buchholzi from an unknown site in the lower Niger basin (AB04368), Hiodon alosoides (AP004356), Scleropages formosus (DQ023143), Osteoglossum bicirrhosum (AB043025) and Elops hawaiensis (the outgroup, NC005798). To confirm inferences from mitogenomic sequences of Pantodon, we also sequenced the nuclear MLL gene and mitochondrial cytochrome b gene for additional specimens of Pantodon from both populations (electronic supplementary material, figures S3 and S4). Additional information on taxonomic sampling and protocols for sequencing are provided in the electronic supplementary material, tables S1 and S2 and text S1.
(b). Inferring phylogenetic trees and divergence times
We aligned protein-coding genes (excluding the heterogeneous base-composition ND6 gene) by eye, and we aligned the 12S and 16S ribosomal RNA (rRNA) sequences and the 22 transfer RNA (tRNA) sequences with Proalign v 0.5a3 [13]. Phylogenetic trees and divergence times with 95 per cent highest posterior density (HPD) intervals were inferred using a partitioned Bayesian method with a relaxed molecular clock and an uncorrelated lognormal rate heterogeneity in BEAST v 1.4.8 [14]. We analysed two different data matrices crossed with two different fossil constraint distributions. The first constraint distribution (lognormal) assumes that the fossil record is sufficiently informative to provide minimum and maximum estimated ages for at least specific clades, whereas the second distribution (uniform) assumes that the fossil record of Teleostei is too incomplete to provide any maximum estimated ages (see details in the electronic supplementary material, text S1). The first data matrix (‘12RT’; 10 695 positions) included concatenated nucleotide sequences from the rRNA genes, tRNA genes and protein-coding genes excluding third codon positions. The second data matrix (‘123RT’; 14 326 nucleotides) included all positions. These data matrices were partitioned according to codon positions and non-protein coding sequences. For each matrix, we used the GTR + I + Γ model of sequence evolution, a fossil-based calibration constraint scheme and four independent runs of 5 × 107 generations. Additional BEAST parameters and details of the fossil-based calibration are presented in the electronic supplementary material, text S1.
(c). Estimating morphological divergence
We compared phenotypic similarity among six osteoglossomorph taxon pairs based on 15 morphometric measurements (electronic supplementary material, table S3, text S2 and figure S5). Log10-transformed measurements from all taxa were analysed by principal component analysis (PCA). We quantified morphological dissimilarity as Mahalanobis generalized distance (D) between each pair of taxa in the PCA-based morphospace. We discarded the first principal component (PC1, 79.89%), which reflected only body size, and focused on the second (PC2, 9.88%) and third (PC3, 5.74%) principal components that represented body shape and captured 49.16 and 28.54% of the remaining morphological variation, respectively. To further test whether Pantodon populations could be distinguished using size-standardized morphometric variation, we used jack-knifed multivariate assignment tests from discriminant function analysis, and trait-specific ANOVAs (electronic supplementary material, text S2).
3. Results
Gene content and gene order of the 10 newly sequenced mitogenomes are typical of osteoglossomorphs and other teleosts. Bayesian phylogenetic trees derived from mitogenomic sequences (described in §2) strongly support the monophyly of Pantodon within the Osteoglossomorpha. Congruent with previous results [15], Pantodon is sister to a large clade comprising the Osteoglossidae, Notopteridae, Gymnarchidae and Mormyridae (figure 2). However, the genetic divergence between the two Pantodon mitogenomes was unexpectedly large (uncorrected p-distance excluding the control region = 15.2%). Similar mitogenomic divergence was observed between the South American O. bicirrhosum and the southeast Asian S. formosus (16.0%) and throughout the entire, morphologically diverse family of mormyrid electric fishes (14.5%).
Figure 2.
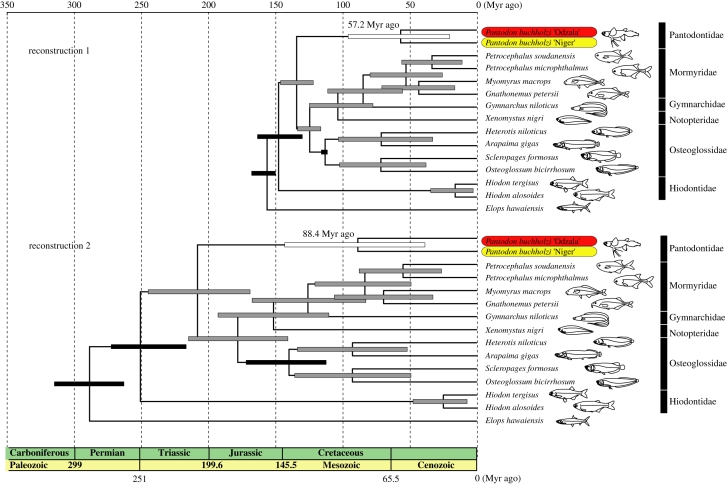
Molecular phylogenetic chronograms of osteoglossomorphs based on a Bayesian relaxed-clock analysis. We used the 12RT data matrix and two different fossil-based constraint distributions (lognormal (reconstruction 1) versus uniform (reconstruction 2)). Elops hawaiensis was used to root the trees. All internal nodes are supported by posterior probabilities of 1.0. Horizontal time scale is in millions of years before present (Myr ago). Horizontal bars show 95% HPD intervals: black, fossil-based calibrated nodes; white, divergence between Pantodon populations; and grey, all other nodes. Numbers next to the Pantodon nodes are divergence time estimates in Myr ago.
Estimated divergence times between Pantodon populations depend on the fossil-based calibration method employed (figure 2 and electronic supplementary material, figure S1). Among the four analytical approaches we tested, the most recent estimate for the divergence of Pantodon populations was 57.2 Myr ago (95% HPD = 23.3–91.3 Myr ago; figure 2, reconstruction 1), placing their separation in the Late Paleocene. The oldest divergence time we estimated between populations was 102.5 Myr ago (95% HPD = 34.5–161.3 Myr ago; electronic supplementary material, figure S1, reconstruction 4), placing their separation in the Mid-Cretaceous.
The nuclear gene MLL similarly showed that Pantodon populations exhibit a pattern of deep genetic divergence (5.6%) comparable to the pattern observed in the family Mormyridae (5.9%) and larger than the divergence observed between two distinctive genera of knifefishes (Notopteridae), Notopterus and Chitala (2.9%). Moreover, we detected the presence of two diagnostic indels in the MLL coding sequence that distinguish the two Pantodon populations (electronic supplementary material, figures S2 and S4). Both the absence of key taxa needed to calibrate the tree using fossils and the short sequence length (751 bp) precluded reliable estimation of divergence times using the MLL data.
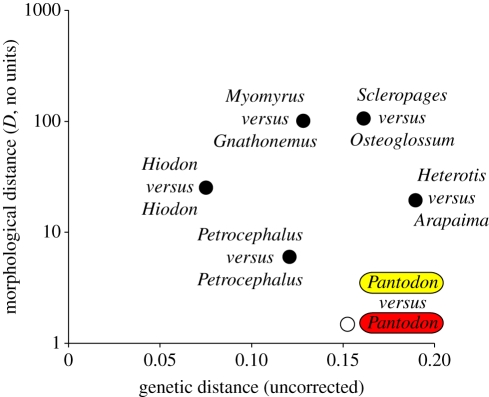
Pantodon populations exhibit the smallest morphological differentiation among available pairs of osteoglossomorph species as well as the lowest rate of morphological evolution. We found no meristic differences between the two Pantodon populations (electronic supplementary material, text S2), and we were unable to consistently distinguish them using morphometrics. Discriminant function analysis of morphometric data allowed correct assignment of individuals to populations in only 64–67% of cases. Size-standardized caudal peduncle length (F1,12 = 4.98, p = 0.048) and head length (F1,12 = 4.61, p = 0.053) were the only traits in which modest statistical differences were apparent. Morphological divergence within other pairs of osteoglossomorph species varied widely in the taxa we examined, indicating that some clades have changed considerably in shape while others show little differentiation (yet more than in Pantodon) over periods of 16.3–72.0 Myr (figure 3).
Figure 3.
Semi-log plot of morphological distances versus genetic distances for six terminal pairs of Osteoglossomorpha. Morphological distances are estimated as Mahalanobis distances (shown on a logarithmic scale). Genetic distances are uncorrected p-distances (number of substitutions/total number of characters) based on complete mitogenomic data (shown on a linear scale).
4. Discussion
Pantodon populations from the Congo and Niger basins offer a striking case of phenotypic stasis over a very long period of evolutionary time. This result is particularly noteworthy in the context of osteoglossomorph fishes, which are generally known for their morphological conservatism [9]. Indeed, Pantodon's morphological constancy despite at least 57.2 Myr of divergence is remarkable in comparison to most invertebrate and all vertebrate examples in the fossil record [3,6]. Morphological stasis in Pantodon appears to surpass other known examples of morphological conservatism in extant vertebrates—including plethodontid salamanders, lampreys and coelacanths—in which slight but consistent morphological differences invariably allow the recognition of distinct taxa [7,8,16]. In addition to extremely deep mitogenomic divergence, we also found striking divergence in nuclear MLL sequences, implicating widespread genetic divergence across both the nuclear and mitochondrial genomes. Genetic differentiation between these two Pantodon populations is so great that they might even be reproductively isolated owing to genetic incompatibilities. Clearly, the possibility of two or more cryptic species within P. buchholzi deserves further study.
Eldredge et al. [4] emphasize the significance of geographical population structure and population expansion when attempting to understand morphological stasis. Over the last 50 Myr, tropical Africa has experienced major climatic oscillations resulting in repeated cycles of forest expansion and fragmentation [17,18]. These fluctuating environmental conditions may have contributed to genetic divergence in Pantodon by repeatedly isolating populations in refugia, as suggested by their current disjunct distribution [19]. However, our data show that genetic isolation in Pantodon began long before the Quaternary arid period, which has been implicated in refugia-driven allopatric divergence of other African species [17]. Geographically fine-scale collections and additional mitochondrial and nuclear sequence data are needed to refine understanding of population structure and divergence times within Pantodon.
Proposed mechanisms of morphological stasis include stabilizing selection, ecological niche conservatism and genetic and developmental constraints [5]. Persistent stabilizing selection could be maintained in habitat refugia, but the lengthy period of morphological conservatism in Pantodon would require the selective milieu to have remained consistent over several major climatic fluctuations, or to have converged recently to an extraordinary degree. Perhaps the many highly specialized aspects of form, sensory biology and behaviour in Pantodon are so tightly interconnected that selection has constrained phenotypic divergence in this lineage. Nevertheless, there are many mechanisms by which genetic and developmental constraints are counteracted over long periods of evolutionary time.
Regardless of which specific mechanisms of phenotypic stasis are at work, Pantodon offers the most extreme example known from extant vertebrates of morphological conservatism over geological time scales. Moreover, the existence of allopatric, genetically divergent Pantodon populations offers rare opportunities to assess the relative roles of intrinsic or extrinsic constraints in stabilizing a complex and unusual morphology. These investigations will complement evolutionary inferences from taxa that exhibit phenotypic lability with minimal genetic differentiation, as well as providing insights into the underpinnings of the long-term phenotypic stasis often observed in the fossil record.
Acknowledgements
Collecting permits were provided by la Direction de la Faune et des Aires Protégées and la Délégation à la Recherche Scientifique et Technologique of the Republic of Congo, and ECOFAC-Congo. All of our fish collection and handling methods conformed to protocols approved by Cornell's Center for Research Animal Resources.
We thank Gerald Smith, Amy McCune, William Fink, Peter Konstantinidis and Katie Peichel for providing helpful comments on the study. John Sullivan, Valentin Mbossi, John Friel and Eric Kinzonzi assisted with specimen collection, and Didier Paugy created the distribution map. John Friel (CUMV) curated the voucher specimens, and Frank Kirschbaum and Li Chenhong donated tissue samples. Funding was provided by the National Geographic Society (7879-05) and by JSPS Research grants (16570082, 15380131, 17207007 and 19207007).
References
- 1.Avise J. C. 2004. Molecular markers, natural history and evolution, 2nd edn. Sunderland, MA: Sinauer Associates [Google Scholar]
- 2.Hoekstra H. E., Coyne J. A. 2007. The locus of evolution: evo devo and the genetics of adaptation. Evolution 61, 995–1016 10.1111/j.1558-5646.2007.00105.x (doi:10.1111/j.1558-5646.2007.00105.x) [DOI] [PubMed] [Google Scholar]
- 3.Stanley S. M. 1998. Macroevolution: pattern and process. Baltimore, MD: Johns Hopkins University Press [Google Scholar]
- 4.Eldredge N., et al. 2005. The dynamics of evolutionary stasis. Paleobiology 31, 133–145 10.1666/0094-8373(2005)031[0133:TDOES]2.0.CO;2 (doi:10.1666/0094-8373(2005)031[0133:TDOES]2.0.CO;2) [DOI] [Google Scholar]
- 5.Erwin D. H. 2007. Disparity: morphological pattern and developmental context. Palaeontology 50, 57–73 10.1111/j.1475-4983.2006.00614.x (doi:10.1111/j.1475-4983.2006.00614.x) [DOI] [Google Scholar]
- 6.Avise J. C., Nelson W. S., Sugita H. 1994. A speciational history of ‘living fossils’: molecular evolutionary patterns in horseshoe crabs. Evolution 48, 1986–2001 10.2307/2410522 (doi:10.2307/2410522) [DOI] [PubMed] [Google Scholar]
- 7.Min M. S., Yang S. Y., Bonett R. M., Vieites D. R., Brandon R. A., Wake D. B. 2005. Discovery of the first Asian plethodontid salamander. Nature 435, 87–90 10.1038/nature03474 (doi:10.1038/nature03474) [DOI] [PubMed] [Google Scholar]
- 8.Kuraku S., Kuratani S. 2006. Time scale for cyclostome evolution inferred with a phylogenetic diagnosis of hagfish and lamprey cDNA sequences. Zool. Sci. 23, 1053–1064 10.2108/zsj.23.1053 (doi:10.2108/zsj.23.1053) [DOI] [PubMed] [Google Scholar]
- 9.Hilton E. J. 2003. Comparative osteology and phylogenetic systematics of fossil and living bony-tongue fishes (Actinopterygii, Teleostei, Osteoglossomorpha). Zool. J. Linn. Soc. 137, 1–100 10.1046/j.1096-3642.2003.00032.x (doi:10.1046/j.1096-3642.2003.00032.x) [DOI] [Google Scholar]
- 10.Käse R. H., Bleckmann H. 1987. Prey localization by surface wave ray-tracing: fish track bugs like oceanographers track storms. Experientia 43, 290–293 [DOI] [PubMed] [Google Scholar]
- 11.Saidel W. M. 2000. Coherence in nervous system design: the visual system of Pantodon buchholzi. Phil. Trans. R. Soc. Lond. B 355, 1177–1181 10.1098/rstb.2000.0662 (doi:10.1098/rstb.2000.0662) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Starosciak A. K., Kalola R. P., Perkins K. P., Riley J. A., Saidel W. M. 2008. Fast and singular muscle responses initiate the startle response of Pantodon buchholzi (Osteoglossomorpha). Brain Behav. Evol. 71, 100–114 10.1159/000111457 (doi:10.1159/000111457) [DOI] [PubMed] [Google Scholar]
- 13.Löytynoja A., Milinkovitch M. C. 2003. A hidden Markov model for progressive multiple alignment. Bioinformatics 19, 1505–1513 10.1093/bioinformatics/btg193 (doi:10.1093/bioinformatics/btg193) [DOI] [PubMed] [Google Scholar]
- 14.Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lavoué S., Sullivan J. P. 2004. Simultaneous analysis of five molecular markers provides a well-supported phylogenetic hypothesis for the living bony-tongue fishes (Osteoglossomorpha: Teleostei). Mol. Phylogenet. Evol. 33, 171–185 10.1016/j.ympev.2004.04.021 (doi:10.1016/j.ympev.2004.04.021) [DOI] [PubMed] [Google Scholar]
- 16.Holder M. T., Erdmann M. V., Wilcox T. P., Caldwell R. L., Hillis D. M. 1999. Two living species of coelacanths? Proc. Natl Acad. Sci. USA 96, 12 616–12 620 10.1073/pnas.96.22.12616 (doi:10.1073/pnas.96.22.12616) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamilton A. C., Taylor D. 1991. History of climate and forests in tropical Africa during the last 8 million years. Climate Change 19, 65–78 10.1007/BF00142215 (doi:10.1007/BF00142215) [DOI] [Google Scholar]
- 18.Weijers J. W. H., Schefuß E., Schouten S., Sinninghe Damsté J. S. 2007. Coupled thermal and hydrological evolution of tropical Africa over the last deglaciation. Science 315, 1701–1704 10.1126/science.1138131 (doi:10.1126/science.1138131) [DOI] [PubMed] [Google Scholar]
- 19.Moritz C., Patton J. L., Schneider C. J., Smith T. B. 2000. Diversification of rainforest faunas: an integrated molecular approach. Annu. Rev. Ecol. Syst. 31, 533–563 10.1146/annurev.ecolsys.31.1.533 (doi:10.1146/annurev.ecolsys.31.1.533) [DOI] [Google Scholar]